Abstract
Intraepithelial lymphocytes expressing the gamma delta T cell receptor (gamma delta IELs) serve as a first line of defense against luminal microbes. Although the presence of an intact microbiota is dispensable for gamma delta IEL development, several microbial factors contribute to the maintenance of this sentinel population. However, whether specific commensals influence population of the gamma delta IEL compartment under homeostatic conditions has yet to be determined. We identified a novel gamma delta IEL hyperproliferative phenotype that is characterized by expansion of multiple Vgamma subsets. Horizontal transfer of this hyperproliferative phenotype to mice harboring a phenotypically normal gamma delta IEL compartment was prevented following antibiotic treatment, thus demonstrating that the microbiota is both necessary and sufficient for the observed increase in gamma delta IELs. Further, we identified two guilds of small intestinal or fecal bacteria represented by 12 amplicon sequence variants (ASV) that are strongly associated with gamma delta IEL expansion. Using intravital microscopy, we find that hyperproliferative gamma delta IELs also exhibit increased migratory behavior leading to enhanced protection against bacterial infection. These findings reveal that transfer of a specific group of commensals can regulate gamma delta IEL homeostasis and immune surveillance, which may provide a novel means to reinforce the epithelial barrier.
Introduction
Intraepithelial lymphocytes (IEL) are located within the intestinal epithelium and provide the first line of defense against luminal microorganisms1. Nearly half of murine IELs express the γδ T cell receptor (TCR), which exhibit a largely protective response to dampen acute inflammation2, 3 and promote mucosal barrier integrity4, 5. These protective functions have largely been attributed to IELs bearing the Vγ7 TCR, which comprise the majority of the γδ IEL population6. We have shown that γδ IELs limit microbial translocation by migrating into lateral intercellular space (LIS) between adjacent epithelial cells to provide surveillance of the barrier7, 8. However, our understanding of the factors involved in regulating γδ IEL migratory behavior and activation remains limited.
γδ IELs are maintained in a partially-activated state to provide an immediate defense against invasive microbes, while limiting the potential for autoimmunity. Recently, activation of γδ IELs was shown to induce the production of interferons (IFN)4, which are potent immunomodulatory cytokines that are rapidly induced in response to viral or bacterial infection9. Type I IFN activates the IFNα/β receptor that is comprised of IFNAR1 and IFNAR2 (IFNAR). The presence of commensal bacteria at birth induces tonic type I IFN production by myeloid cells in the intestinal mucosa10, which contributes to the development of mucosal immunity11, 12. Although tonic IFNAR signaling is critical for the maintenance of lamina propria lymphocyte populations13, its effect on IELs, and γδ T cells in general, remains unclear.
Gnotobiotic, or germ-free (GF), mice exhibit an overall reduction in IELs, yet the number of γδ IELs remains largely intact14, 15. While these findings indicate that microbiota is not required for γδ IEL development, four-week-old GF mice exhibit a delay in population of the IEL compartment compared to SPF mice16. Further, administering antibiotics to wildtype (WT) mice immediately after birth significantly reduces γδ IEL number in the small intestine (SI) without affecting other peripheral lymphocytes17. Consistent with this, signaling through pattern recognition receptors and the aryl hydrocarbon receptor contribute to IEL homeostasis17–19. Whereas Lactobacillus reuteri promotes the development of CD4+ CD8αα+ IELs20, the extent to which specific commensal bacteria influence γδ IEL number and function has yet to be determined.
In this study, we have serendipitously discovered a novel γδ IEL hyperproliferative phenotype that arises early and persists throughout life. This γδ IEL expansion is driven by active proliferation of all Vγ subsets; however, the overall composition of this population skews toward Vγ7− lymphocytes. We find that the microbiota is both necessary and sufficient to transfer the γδ IEL hyperproliferative phenotype to non-phenotypic mice, and further, we identified 12 amplicon sequence variants (ASV) that are closely associated with the expansion of γδ IELs. Interestingly, the hyperproliferative γδ IELs also exhibit enhanced migratory behavior at steady-state and confer protection against systemic Salmonella infection. These findings highlight the contribution of a specific group of commensals in the regulation of γδ IEL homeostasis and surveillance behavior, which may provide a novel means to reinforce the mucosal barrier in response to injury or infection.
Results
Commensal bacteria promote γδ IEL surveillance behavior.
Previous studies have demonstrated that the microbiota is dispensable for population of the γδ IEL compartment; however, in the absence of commensal microbiota γδ IEL surveillance was reduced along the crypt-villus axis21. To determine the effect of the microbiota on the kinetics of γδ IEL migratory behavior within the epithelial compartment, GFP γδ T cell reporter mice (TcrdEGFP) were treated with broad-spectrum antibiotics and intravital microscopy was performed. We find that depletion of commensal bacteria results in reduced γδ IEL migratory speed, the frequency of migration into, and dwell time within the lateral intercellular space (LIS) (Fig. S1). These data support previous findings that commensal-derived signals contribute to γδ IEL motility patterns within the intestinal mucosa.
Identification of a γδ IEL hyperproliferative phenotype that is accompanied by skewed Vγ subset composition.
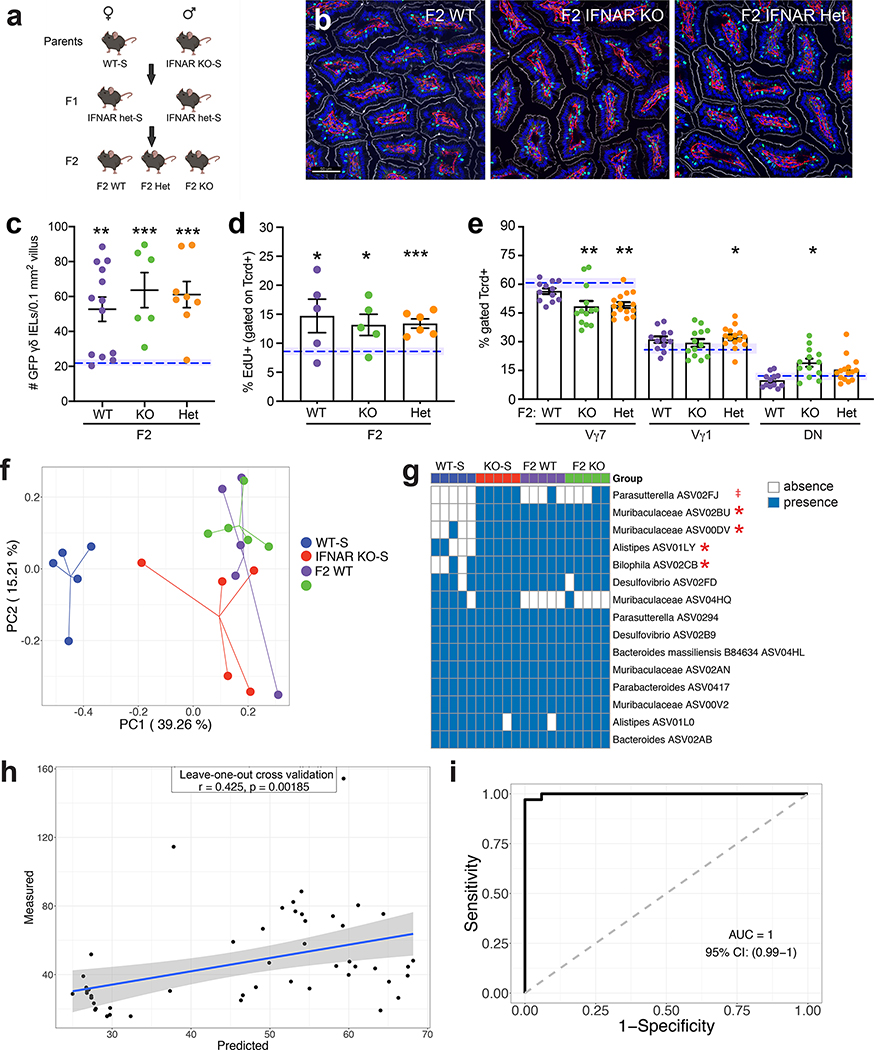
We and others have shown that epithelial-immune crosstalk is required to promote γδ IEL homeostasis and enable a rapid response to microbial pathogens. For example, epithelial MyD88 signaling promotes increased γδ IEL migration in response to bacterial infection21. Based on the known roles for tonic type I IFN in priming the host response to enteric infection10–12, we investigated the contribution of type I IFN signaling to γδ IEL homeostasis. To this end, we crossed IFNAR-deficient mice to those expressing the TcrdEGFP reporter. Morphometric analysis of the jejunum revealed a substantial increase in the number of GFP+ γδ IELs in adult IFNAR KO mice compared to TcrdEGFP (WT) (Fig. 1a,b). We find that this increase is due to enhanced γδ IEL proliferation as IFNAR KO mice exhibit a 50% increase in EdU+ γδ IELs relative to WT (Fig. 1c). Based on these findings, we next asked whether this enhanced proliferation could be attributed to a specific Vγ subset within the IEL compartment. Compared with WT, we were surprised to find that the relative proportion of γδ IELs skewed toward Vγ7− subsets in IFNAR KO mice (Fig. 1d, S2a,b). Further analysis of each Vγ subset revealed that IFNAR KO mice exhibit a 22% and 66% increase in proliferation in Vγ7+ IEL and Vγ7− IEL populations, respectively, compared to WT counterparts (Fig. 1e, S2c). Although the overall number is increased, the relative proportion of IEL subsets remained similar between WT and IFNAR KO mice (Fig. S2d). Interestingly, we found that both the frequency and the total number of γδ T cells from the spleen and mesenteric lymph nodes (MLN) were similar between the two genotypes (Fig. S2e,f).
Figure 1. IFNAR-deficient mice exhibit a γδ IEL hyperproliferative phenotype and skewed Vγ composition early in life.
a Immunofluorescent micrographs of jejunum from TcrdEGFP WT and IFNAR KO mice. Scale bar = 50μm. Nuclei, blue; laminin, red; γδ T cell, green; F-actin, white. b Quantification of jejunal GFP+ γδ T cells. c Percentage of EdU+ γδ IELs and d frequency of SI Vγ IEL subsets in WT and IFNAR KO mice (gated on GFP). e Percentage of EdU+ cells (gated on individual Vγ subsets). f Immunofluorescent micrographs of jejunum from 1-week old TcrdEGFP WT and IFNAR KO mice. Scale bar = 50μm. g Number of GFP+ γδ T cells in the SI of WT or IFNAR KO mice at various ages, n=4–10. Frequency of h CD3+ cells (gated on live cells), i Tcrd+ and Tcrb+ IELs (gated on CD3) and j Vγ subsets (gated on GFP) in WT and IFNAR KO postnatal mice, n=3–6. Data represent mean (±SEM) and are from at least two independent experiments. Each data point represents an individual mouse. Statistical analysis: b,c,j: unpaired t-test; d,e,g,h: two-way ANOVA with Sidak’s post hoc test; i: two-way ANOVA with Tukey’s post hoc test. *P<0.05, **P<0.01, *** P<0.001, # P<0.0001. ns, not significant.
To determine when this γδ IEL hyperproliferative phenotype is established, we performed morphometric analysis of intestinal tissue sections obtained from neonatal and weanling mice (Fig. 1g). As early as one week after birth, we observed a substantial increase in γδ IELs in IFNAR KO mice compared to WT (Fig. 1f,g). At one week of age, the emerging IEL compartment in IFNAR KO mice was heavily skewed toward TCRγδ+ IELs (Fig. 1h,i). However, the relative frequency among Vγ subsets was similar between the two genotypes at these early timepoints (Fig. 1j), suggesting that the differential expansion of Vγ subsets that we observed in IFNAR KO mice occurs post-weaning. These data demonstrate that the expansion of the γδ IEL compartment occurs early in life and continues throughout adulthood.
We next investigated the potential factors that could drive the expansion of and/or skewing of Vγ IEL subsets. Butyrophilin-like (Btnl) −1 and −6 expressed in the murine SI epithelium jointly regulate the maturation and expansion of Vγ7+ IELs in the gut22. The expression of three major Btnl genes (−1, −4, −6) was similar in SI of neonatal and adult WT and IFNAR KO mice (Fig. S2g). IL-7 and IL-15 have known roles in the development and proliferation of intestinal γδ T cells23, 24, yet no change in expression was detected in the SI of adult mice (Fig. S2h,i). Although the ontogeny of γδ IELs is somewhat controversial with studies demonstrating that IELs develop both extrathymically and from thymic precursors22, we found no difference in DN2 and DN3 T cell precursors or γδ T cells in E18.5 fetal thymus (Fig. S2j,k). Taken together, these data indicate that the γδ IEL hyperproliferative phenotype develops after birth but cannot be attributed to altered butyrophilin or cytokine expression, or thymic γδ T cell development.
We next asked whether the increase in the γδ IEL compartment in IFNAR KO mice could be attributed to clonal expansion. Analysis of the γδ TCR repertoire revealed both WT and IFNAR KO γδ IELs are comprised of heterogeneous populations (Fig. S3). Consistent with our flow cytometric analysis (Fig. 1d), Vγ7, Vγ1, and Vγ4 subsets were the main TCRs expressed and Vγ7− subpopulations were predominant in IFNAR KO mice (Fig. S3d). From these data, we conclude that under these highly proliferative γδ IELs maintain their characteristic heterogeneity.
Horizontal transfer of the microbiota is necessary and sufficient to induce the γδ IEL hyperproliferative phenotype.
The experiments described above were performed in a standard SPF barrier facility (S); however, a colony of IFNAR KO mice were also maintained in an enhanced barrier facility (E) in which murine norovirus and Helicobacter species are excluded. In this enhanced facility, TcrdEGFP (WT)-(E) and IFNAR KO-E mice were crossed resulting in the generation of WT, IFNAR+/− and IFNAR-deficient littermates expressing the GFP γδ T cell reporter. To our surprise, morphometric analysis and flow cytometry revealed that IFNAR KO-E mice did not display the γδ IEL hyperproliferative phenotype; these mice exhibited a similar number of γδ IELs and proportion of Vγ subsets compared to WT-S and WT-E mice (Fig. 2a, S4). These findings provided the first evidence that loss of type I IFN signaling alone was not sufficient to induce the γδ IEL hyperproliferative phenotype.
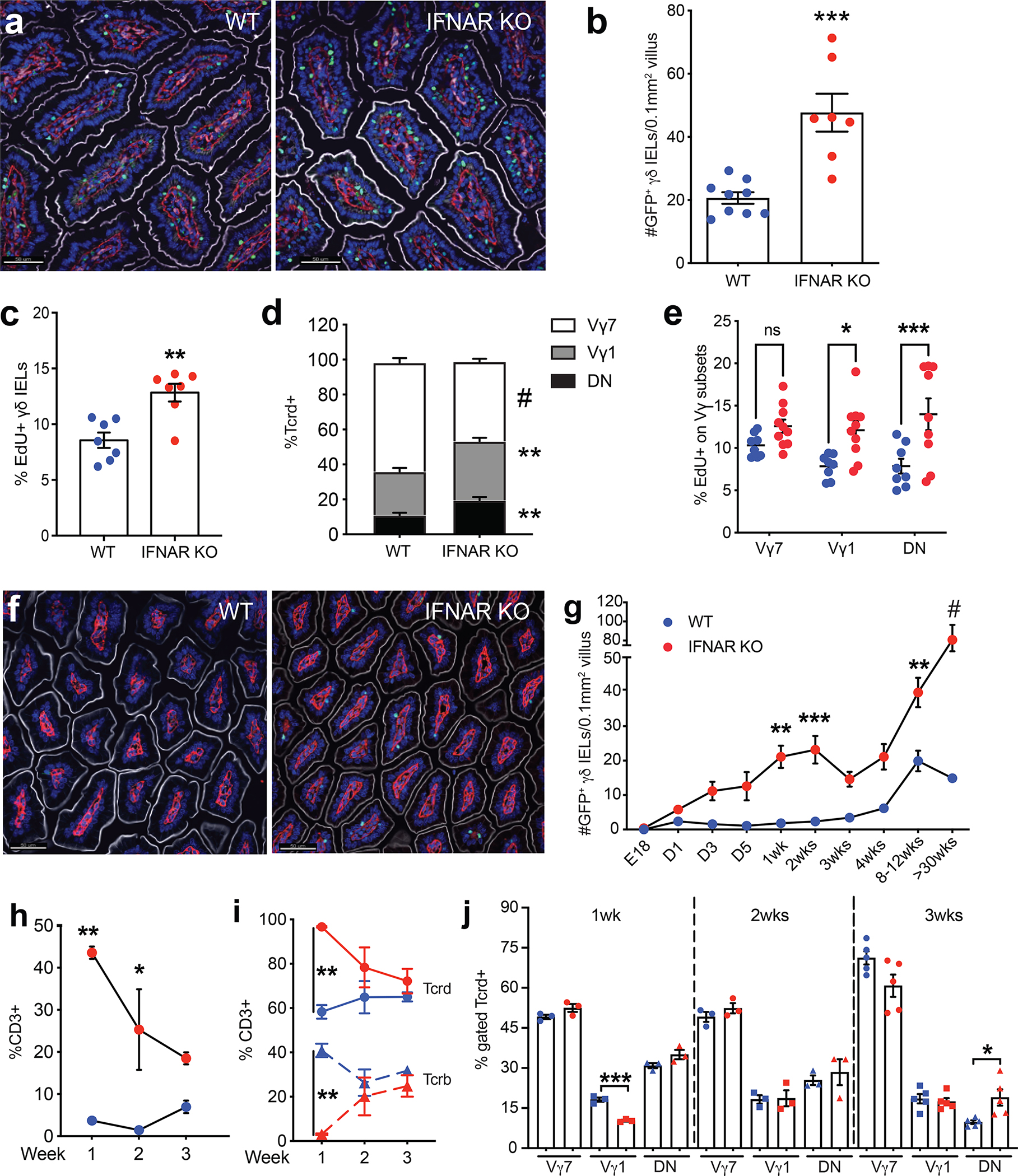
Figure 2. Horizontal transfer of the microbiota is necessary and sufficient to induce the γδ IEL hyperproliferative phenotype.
a Morphometric analysis of the number of GFP+ γδ T cells in untreated WT-E, IFNAR KO-E mice; WT-E or IFNAR KO-E breeders and offspring following IFNAR KO-S bedding transfer in the presence or absence of antibiotic (Abx) treatment. Dashed line indicates the number of γδ IELs in IFNAR KO-S donor mice. n=5–15. Principal coordinates analysis of 16S rRNA sequencing of fecal microbiota from donor and recipient breeders b and offspring c n=4–6. d Binary heatmap of 69 ASVs shared in recipient mice (breeders and offspring) following horizontal transfer of microbiota. The red asterisk highlights the 15 ASVs which had significantly lower prevalence in pre-transfer breeders compared with donors, post-transfer breeders and post-transfer breeders’ offspring. Each data point represents an individual mouse. Data represent mean (±SEM). Statistical analysis: a: one-way ANOVA with Dunnett’s post hoc test **P<0.01, *** P<0.001, # P<0.0001; d: Fisher’s exact test *P<0.05
It is well-established that the microbiome can influence the immune phenotype of mice maintained in separate facilities25. Thus, we next investigated the contribution of the microbiota to the observed changes in the γδ IEL compartment. First, we asked whether horizontal transfer of the microbiota is sufficient to induce this γδ IEL hyperproliferative phenotype. Since mice are coprophagic, dirty bedding was transferred from cages of IFNAR KO-S mice into those housing breeding pairs of WT-E or IFNAR KO-E mice. After bedding transfer, the number of γδ IELs in WT-E or IFNAR KO-E breeders and their adult offspring resembled that of IFNAR KO-S mice (Fig. 2a). Next, to determine whether the microbiota is necessary for the transfer of the γδ IEL hyperproliferative phenotype, broad-spectrum antibiotics were administered in parallel to the bedding transfer in a subset of cages. Antibiotic treatment was able to prevent the horizontal transfer of the γδ IEL phenotype to both WT-E and IFNAR KO-E breeders and their adult offspring (Fig. 2a). Pre-treatment with antibiotics was required to successfully transfer the phenotype to WT-S mice (Fig. S5), indicating that the absence of certain commensals in the enhanced (E) barrier may be more permissive to microbial transfer. Collectively, these data demonstrate that the whole microbiota play a causal role in the establishment of the γδ IEL hyperproliferative phenotype.
To explore the changes to gut microbiota following horizontal transfer and identify the members associated with the γδ IEL hyperproliferative phenotype, we next performed 16S rRNA gene V4 sequencing on fecal samples collected from bedding transfer donors (IFNAR KO-S) and recipients (WT-E and IFNAR KO-E breeders and offspring). Prior to bedding transfer, the alpha diversity of WT-E and IFNAR KO-E breeders differed from that of IFNAR KO-S mice (Fig. S6a,b) which had higher richness. After bedding transfer, the alpha diversity of WT-E and IFNAR KO-E breeders was significantly higher than prior to transfer and more similar to that of IFNAR KO-S mice. Principal coordinates analysis (PCoA) based on Bray-Curtis distance showed a clear separation of the overall gut microbiota structure between donors and pre-transfer breeders along PC1 (Fig. 2b). Following bedding transfer, the gut microbiota of recipients became more similar to donors along PC1 though there was a separation along PC2. PERMANOVA test showed significant difference (P<0.001) between donors, pre-transfer and post-transfer breeders when compared in a pairwise fashion. Using redundancy analysis (RDA) with the group information as constraining variable, 101 ASVs were identified as differential variables between donors, pre- and post-transfer breeders (Fig. S7a, S8a). Next, we performed the same analysis between donors, pre-transfer breeders and the offspring of post-transfer breeders. The alpha diversity of the gut microbiota in the offspring was significantly lower than that of the other two groups (Fig. S6c,d) and their overall gut microbiota structure was more similar to donors compared to their parents prior to transfer (Fig. 2c). PERMANOVA test showed a significant difference between the three groups (P<0.001) and 108 ASVs were identified as differential variables using RDA (Fig. S7b, S8b). After combining the results from the two analyses, 69 ASVs were commonly identified by RDA (Fig 2d, S7c), from which 15 ASVs showed significantly lower prevalence in pre-transfer breeders as compared with donors, post-transfer breeders and their offspring. There was no marked difference between the latter three groups. Taken together, these analyses identified 15 fecal ASVs that are associated with the horizontal transfer of the γδ IEL hyperproliferative phenotype.
The γδ IEL hyperproliferative phenotype can be transmitted vertically to WT offspring.
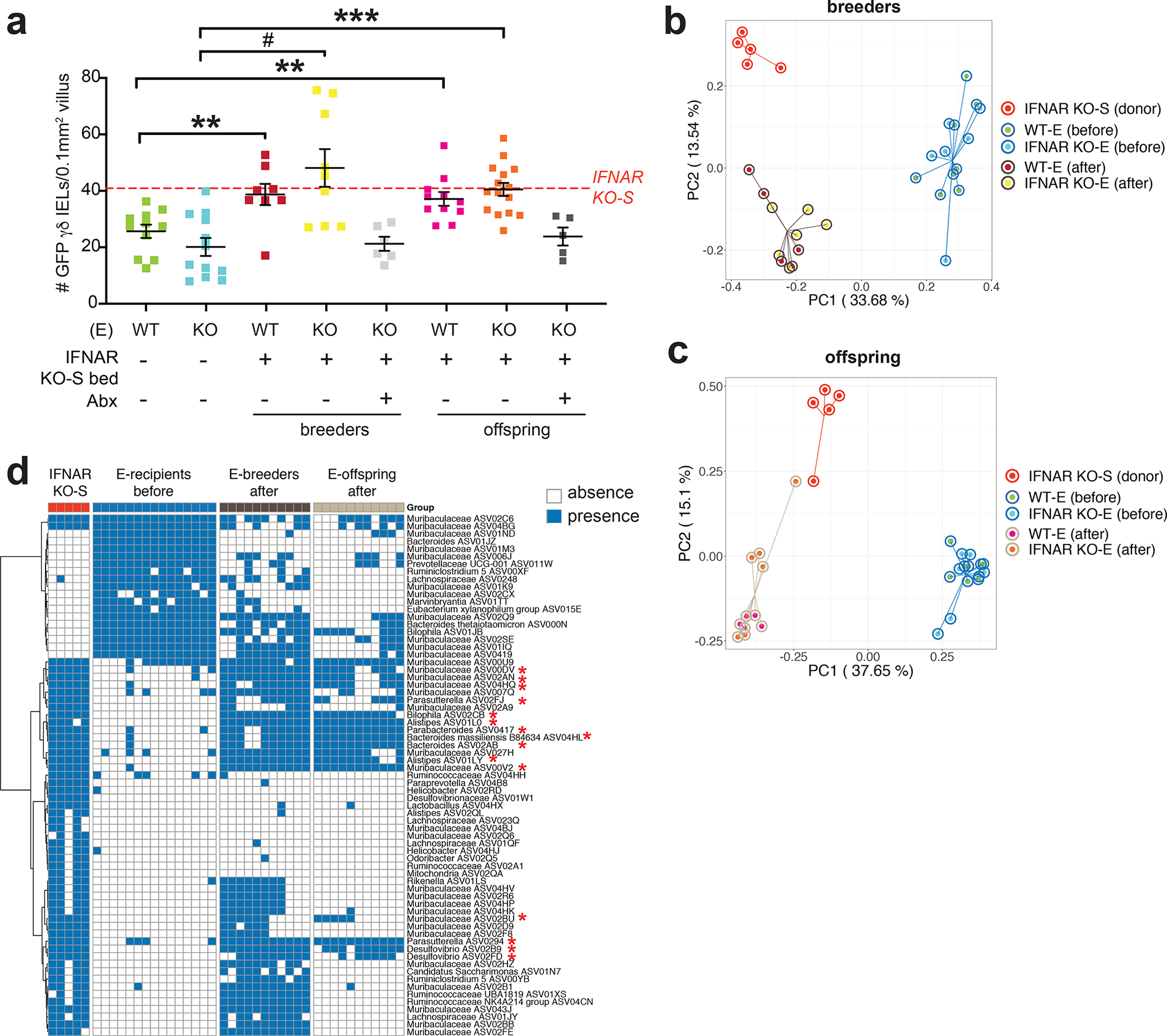
Our findings indicate that the microbiota, and not loss of IFNAR signaling, may be the primary factor leading to the expansion of the γδ IEL compartment. When initially generating the IFNAR KO-S line, we crossed the parental TcrdEGFP (WT-S) to IFNAR KO-S mice and subsequently housed the two strains separately. Housing mice of different genotypes separately can induce a confounding variable when evaluating the effect of the microbiome on a given phenotype. Thus, to control for the maternal effect of the microbiome, we crossed separately housed WT-S dams with IFNAR KO-S sires to generate F2 littermates (Fig. 3a). We observed that adult F2 WT mice exhibited a 2-fold increase in the number of γδ IELs, similar to parental IFNAR KO-S mice (Fig. 3b,c). Further, F2 WT γδ IELs exhibit enhanced proliferation compared to WT-S (Fig. 3d). Vγ1+ cells were increased and Vγ7+ cells were largely decreased in all three genotypes of F2 littermates relative to WT-S mice (Fig. 3e). The reciprocal cross was performed (Fig. S9a) and the resultant F2 WT mice showed a similar expansion of the γδ IEL compartment (Fig. S9b–d), indicating that the genotype of the dam was not a contributing factor. Together, these data demonstrate that the observed phenotype is vertically transmissible and confirms our earlier observation that γδ IEL hyperproliferation occurs independently of IFNAR signaling.
Figure 3. The γδ IEL hyperproliferative phenotype is transmitted vertically independently of genotype.
a Breeding strategy used to generate F2 WT and IFNAR KO littermates. b Representative images and c morphometric analysis of GFP+ γδ T cells in the jejunum of adult F2 littermates. Scale bar = 50μm. n=6–13 mice in three independent experiments. d Percentage of EdU+ γδ IELs and e proportion of Vγ IEL subsets in adult F2 littermates. Dashed lines indicate separately housed WT-S mean values, shaded area (±SEM). f Principal coordinates analysis of 16S rRNA sequencing of fecal microbiota from WT-S, IFNAR KO-S and F2 WT and IFNAR KO littermates. g Occurrence heatmap of the 15 ASVs identified in the horizontal transfer dataset. Blue and white show the presence or absence of ASVs, respectively. h and i Associations between the 5 ASVs (highlighted in g) and the γδ IEL hyperproliferative phenotype. Data represent mean (±SEM) from two independent experiments. Each data point represents an individual mouse. Statistical analysis: c,d: unpaired t test compared to separately housed WT-S values; e: one-way ANOVA with Dunnett’s post hoc test; g: Fisher’s exact test was performed to compared prevalence of ASVs between WT-S and the combination of the other 3 groups. h: Random Forest model with leave-one-out cross-validation was applied to use the ASVs abundance to regress the γδ IEL cell number or i: to classify mice with or without γδ IEL hyperproliferative phenotype. Pearson correlation was used to compare the predicted and measured values for regression model. (AUC) area under the curve of ROC (Receiver Operating Characteristics) was used to assess the classification model. ‡P<0.1 *P<0.05, **P<0.01, *** P<0.001.
Microbiome analysis of fecal samples collected from WT-S, IFNAR KO-S, F2 WT and F2 IFNAR KO mice revealed different alpha diversity of gut microbiota among the 4 groups (Fig. S10). Specifically, WT-S had significantly lower richness compared to the mice exhibiting the γδ IEL hyperproliferative phenotype, and the alpha diversity of the gut microbiota in F2 littermates was similar to IFNAR KO-S mice. PCoA plot based on Bray-Curtis distance showed a clear separation between WT-S and the phenotypic groups along PC1 (Fig. 3f) (PERMANOVA test, P < 0.001 between WT-S and the other 3 groups). The F2 littermates of both genotypes had a similar gut microbiota composition (PERMANOVA test, P = 0.86) close to IFNAR KO-S along PC1, but still showed a significant difference along PC2 (PERMANOVA test, F2 IFNAR KO vs IFNAR KO-S: P = 0.02; F2 WT vs IFNAR KO-S: P = 0.14). We next assessed the prevalence of the 15 bacterial ASVs identified in the horizontal transfer datasets (Fig. 2d) in this experimental group. Among the initial 15 ASVs, we found 5 (one from Parasutterella, two from Muribaculaceae, one from Alistipes and one from Bilophila) with significantly higher prevalence in the phenotypic mice compared to WT-S (Fig. 3g). These 5 ASVs also increased and decreased together across the samples (pairwise spearman correlation: all ρ > 0), which showed co-abundance behavior and may work as a guild26 (hereafter referred to as “fecal guild”). We further explored the associations between the fecal abundance of the 5 ASVs in the fecal guild and the number of γδ IELs using Random Forest regression with leave-one-out cross-validation. The predicted values showed a significantly positive correlation with the measured values indicating that these commensal bacteria likely contribute to the γδ IEL hyperproliferative phenotype (Fig. 3h). Furthermore, the abundances of the 5 ASVs in feces were able to accurately classify phenotypic and non-phenotypic mice using a Random Forest classification model (AUC =1, 95% CI: (0.99 – 1)) (Fig. 3i).
Small intestinal and fecal microbiota signatures are associated with the local expansion of γδ IELs, and in combination, can accurately classify the hyperproliferative phenotype.
We next assessed the regional effect of the microbiota to the γδ IEL hyperproliferative phenotype. Morphometric analysis revealed that γδ IELs were expanded throughout the length of the SI in phenotypic mice, yet not in the cecum or colon (Fig. 4a). Microbiome analysis of luminal contents in various segments of the gut revealed that the overall microbial composition of phenotypic mice was significantly different from those without the phenotype in each SI segment (Fig. 4b and Table S1). Since there was no significant difference in the microbial composition between the different SI segments within each group (Table S2), we combined the samples from duodenum, ileum and jejunum together to explore the differential ASVs between phenotypic and non-phenotypic mice in the SI. In total, we found 7 ASVs were significantly (BH adjusted P < 0.05) enriched, with a fold change from 3 to 10, in phenotypic mice as compared with non-phenotypic ones (Fig. 4c and Fig. S11). Four of these 7 ASVs were from Lactobacillus, 1 from Enterococcus, 1 from Faecalibaculum and 1 from Muribaculaceae. Notably, the four Lactobacillus ASVs were also dominant members in the phenotypic mice as ASV00U8, ASV01JJ, ASV00UH and ASV01NA accounted for 28.64%, 9.96%, 5.96% and 1.91% of the total abundance respectively. These 7 ASV also increased and decreased together across the samples (pairwise spearman correlation: all ρ > 0) and may work as a guild26 (hereafter referred to as the “SI guild”). We further explored the associations between the SI abundance of these 7 ASVs in the SI guild and the number of γδ IELs using Random Forest regression with leave-one-out cross-validation. The predicted values showed a significantly positive correlation with the measured values indicating that these 7 bacteria likely contribute to the γδ IEL hyperproliferative phenotype in the SI (Fig. 4d). In addition, the abundance of the 7 ASVs in the SI were able to accurately classify phenotypic and non-phenotypic mice by using a Random Forest classification model (AUC = 0.97, 95% IC: (0.91–1)) (Fig. 4e).
Figure 4. Small intestinal microbiota signature is associated with the local expansion of the γδ IEL compartment.
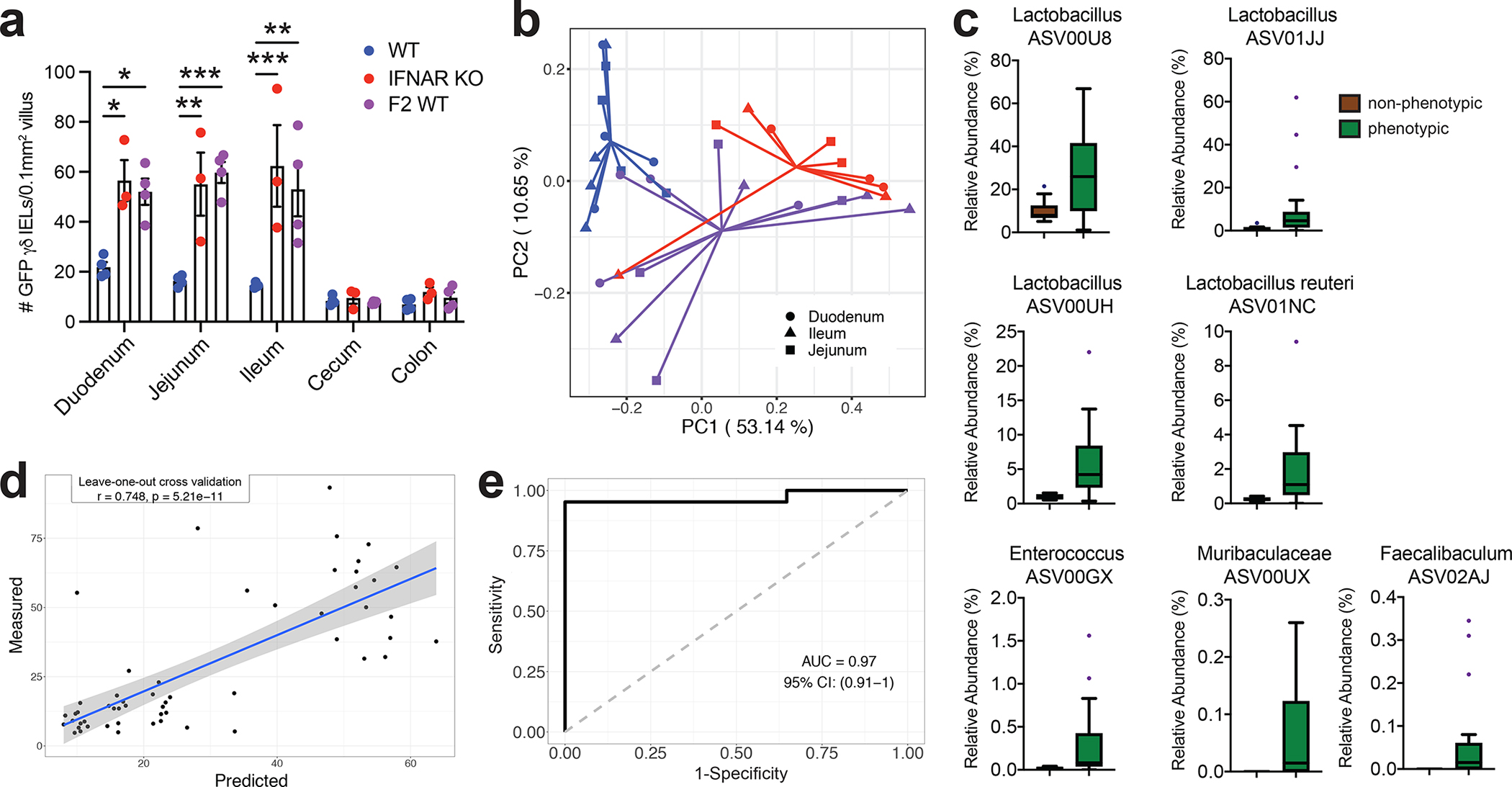
a Morphometric analysis of the number of GFP+ γδ T cells in untreated WT-S, IFNAR KO-S or F2 WT mice. b Principal coordinates analysis of 16S rRNA sequencing of SI luminal microbiota from WT-S, IFNAR KO-S and F2 WT mice. Bray Curtis distance was applied. c Seven ASVs in the SI were enriched in the mice with γδ IEL hyperproliferative phenotype. d and e Associations between the 7 ASVs in the SI enriched by the mice with γδ IEL hyperproliferative phenotype. Statistical analysis: a: one-way ANOVA with Tukey’s post hoc test; c: MaAsLin2 was applied to explore the differential ASVs in SI samples between mice with (IFNAR KO-S and F2 WT) and without the γδ IEL hyperproliferative phenotype (WT-S). BH-adjusted p values < 0.05 considered as significant. Boxes show the medians and the interquartile ranges (IQRs), the whiskers denote the lowest and highest values that were within 1.5 times the IQR from the first and third quartiles, and outliers are shown as individual points; d: Random Forest model with leave-one-out cross-validation was applied to use the ASVs abundance in the intestine to regress the intestinal γδ IEL cell number or e: to classify intestinal segments based on the γδ IEL hyperproliferative phenotype. Pearson correlation was used to compare the predicted and measured values. *P<0.05, **P<0.01, *** P<0.001.
We then explored the association between the γδ IEL hyperproliferative phenotype and the combination of signatures from the SI and fecal guilds. For the SI samples, using the abundance of the 12 ASVs from both guilds, we built a Random Forest regression model against γδ IEL number and a Random Forest classification model to classify phenotypic and non-phenotypic mice (Fig. 5a,b). The models using the combined guilds were similar to those based on the SI guild alone (Fig. 4d, 5a). We performed same analysis in the fecal samples (Fig. 5c,d), and found that the two guild combined regression model performed better than the model using only the fecal guild (Fig. 3h, 5c).
Figure 5. Combining the fecal and small intestinal microbiota signatures accurately predicts the γδ IEL hyperproliferative phenotype.
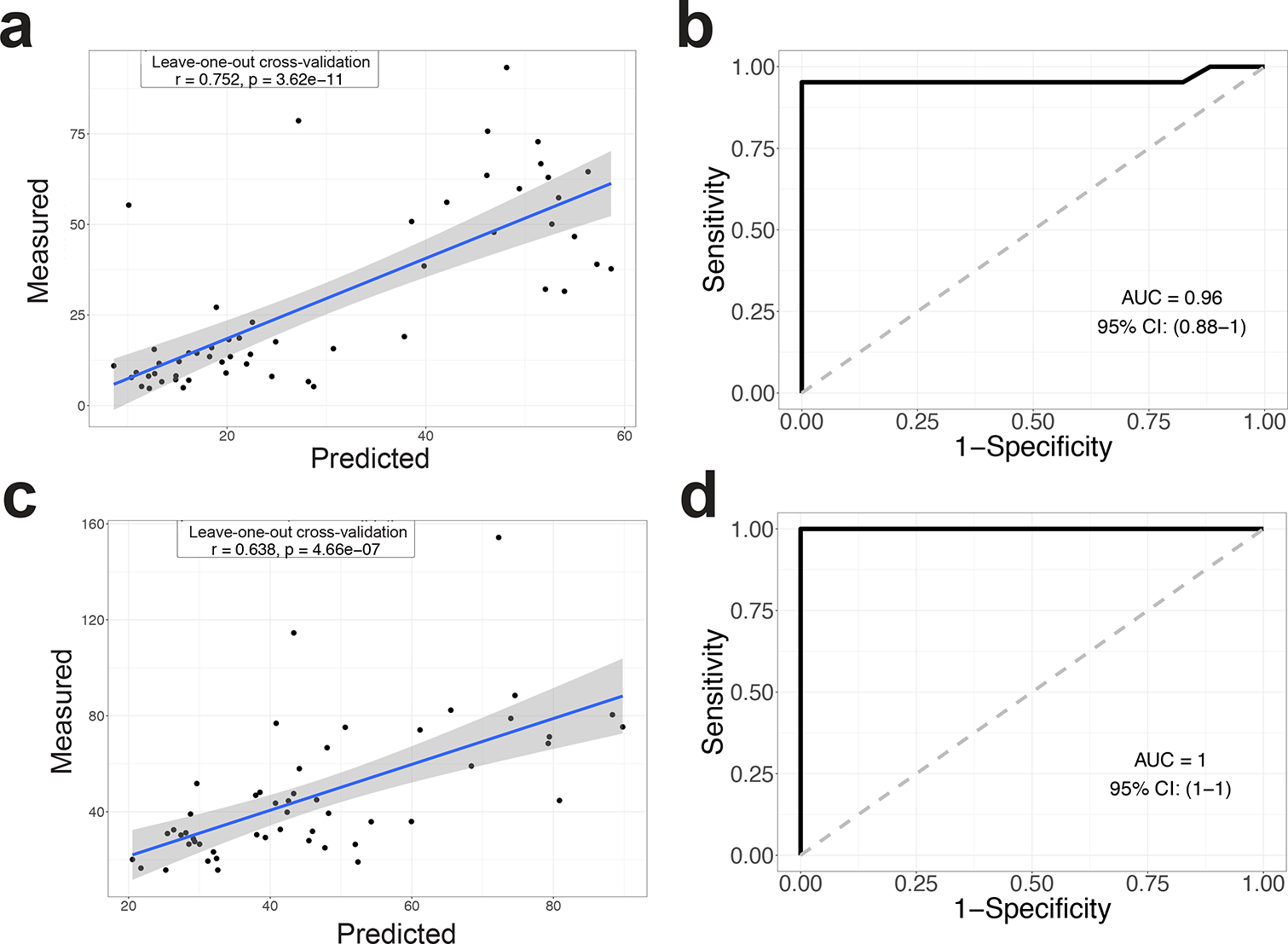
Random Forest model with leave-one-out cross-validation was applied to use the 12 ASVs abundance in the intestine (a) and feces (c) to regress the intestinal γδ IEL cell number. b Random Forest model with leave-one-out cross-validation was applied to use the 12 ASVs abundance in the intestine (b) and feces (d) to classify intestinal segments with or without the γδ IEL hyperproliferative phenotype. Pearson correlation was used to compare the predicted and measured values for regression model. (AUC) area under the curve of ROC (Receiver Operating Characteristics) was used to assess the classification model.
Hyperproliferative γδ IELs exhibit enhanced surveillance behavior to confer protection against bacterial infection.
We previously reported that γδ IELs provide continuous surveillance of the intestinal epithelium to limit acute pathogen invasion8, 27. Thus, to determine whether the expansion and/or alteration of γδ IEL subsets affects their overall surveillance behavior, we performed intravital microscopy of WT-S and F2 WT SI mucosa under homeostatic conditions. Image analysis of time lapse videos revealed that γδ IELs in WT mice with the hyperproliferative phenotype migrated into the LIS more frequently than those in WT-S mice (Fig. 6a,b, Video S1). This increase in flossing behavior was accompanied by increased average track speed and reduced dwell time in the epithelium of F2 WT mice compared to WT-S. (Fig. 6c,d). As expected, the frequency with which an enterocyte was contacted by a γδ IEL was increased due to the expansion of γδ IELs within the epithelial compartment (Fig. 6e).
Figure 6. γδ IEL surveillance behavior is enhanced in WT mice exhibiting the hyperproliferative phenotype and confers protection against enteric infection.
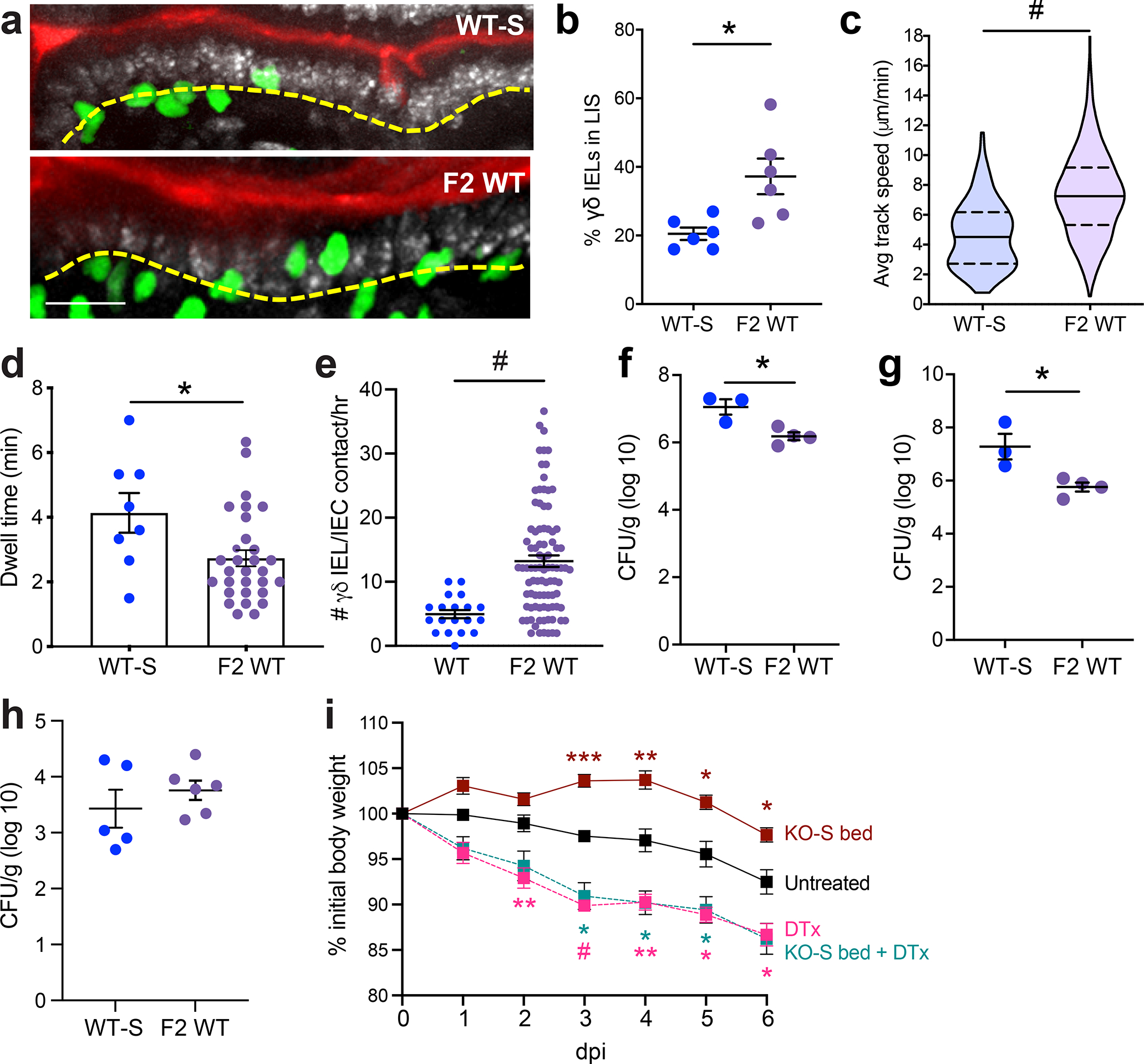
a Maximum projection images taken from intravital microscopy of γδ IELs (green) within the jejunal epithelium of WT-S and F2 WT mice. Scale bar = 30μm. Nuclei, white; lumen, red. Yellow dashed line approximates the basement membrane. b Frequency of γδ IELs in the lateral intercellular space (LIS), c average track speed, d dwell time and e γδ IEL/IEC contacts/hr. WT-S and F2 WT mice were infected orally with Salmonella Typhimurium and CFU were measured at 6 dpi in f spleen and g liver homogenates. h CFU from ileal homogenate 24 h post-infection i Weight loss of TcrdGDL mice receiving IFNAR KO-S bedding and/or DTx in response to Salmonella infection. Each data point represents an individual mouse, except c,d,e in which each data point represents an individual track, γδ IEL or IEC, respectively, and i in which the data is grouped. Data represent mean (±SEM). Statistical analysis: a-h: unpaired t-test; i: two-way ANOVA with Tukey’s post hoc test. *P<0.05, **P<0.01, *** P<0.001, # P<0.0001.
We previously demonstrated that migration of γδ IELs into the LIS is critical to limit Salmonella Typhimurium invasion and subsequent spread to peripheral sites8. To address whether the expansion and increased motility of γδ IELs contributes to the host response to infection, WT-S and F2 WT mice were infected orally with Salmonella. Six days post-infection, F2 WT mice showed reduced bacterial load in the spleen and liver compared to WT-S mice (Fig. 6f,g). We found that the differences in microbiota did not affect the initial colonization of Salmonella (Fig. 6h), but instead that the presence of γδ IELs was central to the protection afforded by microbiota transfer (Fig. 6i). Together, these data demonstrate that the transfer of this hypermotile and hyperproliferative γδ IEL phenotype to WT mice results in enhanced γδ IEL surveillance capacity to effectively reduce the severity of systemic salmonellosis.
Discussion
In this study, we serendipitously identified a novel group of commensals that correlate with the expansion of the γδ IEL compartment. This increase in proliferation is polyclonal and not restricted to one Vγ subset, although Vγ7− IELs expand to a greater extent than Vγ7+ IELs. While we initially attributed this finding to a loss of tonic IFNAR signaling, differences in the IEL compartment between mice housed in different facilities pointed to an environmental factor as the potential cause. Through horizontal and vertical transfer experiments, we determined that the microbiota was required for this phenotype, and further identified 12 ASVs that strongly correlate with the expansion of γδ IELs. In addition to increased proliferation, we found that these lymphocytes exhibit enhanced surveillance behavior within the epithelium. Consistent with our previous reports8, 27, we showed that transfer of the γδ IEL hyperproliferative phenotype to WT mice results in protection against systemic salmonellosis.
Microbial colonization early in life plays a key role in the development of mucosal immunity. Unlike mucosal-associated invariant T (MAIT) cells, antigen-specific effector and regulatory T cells that develop during early life in response to commensal exposure28–31, the microbiota is dispensable for γδ IEL development14, 15. However, through horizontal transfer and antibiotic treatment, we found that the whole gut microbiota is both necessary and sufficient to induce the expansion of γδ IELs in both neonatal and adult mice. Moreover, we identified two different microbial guilds associated with the γδ IEL hyperproliferative phenotype from fecal and SI habitats separately. Compared with the abundance of the identified guild in fecal samples, the SI guild showed a stronger association with the phenotype. This indicates that local alterations of the guild in the SI may more likely directly contribute to the γδ IEL hyperproliferative phenotype. However, the identification of the fecal guild shows that the fecal gut microbiota remained different between phenotypic and non-phenotypic mice.
Among all the 12 ASVs identified in association with the expansion of γδ IELs, the 4 ASVs of the Lactobacillus genus are the most likely to play a causal role. To date, L. reuteri is the only commensal that has been identified to be associated with the development of a specific IEL subpopulation, namely CD4+ CD8αα+ (DP) IELs20. L. reuteri and other Lactobacillus spp. colonize the SI mucosa and are considered to play a beneficial role in regulating host immunity32. Cervantes-Barragan et al. showed that L. reuteri promoted the development of DP IELs via T-cell-intrinsic aryl hydrocarbon receptor (AhR) signaling by increasing tryptophan metabolism20, 33, which is also required for the maintenance of γδ IELs19. AhR-deficient mice exhibit reduced γδ IEL number and increased susceptibility to intestinal injury due to impaired microbial control19. A similar reduction in γδ IELs was also observed in mice deficient in TL1A, which was accompanied by a reduction in Lactobacillus spp. in the ileal mucosa34. Interestingly, administration of the probiotics L. acidophilus and B. longum in conjunction with trinitrobenzenesulfonic acid (TNBS) treatment resulted in increased γδ IEL number and protection against colitis35. Administration of an AhR agonist during challenge with DSS increased the number of CD8αα+ TCRαβ+ IELs, yet had no effect on γδ IELs36. Thus, further studies are required to determine the precise mechanism by which these commensals promote γδ IEL proliferation and whether this is AhR-dependent.
Although our data demonstrate that the γδ IEL hyperproliferative phenotype occurs independently of IFNAR signaling, we cannot rule out the possibility that global loss of type I IFN signaling contributed to the expansion of specific ASVs. The results from our microbiome analyses are consistent with previous reports indicating that loss of global IFNAR signaling (IFNAR KO-E mice) does not induce significant changes to the fecal microbial community37. However, IEC-specific IFNAR KO mice exhibit increased numbers of Paneth cells and lysozyme, suggesting that IFNAR-deficiency may promote local changes in the microbiota38.
We find that the microbiota is both necessary and sufficient to enhance γδ IEL proliferation; however, we cannot exclude the effect of the microbiota on commensal viruses, which also contribute to IEL maintenance18. Murine norovirus (MNV) is present in our standard (S) but not enhanced (E) barrier facility. Whereas MNV can induce IFNAR signaling in the absence of an intact microbiota to maintain in lamina propria lymphocyte populations13, the role of MNV in the regulation of γδ IELs is unknown. Thus, the extent to which type I IFN signaling modulates the IEL compartment and the potential contribution of commensal viruses to this phenotype would be of interest for future investigation.
Several factors contribute to the selection and maintenance of γδ T cell subsets at barrier surfaces. Interestingly, we did not observe altered Vγ populations in the SI in neonatal or weanling mice (Fig. 1j), indicating that the bias towards Vγ7− IELs likely occurs post-weaning. The local microbiota drives the proliferation of Vγ6+ T cells in the oral mucosa, lung and reproductive tract39–41; therefore, we speculate that the continued expansion and stabilization of the local microbial community may induce the preferential proliferation of Vγ7− IELs. While the reciprocal interactions between γδ17 cells and the microbiota have begun to be elucidated39–41, how commensals influence γδIFN populations remains poorly understood.
Dysregulation or aberrant expansion of the IEL compartment is associated with disease states such as celiac disease or inflammatory bowel disease42; however, we do not observe overt intestinal pathology in phenotypic adult mice. Although further study is required to better understand how this microbiota affects other aspects of mucosal immunity, our findings open new avenues to explore how the microbiota or microbial-derived products can be manipulated to modulate γδ IEL proliferation and migratory behavior as a means to reinforce the epithelial barrier in the context of gastrointestinal infection and inflammation.
Methods
Animals
All mice were maintained on a C57BL/6 background and unless otherwise noted, mice of both sexes were analyzed between 8–12 weeks of age. Mice were housed under SPF barrier conditions, with colonies maintained in a standard barrier (S) or enhanced barrier facility (E) which is Helicobacter- and murine norovirus-free. Mice were fed an autoclaved commercial rodent 5010 diet and tap water. All mice were kept in the room with standard 12 hours light-dark cycle and humidity and temperature were monitored. TcrdEGFP mice43 were crossed to IFNAR1 KO mice (provided by Sergei Kotenko and Joan Durbin, Rutgers NJMS). F2 littermates were generated in the SBF by crossing separately housed WT-S and IFNAR KO-S mice. TcrdGDL mice44 were provided by Immo Prinz and Inga Sandrock (UKE/Hannover). All studies were conducted in an Association of the Assessment and Accreditation of Laboratory Animal Care–accredited facility using protocols approved by Rutgers New Jersey Medical School Comparative Medicine Resources.
Intravital microscopy
Time lapse intravital microscopy of the jejunal mucosa was performed as previously described45. Image analysis was performed using Imaris (v.9.7; Bitplane), in which surface-to-surface distance was calculated between GFP+ γδ T cells and the lumen. γδ T cells within 10–12 μm from the lumen were considered within the LIS. γδ IEL track speed was calculated by an autoregressive tracking algorithm confirmed by manual verification of individual tracks. Dwell time and the frequency of γδ IEL/epithelial interactions were quantified manually.
In vivo treatments
Vancomycin (400 μg/mL, Hospira) meropenem (200 μg/mL, Bluepoint Laboratories) were administered in the drinking water for two weeks. For bedding transfer experiments, WT-E and IFNAR KO-E breeding pairs that were transferred into the SBF (S) facility were housed with dirty bedding obtained from IFNAR KO-S cages, which was replenished biweekly. Some breeding cages were maintained on Abx while exposed to dirty bedding (Fig. 2). All offspring were weaned into clean cages with tap water. WT-S or TcrdGDL mice were housed with dirty bedding obtained from IFNAR KO-S cages for 3 weeks with or without 2 week pre-treatment with Abx. Mice were injected intraperitoneally with 0.5 mg 5-Ethynyl-2’-deoxyuridine (EdU, TCI America) daily for five days. Mice were gavaged with 200 mg/ml streptomycin (Sigma-Aldrich) 24 h prior to gavage with 108 colony-forming units (CFU) of DsRed-labeled Salmonella Typhimurium (strain SL3201; Andrew Neish, Emory). TcrdGDL mice were administered 15 ng of diphtheria toxin (List Biological, Campbell, CA) per gram body weight i.p. 24 and 48 h prior to Salmonella infection. Mice were euthanized six days post-infection after which spleen and liver homogenates were plated and colonies counted.
Immunofluorescence and image analysis
Neonatal and post-neonatal (0–3 weeks old) intestine was fixed in 10% formalin and embedded in paraffin, whereas post-weanling and adult intestine were fixed and embedded as described previously27. Briefly, tissue was fixed in 1% paraformaldehyde, washed with 50 mM NH4Cl, cryoprotected in 30% sucrose (wt/vol) and embedded in Optimal Cutting Temperature (OCT, Tissue-Tek) medium. Immunostaining of frozen or FFPE sections (5–7 μm) was performed using the rabbit anti-laminin (Sigma-Aldrich) or biotin-labeled anti-GFP (Abcam), followed by Alexa Fluor 594 goat anti-rabbit IgG (H+L), Alexa Fluor 647 phalloidin, Alexa Fluor 647 Streptavidin (Invitrogen) and/or Hoechst 33342 dye (Invitrogen). Slides were mounted with ProLong Glass (Invitrogen) and images were acquired on an inverted DMi8 microscope (Leica) equipped with a CSU-W1 spinning disk, ZYLA SL150 sCMOS camera (Andor), PL APO 40x/0.85 dry objectives, and iQ3 acquisition software (Andor). The number of GFP+ cells per 0.1 mm2 villus was quantified by an observer blinded to the condition.
IEL isolation and flow cytometric analysis
SI IELs were isolated as previously described7 and GFP+ γδ IELs were sorted to 98% purity using a BD FACSAria II. IELs were stained with viability dye (eFluor 450 or eFluor 780), anti-CD3 (2C11), anti-CD8α (53–6.7), anti-CD8β (H35–17.2), anti-CD4 (GK1.5), anti-TCRβ (H57-597), anti-TCRγδ (GL3) (eBioscience), anti-Vγ7 (clone GL1.7; Rebecca O’Brien, National Jewish Health, Denver, CO), and anti-Vγ1 (clone 2.11, BioLegend) or a Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Invitrogen). Flow cytometry was performed on an LSR Fortessa (BD Biosciences) in the New Jersey Medical School Flow Cytometry and Immunology Core Laboratory. Data were analyzed by FlowJo (v.10.4.0; Tree Star).
TCR repertoire data analysis
RNA was isolated from 3×105 γδ IELs, extracted using TRIzol (Invitrogen) and purified by RNeasy Mini Kit (Qiagen). Library construction and sequencing were performed by iRepertoire (Huntsville, AL, USA). The usage of V, D and J gene and complementarity-determining region 3 (CDR3) sequences were determined, and tree maps were generated using iRweb tools (iRepertoire). Datasets were processed using the MiXCR software package (v3.0.13) to further correct for PCR and sequencing errors. Diversity metrics, clonotype overlap and gene usage were plotted in R, by VDJTools (v1.2.1) using standard packages.
qPCR assays
RNA was extracted from homogenized tissue as described above. Total RNA was reverse transcribed using an iScript cDNA Synthesis Kit (Bio-Rad) and qPCR was performed using SYBR Green (Fisher Scientific) using QuantStudio 6 platform (ThermoFisher Scientific). Ct values were normalized to the Ct values for Ecad (Btln1,4,6,Il15) and Gapdh (Il7), respectively.
Gut microbiome analysis
Genomic DNA was extracted using protocol Q46 with minor modification. Hypervariable region V4 of the 16S rRNA gene was amplified using the 515F and 806R primers modified by Parada et al.47 and Apprill et al.48, respectively and sequenced using the Ion GeneStudio S5 (ThermoFisher Scientific). Primers were trimmed from the raw reads using cutadapt via QIIME 2. Amplicon sequence variants (ASVs) were obtained by denoising using the dada2 denoise-single command in QIIME 2 with parameters --p-trim-left 0 --p-trunc-len 215. Spurious ASVs were further removed by abundance filtering49. A phylogenetic tree of ASVs was built using the QIIME 2 commands alignment mafft, alignment mask, phylogeny fastree and phylogeny midpoint-root. Taxonomy assignment was performed by the q2-feature-classifier plugin in QIIME 2 based on the silva database (release 132)50. The data were then rarified to 38,000 reads/sample for subsequent analyses. Alpha diversity indices (Shannon Index, observed ASVs, Faith’s phylogenetic diversity and evenness) and Bray-Curtis distance were used to evaluate the overall microbiota structure. Principal coordinates analysis (PCoA) was performed by the R packages “ape”. Redundancy analysis (RDA) which describes the relationships between the grouping and ASVs in microbial communities, was performed with R package “vegan” based on Hellinger transformed abundance of the ASVs. ASVs, which had more than 50% of the variation can be explained by the grouping, were selected. Random Forest regression and classification model were performed and cross-validated using the R “randomForest” and “caret” package to test for correlations between ASVs and γδ IEL hyperproliferative phenotype.
Statistical analyses
Data were shown as the mean ± SEM. Statistical analysis was conducted in GraphPad Prism8. The significance between two independent samples was determined by unpaired t-test or for multiple independent variables one-way or two-way ANOVA was performed.
Data and code availability
All raw 16S rRNA sequencing data is accessible in NCBISRA with accession number: PRJNA744534. The accession number of γδ TCR sequencing data is PRJNA744491. Scripts and command lines related to the current study are freely available from the corresponding author upon request.
Supplementary Material
Supplementary Video 1. Intravital microscopy of γδ T cells (green), luminal Alexa Fluor 633 (red) and nuclei (white) in jejunum of TcrdEGFP (WT) or F2 WT mice. Frames were collected approximately every 20 s.
Acknowledgments
The authors would like to thank Sergei Kotenko and Joan Durbin for providing the IFNAR KO mice. Cell sorting was performed at the NJMS Flow Cytometry and Immunology Core Laboratory and supported by National Institute for Research Resources Grant S10RR027022. This work was supported by Busch Biomedical Research Grant, New Jersey Commission on Cancer Research Bridge Grant (DCHS19CRF009), National Institute of Health Grant DK119349 (K.L.E.).
Footnotes
Disclosure: Liping Zhao is a co-founder of Notitia Biotechnologies Company.
References
- 1.Hu MD, Jia L, Edelblum KL. Policing the intestinal epithelial barrier: Innate immune functions of intraepithelial lymphocytes. Curr Pathobiol Rep 2018; 6(1): 35–46. [PMC free article] [PubMed] [Google Scholar]
- 2.Komano H, Fujiura Y, Kawaguchi M, Matsumoto S, Hashimoto Y, Obana S et al. Homeostatic regulation of intestinal epithelia by intraepithelial gamma delta T cells. Proc Natl Acad Sci U S A 1995; 92(13): 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc Natl Acad Sci U S A 2002; 99(22): 14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swamy M, Abeler-Dorner L, Chettle J, Mahlakoiv T, Goubau D, Chakravarty P et al. Intestinal intraepithelial lymphocyte activation promotes innate antiviral resistance. Nat Commun 2015; 6: 7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL et al. {gamma}{delta} intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci U S A 2011; 108(21): 8743–8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nat Rev Immunol 2002; 2(5): 336–345. [DOI] [PubMed] [Google Scholar]
- 7.Edelblum KL, Shen L, Weber CR, Marchiando AM, Clay BS, Wang Y et al. Dynamic migration of gammadelta intraepithelial lymphocytes requires occludin. Proc Natl Acad Sci U S A 2012; 109(18): 7097–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edelblum KL, Singh G, Odenwald MA, Lingaraju A, El Bissati K, McLeod R et al. gammadelta Intraepithelial Lymphocyte Migration Limits Transepithelial Pathogen Invasion and Systemic Disease in Mice. Gastroenterology 2015; 148(7): 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H, Kelsall BL. The role of type I interferons in intestinal infection, homeostasis, and inflammation. Immunol Rev 2014; 260(1): 145–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaupp L, Muth S, Rogell L, Kofoed-Branzk M, Melchior F, Lienenklaus S et al. Microbiota-Induced Type I Interferons Instruct a Poised Basal State of Dendritic Cells. Cell 2020; 181(5): 1080–1096 e1019. [DOI] [PubMed] [Google Scholar]
- 11.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity 2012; 37(1): 171–186. [DOI] [PubMed] [Google Scholar]
- 12.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012; 37(1): 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature 2014; 516(7529): 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandeira A, Mota-Santos T, Itohara S, Degermann S, Heusser C, Tonegawa S et al. Localization of gamma/delta T cells to the intestinal epithelium is independent of normal microbial colonization. J Exp Med 1990; 172(1): 239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki H, Jeong KI, Itoh K, Doi K. Regional variations in the distributions of small intestinal intraepithelial lymphocytes in germ-free and specific pathogen-free mice. Exp Mol Pathol 2002; 72(3): 230–235. [DOI] [PubMed] [Google Scholar]
- 16.Jung J, Surh CD, Lee YJ. Microbial Colonization at Early Life Promotes the Development of Diet-Induced CD8alphabeta Intraepithelial T Cells. Mol Cells 2019; 42(4): 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang W, Wang X, Zeng B, Liu L, Tardivel A, Wei H et al. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J Exp Med 2013; 210(11): 2465–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Gong T, Tao W, Lin B, Li C, Zheng X et al. Commensal viruses maintain intestinal intraepithelial lymphocytes via noncanonical RIG-I signaling. Nature Immunology 2019; 20(12): 1681–1691. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 2011; 147(3): 629–640. [DOI] [PubMed] [Google Scholar]
- 20.Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science 2017; 357(6353): 806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoytema van Konijnenburg DP, Reis BS, Pedicord VA, Farache J, Victora GD, Mucida D. Intestinal Epithelial and Intraepithelial T Cell Crosstalk Mediates a Dynamic Response to Infection. Cell 2017; 171(4): 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Marco Barros R, Roberts NA, Dart RJ, Vantourout P, Jandke A, Nussbaumer O et al. Epithelia Use Butyrophilin-like Molecules to Shape Organ-Specific gammadelta T Cell Compartments. Cell 2016; 167(1): 203–218 e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baccala R, Witherden D, Gonzalez-Quintial R, Dummer W, Surh CD, Havran WL et al. Gamma delta T cell homeostasis is controlled by IL-7 and IL-15 together with subset-specific factors. J Immunol 2005; 174(8): 4606–4612. [DOI] [PubMed] [Google Scholar]
- 24.Ma LJ, Acero LF, Zal T, Schluns KS. Trans-presentation of IL-15 by intestinal epithelial cells drives development of CD8alphaalpha IELs. J Immunol 2009; 183(2): 1044–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stappenbeck TS, Virgin HW. Accounting for reciprocal host-microbiome interactions in experimental science. Nature 2016; 534(7606): 191–199. [DOI] [PubMed] [Google Scholar]
- 26.Wu G, Zhao N, Zhang C, Lam YY, Zhao L. Guild-based analysis for understanding gut microbiome in human health and diseases. Genome Med 2021; 13(1): 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu MD, Ethridge AD, Lipstein R, Kumar S, Wang Y, Jabri B et al. Epithelial IL-15 Is a Critical Regulator of gammadelta Intraepithelial Lymphocyte Motility within the Intestinal Mucosa. J Immunol 2018; 201(2): 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S et al. Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci Immunol 2017; 2(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139(3): 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 2019; 366(6464). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Dejardin F et al. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity 2019; 50(5): 1276–1288 e1275. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Zhou C, Huang J, Kuai X, Shao X. The potential therapeutic role of Lactobacillus reuteri for treatment of inflammatory bowel disease. Am J Transl Res 2020; 12(5): 1569–1583. [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Tian X, He B, Hoang TK, Taylor CM, Blanchard E et al. Lactobacillus reuteri DSM 17938 feeding of healthy newborn mice regulates immune responses while modulating gut microbiota and boosting beneficial metabolites. Am J Physiol Gastrointest Liver Physiol 2019; 317(6): G824–G838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tougaard P, Skov S, Pedersen AE, Krych L, Nielsen DS, Bahl MI et al. TL1A regulates TCRgammadelta+ intraepithelial lymphocytes and gut microbial composition. Eur J Immunol 2015; 45(3): 865–875. [DOI] [PubMed] [Google Scholar]
- 35.Roselli M, Finamore A, Nuccitelli S, Carnevali P, Brigidi P, Vitali B et al. Prevention of TNBS-induced colitis by different Lactobacillus and Bifidobacterium strains is associated with an expansion of gammadeltaT and regulatory T cells of intestinal intraepithelial lymphocytes. Inflamm Bowel Dis 2009; 15(10): 1526–1536. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Pu A, Sheng B, Zhang Z, Li L, Liu Z et al. Aryl hydrocarbon receptor activation modulates CD8alphaalpha(+)TCRalphabeta(+) IELs and suppression of colitis manifestations in mice. Biomed Pharmacother 2017; 87: 127–134. [DOI] [PubMed] [Google Scholar]
- 37.Rauch I, Hainzl E, Rosebrock F, Heider S, Schwab C, Berry D et al. Type I interferons have opposing effects during the emergence and recovery phases of colitis. Eur J Immunol 2014; 44(9): 2749–2760. [DOI] [PubMed] [Google Scholar]
- 38.Tschurtschenthaler M, Wang J, Fricke C, Fritz TM, Niederreiter L, Adolph TE et al. Type I interferon signalling in the intestinal epithelium affects Paneth cells, microbial ecology and epithelial regeneration. Gut 2014; 63(12): 1921–1931. [DOI] [PubMed] [Google Scholar]
- 39.Wilharm A, Tabib Y, Nassar M, Reinhardt A, Mizraji G, Sandrock I et al. Mutual interplay between IL-17-producing gammadeltaT cells and microbiota orchestrates oral mucosal homeostasis. Proc Natl Acad Sci U S A 2019; 116(7): 2652–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilharm A, Brigas HC, Sandrock I, Ribeiro M, Amado T, Reinhardt A et al. Microbiota-dependent expansion of testicular IL-17-producing Vgamma6(+) gammadelta T cells upon puberty promotes local tissue immune surveillance. Mucosal Immunol 2021; 14(1): 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell 2019; 176(5): 998–1013 e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu MD, Edelblum KL. Sentinels at the frontline: the role of intraepithelial lymphocytes in inflammatory bowel disease. Curr Pharmacol Rep 2017; 3(6): 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prinz I, Sansoni A, Kissenpfennig A, Ardouin L, Malissen M, Malissen B. Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nat Immunol 2006; 7(9): 995–1003. [DOI] [PubMed] [Google Scholar]
- 44.Sandrock I, Reinhardt A, Ravens S, Binz C, Wilharm A, Martins J et al. Genetic models reveal origin, persistence and non-redundant functions of IL-17-producing gammadelta T cells. J Exp Med 2018; 215(12): 3006–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia L, Edelblum KL. Intravital Imaging of Intraepithelial Lymphocytes in Murine Small Intestine. J Vis Exp 2019; (148). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costea PI, Zeller G, Sunagawa S, Pelletier E, Alberti A, Levenez F et al. Towards standards for human fecal sample processing in metagenomic studies. Nature Biotechnology 2017; 35(11): 1069–1076. [DOI] [PubMed] [Google Scholar]
- 47.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environmental Microbiology 2016; 18(5): 1403–1414. [DOI] [PubMed] [Google Scholar]
- 48.Apprill A, McNally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecology 2015; 75(2): 129–137. [Google Scholar]
- 49.Wang J, Zhang Q, Wu G, Zhang C, Zhang M, Zhao L. Minimizing spurious features in 16S rRNA gene amplicon sequencing: PeerJ Preprints; 2018. Report no. 2167–9843. [Google Scholar]
- 50.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research 2012; 41(D1): D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1. Intravital microscopy of γδ T cells (green), luminal Alexa Fluor 633 (red) and nuclei (white) in jejunum of TcrdEGFP (WT) or F2 WT mice. Frames were collected approximately every 20 s.
Data Availability Statement
All raw 16S rRNA sequencing data is accessible in NCBISRA with accession number: PRJNA744534. The accession number of γδ TCR sequencing data is PRJNA744491. Scripts and command lines related to the current study are freely available from the corresponding author upon request.