Abstract
We analyzed the composition of aggregate (lake snow)-associated bacterial communities in Lake Constance from 1994 until 1996 between a depth of 25 m and the sediment surface at 110 m by fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes of various specificity. In addition, we experimentally examined the turnover of dissolved amino acids and carbohydrates together with the microbial colonization of aggregates formed in rolling tanks in the lab. Generally, between 40 and more than 80% of the microbes enumerated by DAPI staining (4′,6′-diamidino-2-phenylindole) were detected as Bacteria by the probe EUB338. At a depth of 25 m, 10.5% ± 7.9% and 14.2% ± 10.2% of the DAPI cell counts were detected by probes specific for α- and β-Proteobacteria. These proportions increased to 12.0% ± 3.3% and 54.0% ± 5.9% at a depth of 50 m but decreased again at the sediment surface at 110 m to 2.7% ± 1.4% and 41.1% ± 8.4%, indicating a clear dominance of β-Proteobacteria at depths of 50 and 110 m, where aggregates have an age of 3 to 5 and 8 to 11 days, respectively. From 50 m to the sediment surface, cells detected by a Cytophaga/Flavobacteria-specific probe (CF319a) comprised increasing proportions up to 18% of the DAPI cell counts. γ-Proteobacteria always comprised minor proportions of the aggregate-associated bacterial community. Using only two probes highly specific for clusters of bacteria closely related to Sphingomonas species and Brevundimonas diminuta, we identified between 16 and 60% of the α-Proteobacteria. In addition, with three probes highly specific for close relatives of the β-Proteobacteria Duganella zoogloeoides (formerly Zoogloea ramigera), Acidovorax facilis, and Hydrogenophaga palleroni, bacteria common in activated sludge, 42 to 70% of the β-Proteobacteria were identified. In the early phase (<20 h) of 11 of the 15 experimental incubations of aggregates, dissolved amino acids were consumed by the aggregate-associated bacteria from the surrounding water. This stage was followed by a period of 1 to 3 days during which dissolved amino acids were released into the surrounding water, paralleled by an increasing dominance of β-Proteobacteria. Hence, our results show that lake snow aggregates are inhabited by a community dominated by a limited number of α- and β-Proteobacteria, which undergo a distinct succession. They successively decompose the amino acids bound in the aggregates and release substantial amounts into the surrounding water during aging and sinking.
Macroscopic organic aggregates, known as marine snow and lake snow aggregates, have been identified as microcenters of the recycling of nutrients in pelagic ecosystems (21, 22, 28, 54, 59) and as important components of the sinking flux of particulate organic matter (3, 18, 23). They consist of living, senescent, and dead algae, zooplankton molts, and carcasses and of unidentifiable organic and inorganic debris. The numbers of microbial cells and substrate concentrations on aggregates are greatly enhanced compared to the surrounding water (21, 22, 40, 50, 55), reflecting the high microbial activity of these hot spots in an otherwise often nutrient-poor pelagic environment. The high microbial activity may even lead to anoxic conditions in the center of the aggregates (2, 43).
Despite many studies on the microbial colonization and nutrient dynamics of marine snow and lake snow aggregates, surprisingly little is known about the composition of the aggregate-associated microbial community. On the basis of 16S rRNA gene sequences, fairly high diversities differing also from that of the bacterial community in the surrounding water were found on particle- and aggregate-associated bacterial communities in various marine systems and an estuary (1, 14, 16, 41, 44). Weiss et al. (62) and Grossart and Simon (21), applying fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes, found that bacterial communities on lake snow aggregates are largely dominated by β-Proteobacteria.
Examining the composition of microbial communities with phylogenetically based oligonucleotide probes, however, indicates only in rare cases specific physiological traits of the population of interest, e.g., ammonia oxidation by Nitrosomonas/Nitrosospira spp. within the β-Proteobacteria (41). To better understand the functional response of the dominating members of the heterotrophic microbial communities on macroscopic aggregates, it would be of great importance to study their growth dynamics simultaneously with the turnover of the major labile organic substrates on aggregates, dissolved amino acids and carbohydrates.
The aim of this study was to examine the microbial colonization of lake snow aggregates in mesotrophic Lake Constance, Germany, by FISH together with dynamics of the turnover of dissolved amino acids and carbohydrates. The results show that a specialized bacterial community, dominated only by a few closely related phylogenetic clusters of α- and β-Proteobacteria, and later on of Cytophaga/Flavobacteria, established on the aggregates. High and increasing proportions of β-Proteobacteria were usually associated with a release of dissolved amino acid into the surrounding water.
MATERIALS AND METHODS
Study site and sampling.
Upper Lake Constance is a mesotrophic and warm monomictic prealpine lake with a surface area of 472 km2 and maximum and mean depths of 253 and 101 m, respectively. The lake has been studied intensively in the recent past (20, 25, 27, 51) and also with respect to the significance of lake snow aggregates (21, 22, 23, 24, 62). The study was carried out between June 1994 and December 1996. In 1995 and 1996 the abundance of lake snow aggregates at depths between 10 and 30 m was determined on photographs taken in situ with a Nikonos V (Nikon) underwater camera system as described by Grossart et al. (24). Photographs were also used to measure the size of the aggregates from which their volumes were calculated assuming spherical shape. From June to August 1994 and from May to September 1996, lake snow aggregates were sampled at depths of 10 to 30 m in the center of Lake Überlingen, a northwestern fjord-like arm of Lake Constance with maximum and mean depths of 147 and 90 m, respectively. Between 10 April and 20 August 1995, lake snow aggregates were sampled at a station in the southern area of Lake Constance near Romanshorn, Switzerland, at depths of 10 to 30 m and 50 m and at the sediment surface at 110 m. We chose this site because of its low allochthonous impact and low advective water movement (7). Lake snow aggregates at depths between 10 and 30 m were collected by scuba divers with plastic syringes (59). Samples of sinking lake snow aggregates at a depth of 50 m were collected by a conical sediment trap (Aquatec Kiel) exposed for 6 h. This short period ensured that freshly collected particulate organic matter (POM), decomposed only slightly in the sediment trap, was sampled. It also ensured that the bacterial community on the settled material still resembled closely the community found on aggregates at this depth. Grossart and Simon (23) estimated that lake snow aggregates contributed 40 to 60% to the sinking POM at 25 m. Therefore, it was justified to assume that the majority of POM collected in the sediment trap was of lake snow origin. Surface sediment samples were collected from a depth of 110 m with a Plexiglas sediment corer. Based on the assumption that the uppermost layer of the sediment core consists of very recently settled lake snow aggregates, we carefully sampled the top layer of the sediment core with a pipette. All samples were stored in a cooler, brought to the lab, and frozen at −20°C until further processing.
Experimental design.
Experiments were carried out to examine the microbial colonization of aggregates over time, in most cases together with dynamics of dissolved free and combined amino acids (DFAA and DCAA) and/or free and combined carbohydrates (DFCHO and DCCHO). Therefore, lake snow aggregates were formed from fresh samples of a depth of 6 m transferred into 1.2-liter Plexiglas cylinders rolling horizontally at 2 rpm according to the technique of Shanks and Edmondson (49). The samples were incubated at in situ temperature at ambient light conditions until aggregates of 3 mm or larger had formed, usually within 24 h. In total 16 experiments were carried out between 1994 and 1996, but bacterial production, uptake of DFCHO and DFAA, and aminopeptidase activity (see below) were measured only in two experiments. To study the release of DFAA, DCAA, DFCHO, and DCCHO into the surrounding water or consumption of these substrates from it, the aggregates were transferred to 50-ml glass syringes filled with particle-free (0.2-μm [pore-size]- filtered) lake water. Eight or nine syringes were filled with one aggregate each. One syringe filled only with particle-free water was used as a control. The syringes were rotated vertically at 5 rpm and were incubated in the dark at the in situ temperature at 25 m depth (5 to 10°C) to simulate the conditions of aggregates sinking through the upper hypolimnion. Periodically, 5-ml water samples were withdrawn from the control syringe and from one syringe with an aggregate to measure the change of concentration of dissolved amino acids (DAA) and carbohydrates (DCHO) over time. At each subsampling, one aggregate from another syringe was also collected to examine the composition of the aggregate-associated bacterial community. This sampling design was only valid under the assumption that the size and composition of the aggregates and the composition of the aggregate-associated bacterial community was similar on all aggregates at a given time. In order to test this assumption, we examined the composition of the bacterial community by FISH and group-specific probes on three aggregates of the same origin but incubated separately in syringes over 41 h. In seven subsamples collected over this period the proportions of α-, β-, and γ-Proteobacteria of the triplicate samples agreed within 10% of the mean even though the proportions of α- and β-Proteobacteria varied from 15 to 28% and from 33 to 62% of the DAPI (4′,6′-diamidino-2-phenylindole)-stainable cells, respectively.
In situ hybridization.
For the analysis by FISH (6), we used probes specific for Bacteria, the α-, β-, and γ-subclasses Proteobacteria (37), and the cluster Cytophaga/Flavobacteria (36) and probes targeting 16S ribosmal DNA (rDNA) clones from lake snow aggregates (Table 1) (30). The probes were linked to either of the fluorochromes 5,(6)-carboxyfluorescein-N-hydroxysuccinimidester (Fluos), tetramethyl rhodamine-5,6-isothiocyanate (TRITC), or Cy3 (derivate of succinimidester Cy3 of a cyanine). For in situ hybridization, samples were dried and fixed with a 4% paraformaldehyde solution on Teflon-coated glass slides and processed as described previously (62). In situ hybridization was performed at 46°C for 90 min. The hybridization buffer contained 0.9 M NaCl, either 20 or 35% formamide depending on the stringency (Table 1), 20 mM Tris-HCl (pH 7.4), and 0.01% sodium dodecyl sulfate (SDS). The concentration of each probe was 50 ng μl−1. Probes for BET42a and GAM42a have to be used with a competitor oligonucleotide (37, 57). To stop hybridization, the slides were rinsed and incubated at 46°C for 15 min in washing buffer containing 180 or 40 mM NaCl for 20 and 35% formamide hybridization, respectively; 20 mM Tris-HCl (pH 7.4); and 0.01% SDS. After being rinsed with distilled water, the slides were dried and stained with DAPI (0.01%). Finally, the samples were embedded in Citifluor (Citifluor, Ltd., Canterbury, United Kingdom). The samples were visualized by an epifluorescence microscope (Labophot 2A; Nikon) equipped with the Nikon filter sets UV-2A (DAPI), B-2A (Fluos), G-2A (TRITC), and XF32 NM198 (Omega Optical, Inc.) for Cy3. At least 500 to 600 bacteria on 10 viewfields per sample were counted in triplicates. The average coefficient of variation ranged between 2 and 15%.
TABLE 1.
Description of oligonucleotide probes used to examine the composition of bacterial communities on lake snow aggregates
| Probe | Target group | Sequence (5′ → 3′)a | % Formamideb | Reference |
|---|---|---|---|---|
| EUB338 | Bacteria | GCT GCC TCC CGT AGG AGT | 20 | 5 |
| ALF1b | α-Proteobacteria | CGT TCG (C/T)TC TGA GCC AG | 20 | 37 |
| BET42a | β-Proteobacteria | GCC TTC CCA CTT CGT TT | 35 | 37 |
| GAM42a | γ-Proteobacteria | GCC TTC CCA CAT CGT TT | 35 | 37 |
| CF319a | Cytophaga/Flavobacteria | TGG TCC GTG TCT CAG TAC | 35 | 36 |
| LSA67 | Clones and isolates from lake snow closely related to B. diminuta | GGA CCT TTC GGG GTT AGT | 35 | 30 |
| LSA225 | Sphingomonas sp. and relatives | TCC TAC GCG GGC TCG TCC | 35 | 30 |
| LSA644 | Clones and isolates from lake snow within the genus Sphingomonas | CCA GGA TTC AAG CAA TCC | 35 | 30 |
| SNA23a | S. natans and relatives | CAT CCC CCT CTA CCG TAC | 35 | 61 |
| LSB65 | D. zoogloeoides and relatives | GTT GCC CCG CGC TGC CGT | 35 | 30 |
| LSB70 | Hydrogenophaga sp. and relatives | AGC ACC TTG CGG CCT GTT | 35 | 30 |
| LSB145 | A. facilis and relatives | CTT TCG CTC CGT TAT CCC | 35 | 30 |
Nucleotide sequences according to the work of Brosius et al. (12).
Concentrations of formamide used for hybridization.
Substrate analysis.
Dissolved amino acids and carbohydrates were measured during the lab experiments in which the bacterial colonization of aggregates over time was examined. DFAA were analyzed by high-performance liquid chromatography (HPLC) after precolumn derivatization with ortho-phthaldialdehyde according to the method of Lindroth and Mopper (33) as modified by Simon and Rosenstock (52). DCAA were analyzed as the DFAA after hydrolysis with 6 N HCl for 1 h at 155°C (35).
DFCHO were analyzed by HPLC and pulsed amperometric detection according to the method of Mopper et al. (39) on a Carbopac PA-1 column (Dionex) slightly modified using 16 N NaOH as the eluent and at 10°C. (For further details, see the study by Bunte and Simon [12]). DCCHO were hydrolyzed by 0.1 M HCl for 20 h (26) and analyzed as monomers.
Bacterial biomass production and substrate uptake.
Biomass production of the aggregate-associated bacteria was measured by determining the uptake of 14C-labeled leucine according to the methods of Kirchman et al. (32) and Simon and Azam (53). One lake snow aggregate with a minimum volume of surrounding water was collected by a pipette with a cutoff 1-ml plastic tip, diluted in 400 μl of particle-free lake water, and sonicated for 5 s. Four 100-μl aliquots were transferred into glass test tubes filled with 2.9 ml of 0.2-μm (pore-size)-filtered water. The samples were labeled with [14C]-leucine (310 mCi mmol−1; Amersham) at a final concentration of 60 nM (21). One sample served as a blank and was fixed immediately with Formalin (2% final concentration). The three other samples were incubated for 1 h in the dark at in situ temperature and fixed thereafter. The samples were filtered onto 0.45-μm (pore-size) nitrocellulose filters (Sartorius), extracted with ice-cold 5% trichloroacetic acid (TCA) for 5 min, rinsed once with 5% TCA and twice with 80% ethanol, and radioassayed by liquid scintillation counting. Bacterial production was calculated according to the method of Simon and Azam (53) assuming a twofold isotope dilution of leucine and a partitioning in the protein fraction of 86% of the total macromolecular fraction (52).
The net uptake of DFCHO and DFAA by aggregate-associated bacteria was measured by determining the uptake of a 1:1 mixture of [3H]glucose (specific activity 15 Ci/mmol) and [3H]galactose (specific activity, 31 Ci/mmol) and by a mixture of 15 3H-labeled amino acids (mean specific activity, 34.8 Ci/mmol). All radiochemicals were from Amersham. The radiolabeled DFCHO and DFAA were added at a final concentration of 1 nM to separate the sets of samples. Incubations, filtrations, and radioassays were done in the same way as for leucine incorporation (see above).
Aminopeptidase activity.
The aminopeptidase activity of aggregate-associated bacteria was measured with the fluorogenic substrate analog l-leucine-4-methyl-7-coumarinylamide (Leu-MCA; Fluka, Geneva, Switzerland) according to the method of Hoppe (29) but slightly modified. Triplicates of 100 μl of the sonicated aggregates were added to 900 μl of 0.2-μm-filtered water and incubated at in situ temperature with 100 μM Leu-MCA (final concentration) for 60 min. Then, 1 ml of 0.2-μm-filtered water served as a blank. The fluorescence of the peptidase-cleaved MCA was read at a 365-nm excitation and a 455-nm emission wavelength and translated into units of MCA concentration with a calibration curve.
RESULTS
Composition of the microbial community on natural lake snow aggregates.
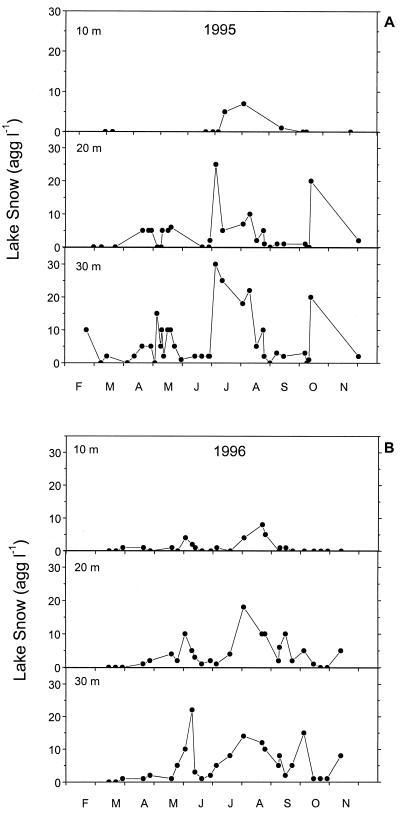
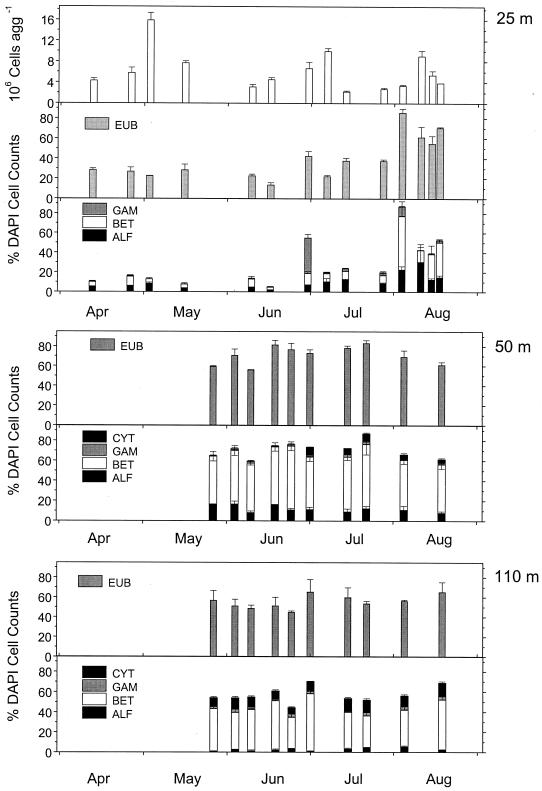
The highest abundances of lake snow aggregates, ranging from <5 to 30 aggregates liter−1, were found during phytoplankton bloom periods such as in May, July, August, and October (Fig. 1). The numbers of aggregates generally increased with depth. The maximum during the spring bloom in May and early June lasted longer in 1995 than in 1996 but was higher in the latter year. In July 1995, the abundance of lake snow aggregates at 30 m was higher than in 1996. Natural lake snow aggregates were densely colonized by microbes. The numbers ranged from 2 × 106 to 16 × 106 cells aggregates liter −1 (Fig. 2). On the basis of the mean volume of lake snow aggregates in Lake Constance of 65 μl, these numbers translate into a volume-specific colonization of 30 × 106 to 246 × 106 cells per ml of aggregate, a 30- to 60-fold denser colonization than in the surrounding water (1 × 106 to 4 × 106 cells ml−1). Between 13 and 100% of the microbes enumerated by DAPI staining were detected by the probe EUB338 specific for Bacteria. In 1994 when we collected only samples at six dates between June and August, this proportion ranged from 55 to 100% of the DAPI cell counts. From April until July 1995 it did not exceed 40% at a depth of 25 m, whereas later in this year and in 1996 proportions of 60 to 82% were detected at this depth (Fig. 2 and 3B). On POM collected in the sediment trap at a depth of 50 m and dominated by lake snow aggregates, we usually detected highest proportions of Bacteria by the EUB338 probe (Table 2), i.e., between 59 and 81% of the DAPI cell counts (Fig. 2). On the sediment surface layer at a 110-m depth proportions of 48 to 62% of the DAPI cell counts were detected by the EUB338 probe (Fig. 2).
FIG. 1.
Abundance of lake snow aggregates in Lake Constance in 1995 at the Romanshorn site and in 1996 at the Lake Überlingen site at 10, 20, and 30 m.
FIG. 2.
Total DAPI cell counts, proportions of Bacteria detected by probe EUB338, proportions of α-, β-, and γ-Proteobacteria detected by probes ALF1b, BET42a, and GAM42a, and proportions of Cytophaga/Flavobacteria detected by probe CF319a on lake snow aggregates in Lake Constance at the Romanshorn site in 1995 (percentage of DAPI cell counts). Samples at a 25-m depth were collected by scuba divers, at a 50-m depth in sediment traps, and at a 110-m depth by a sediment corer.
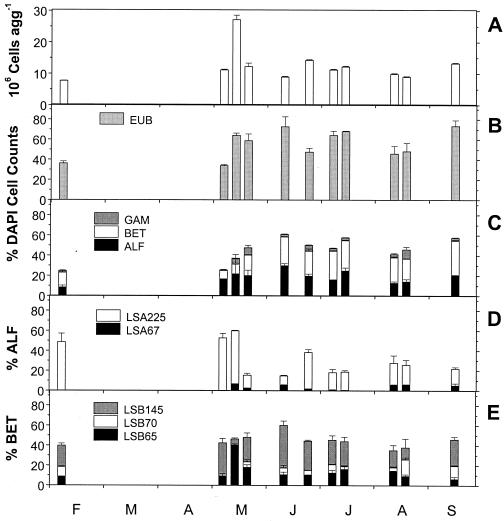
FIG. 3.
Composition of the bacterial community on lake snow aggregates at 25 m in Lake Constance at the Lake Überlingen site in 1996. (A) Total cell numbers per aggregate. (B) Proportions of Bacteria detected by probe EUB338. (C) Proportions of α-, β-, and γ-Proteobacteria detected by probes ALF1b, BET42a, and GAM42a. (D) Percentages of α-Proteobacteria detected by probes LSA67 (several clones of B. diminuta and M. bullata) and LSA225 (specific for Sphingomonas spp., C. subvibrioides, and R. suberifaciens). (E) Percentages of β-Proteobacteria detected by probes LSB65 (D. zoogloeoides and relatives), LSB70 (Hydrogenophaga sp. and relatives), and LSB145 (A. facilis and relatives).
TABLE 2.
Mean proportions of Bacteria, various subclasses of Proteobacteria, and Cytophaga/Flavobacteria detected by FISH on lake snow aggregates in Lake Constance between April and November 1995 at the Romanshorn site at 25 and 50 m and on the surface sediment layer at 110 ma
| Depth (m) | % DAPI cell counts ± SD of:
|
||||
|---|---|---|---|---|---|
| Bacteria | α-Proteobacteria | β-Proteobacteria | γ-Proteobacteria | Cytophaga | |
| 25 | 39.6 ± 21.0 | 10.5 ± 7.9 | 14.2 ± 10.2 | 4.2 ± 9.5 | ND |
| 50 | 70.9 ± 9.4 | 12.0 ± 3.3 | 54.0 ± 5.9 | 1.9 ± 0.7 | 5.8 ± 2.0 |
| 110 | 55.3 ± 6.8 | 2.7 ± 1.4 | 41.1 ± 8.4 | 2.5 ± 0.8 | 10.4 ± 2.1 |
Aggregates at 25 m were collected by scuba divers, at 50 m by a sediment trap, and at 110 m by a sediment corer. For a description and identification of the probes used for FISH, see Table 1. ND, not detected.
In 1995, we examined the composition of the bacterial community on lake snow aggregates at the Romanshorn site by the probes specific for the α-, β-, and γ-Proteobacteria and the Cytophaga/Flavobacteria cluster between a 25-m depth and the sediment surface at 110 m. At 25 m, proportions of α- and β-Proteobacteria were fairly similar until July and ranged between 5 and 15% of the DAPI cell counts, respectively (Fig. 2, Table 2). In August, β-Proteobacteria dominated and comprised proportions of 15 to 60%. Proportions of γ-Proteobacteria remained below 5% except at the end of June. No cells at this depth were detected by the probe specific for the Cytophaga/Flavobacteria cluster. The cumulative proportions of all cells detected by the group-specific probes at a 25-m depth ranged between 8 and 84% of the DAPI cell counts, with a mean of 29%, and between 43 and 100% of cells detected by the Bacteria-specific probe, with a mean of 73%.
At 50 m, β-Proteobacteria always dominated the aggregate-associated bacterial community and ranged between 38 and 64% of the DAPI cell counts (Fig. 2, Table 2), whereas proportions of α-Proteobacteria remained below 20% of the DAPI cell counts, with a mean of 12%. Also, γ-Proteobacteria were detected on aggregates at this depth but comprised only minor proportions (Fig. 2, Table 2). Cells of the Cytophaga/Flavobacteria cluster did not appear until the beginning of July but their proportion remained below 8% of the DAPI cell counts and comprised 5.8% as a mean (Fig. 2, Table 2). The cumulative proportion of bacteria detected by the group-specific probes at a 50-m depth ranged between 58 and 83% of the DAPI cell counts, with a mean of 74%. This proportion matched 100% of the counts by the Bacteria-specific probe.
On the surface sediment layer at a 110-m depth the bacterial community was also largely dominated by β-Proteobacteria, which comprised 31 to 56% of the DAPI cell counts, but cells of the Cytophaga/Flavobacteria cluster comprised a proportion twice as high as at 50 m, up to 18% of the DAPI cell counts (Fig. 2, Table 2). In contrast to the findings at 50 m, cells of this cluster were found on the sediment surface throughout the study period. α- and γ-Proteobacteria were also detected at this depth but comprised proportions of <3% of the DAPI cell counts each (Fig. 2, Table 2). The cell counts of all group-specific probes together comprised 57% of the DAPI cell counts which, as at 50 m, matched 100% of the cell counts by the Bacteria-specific probe.
In 1996, we analyzed the bacterial community on lake snow aggregates collected at 25 m in Lake Überlingen by probes whose nucleotide sequences were derived from 16S rDNA clones amplified directly from lake snow aggregates by PCR (Table 1) (30). These probes are specific for narrow clusters of sequences derived from lake snow aggregates with similarities of >97.5% within β-Proteobacteria closely related to Acidovorax facilis (LSB145), Hydrogenophaga palleroni (LSB70), Duganella zoogloeoides (formerly Zoogloea ramigera ATCC25935; LSB65), and of >96% within α-Proteobacteria closely related to different Sphingomonas species, Caulobacter subvibrioides, Rhizomonas suberifaciens (LSA225), and several clones closely related to Brevundimonas diminuta and Mycoplasma bullata (LSA67). The patterns of aggregate colonization by Bacteria and the α-, β-, and γ-Proteobacteria were similar to those of the previous year, showing also the dominance of α-and β-Proteobacteria, of the latter particularly from June to September (Fig. 3B and C).
The highly specific probes together detected between 14.3 and 52% of the DAPI cell counts with a mean of 30.6%. Probes LSA225 and LSA67 together detected 4.3 to 15.6% of the DAPI cell counts, with a mean of 8.3%, and 16 to 60% of the α-Proteobacteria, with a mean of 34% (Fig. 3D, Table 3). LSA225 always comprised a much larger proportion than LSA67, ranging from 9.2 to 52.3% of the α-Proteobacteria, with a mean of 27.6%. Proportions of LSA67 remained always below 4.1%. The probe LSA644, specific for a subgroup of isolates and clones from lake snow aggregates targeted by probe LSA225, detected 11 to 71% of the bacteria identified by the latter probe (Table 3).
TABLE 3.
Proportions of bacteria detected by various oligonucleotide probes specific for distinct subpopulations of the α-Proteobacteria (LSA67, LSA225, and LSA644) and β-Proteobacteria (LSB65, LSB70, and LSB145) on lake snow aggregates collected at a 25-m depth in Lake Constance (Lake Überlingen) in 1996a
| Date (1995) | α- and β-Proteobacteria, LSB65 + LSB70 + LSB145 + LSA67 + LSA225 (% DAPI cell counts) | α-Proteobacteria, LSA67 + LSA225 (% ALF1b) | α-Proteobacteria, LSA644 (% LSA225) | β-Proteobacteria, LSB65 + LSB70 + LSB145 (% BET42a) |
|---|---|---|---|---|
| 7 April | 18.4 | 57.2 | NA | 42.4 |
| 10 May | 16.4 | 57.4 | NA | 49.6 |
| 17 May | 14.4 | 60.4 | 29.9 | 48.8 |
| 24 May | 26.4 | 17.7 | 10.8 | 57.7 |
| 14 June | 38.2 | 16.1 | 71.0 | 69.7 |
| 28 June | 30.3 | 41.3 | 12.4 | 46.1 |
| 12 July | 33.6 | 21.9 | 41.9 | 53.0 |
| 19 July | 40.9 | 20.3 | 46.0 | 51.6 |
| 16 July | 33.4 | 35.7 | 39.4 | 41.9 |
| 23 July | 32.5 | 31.2 | 40.0 | 50.3 |
| 20 September | 52.5 | 25.6 | 42.2 | 51.7 |
| Mean (SD) | 30.6 (10.9) | 35.0 (16.0) | 37.1 (17.2) | 51.2 (7.3) |
Probes LSB145 (A. facilis and relatives), LSB65 (D. zoogloeoides and relatives), and LSB70 (H. palleroni and relatives) individually detected up to 15.5, 12.4, and 4.5% of the DAPI cell counts, respectively, with means of 9.3, 8.2, and 2.1%. On single aggregates, probes LSB145 and LSB65 together detected up to 23% of the DAPI cell counts and as a mean 13%. Based on the numbers of total β-Proteobacteria, probes LSB145 and LSB65 individually comprised proportions of up to 42.5 and 40.3, respectively, with means of 23.8 and 14.7. All three probes together accounted for 42 to 69.7% of β-Proteobacteria (Fig. 3E, Table 3).
Bacterial colonization and substrate dynamics of lab-made lake snow aggregates.
We examined functional relationships between the structure of the heterotrophic bacterial community on lake snow aggregates and the turnover of the most important labile substrates in experiments with aggregates produced in rolling tanks. Therefore, we determined the composition of the aggregate-associated bacterial community together with the consumption from and release into the surrounding water of dissolved amino acids and carbohydrates. Lab-made aggregates usually were more compact and tended to be larger than natural aggregates. However, as tested in six comparisons made between June and August 1994, the bacterial colonization of the lab-made aggregates that were 1 to 2 days old was not statistically different from that of aggregates collected at a 25-m depth within a few days of the date at which samples were collected at 6 m for forming lab-made aggregates. The mean percentages ± standard deviations of DAPI cell counts of α-, β-, and γ-Proteobacteria on natural aggregates were 16.9 ± 7.1, 41.5 ± 11.9, and 10.5 ± 8.9 compared to 16.5 ± 7.1, 41.8 ± 14.6, and 8.6 ± 2.3 on lab-made aggregates.
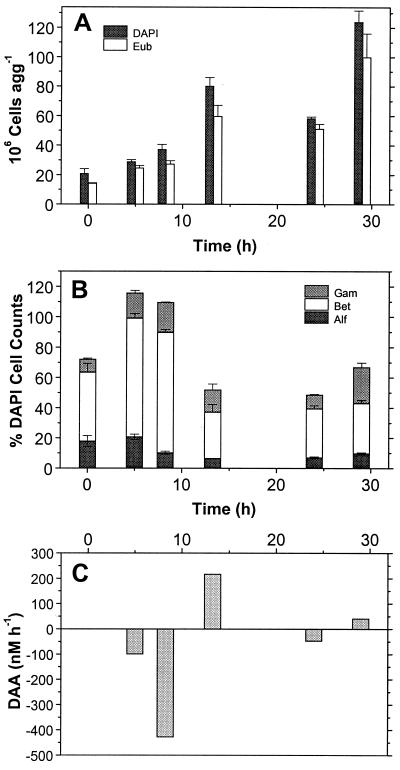
During the course of the experiments, the numbers of DAPI cell counts varied substantially but there was no consistent trend of increasing or decreasing numbers (Fig. 4A, 5A, and 6A). However, in most experiments consistent temporal trends with respect to the relative proportions of α- and β-Proteobacteria occurred. Even though the proportions of α- and β-Proteobacteria during the early phase varied among individual experiments (Fig. 4B, 5B, and 6B), the proportion of α-Proteobacteria decreased in 8 and remained constant in 5 of the 16 experiments and reached average values of <27% toward the end of the experiments, after 20 to 96 h (Table 4). The decrease was statistically significant (t test, P < 0.05) when we compared the mean proportions of the early time points (<20 to 24 h) with those of the late time points (>20 to 24 h). In contrast, the proportions of β-Proteobacteria significantly increased in 11 of 16 experiments (t test, P < 0.05) to proportions of >27% and in 10 experiments even to >36% toward the end of the experiment, irrespective of the dynamics of their absolute numbers (Fig. 4B, 5B, and 6B; Table 4). In the experiment of 9 June 1995, the proportions were already 66% at the beginning and remained constantly high (Fig. 4B, Table 4). In three of the four experiments with decreasing proportions of β-Proteobacteria (9 June 1994, 22 May 1996, 10 August 1996; Fig. 5B), the proportions of α-Proteobacteria also decreased to levels not exceeding those of the former. The temporal dynamics of γ-Proteobacteria, comprising occasionally higher but in most cases lower proportions than α- and β-Proteobacteria, did not show a consistent trend (Table 4). Bacteria of the Cytophaga/Flavobacteria cluster, not enumerated in all experiments, occurred after 20 h and reached up to 15% of the DAPI cell counts after 48 h (data not shown).
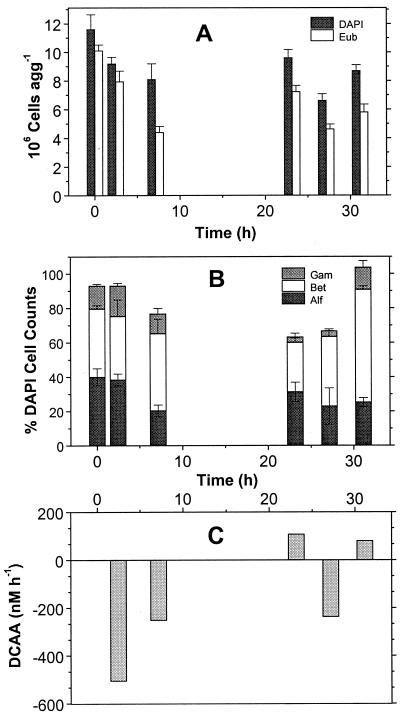
FIG. 4.
Temporal dynamics of aggregate-associated bacteria and DCAA on lab-made aggregates produced in rolling tanks on 9 May 1995 of water from a 6-m depth. (A) Numbers of total cells agg−1 and of Bacteria detected by probe EUB338. (B) Percentages of α-, β-, and γ-Proteobacteria detected by probes ALF1b, BET42a, and GAM42a. (C) Consumption by aggregate-associated bacteria (negative scale) and release of DCAA from aggregates into the surrounding water (positive scale).
FIG. 5.
Temporal dynamics of aggregate-associated bacteria and DCAA on lab-made aggregates produced in rolling tanks on 22 May 1996 of water from 6 m. For definitions, see the legend to Fig. 4.
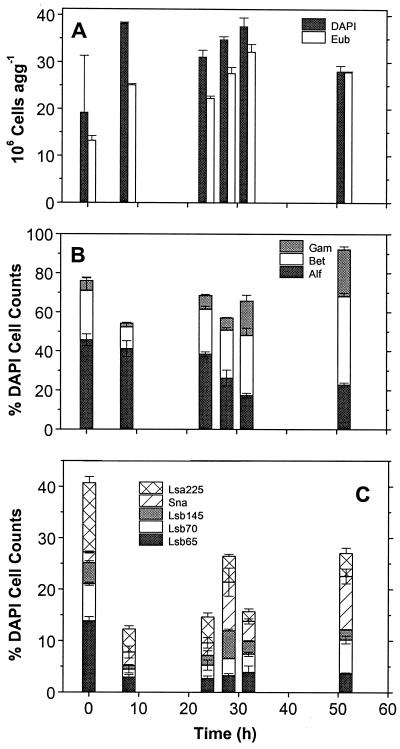
FIG. 6.
Temporal dynamics of aggregate-associated bacteria on lab-made aggregates produced in rolling tanks on 24 November 1996 of water from 6 m. (A) Numbers of total cells per aggregate and of Bacteria detected by the probe EUB338. (B) Percentages of α-, β-, and γ-Proteobacteria detected by probes ALF1b, BET42a, and GAM42a. (C) Percentages of α-Proteobacteria detected by probe LSA225 and of β-Proteobacteria detected by probes SNA23, LSB65, LSB70, and LSB145.
TABLE 4.
Temporal dynamics of the colonization of laboratory-made lake snow aggregates by α-, β-, and γ-Proteobacteria detected by probes ALF1b, BET42a, and GAM 42a
| Date | Time (h) | No. of time points | % DAPI counts ± SDa of:
|
||
|---|---|---|---|---|---|
| α-Proteobacteria | β-Proteobacteria | γ-Proteobacteria | |||
| 1 June 1994 | 0 | 3 | 42.0 ± 8.8 | 21.1 ± 5.5 | 34.0 ± 8.2 |
| 72–96 | 6 | 21.5 ± 4.8 | 26.8 ± 7.9 | 32.8 ± 6.6 | |
| 9 June 1994 | 12–24 | 6 | 24.7 ± 1.8 | 57.3 ± 3.1 | 26.3 ± 2.3 |
| 48–72 | 6 | 16.8 ± 0.8 | 23.2 ± 3.1 | 14.3 ± 2.7 | |
| 15 June 1994 | 0–4 | 9 | 23.6 ± 4.6 | 40.4 ± 3.4 | 20.9 ± 3.7 |
| 22.6 | 3 | 22.7 ± 3.4 | 85.1 ± 0.5 | 37.9 ± 3.3 | |
| 9 August 1994 | 25.5 | 3 | 23.6 ± 4.6 | 25.7 ± 3.4 | 5.0 ± 0.4 |
| 73–96 | 6 | 22.7 ± 3.4 | 39.9 ± 2.5 | 2.0 ± 0.3 | |
| 19 August 1994 | 24.5 | 3 | 14.1 ± 3.2 | 36.1 ± 2.2 | 3.3 ± 0.7 |
| 49–73 | 6 | 11.1 ± 2.2 | 66.6 ± 4.0 | 6.3 ± 1.1 | |
| 9 May 1995 | 0–2.5 | 6 | 39.2 ± 4.2 | 38.2 ± 5.8 | 15.6 ± 1.2 |
| 27–31 | 6 | 24.1 ± 6.6 | 52.9 ± 2.8 | 8.2 ± 1.4 | |
| 16 May 1995 | 0–17 | 6 | 25.4 ± 0.5 | 17.2 ± 2.1 | 4.5 ± 0.7 |
| 25–40 | 6 | 18.7 ± 3.4 | 39.4 ± 1.5 | 8.2 ± 1.4 | |
| 9 June 1995 | 5.5–10 | 6 | 7.2 ± 1.0 | 66.1 ± 4.8 | 15.3 ± 3.3 |
| 24–30 | 6 | 15.8 ± 3.0 | 66.0 ± 7.2 | 1.6 ± 0.4 | |
| 5 July 1995 | 0–4 | 9 | 27.4 ± 2.4 | 12.4 ± 1.0 | 1.4 ± 0.5 |
| 17–20 | 6 | 26.9 ± 4.7 | 72.1 ± 2.9 | 12.8 ± 4.0 | |
| 8 August 1995 | 0–24 | 6 | 23.0 ± 2.8 | 13.5 ± 4.2 | 2.4 ± 3.3 |
| 48–72 | 6 | 30.9 ± 12.1 | 36.3 ± 10.9 | 3.5 ± 2.3 | |
| 22 May 1996 | 0–8 | 9 | 16.3 ± 2.1 | 67.8 ± 3.7 | 15.1 ± 0.8 |
| 24–29 | 6 | 8.4 ± 0.7 | 32.8 ± 2.2 | 16.6 ± 1.6 | |
| 10 August 1996 | 0–18 | 6 | 30.1 ± 2.7 | 54.9 ± 7.5 | 19.3 ± 1.1 |
| 42–48 | 6 | 25.9 ± 3.0 | 21.9 ± 0.5 | 7.8 ± 1.8 | |
| 24 October 1996 | 0–16 | 6 | 26.2 ± 4.3 | 57.7 ± 4.9 | 5.9 ± 0.8 |
| 40–48 | 6 | 35.1 ± 5.1 | 36.1 ± 6.7 | 6.7 ± 0.7 | |
| 31 October 1996 | 0–21 | 9 | 35.2 ± 6.0 | 47.0 ± 4.2 | 3.9 ± 0.8 |
| 49–68 | 6 | 23.2 ± 4.9 | 55.3 ± 4.8 | 5.8 ± 0.6 | |
| 24 November 1996 | 0–8 | 6 | 43.4 ± 3.5 | 18.2 ± 4.3 | 3.6 ± 1.1 |
| 32–51 | 6 | 20.1 ± 1.1 | 38.0 ± 2.8 | 21.0 ± 2.2 | |
| 5 December 1996 | 0–18 | 9 | 35.2 ± 2.5 | 19.8 ± 3.2 | 5.0 ± 0.9 |
| 34–42 | 6 | 12.6 ± 0.7 | 44.1 ± 4.4 | 19.4 ± 1.9 | |
Mean values ± the standard deviations from three to nine time points during early and late phases of the incubation period are shown.
We also examined the composition of the α- and β-proteobacterial communities on lab-made aggregates over time by using clone-specific probes (Fig. 6C). In the six experiments analyzed, bacterial cells detected by probe LSA225 (specific for Sphingomonas spp., C. subvibrioides, R. suberifaciens) ranged from 9 to 95% of the total α-Proteobacteria. Cells detected by probes LSB65 (D. zoogloeoides and relatives) and LSB145 (A. facilis and relatives) ranged from 8 to 74% and 5 to 56% of the total β-Proteobacteria, respectively. In addition to these specific probes, in four experiments we applied probe SNA23a which detects Sphaerotilus natans and relatives. Its proportion ranged from 5 to 38% of the total β-Proteobacteria. Together, the highly specific probes detected between 13 and 80% of the DAPI-stainable cells, and in 68% of the analyses they detected at least 30%.
In 15 experiments we simultaneously measured the temporal dynamics of the composition of the aggregate-associated bacterial community with that of DAA in the surrounding water and calculated the consumption and release rates of DAA by the aggregate-associated microbial community (Table 5). These rates were calculated from the difference in concentrations over time for the same periods for which mean proportions of α- and β-Proteobacteria were determined (Table 4). The dynamics of DCAA were much more pronounced than that of DFAA (Table 5, Fig. 7A). Three experiments exhibited a continuous DAA release. In 11 experiments, DAA were initially consumed from the surrounding water and later released into it. In 7 of the 11 experiments the transition from consumption to release of DAA covaried with increasing proportions of β-Proteobacteria by at least a factor of 1.3 and in 5 cases by a factor of >2, reaching final values between 32 and 85% of the DAPI-stainable cells (Table 4). In the experiments with a continuous DAA release (1 June 1994, 16 May 1995, 31 October 1996) the proportions of β-Proteobacteria were either permanently high or increased as well (Tables 4 and 5). In the only experiment with an initial release of DAA and a later consumption (9 June 1994), the proportions of β-Proteobacteria significantly decreased. α-Proteobacteria did not show such a consistent trend with the dynamics of DAA, but in 11 experiments with an initial DAA consumption and later release their proportions decreased or remained constant. Because the numbers of free-living bacteria in the surrounding water remained fairly constant and below 2 × 105 ml−1, we attribute the changes in concentrations of DAA to the activities of the aggregate-associated bacteria.
TABLE 5.
Consumption from the surrounding water (negative values) and release (positive values) of DFAA, DCAA, DFCHO, and DCCHO into it by microbial communities associated with lake snow aggregates over timea
| Date | Time (h) | Consumption or release (nmol liter−1 h−1)
|
|||
|---|---|---|---|---|---|
| DFAA | DCAA or DAA | DFCHO | DCCHO or DCHO | ||
| 1 June 1994 | 0–24 | −1.0 | 75.8 | – | – |
| 72–96 | – | 38.3 | – | – | |
| 9 June 1994 | 12–24 | – | 30.7 | – | – |
| 48–72 | – | −44.7 | – | – | |
| 15 June 1994 | 0–4 | – | −334.8 | – | – |
| 22.6 | – | 45.2 | – | – | |
| 9 August 1994 | 25.5 | – | −45.7 | – | – |
| 73–96 | – | 48.2 | – | – | |
| 19 August 1994 | 24.5 | – | −63.2 | – | – |
| 49–73 | – | 27.1 | – | – | |
| 9 May 1995 | 0–2.5 | 18.2 | −506.1 | – | – |
| 27–31 | −3.3 | 80.3 | – | – | |
| 16 May 1995 | 0–17 | 226.3 | 1848.3 | – | −0.4 |
| 25–40 | 83.8 | 1779.2 | – | 0.3 | |
| 9 June 1995 | 5.5–10 | −6.3 | −172.7 | – | – |
| 24–30 | 11.5 | −73.1 | – | – | |
| 5 July 1995 | 0–4 | −25.3 | −436.4 | – | – |
| 17–20 | −1.0 | 33.5 | – | – | |
| 22 May1996 | 0–8 | – | −263.7 | – | −148.4 |
| 24–29 | – | 40.7 | – | 323.3 | |
| 10 August 1996 | 0–18 | – | −250.1 | – | 31.0 |
| 42–48 | – | 22.4 | – | 40.7 | |
| 24 October 1996 | 0–16 | – | −27.3 | – | 246.8 |
| 40–48 | – | 66.7 | – | −39.1 | |
| 31 October 1996 | 0–21 | – | 4.3 | – | – |
| 49–68 | – | 22.2 | – | – | |
| 24 November 1996 | 0–8 | 0.3 | −72.8 | −18.0 | −153.3 |
| 32–51 | 1.9 | 50.0 | 4.2 | 48.7 | |
| 5 December 1996 | 0–18 | 2.2 | −1.8 | – | – |
| 34–42 | 4.4 | 63.1 | – | – | |
The aggregates were formed in the lab from natural samples of Lake Constance and incubated in rotating 50-ml syringes in the dark at 5 to 10°C (in situ temperature of the hypolimnion). Consumption and release rates were calculated from the differences in concentration over time in the early and late phases of the incubations. If DFAA and DFCHO were not analyzed separately (–), the rates include the total dissolved amino acids (DAA) and the total dissolved carbohydrates (DCHO).
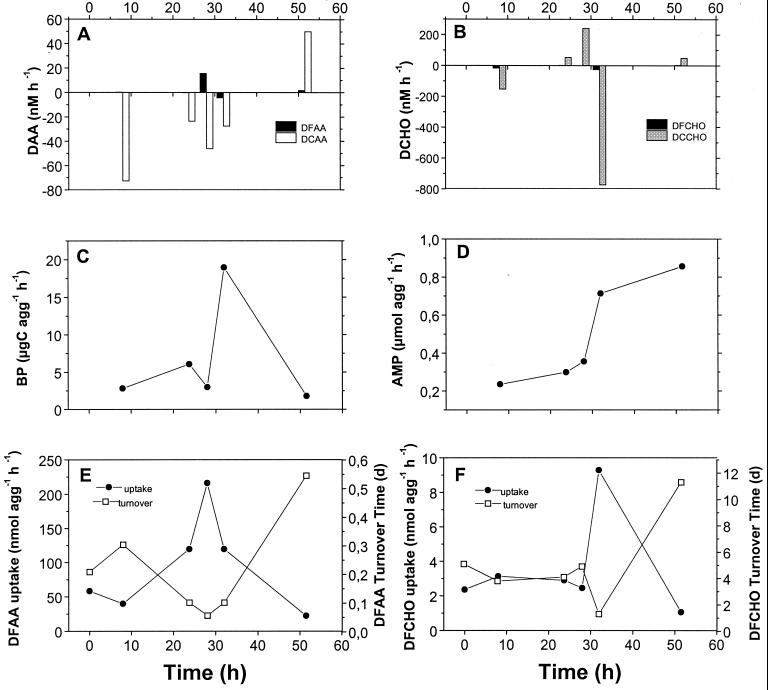
FIG. 7.
Temporal dynamics of the turnover DAA and DCHO by aggregate-associated bacteria on lab-made aggregates produced in rolling tanks on 24 November 1996 of water from 6 m. (A and B) Consumption by aggregate-associated bacteria (negative scale) and release (positive scale) of dissolved amino acids (DAA; DFAA, DCAA) and dissolved carbohydrates (DCHO; DFCHO, DCCHO) from aggregates into the surrounding water. (C) Bacterial biomass production (BP) of aggregate-associated bacteria. (D) aminopeptidase (AMP) activity of aggregate-associated bacteria. (E and F) Uptake rates and turnover times of DFAA and DFCHO.
In five experiments we measured the dynamics of DCHO and calculated rates of consumption from and release into the surrounding water in the same way as for DAA (Fig. 7B, Table 5). Two experiments exhibited an initial consumption and later release, whereas two other experiments showed either no change over time or a continuous release. There was no consistent temporal covariation with the proportions of either α- or β-Proteobacteria.
In two experiments (24 November 1996, 5 December 1996) we measured rates of substrate hydrolysis and uptake. In both experiments, DCAA were first consumed and later released and the proportions of β-Proteobacteria increased (Tables 4 and 5). As illustrated by the experiment of 24 November (Fig. 6 and 7), the shift from the relative dominance of α-Proteobacteria to β-Proteobacteria and from the consumption to the release of DCAA covaried with substantially enhanced activities of the aminopeptidase (Fig. 6B and 7D). During the transitional phase at around 30 h, the rates of bacterial production and uptake of DFAA and DFCHO also reached their maxima but strongly decreased thereafter (Fig. 7C, E, and F).
DISCUSSION
Our results show that the community of lake snow-associated bacteria in Lake Constance was dominated by α- and β-Proteobacteria, 20 to >50% of which comprised cells of a few closely related clusters which we detected with only five oligonucleotide probes of high specificity. Members of both subclasses of Proteobacteria constituted roughly similar proportions of the aggregate-associated bacterial community at a 25-m depth from spring until July 1995 and June 1996, respectively, whereas later on in the season of both years and at depths of 50 and 110 m the β-Proteobacteria largely dominated. With increasing depth, bacteria of the Cytophaga/Flavobacteria cluster occurred in increasing proportions, constituting up to 18% of the DAPI cell counts. Further, the majority of our experiments showed evidence that during the first day after aggregate formation the aggregate-associated bacterial community consumed DAA from the surrounding water. Thereafter, when β-Proteobacteria increasingly dominated and the proportions of α-Proteobacteria decreased, DAA were released into the surrounding water.
Lake snow aggregates in Lake Constance are formed in the epilimnion in the upper 10 m from senescent algae, molts, and dead zooplankton by various processes, including wind-induced shear and aggregation by transparent exopolymer particles (24, 34). Hence, the formation and abundance of lake snow aggregates is a function of the POM concentration. The numbers of aggregates and their seasonal dynamics in 1995 and 1996 are similar to those reported by Grossart et al. (24) for 1993. Lake snow contributes 40 to 60% to the sinking flux and settles through the water column with estimated mean sinking rates of 10 to 15 m day−1, even though individual aggregates may have higher sinking rates (23). Therefore, aggregates have an estimated age of 1.5 to 3 days at depths of 25 to 30 m, of 3 to 5 days at 50 m, and of 8 to 11 days when they reach the sediment surface at a 110-m depth. Thus, in our experiments we simulated the initial days of the aggregate decomposition that took place in the lake at depths of between 10 and 50 m. The results of the majority of these experiments indicated that consumption of DAA from the surrounding water occurred mainly at 10 to 25 m, whereas further below in the hypolimnion, DAA were released from the aggregates into the surrounding water. Some experiments even showed a continuous release. Because we did not run replicates of the release assays during individual experiments the results are not statistically significant but indicate a trend. This trend from an early consumption to a later release of DAA was consistent in 73% of our experiments. Hence, we assume that this trend documents findings which can be generalized and are of general significance for the decomposition of amino acid components of POM on lake snow aggregates. This trend is also consistent with the results of Grossart and Simon (22, 23), who showed that aminopeptidase activities of aggregate-associated bacteria in Lake Constance are highest at depths of 15 to 25 m and that aggregates and settled POM in sediment traps at 50 m directly release dissolved amino acids.
With the Bacteria-specific probe EUB338, we detected 50 to more than 80% of the DAPI cell counts except from April to July 1995, indicating that during most of our study we were able to identify and quantify the majority of the microbial cells on aggregates. These results are consistent with previous studies on the microbial colonization of lake snow aggregates in Lake Constance (22, 62). Usually, α- and in particular β-Proteobacteria dominated the bacterial community on aggregates and more than 80 and often 100% of Bacteria were detected by probes specific for these subclasses of Proteobacteria and for the Cytophaga/Flavobacteria cluster. This observation indicates that nearly all of the cells identified on the aggregates by FISH belonged to either of these phylogenetic lineages and that we did not miss any further potentially important lineages. It is noteworthy that we were able to identify 16 to 60% of the α-Proteobacteria by only two probes specific for distinct clusters from lake snow aggregates belonging to Sphingomonas and relatives (LSA67, LSA225), and 42 to 70% of the β-Proteobacteria by three probes specific for distinct clusters of the β1-subclass of Proteobacteria closely related to D. zoogloeoides (LSB65), H. palleroni (LSB70), and A. facilis (LSB145). Obviously, the microenvironment of lake snow aggregates selects for a bacterial community specialized in decomposing these polymer-rich aggregates because it is highly enriched in labile organic and inorganic nutrients (22, 23) irrespective of the source POM and season. This α- and β-Proteobacteria-dominated community was already established on all of the aggregates we analyzed, including the natural ones and the youngest formed in rolling tanks. It was even present on precursor diatom microaggregates of an age of a few hours (S. Knoll, W. Zwisler, and M. Simon, submitted for publication). On naturally occurring microaggregates in Lake Constance, however, this highly specific bacterial community was never detected (T. Brachvogel and M. Simon, unpublished data). On these microaggregates, the associated bacterial community was dominated by β-Proteobacteria and members of the Cytophaga/Flavobacteria cluster, whereas α-Proteobacteria were not detected (T. Brachvogel, B. Schweitzer, and M. Simon, submitted for publication). In the community of free-living bacteria in Lake Constance, even though also dominated by β-Proteobacteria, the number of bacteria detected by the probes specific for 16S rDNA clones from lake snow aggregates was below the detection limit (W. Zwisler and M. Simon, unpublished data). Hence, the bacterial communities on different types of micro- and macroaggregates and those in the surrounding water exhibit pronounced differences. Similar observations based, however, mainly on qualitative differences, were made for bacterial communities on particles and marine aggregates compared to the surrounding water in estuaries, in Californian coastal waters, and in the Mediterranean Sea (1, 8, 14, 16, 41). Brümmer et al. (9) analyzing the bacterial community on river biofilms also found that α- and β-Proteobacteria were its dominant components.
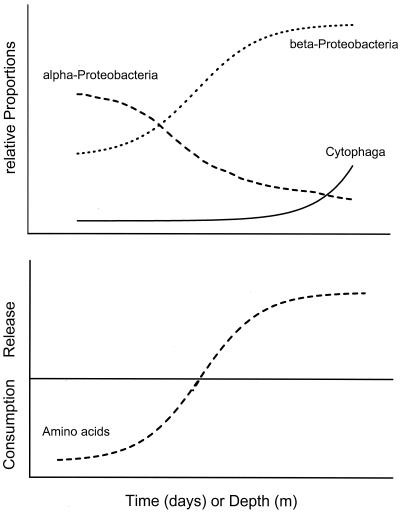
According to the majority of our experimental results, consumption of DAA by the aggregate-associated bacterial community took place on aggregates of an age of 2 to 3 days, equivalent to aggregates occurring at a depth of 15 to 30 m. During this phase, the relative proportions of α- and β-Proteobacteria were rather variable. In most cases consumption of DAA by the aggregate-associated bacterial community exceeded the supply by the aggregate-associated POM such that DAA from the surrounding water were utilized as well. Later on, when proportions of β-Proteobacteria increasingly dominated and proportions of α-Proteobacteria decreased and constituted <27% of the DAPI cell counts, DAA were released into the surrounding water. At this stage, occurring after 3 to 5 days and at depths of 25 to 50 m, the intense hydrolysis of aggregate-associated combined amino acids predominantly by β-Proteobacteria obviously became uncoupled from consumption such that net release into the surrounding water occurred. The closest known relatives of the β-Proteobacteria that we identified (A. facilis, H. palleroni, D. zoogloeoides) are well known for a high potential of hydrolytic enzyme activities and for producing mucopolysaccharides (see below). Hence, the physiology of these bacteria is consistent with our experimental and field observations. At later stages occurring in the deep hypolimnion of the lake, when the relative abundance of β-Proteobacteria decreased again, bacteria of the Cytophaga/Flavobacteria cluster became more abundant on aggregates. These bacteria are known to have a high potential for hydrolyzing complex polysaccharides of various compositions including cellulose, which are rather refractory to decomposition by other aerobic bacteria such as those mentioned above (45). Hence, the greater abundance of cells of the Cytophaga/Flavobacteria cluster in later stages of lake snow aggregates suggests that in these stages labile organic matter was rather depleted and that refractory components dominated more and more. We did not find such clear temporal patterns of DCHO consumption and release in relation to the bacterial colonization of aggregates, presumably because utilization and hydrolysis of dissolved and aggregate-associated polysaccharides is not related as closely to the growth dynamics of the aggregate-associated bacterial community. Figure 8 synthesizes our findings on the bacterial colonization and substrate dynamics on lake snow aggregates into a generalized scheme.
FIG. 8.
Generalized scheme of the microbial colonization and turnover of dissolved amino acids on lake snow aggregates. (Upper panel) Succession of α- and β-Proteobacteria and Cytophaga/Flavobacteria over time (days) or depth (meters). (Lower panel) Consumption and release of dissolved amino acids over time (days) or depth (meters).
The closest identified relatives of the bacteria dominating the community on lake snow aggregates are the α-Proteobacteria Sphingomonas spp., S. capsulata, and B. diminuta and the β-Proteobacteria A. facilis, Hydrogenophaga spp., and D. zoogloeoides. Sphingomonas spp. are gram-negative chemo-organoheterotrophic bacteria that are widely distributed in soil, sediments, and water. They are capable of degrading various contaminants and occur on activated sludge (see references 17, 38, and 60 and references therein). A. facilis and Hydrogenophaga spp. are also gram-negative chemo-organoheterotrophic bacteria that utilize various carbon sources and occur on activated sludge as well (15, 48, 57). D. zoogloeoides metabolizes labile DOM such as amino acids, proteins, and carbohydrates and produces mucopolysaccharides. It has been found on the mucilage of filamentous cyanobacteria and occurs on activated sludge flocs in sewage treatment plants (13, 17, 31). Interestingly, D. zoogloeoides, formerly assigned to the polyphyletic species Zoogloea ramigera, could not be detected in various sewage treatment plants even though it exhibits the greatest physiological diversity of the three strains known (46, 47). According to this comparison, the composition and density of the bacterial community on 1 to 5-day-old lake snow aggregates is not identical to but resembles that found on activated sludge flocs. Previous observations showed that activated sludge flocs are also dominated by β-Proteobacteria closely related to those we found (10, 60, 61). Hence, lake snow aggregates of an age of a few days that occur in the upper 50 m of a lake such as Lake Constance can be considered as activated sludge flocs which occur in dilute concentrations but exhibit similar functions as those found in sewage treatment plants.
There are quite a few studies showing that lake snow and marine snow aggregates are similar with respect to their significance in the microbial solubilization, recycling, and sedimentation of organic matter (3, 21, 22, 23, 28, 39, 50, 54). However, so far little is known about the quantitative composition of the microbial community of marine aggregates. On the basis of our results, we hypothesize that marine aggregates are also colonized by a specialized and well-adapted bacterial community of limited diversity. Smith et al. (55) showed that the hydrolytic activities of marine snow-associated microbial communities led to a net release of DCAA into the surrounding water. On the basis of our results we hypothesize that this microbial community was dominated by microbes similar in function to the β-Proteobacteria which dominated on our aggregates of an age of a few days. During an experimentally induced diatom bloom, Smith et al. (56) found an aggregate-associated microbial community which consumed to a great extent the organic matter hydrolyzed on the aggregates and did not release it into the surrounding water. Hence, we hypothesize on the basis of our results that in this case a community dominated on the aggregates which was functionally more similar to the ones we found to be dominated by α-Proteobacteria. To better understand the functional relationship between microbial colonization and decomposition of marine snow aggregates, it would be of great importance to examine what the counterparts of α- and β-Proteobacteria on marine snow are.
Ploug et al. (42) reported that bacteria of the Cytophaga/Flavobacteria cluster comprised 13 to 74% of the DAPI cell counts on marine snow aggregates in the Southern California Bight. On naturally derived marine snow aggregates at the polar front in the Southern Ocean, the associated bacterial community was dominated by γ-Proteobacteria and also by members of the Cytophaga/Flavobacteria cluster, whereas α-Proteobacteria were of minor importance (M. Simon, unpublished data). Two reports of highly diverse bacterial communities on marine snow aggregates (16, 44) seem to contradict our finding of a limited diversity of lake snow-associated microbial communities. A diverse particle-associated community of bacterial phylotypes was also found in Mediterranean waters at various depths that showed close similarities to that of the surrounding water at 400-m a depth but not in the mixed layer (1). This report, however, only presented sequence data derived from PCR-amplified 16S rDNA clones which merely show the qualitative diversity of microbes on marine aggregates but do not give any indication of their quantitative occurrence. In fact, a qualitative diversity at least as great as that reported for marine snow-associated microbial communities also exists on lake snow aggregates in Lake Constance. Huber (30), sequencing 16S rDNA clones of four samples of natural and lab-made aggregates in Lake Constance at various seasons, found a total of 230 different clones, some of which matched known bacterial species by 100% sequence similarity. Thus, it seems even more surprising that the microbial community on lake snow aggregates was composed of bacteria belonging to narrow phylogenetic clusters. This observation, together with the presented results, demonstrates that the qualitative diversity of a microbial community does not directly represent the quantitative occurrence of their individual members. It shows instead that aggregates and presumably also other habitats harbor a huge pool of very different microbes like seeds which may only propagate when the (micro)environmental and growth conditions become favorable.
In conclusion, our results show that during the formation and microbial decomposition of lake snow aggregates a specialized bacterial community of a small number α- and β-Proteobacteria evolves. During aging and sinking of the aggregates it undergoes a distinct succession and mediates their decomposition. This community is instrumental in key processes of solubilizing POM and recycling of labile DOM in lacustrine environments such as Lake Constance and appears to be closely related to that found on activated sludge flocs.
ACKNOWLEDGMENTS
We are most grateful for the assistance in the field to T. Gries and C. Wünsch and to two anonymous reviewers for their critical suggestions on an earlier version of this article.
This work was supported by grants from the Deutsche Forschungsgemeinschaft awarded to M.S., R.A., and W.L., and by the Special Collaborative Program “Cycling of Matter in Lake Constance” (SFB-248).
REFERENCES
- 1.Acinas S G, Antón J, Rodríguez-Valera F. Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol. 1999;65:514–522. doi: 10.1128/aem.65.2.514-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alldredge A L, Cohen Y. Can microscale chemical patches persist in the sea? Microelectrode study of marine snow, fecal pellets. Science. 1987;235:689–691. doi: 10.1126/science.235.4789.689. [DOI] [PubMed] [Google Scholar]
- 3.Alldredge A L, Silver M L. Characteristics, dynamics and significance of marine snow. Prog Oceanogr. 1988;20:41–82. [Google Scholar]
- 4.Alldredge A L, Cole J J, Caron D A. Production of heterotrophic bacteria inhabiting macroscopic marine aggregates (marine snow) from surface waters. Limnol Oceanogr. 1986;31:68–78. [Google Scholar]
- 5.Amann R I, Krumholz L, Stahl D A. Fluorescent oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amann R I, Ludwig W, Schleifer K H. Identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bäuerle E, Ollinger D, Ilmberger J. Some meteorological, hydrological, and hydrodynamical aspects of upper Lake Constance. Arch Hydrobiol Spec Issues Adv Limnol. 1998;53:31–83. [Google Scholar]
- 8.Bidle K D, Fletcher M. Comparison of free-living and particle-associated bacterial communities in the Chesapeake Bay by stable low-molecular weight RNA analysis. Appl Environ Microbiol. 1995;61:944–952. doi: 10.1128/aem.61.3.944-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brümmer I H M, Fehr W, Wagner-Döbler I. Biofilm community structure in polluted rivers: abundance of dominant phylogenetic groups over a complete annual cycle. Appl Environ Microbiol. 2000;66:3078–3082. doi: 10.1128/aem.66.7.3078-3082.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 12.Bunte C, Simon M. Bacterioplankton turnover of dissolved free monosaccharides in a mesotrophic lake. Limnol Oceanogr. 1999;44:1862–1870. [Google Scholar]
- 13.Caldwell D E, Caldwell S J. A Zoogloea sp. associated with blooms of Anabaena flos-aquae. Can J Microbiol. 1978;24:922–931. doi: 10.1139/m78-154. [DOI] [PubMed] [Google Scholar]
- 14.Crump B C, Armbrust E V, Baross J A. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dangmann E, Stolz A, Kuhm A E, Hammer A, Feigl B, Noisommit-Rizzi N, Rizzi M, Reuss M, Knackmuss H J. Degradation of 4-aminobenzenesulfonate by a two-species bacterial coculture. Physiological interactions between Hydrogenophaga palleroni S1 and Agrobacterium radiobacter S2. Biodegradation. 1996;7:223–229. doi: 10.1007/BF00058181. [DOI] [PubMed] [Google Scholar]
- 16.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 17.Dugan P R, Stoner D L, Pickrum H M. The genus Zoogloea. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 3952–3964. [Google Scholar]
- 18.Fowler S W, Knauer G A. Role of large particles in the transport of elements and organic compounds through the oceanic water column. Prog Oceanogr. 1986;16:147–194. [Google Scholar]
- 19.Frederickson J K, Balkwill D L, Drake G R, Romine M F, Ringelberg D B, White D C. Aromatic-degrading Sphingomonas isolates from the deep surface. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.5.1917-1922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaedke U. Functional and taxonomical properties of the phytoplankton community of large and deep Lake Constance: interannual variability and response to re-oligotrophication (1979–1993) Arch Hydrobiol Spec Issues Adv Limnol. 1998;53:119–141. [Google Scholar]
- 21.Grossart H P, Simon M. Limnetic macroscopic organic aggregates (lake snow): abundance, characteristics, and bacterial dynamics in Lake Constance. Limnol Oceanogr. 1993;38:532–546. [Google Scholar]
- 22.Grossart H P, Simon M. Bacterial colonization and microbial decomposition of limnetic organic aggregates (lake snow) Aquat Microb Ecol. 1998;15:127–140. [Google Scholar]
- 23.Grossart H P, Simon M. Significance of limnetic organic aggregates (lake snow) for the sinking flux of particulate organic matter in a large lake. Aquat Microb Ecol. 1998;15:115–125. [Google Scholar]
- 24.Grossart H P, Simon M, Logan B. Formation of macroscopic organic aggregates (lake snow) in a large lake: the significance of transparent exopolymer particles (TEP), phyto- and zooplankton. Limnol Oceanogr. 1997;42:1651–1659. [Google Scholar]
- 25.Güde H, Gries T. Phosphorus fluxes in Lake Constance. Arch Hydrobiol Spec Issues Adv Limnol. 1998;53:505–544. [Google Scholar]
- 26.Hanisch K, Schweitzer B, Simon M. Use of dissolved carbohydrates by planktonic bacteria in a mesotrophic lake. Microb Ecol. 1996;31:41–55. doi: 10.1007/BF00175074. [DOI] [PubMed] [Google Scholar]
- 27.Häse C, Gaedke U, Seifried A, Beese B, Tilzer M M. Phytoplankton response to re-oligotrophication in large and deep Lake Constance: photosynthetic rates and chlorophyll concentrations. Arch Hydrobiol Spec Issues Adv Limnol. 1998;53:159–178. [Google Scholar]
- 28.Herndl G J. Marine snow in the northern Adriatic Sea: possible causes and consequences for a shallow ecosystem. Mar Microb Food Webs. 1992;6:149–172. [Google Scholar]
- 29.Hoppe H G. Use of fluorogenic model substrates for extracellular enzyme activity (EEA) measurements of bacteria. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publishers; 1993. pp. 423–432. [Google Scholar]
- 30.Huber I. Bacterial diversity on lake snow: phylogenetic and in situ analyses. Ph.D. thesis. Munich, Germany: Technische Universität München; 1997. . (In German.) [Google Scholar]
- 31.Ikeda F, Shuto H, Saito T, Fukui T, Tomita K. An extracellular polysaccharide produced by Zoogloea ramigera 115. Eur J Biochem. 1982;123:437–445. doi: 10.1111/j.1432-1033.1982.tb19787.x. [DOI] [PubMed] [Google Scholar]
- 32.Kirchman D L, K'nees E, Hodson R E. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural systems. Appl Environ Microbiol. 1985;49:599–607. doi: 10.1128/aem.49.3.599-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindroth P, Mopper K. High performance liquid chromatographic determination of subpicomole amounts of amino acids by precolumn fluorescence derivatization with o-phthaldialdehyde. Anal Chem. 1979;51:1667–1674. [Google Scholar]
- 34.Logan B E, Passow U, Alldredge A L, Grossart H-P, Simon M. Mass sedimentation of diatom blooms as large aggregates is driven by coagulation of transparent exopolymer particles (TEP) Deep-Sea Res. 1995;42:203–214. [Google Scholar]
- 35.Lookhart G L, Jones B L, Cooper D B, Hall S B. A method for hydrolyzing and determining amino acid composition of picomole quantities of protein in less than three hours. J Biochem Biophys Methods. 1982;7:15–23. doi: 10.1016/0165-022x(82)90032-x. [DOI] [PubMed] [Google Scholar]
- 36.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 37.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K H. Phylogenetic oligonucleotide probes for the major subclasses of Proteobacteria: problems and solutions. System Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 38.Mohn W M. Bacteria obtained from a sequencing batch reactor that are capable of growth on dehydroabietic acid. Appl Environ Microbiol. 1995;61:2145–2150. doi: 10.1128/aem.61.6.2145-2150.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mopper K, Schultz C A, Chevolot L, Germain C, Revuelta R, Dawson R. Determination of sugars in unconcentrated seawater and other natural waters by liquid chromatography and pulsed amperometric detection. Environ Sci Technol. 1992;26:133–138. [Google Scholar]
- 40.Müller-Niklas G, Schuster S, Kaltenböck E, Herndl G J. Organic content and bacterial metabolism in amorphous aggregations in the Northern Adriatic Sea. Limnol Oceanogr. 1994;39:58–68. [Google Scholar]
- 41.Phillips C J, Smith Z, Embley T M, Prosser J I. Phylogenetic differences between particle-associated and planktonic ammonia-oxidizing bacteria of the β-subdivision of the class Proteobacteria in the northwestern Mediterranean Sea. Appl Environ Microbiol. 1999;65:779–786. doi: 10.1128/aem.65.2.779-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ploug H, Grossart H-P, Azam F, Jorgensen B B. Photosynthesis, respiration, and carbon turnover in sinking marine snow from surface waters of Southern California Bight: implications for the carbon cycle in the ocean. Mar Ecol Prog Ser. 1999;179:1–11. [Google Scholar]
- 43.Ploug H, Kühl M, Buchholz-Cleven B, Jørgensen B B. Anoxic aggregates—an ephemeral phenomenon in the pelagic environment. Aquat Microb Ecol. 1997;13:285–294. [Google Scholar]
- 44.Rath J, Wu K Y, Herndl G J, DeLong E F. High phylogenetic diversity in a marine snow-associated bacterial assemblage. Aquat Microb Ecol. 1998;14:261–269. [Google Scholar]
- 45.Reichenbach H. The order Cytophagales. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokayotes. New York, N.Y: Springer-Verlag; 1992. pp. 3631–3675. [Google Scholar]
- 46.Rosselló-Mora R A, Ludwig W, Schleifer K H. Zoogloea ramigera: a phylogenetically diverse species. FEMS Microbiol Lett. 1993;114:129–134. [Google Scholar]
- 47.Rossellö-Mora R A, Wagner M, Amann R, Schleifer K H. The abundance of Zoogloea ramigera in sewage treatment plants. Appl Environ Microbiol. 1995;61:702–707. doi: 10.1128/aem.61.2.702-707.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulze R, Spring S, Amann R, Huber I, Ludwig W, Schleifer K H, Kämpfer P. Genotypic diversity of Acidovorax strains isolated from activated sludge and description of Acidovorax defluvii sp. nov. System Appl Microbiol. 1999;22:205–214. doi: 10.1016/S0723-2020(99)80067-8. [DOI] [PubMed] [Google Scholar]
- 49.Shanks A L, Edmondson E W. Laboratory-made artificial marine snow: a biological model of the real thing. Mar Biol. 1989;101:463–470. [Google Scholar]
- 50.Shanks A L, Trent J D. Marine snow: microscale nutrient patches. Limnol Oceanogr. 1979;24:43–47. [Google Scholar]
- 51.Simon M, Bunte C, Schulz M, Weiss M, Wünsch C. Bacterioplankton dynamics in Lake Constance (Bodensee): substrate utilization, growth control, and long-term trends. Arch Hydrobiol Spec Issues Adv Limnol. 1998;53:195–221. [Google Scholar]
- 52.Simon M, Rosenstock B. Carbon and nitrogen sources of planktonic bacteria in Lake Constance studied by the composition and isotope dilution of intracellular amino acids. Limnol Oceanogr. 1992;37:1496–1511. [Google Scholar]
- 53.Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- 54.Simon M, Alldredge A L, Azam F. Bacterial carbon dynamics on marine snow. Mar Ecol Prog Ser. 1990;65:205–211. [Google Scholar]
- 55.Smith D C, Simon M, Alldredge A L, Azam F. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature. 1992;359:139–142. [Google Scholar]
- 56.Smith D C, Steward G F, Long R A, Azam F. Bacterial mediation of carbon fluxes during a diatom bloom in a mesocosm. Deep-Sea Res. 1995;42:75–97. [Google Scholar]
- 57.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strunk O, Gross O, Reichel B, May M, Hermann S, Stuckman N, Nonhoff B, Lenke M, Ginhart A, Vilbig A, Ludwig T, Bode A, Schleifer K-H, Ludwig W. ARB: a software environment for sequence data ( http://www.mikro.biologie.tu-muenchen.de). Munich, Germany: Department of Microbiology, Technische Universität München; 1999. [Google Scholar]
- 59.Trent J D, Shanks A L, Silver M W. In situ laboratory measurements on macroscopic aggregates in Monterey Bay, California. Limnol Oceanogr. 1978;23:415–421. [Google Scholar]
- 60.Wagner M, Amann R, Lemmer H, Schleifer K H. Probing activated sludge with oligonucleotides specific for Proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner M, Amann R, Kämpfer P, Assmus B, Hartmann A, Hutzler P, Springer N, Schleifer K H. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. System Appl Microbiol. 1994;17:405–417. [Google Scholar]
- 62.Weiss P, Schweitzer B, Amann R, Simon M. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow) Appl Environ Microbiol. 1996;62:1998–2005. doi: 10.1128/aem.62.6.1998-2005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]