Abstract
Although several studies have identified a distinct gut microbial composition in Parkinson’s disease (PD), few studies have investigated the oral microbiome or functional alteration of the microbiome in PD. We aimed to investigate the connection between the oral and gut microbiome and the functional changes in the PD-specific gut microbiome using shotgun metagenomic sequencing. The taxonomic composition of the oral and gut microbiome was significantly different between PD patients and healthy controls (P = 0.003 and 0.001, respectively). Oral Lactobacillus was more abundant in PD patients and was associated with opportunistic pathogens in the gut (FDR-adjusted P < 0.038). Functional analysis revealed that microbial gene markers for glutamate and arginine biosynthesis were downregulated, while antimicrobial resistance gene markers were upregulated in PD patients than healthy controls (all P < 0.001). We identified a connection between the oral and gut microbiota in PD, which might lead to functional alteration of the microbiome in PD.
Subject terms: Parkinson's disease, Parkinson's disease
Background
Parkinson’s disease (PD) is a chronic progressive neurodegenerative disease affecting more than 6 million people worldwide1. The non-motor symptoms of PD, such as constipation, impaired olfaction, and rapid eye movement sleep behavior disorders, are frequently present before the onset of motor symptoms, which might be explained by the accumulation of alpha-synuclein in the peripheral nervous system before spreading to the substantia nigra2. The progressive spreading of alpha-synuclein from peripheral nerves into the brain was previously proposed in neuropathologic studies and also demonstrated in mouse models, where injection of preformed alpha-synuclein fibrils into the gastric muscular layers resulted in the spread of pathologic alpha-synuclein into the brain, while truncal vagotomy prevented the spread3,4. These findings led to the hypothesis that the pathology of PD may initiate in the gut5.
Animal studies have clarified the role of the gut microbiome in PD. Germ-free mice showed limited alpha-synuclein pathology, while mice with microbiota from PD patients exhibited enhanced motor dysfunction6. Human studies have also revealed distinct gut microbial compositions associated with PD7–11, but little is known about the alteration of the oral microbiome in PD. Dysbiosis of the oral microbiome has been observed in systemic diseases including cancer, autoimmune diseases, and gastrointestinal diseases12. The systemic effects of oral dysbiosis can be explained by local and systemic inflammation as well as the oral-gut connection. In general, the oral bacteria poorly colonize in a healthy digestive system13,14. However, in pathological circumstances, oral pathogens could affect the gut environment, enhancing the oral-gut dysbiosis connection. For example, an increased abundance of the oral-associated microbiome was observed in the gut in patients with inflammatory bowel disease, colon cancer, and liver cirrhosis12,15,16. Oral dysbiosis could also affect the pathogenesis of PD through the oral-gut connection. However, the number of studies on the oral microbiome in PD is limited, especially on its link with the gut microbiome5.
In addition, prior investigations have focused on the analysis of bacterial composition in PD using 16 S rRNA gene-based amplicon sequencing17. This method only targets the 16 S rRNA locus, which is a taxonomically informative marker18. Although this sequencing method is inexpensive and analytically convenient19, it has low taxonomic resolution at the genus level, and only enables indirect assumptions about biological functions20–22. Functional analysis of the microbiome is essential to determine how the bacterial composition affects the development of PD and to use it as a therapeutic target. In addition, the ecological dynamics of the microbiota enable the recovery of initial function, despite compositional changes23. Whole-genome shotgun metagenomic sequencing evaluates DNA from the whole microbial community18, thus providing high-resolution detection at the species level and elucidation of biological functions encoded by the microbial genome18,21,22. The use of whole-genome shotgun sequencing is increasing, but few studies have been conducted in PD patients to date24.
In the present study, we aimed to address the lack of data on the link between the oral and gut microbiome in PD, and on the functional alterations of the PD microbiota. We investigated the taxonomic and functional changes in the oral and gut microbiota in PD patients and healthy controls. In addition, we identified a discriminatory panel of candidate microbial biomarkers using the oral and gut microbiota.
Results
Baseline clinical characteristics of PD patients and healthy controls
The study population included 91 patients with PD and 85 healthy controls (HCs). The mean age at study enrollment was not significantly different between the PD patients and HCs (mean ± standard deviation, 65.1 ± 7.9 vs. 64.6 ± 8.0, P = 0.66) (Table 1). The prevalence of constipation was higher in PD patients than in HCs (47.3% vs. 12.9%, P < 0.001). Sex, body mass index, and dietary intake were not significantly different between two groups. The degrees of swallowing difficulty and olfactory dysfunction were higher in PD patients than in HCs. In PD patients, the median disease duration (IQR) was 2.0 (0.0─6.0) years and the median (IQR) Unified PD Rating Scale (UPDRS) part 3 was 32.0 (25.0─40.0).
Table 1.
Baseline clinical characteristics of patients with Parkinson’s disease (PD) and healthy controls
| PD patients (n = 91) | HCs (n = 85) | P value | |
|---|---|---|---|
| Age, years | 65.1 ± 7.9 | 64.6 ± 8.0 | 0.66 |
| Male | 49 (53.8%) | 40 (47.1%) | 0.45 |
| BMI, kg/m2 | 24.3 (22.3 − 26.4) | 23.9 (21.3 − 25.6) | 0.09 |
| Education, years | 12.0 (12.0 − 16.0) | 12.0 (12.0 − 16.0) | 0.88 |
| Clinical symptoms | |||
| IBS | 3 (3.3%) | (3.5%) | >0.99 |
| Constipation | 43 (47.3%) | 11 (12.9%) | <0.001* |
| Bristol stool scale | 4.0 (2.0 − 4.0) | 4.0 (4.0 − 4.0) | <0.001* |
| Swallowing test | 0.0 (0.0 − 3.0) | 0.0 (0.0 − 0.0) | 0.003* |
| Smell test | 9.0 (6.0 − 10.0) | 10.0 (10.0 − 10.0) | <0.001* |
| Disease duration | 2.0 (0.0 − 6.0) | — | |
| UPDRS | |||
| Part 1 | 5.0 (3.0 − 7.0) | — | |
| Part 2 | 7.0 (4.0 − 11.0) | — | |
| Part 3 | 32.0 (25.0 − 40.0) | — | |
| Part 4 | 0.0 (0.0 − 2.0) | — | |
| Total score | 44.0 (34.0 − 60.0) | — | |
| Hoehn and Yahr stage | 2.0 (2.0 − 3.0) | — | |
| Medication for PD | |||
| Levodopa | 74 (81.3%) | — | |
| Dopamine agonist | 35 (38.5%) | — | |
| COMT inhibitor | 7 (7.7%) | — | |
| MAO-B inhibitor | 25 (27.5%) | — | |
| Amantadine | 19 (20.9%) | — | |
| Levodopa equivalent daily dose, mg | 450.0 (275.0 − 802.5) | — | |
| Daily dietary intake | (n = 83) | (n = 78) | |
| Total energy, kCal | 2168.5 ± 500.5 | 2160.3 ± 566.6 | 0.92 |
| Carbohydrate, g | 295.1 ± 69.9 | 288.4 ± 79.6 | 0.57 |
| Protein, g | 88.2 ± 25.8 | 88.9 ± 25.1 | 0.85 |
| Animal source | 47.2 ± 15.5 | 47.1 ± 16.5 | 0.96 |
| Plant source | 40.9 ± 16.5 | 41.8 ± 16.4 | 0.73 |
| Fat, g | 68.5 ± 26.4 | 67.7 ± 24.2 | 0.84 |
| Animal source | 40.2 ± 20.0 | 38.7 ± 18.4 | 0.63 |
| Plant source | 28.3 ± 12.9 | 29.0 ± 12.8 | 0.75 |
| Fiber, g | 38.4 ± 17.0 | 38.0 ± 17.6 | 0.87 |
| Soluble | 5.0 ± 2.5 | 5.1 ± 2.4 | 0.08 |
| Insoluble | 18.9 ± 8.1 | 19.2 ± 8.6 | 0.80 |
Values are mean ± standard deviation, n (%), or median (interquartile range).
PD Parkinson’s disease, HCs healthy controls, BMI body mass index, IBS irritable bowel syndrome, UPDRS Unified Parkinson’s Disease Rating Scale, COMT catechol-O-methyltransferase, MAO-B monoamine oxidase B.
*P < 0.05.
Previous studies indicated that PD medication such as catechol-o-methyl transferase (COMT) inhibitors and anticholinergics affected the microbiota composition, so we conducted a beta-diversity analysis to detect any effect of PD medication on the microbiota composition. Stool microbiome was related to the use of COMT inhibitor (P = 0.001) and amantadine (P = 0.048), while the oral microbiome was related to the use of dopamine agonist (P = 0.003) and amantadine (P = 0.049).
16 S rRNA gene sequencing of the oral and gut microbiota
Stool samples were analyzed in 88 PD patients and 84 HCs, and oral samples were analyzed in 74 PD patients and 69 HCs after excluding those with low sequence coverage and those who refused to give oral samples (Supplementary Fig. 1). Characteristics of the subset of patients sampled for oral and gut microbiome were similar to those of the total study population (Supplementary Table 1 and Supplementary Table 2).
Beta-diversity analysis showed significant differences in the microbial composition between the two groups in the gut (P = 0.001) and oral samples (P = 0.003) (Fig. 1a, b), while there were no significant differences in alpha diversity (Supplementary Fig. 2).
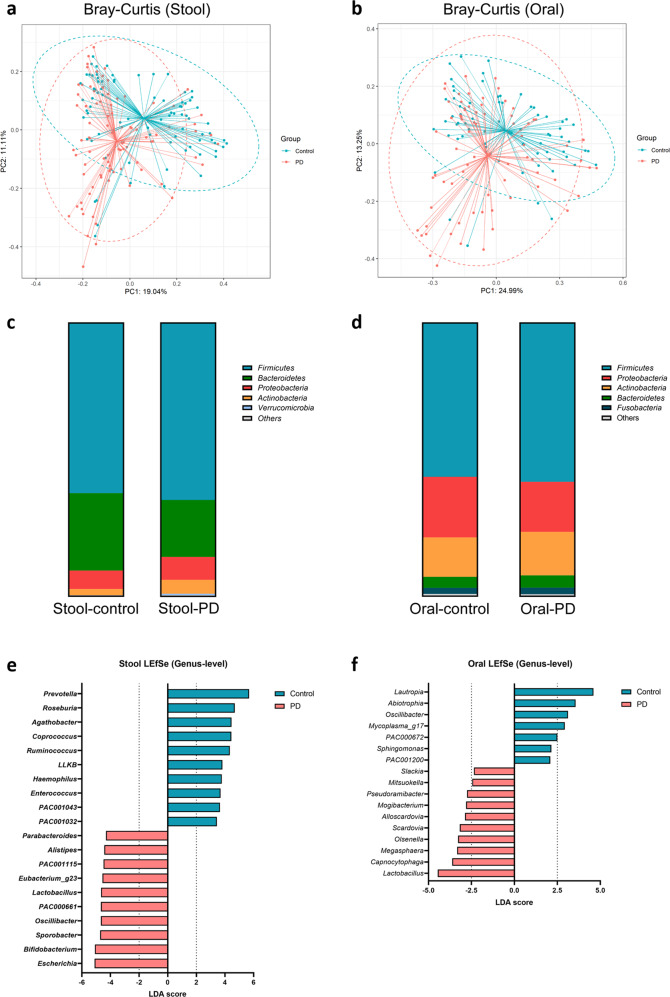
Fig. 1. Gut and oral microbial community structure based on 16 S rRNA gene sequencing.
a, b Principal coordinates analysis based on the Bray-Curtis dissimilarity between Parkinson’s disease (PD) and healthy controls in a stool and b oral samples. c, d Phylum-level bar charts of PD and HCs in the c stool and d oral samples. e, f Comparison of the e gut microbiome and f oral microbiome between PD and HCs using genus-level LEfSe analysis. The top 10 genera based on the LDA score in the PD and control groups are shown. The cut-off value for oral LDA effect size was set at 2.0. LLKB (Lachnospiraceae; LLKB01000001), PAC001043 (Lachnospiraceae, AJ576336), PAC001032 (Lachnospiraceae, unpublished), PAC001115 (Christensenellaceae, HQ716403) PAC000661 (Oscillospiraceae; JN713389) PAC002046 (Lachnospiraceae, GQ897562), and Eubacterium_g23 (Oscillospiraceae, GQ502529) were the phylotype genera (Family, NCBI accession number).
In the gut, the most common phylum was Firmicutes, which had a similar abundance between PD patients and HCs. Bacteroidetes was significantly more abundant in HCs (FDR-adjusted P; Q = 0.007), while Proteobacteria was more abundant in PD patients (Q = 0.048) (Fig. 1c). In the oral samples, the most common phylum was Firmicutes, followed by Proteobacteria and Actinobacteria, all of which did not show significant differences between the two groups (Fig. 1d).
According to the linear discriminant analysis (LDA) effect size (LEfSe) analysis, short-chain fatty acid-producing gut bacteria such as Prevotella, Roseburia, Coprococcus, and Ruminococcus had lower LDA scores in PD patients. Escherichia, Bifidobacterium, Sporobacter, Oscillibacter, and Lactobacillus had higher LDA scores in PD patients than in HCs (Fig. 1e). Aside from the top 20 genera according to the LEfSe analysis, genera that were abundant in patients with PD in previous studies (i.e., Hungatella, Odoribacter, Alistipes, and Collinsella) were also more abundant in PD patients than in HCs (Q < 0.030)25–27, along with the proinflammatory genus Mogibacterium (Q = 0.043). In the oral samples of PD patients, Lautropia, Abiotrophia, and Oscillibacter had lower LDA scores while Lactobacillus, Capnocytophaga, and Megasphaera had higher LDA scores than in HCs (Fig. 1f).
We compared the gut microbiota between patients whose H&Y stage was less than 3 (mild PD) and patients whose H&Y stage was 3 or greater (severe PD). The beta-diversity was not significantly different between mild PD and severe PD (P = 0.147) (Supplementary Fig. 3). LEfSe analysis showed that the PD-associated microbiome in this study, such as Lactobacillus and Bifidobacterium, and gut pathogens such as Klebsiella, were higher in severe PD compared with mild PD. We also compared the oral microbiota between mild PD and severe PD. The beta diversity was significantly different between mild PD and severe PD (P = 0.042). In the severe PD group, oral Hemophilus and Lactobacillus were increased, and Lautropia was decreased.
Functional analysis based on 16 S rRNA gene sequencing
We indirectly compared the function of the oral microbiome using 16 S rRNA gene sequencing between patients with PD and HCs using PICRUSt. Beta-glucoside operon transcriptional antiterminator was higher in the oral samples of PD compared with the healthy controls (Supplementary Table 3). However, the results were not statistically significant after adjusting for multiple comparisons.
Association between the oral and gut microbiome
In the previous four studies that investigated the oral microbiome in PD and in this study, one common oral bacteria consistently found to be increased in PD was Lactobacillus14,28–31. Therefore, we focused on oral Lactobacillus, a facultative anaerobe that could survive in the digestive tract.
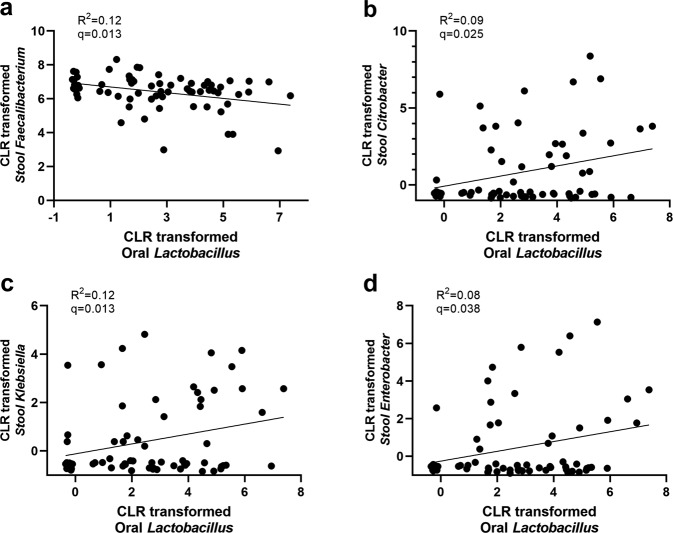
Lactobacillus was significantly more abundant in PD patients, in both the oral cavity (Q = 0.044) and the gut (Q = 0.187 and P = 0.049). We examined the correlation between the abundance of oral Lactobacillus in the mouth with that of gut microbiome. In PD patients, the abundance of oral Lactobacillus was associated with a low abundance of gut Faecalibacterium, which is a representative intestinal commensal bacterium (Q = 0.013), and with a high abundance of gut Citrobacter, Klebsiella, and Enterobacter, all of which belong to the opportunistic pathogenic Enterobacteriaceae (Q ≤ 0.038) (Fig. 2). However, the correlation was not significant in HCs. Oral Lactobacillus and stool Lactobacillus did not show a significant association in both PD and HCs (Q = 0.36).
Fig. 2. Correlation between oral Lactobacillus and stool bacteria in Parkinson’s disease.
a–d Linear regression analysis between oral Lactobacillus and stool. a Faecalibacterium, b Citrobacter, c Klebsiella, and d Enterobacter.
Association between the microbiome and clinical manifestations of PD
Canonical correspondence analysis showed that PD severity index (UPDRS total score) was positively correlated with the disease duration, dysphagia, and the use of COMT inhibitor or amantadine, whereas age, Bristol stool index, IBS, and olfactory function were not significantly correlated with PD severity (Supplementary Fig. 4 and Supplementary Table 4). The total UPDRS score was associated with a higher abundance of Bifidobacterium (Q = 0.002) and Lactobacillus (Q = 0.006) among the top 20 genera from the LEfSe analysis. However, there was no genus correlated negatively with the PD severity indexes. Stool firmness (low Bristol stool scale) was related to a low abundance of Prevotella (Q = 0.003). The use of COMT inhibitor or amantadine was positively associated with disease severity. In contrast, none of the oral microbiota was significantly correlated with the clinical symptoms of PD.
Whole-genome shotgun metagenomics of the gut microbiome
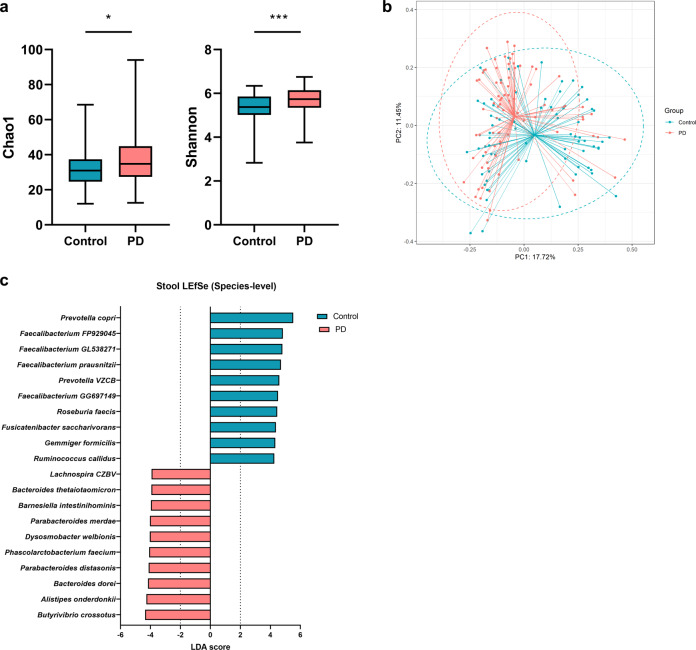
Using stool samples, we conducted high-resolution taxonomic and functional analyses based on the whole-genome shotgun metagenomics data. The alpha diversity was significantly higher in PD patients than in HCs (P = 0.048 for Chao1, and P = 0.0003 for Shannon) (Fig. 3a), and the beta-diversity was also significantly different between the two groups (P = 0.001) (Fig. 3b). We found that Prevotella copri, Prevotella VZCB, and four Faecalibacterium species were less abundant in PD patients than in HCs (Fig. 3c). Conversely, possible pathogens or proinflammatory species, such as Alistipes onderdonkii, Bacteroides dorei, Parabacteroides merdae, and Butyrivibrio crossotus, were more abundant in PD patients. Aside from the top 20 species according to the LEfSe analysis, proinflammatory species (Bacteroides cellulosilyticus, Bacteroides eggerthii, Butyricimonas virosa, and Bilophila wadsworthia) were also significantly more abundant in PD patients (Q < 0.041).
Fig. 3. Gut microbial species-level community structures based on whole-genome shotgun sequencing.
a Alpha diversity between Parkinson’s disease (PD) and healthy controls. b Principal coordinates analysis based on the Bray-Curtis dissimilarity. c Species-level LEfSe analysis. The top 10 genera in the gut based on the LDA score in the Parkinson’s disease and HC groups are shown. Faecalibacterium FP929045 (AJ270470), Faecalibacterium GL538271 (GL538271), Prevotella VZCB (VZCB01000106), Faecalibacterium GG697149 (GG697149) and Lachnospira CZBV (CZBV01000019) were phylotype species (NCBI accession number). Boxplot centerline represents the median (50th percentile). The top and bottom hinges represent 75th and 25th percentiles, respectively. The upper and lower whiskers correspond to the highest and lowest data points. n.s: not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
Functional analysis based on whole-genome shotgun metagenomics
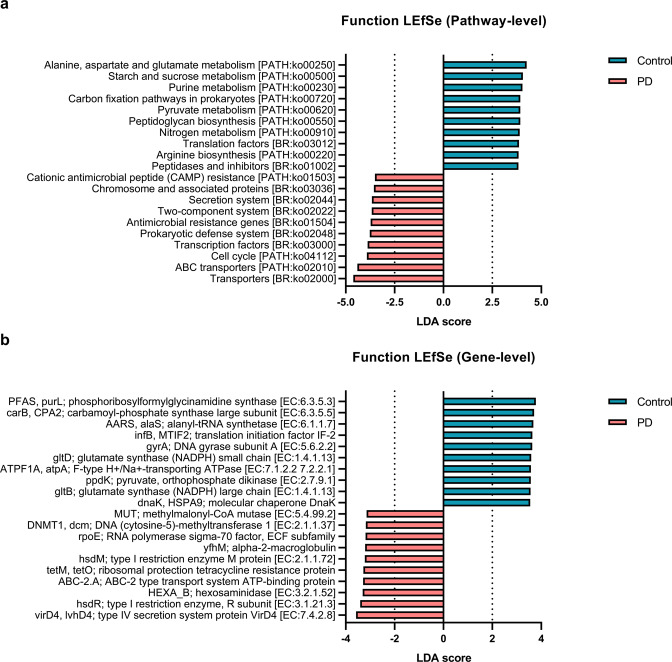
Microbial genes involved in glutamate metabolism (ko00250) and arginine biosynthesis (ko00220) were downregulated in PD patients compared with HCs (all Q < 0.0001) (Fig. 4). Conversely, microbial genes involved in the prokaryotic defense system (ko02048), antimicrobial resistance (ko01504), and cationic antimicrobial peptide (CAMP) resistance (ko01503) were upregulated in PD patients compared with HCs. Interestingly, glutamate decarboxylase, which converts L-glutamate into γ -aminobutyric acid (GABA), was present at a higher frequency in the gut of PD patients than in HCs (Q = 0.004). Conversely, glutamate synthase and phosphoribosylformylglycinamidine synthase, all of which produce L-glutamate, were present at a lower frequency in PD patients than in HCs (all Q < 0.0001) (Supplementary Fig. 5). Furthermore, genes involved in the synthesis of arginine (ornithine carbomoyltransferase, Q = 0.021; argininosuccinate synthase, Q < 0.0001; and argininosuccinate lyase, Q < 0.0001) were present at a lower frequency in PD patients than in HCs, and genes involved in the consumption of arginine (arginase, Q < 0.0001) were present at a higher frequency in PD patients than in HCs.
Fig. 4. Gut microbial functional profiles based on shotgun sequencing.
a, b Comparison of gut microbial functional profiles between patients with Parkinson’s disease (PD) and healthy controls in a pathway-level, and b gene-level.
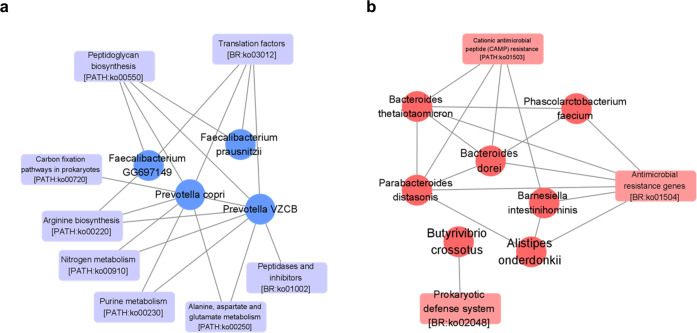
Network analysis showed that alanine, aspartate, and glutamate metabolism (ko00250) and arginine metabolism (ko00220) were strongly correlated with Prevotella copri, Prevotella VZCB, and Faecalibacterium species (all Q < 0.0001) (Fig. 5). Prokaryotic defense system (ko02048), antimicrobial resistance (ko01504), and CAMP resistance (ko01503) genes were significantly correlated with Phascolarctobacterium faecium (Q < 0.0001), Bacteroides thetaiotaomicron (Q < 0.0001), Bacteroides dorei (Q < 0.0001), Parabacteroides distasonis (Q < 0.0001), Barnesiella intestinihominis (Q < 0.0001) and Alistipes onderdonkii (Q < 0.0001).
Fig. 5. Network analysis showing the correlations between gut bacterial species and function.
a, b Network analysis of the 20 selected species and associated functional pathways. The nodes between functions were eliminated and only the edges that have significant correlations are shown. The minimum R value cut-off was 0.35. The red color represents species or functions that were less prevalent in Parkinson’s disease (PD) patients, and the blue colors represent those that were more prevalent in PD patients.
Machine learning analysis for the diagnosis of PD
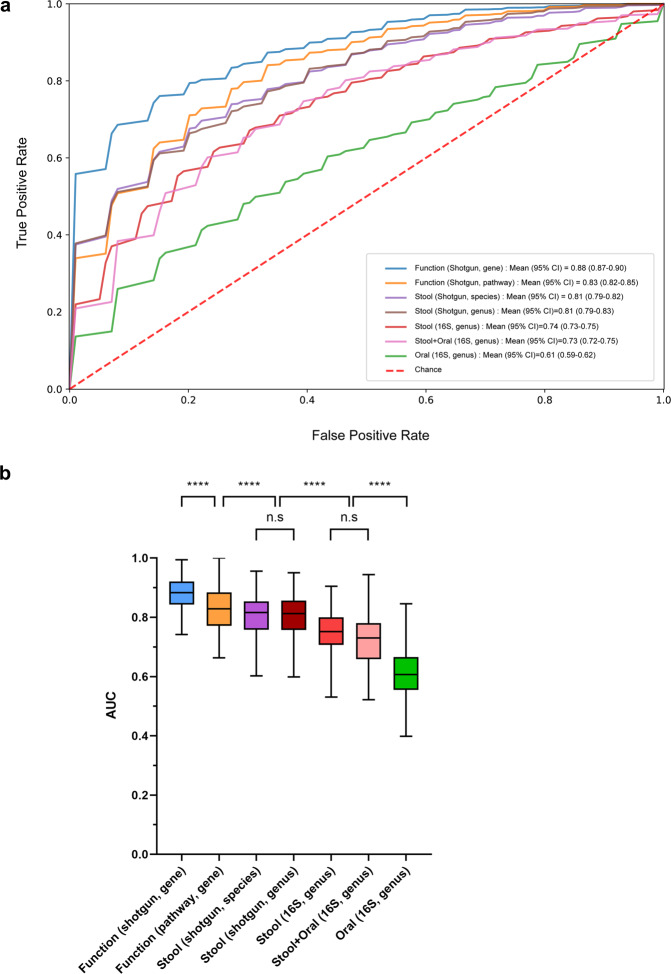
We trained random forest classifiers with bacterial composition (oral, genus level; stool, species and genus level) and function (gene and pathway level). The random forest classifier using gene markers from whole-genome shotgun metagenomic sequencing had the highest area under the curve (AUC) for discriminating PD patients from HCs (0.88, 95% CI 0.87–0.90), followed by the classifiers using functional pathways (AUC = 0.83, 95% CI 0.82–0.85) and species-level taxa (AUC = 0.81, 95% CI 0.79–0.82) from shotgun sequencing (Fig. 6). The most commonly used functional features in the classifier were genes involved in arginine and glutamate biosynthesis, which is consistent with the results of the functional analysis (Supplementary Fig. 6). The classifier using genus-level data from shotgun sequencing had an AUC of 0.81, higher than that using genus-level data from 16 S rRNA gene sequencing (AUC = 0.74, P < 0.0001). Combining the gut and oral microbiome did not significantly improve the discriminatory performance compared to the gut microbiome alone (AUC = 0.73 vs. 0.74, P = 0.63).
Fig. 6. Discriminating Parkinson’s disease from healthy controls based on the oral and gut microbiome.
a Receiver operating characteristic curve of random forest classifier using shotgun or 16 S rRNA gene sequencing. b Comparison of each classifier. Boxplot centerline represents the median (50th percentile). The top and bottom hinges represent 75th and 25th percentiles, respectively. The upper and lower whiskers correspond to the highest and lowest data points. n.s: not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
In the present study, we investigated the connection between the oral and gut microbiome in PD patients, and found that oral Lactobacillus, which was more abundant in PD patients than in HCs, was positively correlated with opportunistic pathogens in the gut. We also identified distinct taxonomic and functional signatures associated with PD. Microbial gene markers for glutamate and arginine biosynthesis were downregulated in PD patients compared with HCs, while antimicrobial resistance genes were upregulated in PD patients. A random forest classifier using the functional gene markers from shotgun metagenomic sequencing was superior to one based on taxonomic markers from 16 S rRNA gene sequencing at distinguishing PD patients from HCs.
The oral microbiome from patients with PD had a distinct taxonomic composition compared with healthy controls. Oral Lactobacillus had the highest LDA score in PD patients than in HCs, and it did not show a significant correlation with confounding factors, including disease duration and dysphagia. In the previous four studies that investigated the oral microbiome in PD and in this study, the one common oral bacteria consistently increased in PD was Lactobacillus14,28–30. The other oral bacteria showed variability among studies; for example, Capnocytophaga was decreased in PD in the study of Pereira et al.28, but it was increased in PD in this study. However, the mechanism by which oral Lactobacillus affects the pathogenesis of PD remains unclear. Commensal bacteria in the oral cavity include Streptococcus, Fusobacterium, Haemophilus, and Prevotella species, while oral Lactobacillus is abundant in dental caries32. Oral pathogens could affect systemic disease through local and systemic inflammation33, or they could reach the stomach from the oral cavity and colonize the intestine34. Interestingly, it was hypothesized that Lactobacillus reuteri might increase the release of alpha-synuclein into the enteric nervous system by increasing the firing rate of mesenteric afferent nerve bundles35,36.
Oral bacteria are poor colonizers in a healthy digestive system13,14. However, oral-associated microbiota were found in the gut of patients with inflammatory bowel disease, colon cancer, and liver cirrhosis12,15,16. We can assume that in pathological circumstances, oral pathogens can colonize the gut, or dead bacteria could affect gut microbiota growth by acting as a nutritional source13,14. In this study, we found that the increase in oral Lactobacillus was associated with an increase in gut pathogens, but only in PD, not in healthy controls. Oral Lactobacillus could migrate to the gut and alter the intestinal environment to promote the colonization of pathogens, as it may do in gastrointestinal disease12,15,16. This implies an oral-gut microbiome connection in pathological conditions involving damage to the enteric nervous system and decreased gastrointestinal motility, as is found in PD. The oral microbiota is an attractive diagnostic and therapeutic target because it is easily accessible and modifiable. Additional studies are required to elucidate the association between oral bacteria, especially Lactobacillus species, and the pathogenesis of PD.
Oral Lactobacillus and stool Lactobacillus did not show a significant association in either PD or HCs. This might be explained by the fact that the main Lactobacillus species in the oral cavity and the intestine are different, even though Lactobacillus is a facultative anaerobe that could survive in the intestine. For example, Lactobacillus rhamnosus, Lactobacillus casei, Lactobacillus fermentum, and Lactobacillus salivarius are oral Lactobacillus species most commonly found in healthy young people32, while Lactobacillus paracaesei, Lactobacillus ruminis, and Lactobacillus casei are gut lactobacillus species most commonly observed in healthy older adults37. Because stool Lactobacillus was higher in PD, and it showed a significant correlation with disease severity, oral and stool Lactobacillus would be expected to show some correlation when using species-level analysis.
The microbiome is an ecosystem in which numerous bacterial taxa exist, and it maintains stability and resilience through functional redundancy23,38,39. Although current approaches to studying the PD microbiome are based on determining the bacterial composition, the function of the ecosystem may not change in responses to differences in the bacterial composition. This is reflected in the result of the random forest classifier in the present study, which showed that gene markers and functional pathways had better discriminating performance than taxonomic composition. Therefore, functional approaches are important when investigating the effect of the microbiome on systematic diseases.
We indirectly compared the function of the oral microbiome using 16 S rRNA gene sequencing between patients with PD and healthy controls, but the results were not statistically significant after adjusting for multiple comparisons. Beta-glucoside operon transcriptional anti-terminator showed higher trends in the oral samples of PD. It is produced by many species in the oral microbiome, and Lactobacillus has high beta-glucosidase activity40,41. Orally secreted beta-glucosidase was associated with halitosis and dental biofilm, possibly mediated through a volatile organic compound.
Whole-genome shotgun metagenomic sequencing enabled precise functional analysis of the gut microbiome in PD patients, which is not possible using traditional 16 S RNA sequencing data. Gut microbial genes in PD patients were related to high consumption and low production of glutamate and arginine, which were also the most frequently used features in the random forest classifier. Decreased glutamate in the serum and stool samples of patients with PD has been observed in previous studies25,42,43. Intestinal cells use glutamate to produce energy and protect the mucosa through glutathione, and decreased glutamate may thus contribute to mucosal damage. In addition, glutamate in the gut environment reduces inflammation by inducing the differentiation of naïve T cells into regulatory T cells44. Arginine reduces gut inflammation and pathology, and is important for normal brain function45. The upregulation of antimicrobial resistance genes observed in PD patients suggests prior infection and the use of antibiotics, and this is consistent with a previous finding that the use of antibiotics increases the risk of PD46. CAMP is an essential defense system for the host against infection47. The upregulation of CAMP resistance genes in the gut microbiome of PD implies increased antimicrobial peptide secretion and inflammation in the host. The gut inflammation promotes alpha-synuclein aggregation in the enteric nerve, or conversely, the accumulation of alpha-synuclein may provoke inflammation and dysbiosis48. Therefore, our data showed functional alteration of the gut microbiome in PD, which could contribute to the pathogenesis of PD.
Shotgun metagenomic sequencing detected PD-associated microbial species that were not found by 16 S rRNA gene sequencing. The PD-associated microbial species included pathogenic or proinflammatory species, such as Alistipes onderdonki, Bacteroides dorei, and Parabacteroides merdae. Alistipes onderdonki was found in the stool of patients with multiple system atrophy, suggesting its role in α-synucleinopathy49. Bacteroides dorei was associated with weight loss and autoimmune disease, and Parabacteroides merdae was enriched in colorectal cancer50. In addition, we found a significant species-level difference in the Prevotella genus between PD patients and HCs using shotgun sequencing. There are more than 40 species in this genus51. In previous studies, Scheperjans et al. found that the Prevotellaceae family was decreased in PD52 and Petrov et al. found that Prevotella copri was decreased in PD53. On the other hand, Wallen et al. reported that the Prevotella_9 genus, including Prevotella copri, was decreased in PD, but the less common genus Prevotella was increased in PD54. Therefore, the analysis accuracy depends on the phylogenetic resolution. We found that Prevotella copri was less abundant in PD patients than in HCs and it was associated with decreased arginine and glutamate metabolism. To date, little is known about whether Prevotella copri significantly affects the pathogenesis of PD. Prevotella is usually abundant in the stool of healthy Asian people, and it benefits the host by helping to digest a high-fiber diet55. The role of Prevotella copri in PD in association with amino acid metabolism requires further investigation, especially in Asian populations.
By directly comparing the discriminatory ability of shotgun and 16 S rRNA gene sequencing, we found that shotgun metagenomic sequencing performs better at discriminating PD patients from HCs. Previous studies have investigated the gut microbiome in PD patients using 16 S rRNA gene sequencing25, and only a small number of recent studies have used shotgun metagenomic sequencing for PD24,56. One study found a mean AUC of 0.92 when discriminating PD patients from HCs using gut metagenomics-derived gene markers24, which is comparable to our maximum AUC of 0.88. We found that gene markers and functional pathways determined using shotgun sequencing had better discriminatory performance than the taxonomic composition, which might be explained by the high levels of functional redundancy in the microbiota23,38,39. In addition, the taxonomic composition derived from shotgun sequencing showed better discriminatory performance than the taxonomic composition derived from 16 S rRNA gene sequencing. These results support the superiority of shotgun metagenomic sequencing over 16 S rRNA gene sequencing for identifying relevant changes to the PD-associated gut microbiota.
The present study has some limitations. First, although our 16 S rRNA gene data revealed significant differences in the oral microbiome between PD patients and HCs, shotgun metagenomic sequencing was performed only on stool samples. Because the gut microbiome showed better discriminating performance for PD than the oral microbiome, whole-genome shotgun metagenomic sequencing was performed on the gut microbiome, considering the cost-effectiveness. Further studies using shotgun metagenomics are therefore required to identify the oral microbial functions and species-level composition. Second, we did not investigate the metabolites associated with the identified gut and oral microbiota. To support the results of our functional analysis on the significant alterations in glutamate and arginine metabolism, these metabolites should be investigated in stool samples.
In conclusion, the present study identified a distinctive connection between the oral and gut microbiota, which might lead to functional alterations of the PD-associated microbiome.
Methods
Study participants
In this case-control study, we prospectively enrolled patients with PD using the UK PD Society brain bank clinical diagnostic criteria57 and their spouses as HCs at Asan Medical Center from 2019 to 2020. The patients’ spouses were selected as HCs because they share common environmental factors. The exclusion criteria were as follows: (1) participants with inflammatory bowel diseases; (2) participants with a history of acute inflammatory or infectious disease within one month prior to participation; (3) participants using antibiotics, steroids, or immunosuppressants; (4) participants who underwent surgery on their gastrointestinal tracts or oral cavity; (5) participants using artificial nutrition; (6) participants who had undergone deep brain stimulation; and (7) participants diagnosed with PD dementia.
Ethics
This study was approved by the Asan Medical Center Institutional Review Board (2019-0929) and was performed in accordance with the relevant guidelines and regulations, including the Declaration of Helsinki. All participants provided written informed consent at study enrollment.
Clinical evaluation
We assessed the baseline characteristics of the cohort, including age, sex, and body mass index. Diet was assessed using a semi-quantitative food frequency questionnaire58. Irritable bowel syndrome and constipation were assessed using the ROME III diagnostic criteria59. Dysphagia was assessed using a swallowing disturbance questionnaire60, and olfactory function was assessed using a scent survey for screening (SSS) test61. Motor function was assessed using the Unified PD Rating Scale (UPDRS) and Hoehn and Yahr (H&Y) stage in the medication-off state.
Preparation of oral and stool samples
Oral swabs and stool samples of patients with PD and their spouses were collected for microbial community analysis. For the oral swab samples, the buccal area was swabbed with an eSwab kit (COPAN Diagnostics Inc., California, USA). A stool sampling kit (CJ Bioscience Inc., Seoul, Korea) was used to collect stool samples. Conventional 16 S rRNA gene sequencing was performed for both the oral and gut microbiome. Because we found that the gut microbiome showed better performance when discriminating PD from controls than the oral microbiome, whole-genome shotgun metagenomic sequencing was only performed on the gut microbiome, considering the cost-effectiveness.
16 S rRNA gene sequencing, taxonomic profiling, and functional profiling
The V3-4 hypervariable region of the 16 S rRNA gene was amplified with primers 341 F and 805 R using the direct PCR method. Libraries were prepared using an NEBNext Ultra II FS DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA). The prepared DNA libraries were sequenced by CJ Bioscience Inc. (Seoul, Korea) using the Illumina Miseq platform (Illumina, San Diego, CA, USA) with 2 × 300 bp kits.
The paired end raw 16 S rRNA sequences data were uploaded to the EzBioCloud and processed using a web-based EzBioCloud microbiome taxonomic profile tool (https://www.ezbiocloud.net/contents/16smtp). High-quality sequence reads were assigned to “species group” at 97% sequence similarity using the PKSSU4.0 database. The prediction of functional biomarkers of the oral microbiota was performed using the PICRUSt with EzBioCloud MTP server62.
Whole-genome shotgun metagenomic sequencing
Whole-genome shotgun metagenomic libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit and the NEBNext Multiplex Oligos for Illumina (New England Biolabs, Ipswitch, USA), according to the manufacturer’s protocols. Fragment size and DNA concentration in the final library were checked using a Bioanalyzer system (Agilent Technologies, Santa Clara, USA) before sequencing using an Illumina NovaSeq 6000 platform (2×150 bp read length) at Macrogen (Seoul, Korea).
Taxonomic profiling of shotgun metagenomics data
A Kraken2 database63 containing bacterial and archaeal species represented in the EzBioCloud database was generated64. For each species, 92 core genes were extracted using the UBCG pipeline65. The total core gene length for each species was stored for further downstream analysis. A Kraken2-compatible taxonomic structure was constructed using EzBioCloud’s taxonomic system, and the core gene sequences were converted into FASTA files using a numerical identifier matching the taxonomic structure file. Finally, the database was compiled with the Kraken2-build command using a k of size 35 and default parameters.
The potential presence of bacterial and archaeal species for each raw metagenomic sample read was initially surveyed using the pre-built Kraken2 core gene database66. After acquiring a list of candidate species, a custom Bowtie2 database was built, utilizing only the core genes from the species found during the first step to reduce the search space and obtain accurate coverage and depth metrics. The raw sample was then mapped against the Bowtie2 database using the very sensitive option and a quality threshold of phred33. Samtools was used to convert and sort the output BAM file. Coverage of the mapped reads against the BAM file was obtained using Bedtools. To avoid false positives, reads that mapped to a given species were only quantified if the total coverage of their core genes was at least 25% according to an in-house script. Finally, species abundance was calculated using the total number of reads counted, and normalized species abundance was calculated using the total length of core genes per species.
Functional profiling based on shotgun metagenomics data
Functional annotations were obtained by matching each read against the KEGG database67 using DIAMOND68. An initial database file was built from the KEGG fasta file containing the ortholog amino acid sequences using DIAMOND’s makedb command with the default parameters. Then, DIAMOND was executed using the blastx parameter, which converts each metagenomic read into multiple amino acid sequences by generating all six open reading frames and then matching these against the pre-built KEGG database. If a read had multiple KEGG hits, the top hit was used. After quantifying all of the KEGG orthologs, minpath was used to predict the presence of KEGG functional pathways69.
Machine learning for discriminating PD
Using the 16 S rRNA-based sequencing data for the oral and gut microbiome and the whole-genome shotgun sequencing data for the gut microbiome, we developed a random forest classifier for discriminating PD from HCs, using custom python scripts employing the Scikit-learn package70. We trained random forest classifiers with the bacterial composition (oral = genus level; gut = species and genus level) and function (gene and pathway level). The model was trained 20 times using a 5-fold cross-validation method, and the average area under the receiver operating characteristic (ROC) curve was calculated.
Statistical analysis
We compared the baseline demographics, dietary intake, and clinical symptoms between PD patients and the HCs using a χ2 test, Student’s t-test, and Mann–Whitney U test, where appropriate. Significance was set at a P value less than 0.05, and all P values were 2-tailed. The species richness was assessed using Chao1, and diversity indices were calculated using the Shannon matrix. The beta-diversity was calculated using the Bray-Curtis metric. The significance of beta-diversity was assessed using PERMANOVA with QIIME271. In the diversity analyses, all features were used.
To examine the taxonomic and functional differences between PD and HC, we performed LEfSe analysis72, which uses effect size to measure phenotypic differences in metagenomic data, as well as statistical significance. Features with less than 0.01% relative abundance in the data were excluded from the analysis to avoid obtaining biologically meaningless results. In addition, we performed beta-diversity and LEfSe analyses to compare the microbiota between patients whose H&Y stage was less than 3 (mild PD) and patients whose H&Y stage was 3 or more (severe PD).
The variation in each taxonomic profile and function between PD patients and HCs was analyzed using a Mann–Whitney U test. The Benjamini–Hochberg method was used to adjust for multiple testing. Statistical significance was set at an adjusted P value (Q-value) of 0.05. Since the oral microbiome could affect the gut microbiome15, the correlation between the oral and gut microbiome composition was determined using linear regression analysis. We imposed a centered log-ratio transformation on the relative abundance data using the ‘phyloseq’ and ‘microbiome’ R packages73–75.
Canonical correspondence analysis (CCA) was performed to identify the bacterial taxa associated with the clinical symptoms of PD. In CCA, we used three demographic features (age, BMI, and disease duration), three drugs that can affect the microbiota (COMT inhibitor, amantadine, and dopamine agonist), and the clinical symptoms of PD (Bristol stool scale indicating stool firmness, IBS symptoms, dysphagia scale, olfaction, H&Y stage and UPDRS). The top 20 strains from LEfSe analysis were used in the analysis. CCA was performed using XLSTAT software (Addinsoft, Paris). Linear regression analysis was also conducted to highlight the clinically meaningful and statistically significant correlation from the CCA.
Network analysis was performed using the top 20 pathway level functions and the top 20 species level taxa based on the LDA Score. Network maps were generated between bacterial species and functional pathways using QIIME2 SCNIC and visualized in Cytoscape version 3.8.276. Hierarchical all-against-all association testing was performed to find multi-resolution associations between the bacterial taxonomic and functional profiles77.
Supplementary information
Acknowledgements
This study was supported by the Korean Healthcare Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (grant number: HI19C0256), and by the Asan Institute for Life Sciences, Asan Medical Center, Republic of Korea (grant number: 2018-0622). The funding sources had no role in the design of the study, the collection, analysis, and interpretation of the data, or in writing the manuscript.
Author contributions
S.J., W.K., J.H.L., and S.J.C. contributed to the study conceptualization and design. All authors contributed to the acquisition and analysis of data. S.J., W.K., H.J.Y., Y.K.P., J.-W.B., and S.J.C. contributed to drafting of the text and preparation of the figures. All authors read and approved the final manuscript. S.J. and W.K. are co-first authors and equally contributed to the manuscript.
Data availability
The datasets generated during the current study are available in the NCBI repository (https://www.ncbi.nlm.nih.gov) and Sequence Read Archive database under the accession numbers PRJNA742875 and PRJNA743718, respectively. PRJNA742875 contains 16S rRNA gene sequencing data from 356 samples, including 172 stool samples (88 PD and 84 HC), 143 oral samples (74 PD, 69 HC), and 41 nasal samples. PRJNA743718 contains shotgun metagenomic sequencing data from 156 stool samples, reflecting the dataset after quality control filtering. The associated metadata include group identifiers in the “host_status” column, indicating whether each sample is from a Parkinson’s Disease (PD) patient or a healthy control (HC).
Code availability
No custom codes were used. All software and packages, their versions, relevant specification and parameters are stated in the Methods section.
Competing interests
W.K., M.C., and J.H.L. declare no competing non-financial interests, but the following competing financial interests: they are employed by CJ Bioscience, Inc., the developer of the EzBioCloud services. S.J., Y.S.H., S.H.L., K.W.P., M.S.K., H.L., H.J.Y., Y.K.P., H.S., J-Y.L., J-W.B., and S.J.C. declared no competing financial and non-financial interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sungyang Jo, Woorim Kang.
Change history
8/25/2025
The Data Availability statement was missing some details about the dataset and should have read “The datasets generated during the current study are available in the NCBI repository (https://www.ncbi.nlm.nih.gov) and Sequence Read Archive database under the accession numbers PRJNA742875 and PRJNA743718, respectively. PRJNA742875 contains 16S rRNA gene sequencing data from 356 samples, including 172 stool samples (88 PD and 84 HC), 143 oral samples (74 PD, 69 HC), and 41 nasal samples. PRJNA743718 contains shotgun metagenomic sequencing data from 156 stool samples, reflecting the dataset after quality control filtering. The associated metadata include group identifiers in the “host_status” column, indicating whether each sample is from a Parkinson’s Disease (PD) patient or a healthy control (HC)". The article has been corrected.
Supplementary information
The online version contains supplementary material available at 10.1038/s41531-022-00351-6.
References
- 1.GBD 2015 Neurological Disorders Collaborator Group. Global, regional, and national burden of neurological disorders during 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Neurol.16, 877–897 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalia, L. V. & Lang, A. E. Parkinson’s disease. Lancet386, 896–912 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Kim, S. et al. Transneuronal propagation of pathologic α-synuclein from the gut to the brain models Parkinson’s disease. Neuron103, 627–641 e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braak, H., de Vos, R. A., Bohl, J. & Del Tredici, K. Gastric α-synuclein immunoreactive inclusions in Meissner’s and Auerbach’s plexuses in cases staged for Parkinson’s disease-related brain pathology. Neurosci. Lett.396, 67–72 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Bell, J. S. et al. From nose to gut - the role of the microbiome in neurological disease. Neuropathol. Appl. Neurobiol.45, 195–215 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Sampson, T. R. et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell167, 1469–1480 e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tremlett, H. et al. The gut microbiome in human neurological disease: a review. Ann. Neurol.81, 369–382 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Chiang, H. L. & Lin, C. H. Altered gut microbiome and intestinal pathology in Parkinson’s disease. J. Mov. Disord.12, 67–83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopfner, F. et al. Gut microbiota in Parkinson disease in a northern German cohort. Brain Res1667, 41–45 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Malkki, H. Parkinson disease: could gut microbiota influence severity of Parkinson disease? Nat. Rev. Neurol.13, 66–67 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Mridula, K. R. et al. Association of Helicobacter pylori with Parkinson’s disease. J. Clin. Neurol.13, 181–186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khor, B. et al. Interconnections between the oral and gut microbiomes: reversal of microbial dysbiosis and the balance between systemic health and disease. Microorganisms9, 496 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seedorf, H. et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell159, 253–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleury, V. et al. Oral Dysbiosis and Inflammation in Parkinson’s Disease. J. Parkinsons Dis.11, 619–631 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas, A. M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med.25, 667–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin, N. et al. Alterations of the human gut microbiome in liver cirrhosis. Nature513, 59–64 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Nishiwaki, H. et al. Meta-analysis of gut dysbiosis in Parkinson’s disease. Mov. Disord.35, 1626–1635 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Sharpton, T. J. An introduction to the analysis of shotgun metagenomic data. Front. Plant Sci.5, 209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poretsky, R. et al. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE9, e93827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, J. S. et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun.10, 5029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranjan, R. et al. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun.469, 967–977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laudadio, I. et al. Quantitative assessment of shotgun metagenomics and 16S rDNA amplicon sequencing in the study of human gut microbiome. OMICS22, 248–254 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Louca, S. et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol.2, 936–943 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Qian, Y. et al. Gut metagenomics-derived genes as potential biomarkers of Parkinson’s disease. Brain143, 2474–2489 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Romano, S. et al. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. NPJ Parkinsons Dis.7, 27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren, T. et al. Gut microbiota altered in mild cognitive impairment compared with normal cognition in sporadic Parkinson’s disease. Front. Neurol.11, 137 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, F. et al. Altered gut microbiota in Parkinson’s disease patients/healthy spouses and its association with clinical features. Parkinsonism Relat. Disord.81, 84–88 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Pereira, P. A. B. et al. Oral and nasal microbiota in Parkinson’s disease. Parkinsonism Relat. Disord.38, 61–67 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Mihaila, D. et al. The oral microbiome of early stage Parkinson’s disease and its relationship with functional measures of motor and non-motor function. PLoS ONE14, e0218252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Z. et al. Oral, nasal, and gut microbiota in Parkinson’s disease. Neuroscience480, 65–78 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Rozas, N. S., Tribble, G. D. & Jeter, C. B. Oral factors that impact the oral microbiota in Parkinson’s disease. Microorganisms9, 1616 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caufield, P. W. et al. Oral lactobacilli and dental caries: a model for niche adaptation in humans. J. Dent. Res.94, 110S–118S (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia, G. et al. The oral microbiota - a mechanistic role for systemic diseases. Br. Dent. J.224, 447–455 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Olsen, I. & Yamazaki, K. Can oral bacteria affect the microbiome of the gut? J. Oral. Microbiol.11, 1586422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunze, W. A. et al. Lactobacillus reuteri enhances excitability of colonic AH neurons by inhibiting calcium-dependent potassium channel opening. J. Cell. Mol. Med.13, 2261–2270 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paillusson, S. et al. Activity-dependent secretion of alpha-synuclein by enteric neurons. J. Neurochem.125, 512–517 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Štšepetova, J. et al. Diversity and metabolic impact of intestinal Lactobacillus species in healthy adults and the elderly. Br. J. Nutr.105, 1235–1244 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Tian, L. et al. Deciphering functional redundancy in the human microbiome. Nat. Commun.11, 6217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moya, A. & Ferrer, M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol24, 402–413 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Teixeira Essenfelder, L. et al. Salivary β-glucosidase as a direct factor influencing the occurrence of halitosis. Biochem. Biophys. Rep.26, 100965 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahajan, P. M., Desai, K. M. & Lele, S. S. Production of cell membrane-bound α- and β-glucosidase by Lactobacillus acidophilus. Food Bioproc. Tech.5, 706–718 (2012). [Google Scholar]
- 42.O’Gorman Tuura, R. L., Baumann, C. R. & Baumann-Vogel, H. Beyond dopamine: GABA, glutamate, and the axial symptoms of Parkinson disease. Front. Neurol.9, 806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vascellari, S. et al. Gut microbiota and metabolome alterations associated with Parkinson’s disease. mSystems5, e00561–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomé, D. The roles of dietary glutamate in the intestine. Ann. Nutr. Metab.73, 15–20 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Qiu, Y. et al. L-Arginine inhibited inflammatory response and oxidative stress induced by lipopolysaccharide via arginase-1 signaling in IPEC-J2 Cells. Int. J. Mol. Sci.20, 1800 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mertsalmi, T. H., Pekkonen, E. & Scheperjans, F. Antibiotic exposure and risk of Parkinson’s disease in Finland: a nationwide case-control study. Mov. Disord.35, 431–442 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Peyrin-Biroulet, L. et al. NODs in defence: from vulnerable antimicrobial peptides to chronic inflammation. Trends Microbiol14, 432–438 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Lema Tomé, C. M. et al. Inflammation and α-synuclein’s prion-like behavior in Parkinson’s disease—is there a link? Mol. Neurobiol.47, 561–574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wan, L. et al. Alterations of the gut microbiota in multiple system atrophy patients. Front. Neurosci.13, 1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng, Q. et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun.6, 6528 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Ashaolu, T. J., Ashaolu, J. O. & Adeyeye, S. A. O. Fermentation of prebiotics by human colonic microbiota in vitro and short-chain fatty acids production: a critical review. J. Appl. Microbiol.130, 677–687 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Scheperjans, F. et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord.30, 350–358 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Petrov, V. A. et al. Analysis of gut microbiota in patients with Parkinson’s disease. Bull. Exp. Biol. Med.162, 734–737 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Wallen, Z. D. et al. Characterizing dysbiosis of gut microbiome in PD: evidence for overabundance of opportunistic pathogens. NPJ Parkinsons Dis.6, 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kovatcheva-Datchary, P. et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab.22, 971–982 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Bedarf, J. R. et al. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med9, 39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hughes, A. J., Daniel, S. E., Kilford, L. & Lees, A. J. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry55, 181–184 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Na, Y. J. & Lee, S. H. Development and validation of a quantitative food frequency questionnaire to assess nutritional status in Korean adults. Nutr. Res. Pract.6, 444–450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rome Foundation. Guidelines–Rome III diagnostic criteria for functional gastrointestinal disorders. J. Gastrointestin. Liver Dis.15, 307–312 (2006). [PubMed] [Google Scholar]
- 60.Manor, Y. et al. Validation of a swallowing disturbance questionnaire for detecting dysphagia in patients with Parkinson’s disease. Mov. Disord.22, 1917–1921 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Yang, Y. et al. Clinical feasibility of scent survey for screening test for olfactory function. Article in Korean. J. Rhinol.25, 14–20 (2018). Korean. [Google Scholar]
- 62.Langille, M. G. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol.31, 814–821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol.20, 257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon, S. H. et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol67, 1613–1617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Na, S. I. et al. UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol.56, 280–285 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Chalita, M. et al. Improved metagenomic taxonomic profiling using a curated core gene-based bacterial database reveals unrecognized species in the genus. Streptococcus. Pathog.9, 204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanehisa, M. et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Ye, Y. & Doak, T. G. A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Comput. Biol.5, e1000465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res.12, 2825–2830 (2011). [Google Scholar]
- 71.Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol.37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol.12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lê Cao, K. A. et al. MixMC: a multivariate statistical framework to gain insight into microbial communities. PLoS ONE11, e0160169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lahti, L. & Shetty, S. Tools for microbiome analysis in R. Microbiome package version 1.15.1. https://github.com/microbiome/microbiome (2017). (Accessed 5 October 2021).
- 76.Kohl, M., Wiese, S. & Warscheid, B. Cytoscape: software for visualization and analysis of biological networks.in Data mining in proteomics (eds Hamacher, M., Eisenacher, M. & Stephan, C.) 291-303 (Humana Press, Totowa, NJ, 2011). [DOI] [PubMed]
- 77.Rahnavard, G. et al. High-sensitivity pattern discovery in large multiomic datasets. https://huttenhower.sph.harvard.edu/halla/ (2017). (Accessed 5 October 2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the NCBI repository (https://www.ncbi.nlm.nih.gov) and Sequence Read Archive database under the accession numbers PRJNA742875 and PRJNA743718, respectively. PRJNA742875 contains 16S rRNA gene sequencing data from 356 samples, including 172 stool samples (88 PD and 84 HC), 143 oral samples (74 PD, 69 HC), and 41 nasal samples. PRJNA743718 contains shotgun metagenomic sequencing data from 156 stool samples, reflecting the dataset after quality control filtering. The associated metadata include group identifiers in the “host_status” column, indicating whether each sample is from a Parkinson’s Disease (PD) patient or a healthy control (HC).
No custom codes were used. All software and packages, their versions, relevant specification and parameters are stated in the Methods section.