Abstract
The human microbiome is comprised of a complex and diverse community of organisms that is subject to dynamic changes over time. As such, cross-sectional studies of the microbiome provide a multitude of information for a specific body site at a particular time, but they fail to account for temporal changes in microbial constituents resulting from various factors. To address this shortcoming, longitudinal research studies of the human microbiome investigate the influence of various factors on the microbiome of individuals within a group or community setting. These studies are vital to address the effects of host and/or environmental factors on microbiome composition as well as the potential contribution of microbiome members during the course of an infection. The relationship between microbial constituents and disease development has been previously explored for skin and soft tissue infections (SSTIs) within congregate military trainees. Accordingly, approximately 25% of the population carries Staphylococcus aureus within their nasal cavity, and these colonized individuals are known to be at increased risk for SSTIs. To examine the evolution of the nasal microbiota of U.S. Army Infantry trainees, individuals were sampled longitudinally from their arrival at Fort Benning, Georgia, until completion of their training 90 days later. These samples were then processed to determine S. aureus colonization status and to profile the nasal microbiota using 16S rRNA gene-based methods. Microbiota stability differed dramatically among the individual trainees; some subjects exhibited great stability, some subjects showed gradual temporal changes and some subjects displayed a dramatic shift in nasal microbiota composition. Further analysis utilizing the available trainee metadata suggests that the major drivers of nasal microbiota stability may be S. aureus colonization status and geographic origin of the trainees. Nasal microbiota evolution within the congregate setting imposed by military training is a complex process that appears to be affected by numerous factors. This finding may indicate that future campaigns to prevent S. aureus colonization and future SSTIs among high-risk military trainees may require a ‘personalized’ approach.
Subject terms: Microbiology, Microbiome
Introduction
The composition of an individual’s microbiome varies widely depending on the sampled body site. Moreover, the microbial constituents that are present at various sites are influenced by a multitude of factors, including geographic location, lifestyle, ethnicity, culture, diet, and physical health1–7. Additionally, it is clear that the presence of particular bacterial species can influence the prevalence of other species that are found within the microbiome4,8–10. These influences can be negative or positive and can occur through a variety of mechanisms. For example, Staphylococcus epidermidis, a commensal component of the human skin microbiome, produces phenol soluble modulins (PSMs) that function in conjunction with host anti-microbial peptides to kill Streptococcus pyrogenes and S. aureus11–13. Similarly, within the nasal cavity, members of the microbial community appear to prevent the establishment of other species; in vitro studies have shown that Corynebacterium pseudodiphtheriticum inhibits the colonization efforts of S. aureus and exhibits bactericidal activity against the pathogen14–16. Additionally, C. accolens produces free fatty acids that inhibit growth of Streptococcus pneumoniae within the nasal cavity15,17. Another skin commensal organism, Corynebacterium striatum, has a demonstrated ability to modulate S. aureus gene expression and elicit the transformation of the microbe from a virulent state to that of a commensal18,19. Conversely, some microbes cooperate with each other to promote colonization and sustained survival. For example, the presence of C. accolens correlates with S. aureus carriage in the nasal cavity and can enhance growth of S. aureus in vitro15–17. Likewise, the gut microbe, Bacteroides ovatus, induces the extracellular digestion of inulin to feed other species, such as B. vulgatus, within the intestinal tract20. This relationship between the species within the intestines has reciprocal benefits and actually increases the fitness of B. ovatus20. Thus, the interplay among the constituents of the microbial community is complex, and each member can positively or negatively influence the presence of other species within the microbial milieu.
While a great deal of information has been gained from cross-sectional studies that define the microbiome of a particular site at a particular time, the composition and relative abundance of various species within the microbiome of an individual can change over time. These changes can be driven by host and/or lifestyle factors1–7 as well as via simple exposure as an individual encounters other bacterial species within an environment and/or through interaction with others2. As such, the dynamic changes that occur within the human microbiome are being examined via longitudinal research studies that seek to examine the impact of environmental factors and/or personal interactions on the microbiome of individuals over a period of time. For example, longitudinal analysis of the gut microbiome revealed the influence of daily dietary changes on microbial composition; stability was linked to a more diverse diet21. Similarly, a longitudinal examination of the skin microbiome in adults revealed relative stability despite exposure to various environmental microbes22. In contrast, analysis of U.S. Air Force Academy cadets revealed an increase in the similarity of skin microbiomes, but not gut microbiota, among roommates upon cohabitation23. Finally, analysis of the nares of healthy individuals indicated a temporal stability in microbial composition as well as a lack of convergence of the nasal microbiomes among cohabiting couples8. Thus, longitudinal analyses of the human microbiome generate results that vary depending on the body site and cohort under examination.
The particular composition of an individual’s microbiome, in combination with host and environmental factors, can precipitate pathogenesis of commensal microorganisms and can result in infections. Furthermore, resident microbes can influence the ability of an encountered pathogen to colonize and cause disease11,18,24–27. The relationship between the presence of particular microbes and disease development has been specifically examined for skin and soft tissue infections (SSTIs), which are frequently encountered maladies within the Military Health System28–30. Due to the close quarters and frequent physical contact associated with military training, the rate of SSTIs (abscesses, cellulitis, folliculitis, etc.) is especially high among military trainees28,30–33. This high prevalence of SSTIs greatly impacts military readiness and creates an economic burden on the Military Health System30.
Even though S. aureus benignly colonizes the nares of approximately 25% of the population at any one time34,35, colonized individuals are known to be at increased risk for subsequent infections35–38. As such, S. aureus is the most frequently identified infectious agent within cutaneous abscesses39,40. Various studies have sought to identify links between the composition of the nasal microbiome, S. aureus colonization, and subsequent SSTI presentation among military recruits32,41–44. For example, among a cohort of military recruits presenting with SSTIs, colonization and infection with methicillin-resistant S. aureus (MRSA) appeared to be associated with a single acquisition event, thus underscoring the potential for host decolonization and environmental disinfection as strategies to prevent SSTIs in high-risk recruits43. In contrast, a separate S. aureus genomic analysis in two cohorts of high-risk recruits identified multiple transmission events of MRSA within platoons45. These two seemingly disparate analyses highlight the need to examine the microbiomes of individual recruits within a platoon to explore the potential exchange of microbes within the congregate setting experienced during military training. Moreover, given that individuals found in other congregate settings (prison inmates, athletic team members, residents of long-term care facilities, children attending daycare) are also at increased risk for SSTI development46–51, a greater understanding of microbiome evolution within the congregate setting may shed light on possible paths to prevent subsequent infections.
Herein we investigated the evolution of the nasal microbiome of a single platoon of male U.S. Army Infantry trainees stationed at Fort Benning, Georgia. Individuals were sampled longitudinally within 24 h of their arrival at Fort Benning until completion of their training 90 days later. Given the congregate setting and uniform diet, schedule, and environmental exposures encountered by these military trainees, we originally hypothesized that we would observe an overall convergence of the individuals’ nasal microbiota to a more similar composition. However, microbiota stability varied dramatically on an individual basis; some subjects showed great stability, some subjects showed gradual temporal changes and some subjects showed dramatic shifts in nasal microbiota composition. Analysis of available patient metadata suggests that S. aureus colonization and geographic origin of the trainees may be key drivers of nasal microbiota stability within this population.
Results
Participant characteristics
A total of 627 male trainees were enrolled from 2015–2016 in an observational cohort study of S. aureus colonization and SSTI52. Participants were recruited from four separate Infantry training companies (two in 2015 and two in 2016), each composed of ~ 200 trainees. To determine the evolution of the nasal microbiota of individuals within the congregate setting imposed by infantry training, 53 male subjects from a single platoon enrolled in 2015 were analyzed. Demographic information for the subjects is provided in Table 1. Briefly, the median age was 19 years (range 17–34), the majority were non-Hispanic whites (37), and the individuals originated from various U.S. locations; approximately half of the subjects were from the South (26), followed by subjects from the Midwest (12), the Northeast (10), and the West (5). Metadata for individual subjects is provided in Supplementary Table S1.
Table 1.
Study subject characteristics.
| Median age (range) | 19 (17–34) |
| Race/ethnicity | |
| Non-Hispanic/Latino | |
| White | 37 |
| Black or African American | 5 |
| Two races (White/Black or African American) | 2 |
| Not reported | 2 |
| Hispanic/Latino | |
| White | 5 |
| Not reported | 2 |
| S. aureus nasal colonization1 | |
| Non-colonized | 20 |
| Intermittent | 22 |
| Persistent | 9 |
| NA2 | 2 |
| Geographic origin3 | |
| Midwest | 12 |
| Northeast | 10 |
| South | 26 |
| West | 5 |
1S. aureus nasal colonization was defined based on culture-positive S. aureus in the nares over the five study visits: non-colonized (positive at 0 study visits), intermittent (positive at ≥ 1 and ≤ 3 study visits), or persistent (positive at ≥ 4 study visits).
2Was not assessed for the two subjects that did not complete the five study visits.
3Based on zipcode listed on intake forms upon arrival. Regions as defined by US Census Bureau.
Nasal swabs and culture analysis
Paired nasal swabs were collected by the study staff upon arrival of subjects at Fort Benning (day 0) and then longitudinally at days 14, 28, 56 and 90. On each day, one swab was immediately used for S. aureus culture, while the second swab was stored at − 80 °C for future microbiota analysis. The day 0 swabs were used to define the baseline S. aureus nasal culture status/nasal microbiota. The subsequent swabs were used to identify any changes in S. aureus colonization status/structure and composition of the nasal microbiota. A total of 12/53 trainees (22.6%) were culture positive for S. aureus at day 0 (Fig. 1). While culture positivity varied across the study, overall colonization was increased at the end of training. A total of 19/51 trainees (37.2%) were culture positive at day 90; two subjects did not complete all 5 study visits and a day 90 sample was not available for these individuals. Overall, of the obtained 261 culture swabs, 81 were S. aureus positive (Fig. 1). S. aureus nasal colonization among individuals is known to fall into three broad categories: persistent carriers, intermittent carriers and persistent non-carriers. Thus, trainees were categorized into these general groupings based on the following definitions: not colonized (no positive cultures), intermittently colonized (≥ 20% and < 80% positive cultures), or persistently colonized (≥ 80% positive cultures) (Fig. 1). The two subjects that did not complete all 5 study visits could not be categorized. A total of 20 subjects (39.2%) were not nasally colonized by S. aureus, 22 subjects (43.1%) were intermittently colonized, and 9 subjects (17.6%) were persistently colonized. These numbers are in line with other studies concerning the prevalence of S. aureus in the nares16,35,53,54. At the individual level, we observed a variety of colonization patterns. Of the 22 intermittently colonized subjects, 12 were positive at only a single study visit. Of these, 7 were positive at one study visit, and negative at all following visits. Even among the 9 persistently colonized subjects, only 3 subjects were positive at each visit (Fig. 1).
Figure 1.

S. aureus culture status in the nares. Nasal swabs were collected upon arrival of subjects at Fort Benning (day 0) and then longitudinally at days 14, 28, 56 and 90. The swab was immediately used for S. aureus culture. Each trainee is listed by subject ID number, and the culture results for each day are shown. Positive culture results are denoted by red boxes, and negative culture results are indicated by white boxes. Results were not obtained for two subjects on different days, and those results are indicated by gray boxes. Trainees are categorized into general groupings based on S. aureus colonization status: not colonized (no positive cultures), intermittently colonized (≥ 20% and < 80% positive cultures), or persistently colonized (≥ 80% positive cultures). The two subjects that did not complete all 5 study visits are not categorized.
Over the course of the 3-month study, 2 subjects developed a purulent SSTI: Subject 2013 on day 94; and Subject 2197 on day 22. Nasal colonization by S. aureus is a known risk factor for SSTI development35,37,38. However, neither subject was colonized by S. aureus in the nares at any of the sample timepoints.
Microbiota sequencing
To analyze temporal changes in the nasal microbiota, the remaining set of paired nasal swabs was subjected to DNA extraction. To reduce the likelihood of bias, the 261 swabs were randomly arrayed across three 96-well plates for the extraction. Additionally, each plate contained one well with a mock community (positive extraction control), and 2 or 3 wells containing no input (negative extraction control). Extracted DNA was used for amplification and MiSeq-based sequencing of the V1-V3 region of the rRNA gene; positive and negative sequencing controls were also included in the sequencing reactions. One sample (Subject 2103 Day 90) yielded no sequence data, even after multiple attempts; the remaining 260 subject samples yielded a total of 18,135,243 raw sequences, with an average of 69,751 sequences per subject. The three positive DNA extraction controls yielded 966,548 sequences (mean, 322,183). Similarly, the three positive sequencing controls generated 368,129 sequences (mean, 122,710) sequences. In contrast, the eight negative DNA extraction and three negative sequencing controls yielded 6299 sequences (mean, 787) and 12,808 sequences (mean, 4269), respectively.
Forward and reverse sequencing reads were trimmed and assembled using FLASH55. Assembled sequences were analyzed using the R Studio implementation of DADA256,57. After quality filtering and chimera removal, a total of 10,265,661 sequences from subject samples remained (56.6%), representing an average of 39,483 sequences per sample. From the control samples, a total of 355,294 (positive extraction controls), 142,401 (positive sequencing controls), 787 (negative extraction controls), and 96 (negative sequencing controls) sequences remained. The sequences assembled into a total of 12,359 amplicon sequence variants (ASVs); these were aligned to the Silva database, with species designations added where possible. The percent abundance of each ASV for each sample is provided in Supplementary Table S2. For downstream analyses, the control sequences were removed. After processing, three trainee samples had fewer than 1000 sequences (Subject 2014 Day 28, Subject 2149 Day 28, and Subject 2198 Day 90) and were removed from the downstream analyses.
Microbiota characterization
The nasal microbiota of the subjects largely consisted of members of the Actinobacteria, Firmicutes, and Proteobacteria phyla. Ten genera averaged ≥ 1% abundance at any time point: Corynebacterium_1, Cutibacterium, and Lawsonella of Actinobacteria; Staphylococcus, Dolosigranulum, Anaerococcus, and Peptoniphilus of Firmicutes; and Moraxella, Haemophilus, and Neisseria of Proteobacteria. The abundance of these genera for each subject at each time point is shown in Supplementary Fig. S1. Based on a visual analysis, the microbiota of some subjects was relatively stable over time (e.g., 2141 and 2207); others changed drastically in composition, often with increases in Proteobacteria (e.g., 2008 and 2220); and other subjects showed steady shifts in their microbiota over time (e.g., 2096 and 2135). At the population level, these changes were visible when the average abundance of the ten genera that averaged ≥ 1% abundance were plotted across time (Fig. 2 and Supplementary Fig. S2). Broadly speaking, the abundance of Actinobacteria and Corynebacterium_1 was unstable, and showed a biphasic pattern of changes; a decrease was observed at day 14, followed by increases at day 28 and 56 and another decrease at day 90. The abundance of Firmicutes was relatively stable across the study, but the contributing genera shifted; a decrease in Staphylococcus and an increase in Dolosigranulum were observed. Finally, the abundance of Proteobacteria represented a mirror image of the Actinobacteria changes; there was an increase at day 14, a decrease at days 28 and 56, and another increase at day 90. Of note, the genera contributing to these increases were different over time: Haemophilus and Moraxella at day 14, but Moraxella and Neisseria at day 90. Statistical analysis revealed that the temporal changes were significant for all of the highest abundance phyla and genera with the exception of Anaerococcus and Peptoniphilus (Supplementary Figs. S3, S4 and Supplementary Tables S3, S4). Overall, while there were observable differences in the nasal microbiota of the trainees, these changes occurred at different times and to a different extent for each trainee.
Figure 2.

Distribution of ten most abundant genera across time. Nasal swabs were collected upon arrival of subjects at Fort Benning (day 0) and then longitudinally at days 14, 28, 56 and 90. The swabs were used to analyze the nasal microbiota of each subject. The sequencing results for the trainees are combined for each time point, and the average abundance of the ten genera that averaged ≥ 1% abundance is plotted across time.
Given that changes differed dramatically across subjects, we hypothesized that host-specific factors influenced the observed stability/instability of the nasal microbiota. To this end, available metadata for each trainee was layered into the analyses. Specifically, we examined the contribution of nasal S. aureus culture (negative or positive), nasal S. aureus colonization status (persistent carriers, intermittent carriers and persistent non-carriers) and subject geographic origin (Midwest, Northeast, South, West). As expected from a prior study41, positive nasal S. aureus culture and persistent colonization status were associated with higher abundance of Firmicutes and Staphylococcus and lower abundance of Actinobacteria and Corynebacterium_1 (Supplementary Figs. S3, S4 and Supplementary Tables S3, S4). The abundance of Corynebacterium_1 was highest in subjects from the Midwest and lowest in subjects from the West, with the opposite observation for Peptoniphilus. Intermittent S. aureus colonization status was associated with an increase in Proteobacteria and Moraxella (Supplementary Fig. S4 and Supplementary Table S4). Further analysis by two-way ANOVA demonstrated that this was specifically due to the increase in Proteobacteria and Moraxella at day 90 among the intermittently colonized subjects (Supplementary Figs. S5, S6 and Supplementary Tables S5, S6).
To determine the genera that were differentially represented in the various groupings by an independent metric, LEfSe analysis was also utilized58. An advantage of this method is that it detects differences among genera of lower abundance. To this end, there were many genera that were differentially represented based on time (Supplementary Fig. S7A). Among the genera that were increased were Lawsonella, Mycobacterium, Sphingomonas, Pantoea, Methylobacterium, and Pseudomonas on Day 14; Cutibacterium and Chryseobacterium at Day 28; and Dolosigranulum and Brachybacterium at Day 90. The genera that were differentially represented based on positive or negative S. aureus culture (Supplementary Fig. S7B) and persistently colonized or not colonized by S. aureus were highly similar (Supplementary Fig. S7C); Staphylococcus, Cutibacterium, Propionibacterium, Anaerococcus, and Peptoniphilus were more highly represented in samples positive for S. aureus culture and in samples from persistently colonized samples. In addition, Corynebacterium_1 and the family Neisseriaceae were more highly represented in trainees that were negative for S. aureus culture and in persistent non-carriers of S. aureus. Moraxella, Dolosigranulum, and Raoultella were highly represented in samples from intermittently colonized subjects. Finally, only two genera were differentially represented based on geographic origin of the subjects (Supplementary Fig. S7D): Corynebacterium_1 in the Midwest and Raoultella in the Northeast.
Alpha and beta diversity analysis
Alpha diversity was measured using three common metrics: observed ASVs to measure richness, and Shannon diversity index and inverse Simpson index to measure diversity. The richness of the samples changed over time and based on the S. aureus nasal colonization category of the subject (Fig. 3 and Supplementary Fig. S8). The number of observed ASVs was lowest at day 0 (mean 63.9 ASVs) and increased over time, with a peak at day 56 (mean 146.6 ASVs) (P < 0.001, Supplementary Table S7). Subjects who were persistently colonized by S. aureus had fewer observed ASVs (77.4 ASVs) compared to those who were not colonized or were intermittently colonized (103.0 and 110 ASVs, respectively, P = 0.033) (Fig. 3). Neither diversity index changed across time or based on metadata (Supplementary Fig. S8 and Supplementary Table S7).
Figure 3.
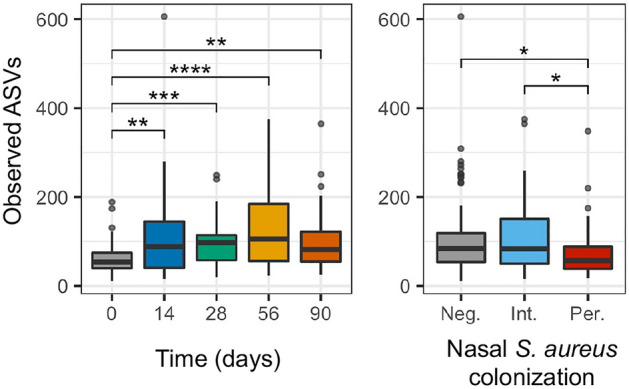
Measurement of alpha diversity. The number of observed ASVs are graphed as a measure of richness. The observed ASVs are displayed over time and based on S. aureus colonization status. S. aureus colonization status is noted as follows: negative (Neg.), intermittent (Int.), or persistent (Per.). For nasal S. aureus colonization, “NA” values are not graphed. Pairwise non-parametric Wilcoxon tests were performed with Benjamini and Hochberg correction for multiple comparisons. Statistical significance is indicated by asterisks (*): *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
To further assess differences between the groups, beta diversity was assessed using the Bray–Curtis metric59, which is calculated based on the structure of the community. For this analysis, beta diversity is measured between each pair of samples, and a matrix of values is returned. The resulting distance matrix was visualized using principal co-ordinate analysis (PCoA) wherein samples that are most dissimilar are plotted the farthest apart, and samples that are more similar are plotted closer together. PCoA visualization of the temporal beta diversity by individual subject is provided in Supplementary Fig. S9. Some subjects were relatively stable over time; the points remained close to each other (e.g., 2145 and 2207). In contrast, many subjects showed substantial temporal changes (e.g., 2012 and 2220); the points were scattered across the PCoA plot. These individualized changes in beta diversity are in keeping with the trainee-specific differences in the abundance of phyla and genera highlighted above (Supplementary Fig. S1).
Beta diversity analysis combined with trainee metadata revealed unexpected interactions. Namely, distinct patterns were observed based on the geographic origin of the subjects; samples from the West and Northeast largely segregated to the upper quadrants and samples from the Midwest and South scattered throughout the plot (Supplementary Fig. S10). This finding was unexpected since only a few significant differences in the abundance of phyla and genera were observed when samples were grouped by geographic origin (Supplementary Figs. S3–S6). Thus, the differences elucidated by the beta diversity analysis are likely driven by the contribution of lower abundance phyla and genera to the structure of the microbiota.
Further examination of the beta diversity plot from the Northeast revealed that the five points located in the lower right-hand quadrant and distant from the other points from the Northeast samples, were from a single subject (2141). Reexamination of the participant questionnaires revealed that subject 2141 was the only subject in the platoon who self-reported the use of antibiotics in the 6 months prior to study enrollment. Thus, antibiotic use may be responsible for diversity differences observed in this individual. To eliminate this potential confounding factor, this subject was removed from downstream analysis. Indeed, upon removal of samples from this subject, the orientation of the PCoA shifted, rotating approximately 180°, with subjects from the Northeast and West now clustered in the lower quadrants of the plot (Fig. 4D).
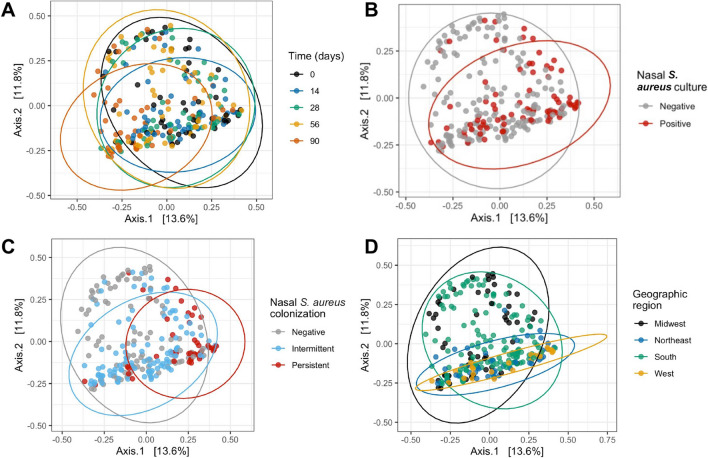
Figure 4.
Principal co-ordinate analysis (PCoA). Bray–Curtis distance of the samples is visualized by PCoA based on time (A), nasal S. aureus culture (B), nasal S. aureus colonization (C), and geographic region (D). For nasal S. aureus colonization, “NA” values are not graphed. Bray–Curtis distances were ordinated by principal co-ordinate analysis (PCoA) using phyloseq, and graphed using phyloseq and ggplot.
Visual analysis of samples based on time, S. aureus culture, S. aureus colonization grouping, and geographic origin elucidated differences within groups (Fig. 4). For example, the day 14 samples were compressed into a smaller area than samples from days 0, 28, and 56. Furthermore, the 90 samples shifted toward the lower left quadrant of the plot (Fig. 4A). These observations suggest time-dependent oscillations in the nasal microbiota structure. While the microbiota at days 0, 28, and 56 were similar, a microbiota shift occurred at day 14. This shift was largely reversed by day 28. An additional shift occurred between days 56 and 90. Next, distribution of samples from S. aureus culture positive trainees were compressed as compared to the culture negative samples (Fig. 4B). Finally, samples from subjects who were intermittently colonized by S. aureus tended to cluster to the lower quadrants, and those from persistently colonized subjects were segregated to a smaller cluster in the lower right quadrant (Fig. 4C).
The statistical significance of these visual observations was next assessed using PERMANOVA analysis: time, S. aureus culture, S. aureus colonization status, geographic origin, and subject were all significant factors (Table 2). Pairwise PERMANOVA analysis further elucidated the relationship between individual factors (Supplementary Table S8). The microbiota at days 0, 28, and 56 were not significantly different from one another. However, day 14 was significantly different from every other time point except day 28, and day 90 was significantly different from every other time point. All samples from subjects that were not colonized, intermittently colonized, and persistently S. aureus colonized were significantly different from each other. Finally, the samples from each geographic origin were significantly different from the others.
Table 2.
Bray–Curtis beta diversity distances were tested by PERMANOVA analysis. One-way comparisons were tested: time, nasal S. aureus culture, nasal S. aureus colonization, geographic region, and subject. For nasal S. aureus colonization, subjects with “NA” status were removed. PERMANOVA was performed using vegan’s adonis function78, using 999 permutations.
| Parameter | Pr (> F) |
|---|---|
| Time | 0.001 |
| Nasal S. aureus culture | 0.001 |
| Nasal S. aureus colonization | 0.001 |
| Geographic region | 0.001 |
| Subject | 0.001 |
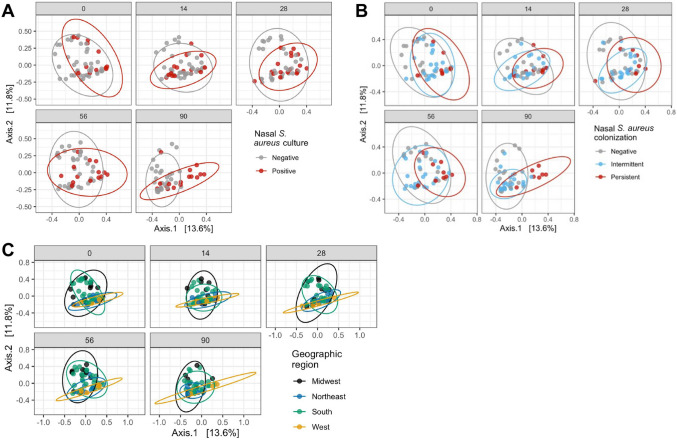
To understand the temporal impact of nasal S. aureus culture, nasal S. aureus colonization status, and geographic origin, each factor was assessed by sample time point (Fig. 5). The difference between samples from S. aureus culture negative or positive trainees were most apparent at the end of the study; S. aureus negative samples scattered throughout the plot at all time points, but samples from S. aureus positive individuals condensed into a smaller area by day 90 (Fig. 5A). Similarly, the difference between samples from subjects not colonized, intermittently colonized or persistently colonized by S. aureus was most apparent at day 90; at this point samples from persistently colonized subjects segregated in the lower quadrants and those from intermittently colonized subjects segregated in the lower left quadrant (Fig. 5B). In contrast, the samples from subjects from different geographic regions were different from one another at all study visits but were temporally consistent at all time points: those from the Northeast and West remained largely within the lower quadrant and those from the Midwest and South were scattered throughout the plot (Fig. 5C).
Figure 5.
Principal co-ordinate analysis (PCoA) over time. Bray–Curtis distance of the samples over time is visualized by PCoA, colored by nasal S. aureus culture (A), nasal S. aureus colonization (B), and geographic region (C). For nasal S. aureus colonization, “NA” values are not graphed. Bray–Curtis distances were ordinated by principal co-ordinate analysis (PCoA) using phyloseq, and graphed using phyloseq and ggplot.
Finally, we examined the contribution of race and ethnicity to the observed beta diversity (Supplementary Fig. S11). Several groups were visually distinct from the others, especially the Non-Hispanic/Latino, White and Black and the Hispanic/Latino, Not reported. However, the low number of subjects that fell into each of these categories (2 subjects each for the categories listed above) limits any conclusions about nasal microbiota difference that may be attributed to race and ethnicity in this study.
Longitudinal analyses
A major goal of this study was to test the hypothesis that the congregate setting would lead to an overall convergence of the individuals’ nasal microbiomes to a more similar composition over time. To measure these changes, the distribution of Bray–Curtis distance values between all pairs of subjects at each time point were plotted (Supplementary Fig. S12). Overall, the dissimilarity between the samples was high, with median values of approximately 0.8. Temporal analysis of the distribution of distance values demonstrated that the distance between the subjects increased over time; the lowest distance (0.77) was seen at day 0 and the highest distance (0.83) was seen at day 90. Thus, instead of converging, the microbiota of the subjects actually appeared to slightly diverge over time; however, the biological significance of a change in diversity from 0.77 to 0.83 units is unclear. Of note, this result was in contrast to the results obtained by visual analysis of the Bray–Curtis PCoA plot over time (Fig. 4); therein the day 90 samples clustered together, implying that they were more similar. The observed differences between these two analyses perhaps demonstrates one of the drawbacks of a purely visual analysis of a PCoA plot, which attempts to represent highly dimensional data in two dimensions.
To further explore the apparent divergence in the microbiota, the dispersivity of the Bray–Curtis distances over time was measured using PERMDISP analysis60 (Fig. 6). This analysis measures the distance of each sample to the group centroid; larger distances from the centroid indicate less similarity in the group, whereas smaller distances from the centroid indicate more similarity in the group. The dispersivity of the samples was larger at day 90 than at other time points; as a factor, time was significant (ANOVA, P = 0.04057). Pairwise analysis determined that the difference was driven by the comparison of Day 0 to Day 90 (Tukey’s correction for multiple comparisons, P = 0.026883), the only significant comparison.
Figure 6.
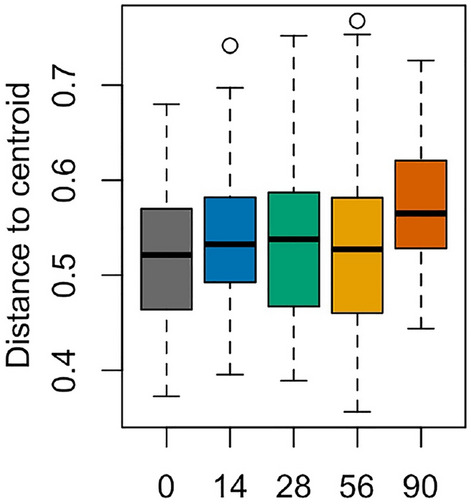
PERMDISP analysis to measure dispersivity. The homogeneity Bray–Curtis distances between the groups were measured using the PERMDISP60 analysis from the vegan package and graphed using ggplot. ANOVA was performed to test for differences, followed by pairwise comparisons with Tukey’s correction for multiple comparisons.
Lastly, to measure the stability of the microbiota over time, the pairwise time point-to-time point variation between same-subject samples was calculated (Fig. 7). This analysis measured the beta diversity of each subjects’ samples over time. Analysis of sequential time points (Day 0 to Day 14, Day 14 to Day 28, etc.) indicated that the dissimilarity of the microbiota between Day 0 and Day 14 was 0.51 units. The microbiota were more similar through Day 56 (0.41 units between Day 14 and Day 28 and 0.39 units between Day 28 and Day 56), and then most dissimilar between Day 56 and Day 90 (0.56 units). Analysis of all pairwise time points showed that the greatest dissimilarities were between Day 90 and any other day. These data suggest that the microbiota changed within the first 2 weeks, partly stabilized during training, and then shifted again before the last sample visit.
Figure 7.
Stability and diversity of the microbiota. The changes in stability and diversity of the microbiota over the course of the study within subjects were evaluated using the Bray–Curtis distance. The pairwise time point-to-time point variation between same-subject samples was calculated and plotted as boxplots. Pairwise Wilcoxon tests with FDR correction for multiple comparisons were performed. Statistical significance is indicated by asterisks (*): *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
Taken together, the temporal analyses indicated that the microbiota of the trainees did change significantly over time. However, contrary to our original convergence hypothesis, the nasal microbiota of the subjects was more similar at day 0 than at day 90. The change in the microbiota of each subject was greatest at the beginning and end of the study, suggesting a biphasic evolution in the nasal microbiota. An early change occurred (between days 0 and 14), followed by some stability in the microbiota for several weeks (between days 14, 28, and 56), and finally another large shift occurred (between days 56 and 90). Thus, nasal microbiota evolution within the congregate setting imposed by military training is a complex process that appears to be affected by numerous factors.
Discussion
To investigate the changes in the nasal microbiota that occur during initial military training, herein, we longitudinally assessed 53 Army trainees from a single platoon at Fort Benning, Georgia. Consistent with previous studies that examined the composition of the nasal microbiome in predominantly healthy individuals, the nasal microbiota of the cohort was dominated by 3 phyla: Actinobacteria, Firmicutes, and Proteobacteria8,16,41,61,62. Similarly, the prevalence of S. aureus nasal colonization of the trainees was comparable to previous reports16,35,53,54, with 39.2% of trainees not colonized, 43.1% of subjects intermittently colonized, and 17.6% of individuals persistently colonized (Fig. 1). As expected63, the number of S. aureus positive cultures increased from 22.6% of the recruits on day 0 to 37.2% of the trainees on day 90. Interestingly, of the two subjects that developed purulent SSTI, neither one was colonized by S. aureus in the nares at any of the sampled timepoints. This was a surprise since S. aureus nasal colonization is a risk factor for SSTI development35,37,38. It is possible that the congregate setting and outdoor activities engaged in by these subjects contributed to the development of the SSTI without a prior nasal colonization event. Alternatively, transient nasal colonization may have occurred outside of the obtained samples.
The microbiota of the individual trainees appeared strongly linked to S. aureus culture results and colonization status. S. aureus positive samples and samples from persistently colonized individuals contained similar genera, while samples that were negative for S. aureus and samples from non-colonized individuals contained a higher representation of Corynebacterium_1 and the Neisseriaceae family (Supplementary Fig. S7B,C). Furthermore, we observed a significant increase in Moraxella at day 90 specifically among intermittently colonized subjects (Fig. 2, Supplementary Figs. S2, S4, and Supplementary Table S4). Of note, some members of Moraxella (Moraxella catarrhalis, specifically) are clinically relevant in respiratory disease64,65. The reasons why some individuals are persistently nasally colonized by S. aureus while others are intermittent carriers or persistent non-carriers remains unclear. However, it is clear that the presence of S. aureus within the nasal niche has a dramatic effect on the nasal microbiome. For example, in subjects that are persistently colonized by S. aureus, the microbiota is dominated by S. aureus (Supplementary Fig. S6). Thus, this bacterium may actively prevent colonization by other opportunistic bacteria. Similarly, in subjects that are persistent non-carriers of S. aureus, perhaps the dominant microbiota is able to prevent S. aureus (and possibly other opportunistic bacteria) from colonizing. In line with this, perhaps the nasal microbiome of intermittently colonized individuals represents a less robust microbiota that is unable to prevent temporary colonization by S. aureus, and, in the case of the trainees described here, Moraxella (Supplementary Fig. S6). This fluctuation may extend to other opportunistic pathogens, preventing or permitting colonization.
A temporal analysis of the stability of the microbiota revealed noteworthy shifts in composition at two distinct time points. The first change appeared to occur between day 0 and day 14, with the microbiota of the individual subjects then remaining stable for several weeks. Another large shift in the trainees’ microbiota then transpired between day 56 and day 90. These particular shifts could be related to activities in which the recruits were engaged during those specific time periods. For example, during week 12, the Infantry training cycle culminates in a week-long field training exercise, wherein soldiers conduct extensive training activities in austere environments and sleep in tents for 7 days before returning to the barracks. Of note, several of the genera identified in the LEfSe analysis are typically located in environmental samples, including soil and water66–71. Detecting these genera in the nares of trainees is not surprising given the amount of time trainees spend outside and the nature of their training that requires intense interaction with their environment.
Due to the small size of the cohort, some metadata could not be reliably included in tests, including ethnicity and race. Additionally, while the contribution of geographic origin to the microbiota is supported elsewhere3,5–7, our observations were complicated by the uneven number of subjects from each geographic region. Our cohort included 12 subjects from the Midwest, 10 from the Northeast, 26 from the South, and only 5 from the West. Furthermore, the assignment of geographic origin was based on the last permanent address provided by each subject in the study intake questionnaire; thus, the amount of time lived at that address and any gaps in which the subject had lived elsewhere were not captured. Consequently, the geographic location that potentially contributed to the nasal microbiome of a given individual may not be the same as the geographic region assigned through the intake questionnaire. Even given these limitations, the individuals’ microbiota still displayed some clustering based on geographic assignment when beta diversity was analyzed.
LEfSe analysis revealed important connections involving the Staphylococcus and Corynebacterium genera. Unsurprisingly, Staphylococcus was highly represented in individuals that were positive for S. aureus culture and in samples from persistently colonized individuals. In contrast, Corynebacterium_1 was more highly represented in samples from trainees that were negative for S. aureus culture and in non-carriers of S. aureus (Supplementary Fig. S7). These data are consistent with our previous nasal microbiome studies that highlight an inverse relationship between colonization with Corynebacterium species and the presence of Staphylococcus species41,44. Furthermore, these data are in agreement with in vitro mechanistic studies that have shown the ability of Corynebacterium to kill pathogenic S. aureus14–16. If these in vitro results actually represent what happens within the nasal environment, this would explain why individuals carrying high levels of Corynebacterium_1 tend to be S. aureus negative or non-carriers. Similarly, it is worth noting that clinical studies show that direct introduction of Corynebacterium species into the nasal cavity can eradicate the S. aureus that is colonizing this niche72. Of note, the LEfSe analysis based on geographic origin revealed the differential representation of Corynebacterium_1 in trainees from the Midwest (Supplementary Fig. S7). In addition, the trainees from this region had fewer S. aureus-positive cultures than trainees from the other regions; after removal of the two individuals that did not complete all study visits, 20% of samples obtained from those from the Midwest were positive, while 26% were positive from those from the Northeast, 31% from those from the South and 60% from those from the West (Supplementary Fig. S10). Given that nasal colonization with S. aureus is a risk factor for SSTI development, this leads to the intriguing possibility that those from the Midwest may be less likely to develop SSTI. To this end, future studies will address the contribution of this and other factors to SSTI development within military trainees.
To complete our longitudinal analysis of the nasal microbiota of platoon members, we analyzed the differences between the individuals’ microbiota at the start of the study and at the end of the study. We expected to see convergence of the microbiota similar to that observed in a study of cohabitating cadets at the U.S. Air Force Academy23. However, we did not find convergence among the microbiota and indeed noted divergence over time. Interestingly, the microbiota of the individual trainees appeared more similar at day 0 than at day 90. Our contrasting observation may result from a difference in the activities of this cohort, especially the outdoor activities performed by the platoon analyzed in this study.
Conclusions
The congregate setting encountered during military training provides numerous, unique environmental and personal interactions for trainees. As such, the numerous encounters with microbes may provide an assault on the existing microbiota of the trainee. Indeed, the longitudinal investigation of the evolution of nasal microbiota presented here reveals a complex process that is affected by numerous factors. The stability of the nasal microbiota varied dramatically among the individuals, and the major driver of nasal microbiota stability appears to be the geographic origin of the trainees. Additionally, the stability of the nasal microbiota seems to correlate with S. aureus colonization status; however, the exact nature of the association between S. aureus colonization and nasal microbiota stability remains unclear. Indeed, a lack of stability within the nasal microbiota may facilitate colonization by S. aureus. These results suggest that broad-spectrum efforts to prevent or eradicate S. aureus colonization and subsequently minimize SSTIs within military settings may not be an effective approach. In contrast, a more individualized approach may ultimately increase the efficacy of these attempts to reduce the burden of SSTIs within the Military Health System.
Methods
Subject recruitment
A longitudinal cohort study was conducted, as described in detail elsewhere52. Trainees from the US Army Infantry at Fort Benning, Georgia, were recruited. The company commenced a 14-week training cycle in September 2015. Infantry training companies are composed of approximately 200 trainees, further segregated into four platoons. Samples from 53 trainees from a single platoon were analyzed for this longitudinal study. Among these individuals, 2 subjects developed a purulent SSTI: Subject 2013 on day 94; and Subject 2197 on day 22. The study was approved by the Uniformed Services University Institutional Review Board (IDCRP-090) and was conducted in accordance with the relevant guidelines and regulations of the institution. Informed consent was obtained from all study participants.
Sample collection
Paired nasal swabs were obtained upon arrival (day 0) and at each study visit (day 14, 28, 56, and 90) by study staff for a total of five study visits. At the initial study visit, trainees completed questionnaires for demographic and MRSA risk factor information. From each set of paired nasal swabs, one swab was utilized to determine the presence of S. aureus by culture at the Benning Martin Army Community Hospital clinical microbiology laboratory as previously described73. The second swab was shipped to Uniformed Services University in Bethesda, MD for DNA extraction and microbiome analysis. Swabs were transported on dry ice and stored at − 80 °C until extraction, as described below.
S. aureus colonization status was defined based on the percent of study visits at which a subject was positive by culture, as follows: persistently colonized (≥ 80% positive); intermittently colonized (≥ 20% and < 80% positive); and not colonized (0% positive). Two subjects did not complete the five study visits, and no colonization status was assigned.
DNA extraction
DNA was extracted from the nasal swabs using the PowerSoil-htp kit (MoBio), which combines chemical and mechanical lysis. A total of 261 sample swabs were arrayed across three 96-well plates. To extract the DNA from the sample, the swab head was broken into the 96-well plate. The location within the plates was randomized, with the exception that no subject could have all five samples in a single 96-well plate to minimize any possible plate-to-plate confounders. As a positive DNA extraction control, a mock microbial community (HM-280, BEI resources) was extracted in parallel. For this, approximately 1 × 108 cells were added to one empty well within each 96-well plate, for a total of three positive controls. As a negative DNA extraction control, eight wells (3 in the first and second plate, and 2 in the third plate) were left empty. The manufacturer’s instructions were used with the following adaptations. PowerSoil bead solution and Solution C1 were mixed, transferred to each well of the plate, and incubated at 65 °C for 15 min prior to mechanical lysis. DNA was eluted from the silica membrane using 100 µL nuclease-free water (IDT).
Microbiome library preparation
The V1–V3 region of the bacterial 16S rRNA gene was amplified using primers modified to include Illumina adapters, 27F (5′-AGAGTTTGATCCTGGCTCAG) and 534R (5′-ATTACCGCGGCTGCTGG) as previously described74–76. The following PCR conditions were used: 2.5 μL 10 × PCR Buffer, 4 μL dNTP Mix, 0.25 μL Takara LA Taq Polymerase (Clontech), 1 μL 27F (10 μM), 1 μL 534R (10 μM), 14.25 μL PCR Water (Qiagen), and 2.0 μL DNA. Reactions were performed in duplicate for 30 cycles, combined, purified using Agencourt AmpureXP (Beckman Coulter), and quantified using Quantifluor dsDNA Kit (Promega). Equivalent amounts of amplicons were pooled together, purified with MinElute PCR purification kit (Qiagen), and sequenced on an Illumina MiSeq along with the positive and negative sequencing controls. Due to a primer synthesis error, eleven of the patient samples yielded no reads. Thus, these samples were separately reamplified with corrected primers and sequenced. The resulting data were combined with the sequencing data from the other samples and then analyzed as described below.
Sequencing analysis
V1–V3 16S rRNA amplicons were sequenced (2 × 300 bp MiSeq read pairs) and demultiplexed. Read pairs were quality trimmed using trimBWAstyle.pl (-q 20), trimmed of primers using tagcleaner.pl (v 0.16; -tag1 ATTACCGCGGCTGCTGG -tag2 AGAGTTTGATCCTGGCTCAG), filtered to remove short reads (< 246 nt) and merged using FLASH (v 1.2.11; default parameters)55.
The 16S rRNA gene sequences were processed using DADA2 v. 1.10.156,57. FilterAndTrim was executed using default parameters for maximum N (maxN), truncation after a specific quality score (truncQ), and removal of phiX; in addition, reads were of length less than 400 base pairs were removed (minLen), and the maxEE score was set to 2. The error model was performed using 2 × 108 bases, with multithread set to true. The data were dereplicated using derepFastq, and this dereplicated dataset was used for the sample inference algorithm. As the reads had already been overlapped, the step to merge forward and reverse reads was skipped. Chimeric sequences represented less than 3% of the amplicon sequence variants (ASVs), and were removed. Taxonomy was assigned using the Silva database (v132), with the optional species assignment.
Processing and analysis were performed using Phyloseq77. Taxonomic filtering was performed to remove the following: ASVs from five phyla that were represented with two or fewer ASVs (Deferribacteres, Dependentiae, Lentisphaerae, Nitrospirae, and Rokubacteria); ASVs from one eukaryotic phylum (Euglenozoa, 10 ASVs); and ASVs that did not classify to a phylum (180 ASVs). After taxonomic analysis, the positive and negative control samples (6 and 11 samples, respectively) were removed from the dataset for the downstream analyses. Post-DADA2 processing, 3 samples contained less than 1000 reads, and were also removed (Subject 2014 Day 28, Subject 2149 Day 28, and Subject 2198 Day 90).
Bar plots of percent abundance of phyla or genera were generated by agglomerating ASVs to the phylum or genus taxonomic level, keeping all taxa that classified to “NA” (NArm = FALSE), transforming read counts to relative abundance, and melting the phyloseq object into a matrix. Agglomerated phyla or genera that represented < 1% of reads at any time point were merged together using the summarize function of dplyr, as “Other”. Scatterplots of phyla or genera were generated using the agglomerated datasets. Alpha diversity (observed ASVs, Shannon diversity index, and Inverse Simpson index) and beta diversity (Bray–Curtis) were measured using phyloseq. Bray–Curtis distances were ordinated by principal co-ordinate analysis (PCoA) using phyloseq, and graphed using phyloseq and ggplot. All remaining graphs were generated using ggplot. Where shown, 95% confidence ellipses were plotted using stat_ellipse (ggplot). Graphs were arranged into grids using plot_grid (cowplots).
LEfSe analysis58 was performed using the Galaxy web application (https://huttenhower.sph.harvard.edu/galaxy). For each metadata set analyzed, the abundance of each agglomerated genus was uploaded as tabular data. Differences in abundance were tested using Kruskal–Wallis (P < 0.01) with the less strict one-against-all analysis. Differential features of 2 LDA were plotted.
Statistics
Statistical analyses were conducted using R. Differences in abundance of genera or phyla were tested using one-way ANOVA across a single grouping or using two-way ANOVA using time and another grouping. Normality of alpha diversity measures was determined using the Shapiro–Wilk test, in which only the Shannon index was found to be normal (W = 0.99161, P = 0.1505). Subsequently, parametric (ANOVA and Student’s t) or non-parametric (Kruskal–Wallis Rank Sum and Wilcoxon Rank Sum) tests were employed. Where significant associations were found, pairwise Student’s t and pairwise Wilcoxon Rank Sum tests were employed, with Benjamini and Hochberg method (FDR) to correct for multiple comparisons. The vegan package was used to test one-way differences in beta diversity (PERMANOVA), for analysis of similarity (ANOSIM), and for beta dispersivity (PERMDISP) with one-way ANOVA to test for differences. Two-way differences in beta diversity were tested using pairwise PERMANOVA, using an adaptation of vegan’s adonis function78. Differences in Bray–Curtis distances were performed by pairwise Wilcoxon test with FDR correction for multiple comparisons.
Ethics approval and consent to participate
The study was approved by the Uniformed Services University Institutional Review Board (IDCRP-090).
Supplementary Information
Acknowledgements
We would like to thank the NISC and NIH High Performance Computing Resources, Sean Conlan, Clayton Deming, and Julie A. Segre for their assistance with microbiome sequencing and bioinformatic analysis. We would also like to thank Cara Olsen from the Biostatistics Consulting Center at USUHS for her assistance with statistical analysis. We thank Ryan Johnson for providing his expertise on microbiome analysis programs. The following reagent was obtained through BEI Resources, NIAID, NIH as part of the Human Microbiome Project: Cells from Microbial Mock Community C in PBS, HM-280.
Disclaimer
The views expressed are the sole responsibility of the author(s) and do not necessarily reflect the views, opinions or policies of Uniformed Services University of the Health Sciences (USU), Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., the National Institute of Health or the Department of Health and Human Services, the Department of Defense (DoD), the Departments of the Army, Navy, or Air Force, or the U.S. Government. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. Government.
Author contributions
D.M., E.M., J.B., M.E., and D.T. conceived and designed the study and experiments. F.B., D.M., and J.W. wrote the manuscript. N.L. and P.C. processed the collected swabs for S. aureus colonization status. F.B. performed the microbiome-related experiments and analyzed the data. E.M., J.B., P.C., N.L., M.E., and C.E. coordinated the sample collection at Fort Benning, Georgia. All authors reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work (IDCRP-090) was conducted by the Infectious Disease Clinical Research Program (IDCRP), a Department of Defense (DoD) program executed by the Uniformed Services University of the Health Sciences (USU) through a cooperative agreement with The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. This project was supported with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), under Inter‐Agency Agreement Y1-Al-5072 (to E. Millar and D. R. Tribble) and from the Defense Health Program, U.S. Department of Defense, under award HU0001190002 (to E. Millar and D. R. Tribble). This study was supported by a U.S. Department of Defense Program project grant (HT9404-12-1-0019 to D. S. Merrell) and a Military Infectious Diseases Research Program award (HU0001-15-2-0031 to D. S. Merrell).
Data availability
The datasets generated and analyzed during the current study are available in the NCBI under BioProject ID: PRJNA780476 [https://dataview.ncbi.nlm.nih.gov/object/PRJNA780476].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-15059-z.
References
- 1.Brooks AW, Priya S, Blekhman R, Bordenstein SR. Gut microbiota diversity across ethnicities in the United States. PLoS Biol. 2018;16(12):e2006842. doi: 10.1371/journal.pbio.2006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat. Med. 2018;24(4):392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez A, Vazquez-Baeza Y, Knight R. SnapShot: the human microbiome. Cell. 2014;158(3):690.e1. doi: 10.1016/j.cell.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 4.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8(1):51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176(3):649–662.e20. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One. 2010;5(5):e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krismer B, Weidenmaier C, Zipperer A, Peschel A. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol. 2017;15(11):675–687. doi: 10.1038/nrmicro.2017.104. [DOI] [PubMed] [Google Scholar]
- 10.Reyes N, Montes O, Figueroa S, Tiwari R, Sollecito CC, Emmerich R, et al. Staphylococcus aureus nasal carriage and microbiome composition among medical students from Colombia: A cross-sectional study. F1000Res. 2020;9:78. doi: 10.12688/f1000research.22035.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen GJ, Brüggemann H. Bacterial skin commensals and their role as host guardians. Benef. Microbes. 2014;5(2):201–215. doi: 10.3920/bm2012.0062. [DOI] [PubMed] [Google Scholar]
- 12.Cogen AL, Yamasaki K, Muto J, Sanchez KM, Crotty Alexander L, Tanios J, et al. Staphylococcus epidermidis antimicrobial delta-toxin (phenol-soluble modulin-gamma) cooperates with host antimicrobial peptides to kill group A Streptococcus. PLoS One. 2010;5(1):e8557. doi: 10.1371/journal.pone.0008557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, MacLeod DT, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Investig. Dermatol. 2010;130(1):192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy BL, Dickey SW, Plaut RD, Riggins DP, Stibitz S, Otto M, et al. Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy. MBio. 2019;10(1):e02491-18. doi: 10.1128/mBio.02491-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardy BL, Merrell DS. Friend or foe: Interbacterial competition in the nasal cavity. J Bacteriol. 2021;203(5):e00480-20. doi: 10.1128/jb.00480-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe. 2013;14(6):631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. MBio. 2016;7(1):e01725–15. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat. Rev. Microbiol. 2018;16(3):143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 19.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front. Microbiol. 2016;7:1230. doi: 10.3389/fmicb.2016.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533(7602):255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, et al. Daily Sampling reveals personalized diet–microbiome associations in humans. Cell Host Microbe. 2019;25(6):789–802.e5. doi: 10.1016/j.chom.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Oh J, Byrd AL, Park M, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell. 2016;165(4):854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma A, Richardson M, Cralle L, Stamper CE, Maestre JP, Stearns-Yoder KA, et al. Longitudinal homogenization of the microbiome between both occupants and the built environment in a cohort of United States Air Force Cadets. Microbiome. 2019;7(1):70. doi: 10.1186/s40168-019-0686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535(7610):85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: Pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018;26(3):229–242. doi: 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumas A, Bernard L, Poquet Y, Lugo-Villarino G, Neyrolles O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell Microbiol. 2018;20(12):e12966. doi: 10.1111/cmi.12966. [DOI] [PubMed] [Google Scholar]
- 27.Tsolis RM, Bäumler AJ. Gastrointestinal host-pathogen interaction in the age of microbiome research. Curr. Opin. Microbiol. 2020;53:78–89. doi: 10.1016/j.mib.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Armed Forces Health Surveillance Center Bacterial skin infections, active component, U.S. Armed Forces, 2000–2012. MSMR. 2013;20(12):2–7. [PubMed] [Google Scholar]
- 29.Landrum ML, Neumann C, Cook C, Chukwuma U, Ellis MW, Hospenthal DR, et al. Epidemiology of Staphylococcus aureus blood and skin and soft tissue infections in the US military health system, 2005–2010. JAMA. 2012;308(1):50–59. doi: 10.1001/jama.2012.7139. [DOI] [PubMed] [Google Scholar]
- 30.Stahlman S, Williams VF, Oh GT, Millar EV, Bennett JW. Skin and soft tissue infections, active component, U.S. Armed Forces, 2013–2016. MSMR. 2017;24(7):2–11. [PubMed] [Google Scholar]
- 31.Campbell KM, Vaughn AF, Russell KL, Smith B, Jimenez DL, Barrozo CP, et al. Risk factors for community-associated methicillin-resistant Staphylococcus aureus infections in an outbreak of disease among military trainees in San Diego, California, in 2002. J. Clin. Microbiol. 2004;42(9):4050–4053. doi: 10.1128/jcm.42.9.4050-4053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millar EV, Rice GK, Elassal EM, Schlett CD, Bennett JW, Redden CL, et al. Genomic characterization of USA300 methicillin-resistant Staphylococcus aureus (MRSA) to evaluate intraclass transmission and recurrence of skin and soft tissue infection (SSTI) among high-risk military trainees. Clin. Infect. Dis. 2017;65(3):461–468. doi: 10.1093/cid/cix327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zinderman CE, Conner B, Malakooti MA, LaMar JE, Armstrong A, Bohnker BK. Community-acquired methicillin-resistant Staphylococcus aureus among military recruits. Emerg. Infect. Dis. 2004;10(5):941–944. doi: 10.3201/eid1005.030604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kluytmans JA, Wertheim HF. Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection. 2005;33(1):3–8. doi: 10.1007/s15010-005-4012-9. [DOI] [PubMed] [Google Scholar]
- 35.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005;5(12):751–762. doi: 10.1016/s1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 36.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N. Engl. J. Med. 2001;344(1):11–16. doi: 10.1056/nejm200101043440102. [DOI] [PubMed] [Google Scholar]
- 37.Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, et al. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet. 2004;364(9435):703–705. doi: 10.1016/s0140-6736(04)16897-9. [DOI] [PubMed] [Google Scholar]
- 38.Davis KA, Stewart JJ, Crouch HK, Florez CE, Hospenthal DR. Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin. Infect. Dis. 2004;39(6):776–782. doi: 10.1086/422997. [DOI] [PubMed] [Google Scholar]
- 39.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010;23(3):616–687. doi: 10.1128/cmr.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ki V, Rotstein C. Bacterial skin and soft tissue infections in adults: A review of their epidemiology, pathogenesis, diagnosis, treatment and site of care. Can. J. Infect. Dis. Med. Microbiol. 2008;19(2):173–184. doi: 10.1155/2008/846453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson RC, Ellis MW, Lanier JB, Schlett CD, Cui T, Merrell DS. Correlation between nasal microbiome composition and remote purulent skin and soft tissue infections. Infect. Immun. 2015;83(2):802–811. doi: 10.1128/iai.02664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson RC, Ellis MW, Schlett CD, Millar EV, LaBreck PT, Mor D, et al. Bacterial etiology and risk factors associated with cellulitis and purulent skin abscesses in military trainees. PLoS One. 2016;11(10):e0165491. doi: 10.1371/journal.pone.0165491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millar EV, Rice GK, Schlett CD, Elassal EM, Cer RZ, Frey KG, et al. Genomic epidemiology of MRSA infection and colonization isolates among military trainees with skin and soft tissue infection. Infection. 2019;47(5):729–737. doi: 10.1007/s15010-019-01282-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh J, Johnson RC, Schlett CD, Elassal EM, Crawford KB, Mor D, et al. Multi-body-site microbiome and culture profiling of military trainees suffering from skin and soft tissue infections at fort Benning, Georgia. mSphere. 2016;1(5):e00232-16. doi: 10.1128/mSphere.00232-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, R. S. M., Eugene, V., Callendrello, A., English, C. E., Elassal, E. M., Ellis, M. W., Bennett, J. W., Hanage, W. P. Genomic epidemiology of methicillin-resistant Staphylococcus aureus in two cohorts of high-risk military trainees (submitted).
- 46.David MZ, Mennella C, Mansour M, Boyle-Vavra S, Daum RS. Predominance of methicillin-resistant Staphylococcus aureus among pathogens causing skin and soft tissue infections in a large urban jail: Risk factors and recurrence rates. J. Clin. Microbiol. 2008;46(10):3222–3227. doi: 10.1128/jcm.01423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dwyer LL, Harris-Kojetin LD, Valverde RH, Frazier JM, Simon AE, Stone ND, et al. Infections in long-term care populations in the United States. J. Am. Geriatr. Soc. 2013;61(3):342–349. doi: 10.1111/jgs.12153. [DOI] [PubMed] [Google Scholar]
- 48.Feldstein D, Sloane PD, Weber D, Ward K, Reed D, Zimmerman S. Current prescribing practices for skin and soft tissue infections in nursing homes. J. Am. Med. Dir. Assoc. 2017;18(3):265–270. doi: 10.1016/j.jamda.2016.09.024. [DOI] [PubMed] [Google Scholar]
- 49.Haysom L, Cross M, Anastasas R, Moore E, Hampton S. Prevalence and risk factors for methicillin-resistant Staphylococcus aureus (MRSA) infections in custodial populations: A systematic review. J. Correct Health Care. 2018;24(2):197–213. doi: 10.1177/1078345818765271. [DOI] [PubMed] [Google Scholar]
- 50.Karanika S, Kinamon T, Grigoras C, Mylonakis E. Colonization with methicillin-resistant Staphylococcus aureus and risk for infection among asymptomatic athletes: A systematic review and metaanalysis. Clin. Infect. Dis. 2016;63(2):195–204. doi: 10.1093/cid/ciw240. [DOI] [PubMed] [Google Scholar]
- 51.Kazakova SV, Hageman JC, Matava M, Srinivasan A, Phelan L, Garfinkel B, et al. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 2005;352(5):468–475. doi: 10.1056/NEJMoa042859. [DOI] [PubMed] [Google Scholar]
- 52.Lee RS, Millar EV, Callendrello A, English CE, Elassal EM, Ellis MW, et al. Genomic epidemiology of methicillin-resistant Staphylococcus aureus in two cohorts of high-risk military trainees. medRxiv. 2019;6:19011445. doi: 10.1101/19011445. [DOI] [Google Scholar]
- 53.Nouwen JL, Ott A, Kluytmans-Vandenbergh MF, Boelens HA, Hofman A, van Belkum A, et al. Predicting the Staphylococcus aureus nasal carrier state: Derivation and validation of a “culture rule”. Clin. Infect. Dis. 2004;39(6):806–811. doi: 10.1086/423376. [DOI] [PubMed] [Google Scholar]
- 54.van Belkum A, Verkaik NJ, de Vogel CP, Boelens HA, Verveer J, Nouwen JL, et al. Reclassification of Staphylococcus aureus nasal carriage types. J. Infect. Dis. 2009;199(12):1820–1826. doi: 10.1086/599119. [DOI] [PubMed] [Google Scholar]
- 55.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Res. 2016;5:1492. doi: 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 1957;27(4):325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 60.Anderson MJ. Distance-based tests for homogeneity of multivariate dispersions. Biometrics. 2006;62(1):245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- 61.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio. 2010;1(3):e00129-10. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012;4(10):77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aamot HV, Eskonsipo PKJ, Jørgensen SB, Blomfeldt A. Staphylococcus aureus colonization during military service: A prospective cohort study. Clin. Microbiol. Infect. 2018;24(7):744–748. doi: 10.1016/j.cmi.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 64.Murphy TF, Parameswaran GI. Moraxella catarrhalis, a human respiratory tract pathogen. Clin. Infect. Dis. 2009;49(1):124–131. doi: 10.1086/599375. [DOI] [PubMed] [Google Scholar]
- 65.Verduin CM, Hol C, Fleer A, van Dijk H, van Belkum A. Moraxella catarrhalis: From emerging to established pathogen. Clin. Microbiol. Rev. 2002;15(1):125–144. doi: 10.1128/cmr.15.1.125-144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gallego V, García MT, Ventosa A. Methylobacterium hispanicum sp. nov. and Methylobacterium aquaticum sp. Nov., isolated from drinking water. Int. J. Syst. Evol. Microbiol. 2005;55(Pt 1):281–287. doi: 10.1099/ijs.0.63319-0. [DOI] [PubMed] [Google Scholar]
- 67.Kim KK, Lee KC, Oh HM, Lee JS. Chryseobacterium aquaticum sp. Nov., isolated from a water reservoir. Int. J. Syst. Evol. Microbiol. 2008;58(Pt 3):533–537. doi: 10.1099/ijs.0.65491-0. [DOI] [PubMed] [Google Scholar]
- 68.Leung KT, Chang YJ, Gan YD, Peacock A, Macnaughton SJ, Stephen JR, et al. Detection of Sphingomonas spp in soil by PCR and sphingolipid biomarker analysis. J. Ind. Microbiol. Biotechnol. 1999;23(4–5):252–260. doi: 10.1038/sj.jim.2900677. [DOI] [PubMed] [Google Scholar]
- 69.Singh H, Du J, Yang JE, Shik Yin C, Kook M, Yi TH. Brachybacterium horti sp. nov., isolated from garden soil. Int. J. Syst. Evol. Microbiol. 2016;66(1):189–195. doi: 10.1099/ijsem.0.000696. [DOI] [PubMed] [Google Scholar]
- 70.Waak MB, LaPara TM, Hallé C, Hozalski RM. Nontuberculous mycobacteria in two drinking water distribution systems and the role of residual disinfection. Environ. Sci. Technol. 2019;53(15):8563–8573. doi: 10.1021/acs.est.9b01945. [DOI] [PubMed] [Google Scholar]
- 71.Walterson AM, Stavrinides J. Pantoea: Insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015;39(6):968–984. doi: 10.1093/femsre/fuv027. [DOI] [PubMed] [Google Scholar]
- 72.Uehara Y, Nakama H, Agematsu K, Uchida M, Kawakami Y, Abdul Fattah AS, et al. Bacterial interference among nasal inhabitants: Eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J. Hosp. Infect. 2000;44(2):127–133. doi: 10.1053/jhin.1999.0680. [DOI] [PubMed] [Google Scholar]
- 73.Ellis MW, Griffith ME, Dooley DP, McLean JC, Jorgensen JH, Patterson JE, et al. Targeted intranasal mupirocin to prevent colonization and infection by community-associated methicillin-resistant Staphylococcus aureus strains in soldiers: A cluster randomized controlled trial. Antimicrob. Agents Chemother. 2007;51(10):3591–3598. doi: 10.1128/aac.01086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2(1):6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jo JH, Deming C, Kennedy EA, Conlan S, Polley EC, Ng WI, et al. Diverse human skin fungal communities in children converge in adulthood. J. Investig. Dermatol. 2016;136(12):2356–2363. doi: 10.1016/j.jid.2016.05.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tirosh O, Conlan S, Deming C, Lee-Lin SQ, Huang X, Su HC, et al. Expanded skin virome in DOCK8-deficient patients. Nat. Med. 2018;24(12):1815–1821. doi: 10.1038/s41591-018-0211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martinez Arbizu, P. PairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. R Package Version 0.4. https://github.com/pmartinezarbizu/pairwiseAdonis (2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available in the NCBI under BioProject ID: PRJNA780476 [https://dataview.ncbi.nlm.nih.gov/object/PRJNA780476].