Abstract
Base editing techniques were developed for precise base conversion on cellular genomic DNA, which has great potential for the treatment of human genetic diseases. The glycosylase base editor (GBE) recently developed in our lab was used to perform C-to-G transversions in mammalian cells. To improve the application prospects of GBE, it is necessary to further increase its performance. With this aim, we replaced the human Ung in GBE with Ung1 from Saccharomyces cerevisiae. The resulting editor APOBEC-nCas9-Ung1 was tested at 17 chromosomal loci and was found to have an increased C-to-G editing efficiency ranging from 2.63% to 52.3%, with an average of 23.48%, which was a significant improvement over GBE, with an average efficiency of 15.54%, but with a decreased purity. For further improvement, we constructed APOBEC(R33A)-nCas9-Rad51-Ung1 with two beneficial modifications adapted from previous reports. This base editor was able to achieve even higher editing efficiency ranging from 8.70% to 72.1%, averaging 30.88%, while also exhibiting high C-to-G purity ranging from 35.57% to 92.92%, and was designated GBE2.0. GBE2.0 provides high C-to-G editing efficiency and purity in mammalian cells, making it a powerful genetic tool for scientific research or potential genetic therapies for disease-causing G/C mutations.
Keywords: base editing, glycosylase base editor
Graphical abstract
The glycosylase base editor (GBE) is able to perform C-to-G transversions in mammalian cells. In this study, Sun et al. develop reconstructed glycosylase base editor GBE2.0 with enhanced C-to-G base editing efficiency and purity, which offers hope for the treatment of human genetic diseases caused by more than 3,000 known C/G SNVs.
Introduction
The CRISPR-Cas9 system can recognize target sequences at single nucleotide resolution, and has been adapted to develop powerful genome-editing techniques.1, 2, 3, 4, 5, 6 Subsequently, base editor techniques were developed for precise cytosine (C) to thymine (T) and adenine (A) to guanine (G) base editing (CBE and ABE, respectively) without the use of a donor DNA template.7, 8, 9 In 2020, our group and the Joung group developed a similar base editor complex comprising Cas9 nickase (nCas9), apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) and uracil DNA glycosylase (Ung). These complexes perform a series of enzymatic functions, including specifically binding to target DNA, cleaving the amine group from cytosine, and excising the resulting uracil to create an apurinic/apyrimidinic (AP) site. Relatively specific C-to-G base conversion was obtained by cellular DNA repair of the AP site.10,11 The glycosylase base editor (GBE) is the first base editor that can perform specific C-to-G transversion in mammalian cells with high purity and a narrow editing window, which offers hope for the treatment of human genetic diseases caused by more than 3,000 known C/G single-nucleotide variants (SNVs).9
Following years of intensive development, generations of CBE and ABE editors were constructed and improved, enabling efficient specific C-to-T and A-to-G conversion.12, 13, 14, 15, 16 Compared with these more mature systems, the performance of GBE still has room for improvement. For example, the editing efficiency of GBE is generally lower than that of ABE and CBE. In our work,10 the editing efficiency of GBE at 30 tested loci varied from 5.3% to 53%, and in the work of Joung et al.,11 the editing efficiency of CGBE1 at 25 tested sites ranged from 4% to 68%. To improve the application prospects of GBE, it is crucial to improve the performance of this technique. In this work, we reconstructed the glycosylase base editor to obtain better GBE editors.
Results
Ung1 from Saccharomyces cerevisiae improved GBE editing efficiency
Ung is a key component of the GBE complex, and it can excise uracils from the DNA single strand to create AP sites. The relatively specific C-to-G base conversion is obtained by cellular DNA repair of the AP site.10,11 Thus, we speculated that different Ung homologs may affect the C-to-G transversion base editing outcomes of GBE, and the enzymatic activity of Ung was positively correlated with the C-to-G editing efficiency of GBE. Previous studies indicated that the differences in C-to-G editing efficiency may be related to the enzymatic activities of Ung orthologs from different species,11,17,18 and an Ung with higher enzymatic activity may be helpful to increase the performance of GBE. Ung with higher enzymatic activity could remove more uracils and create more AP sites, ultimately resulting in an increase in C-to-G conversion efficiency. Therefore, it is necessary to screen the Ung domains from different organisms to obtain better GBE editors.
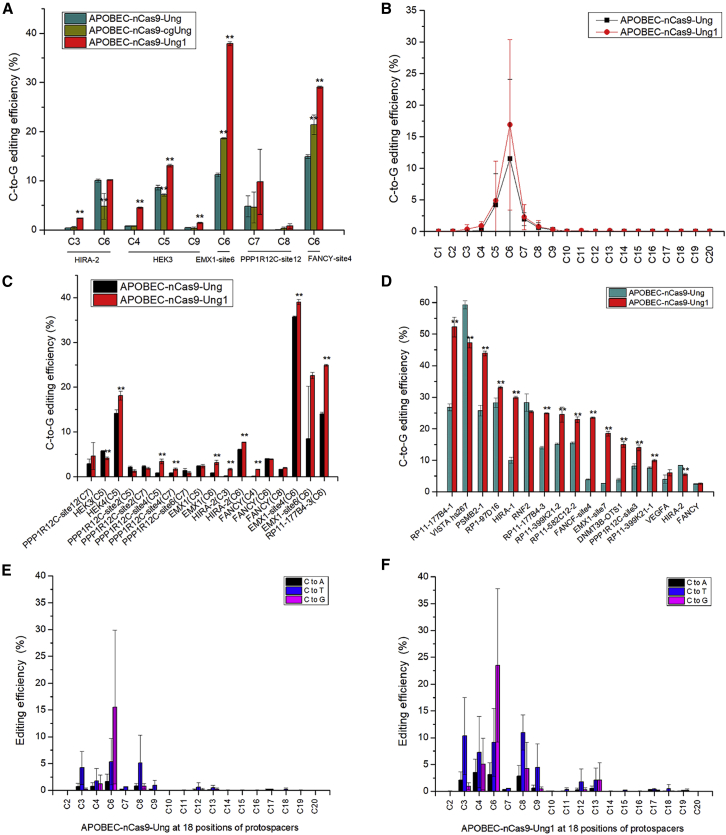
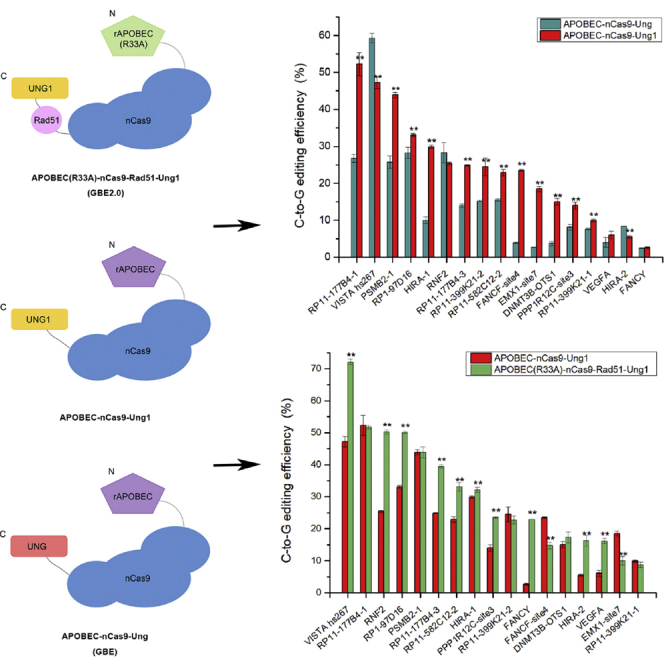
To improve the performance of GBE, we replaced human Ung in GBE with a few other Ung orthologs, such as cgUng from Corynebacterium glutamicum and Ung1 from S. cerevisiae. The C-to-G base editing efficiencies of different GBEs (APOBEC-nCas9-Ung, APOBEC-nCas9-cgUng, APOBEC-nCas9-Ung1) from 5 chromosomal loci in HEK293T cells are shown in Figure 1A. Compared with APOBEC-nCas9-Ung, the APOBEC-nCas9-cgUng editor exhibited a significant reduction in C-to-G base editing efficiency at 2 out of 5 targeted sites (HIRA-2, HEK3) and a significant increase in C-to-G base editing efficiency at 2 out of 5 targeted sites (EMX1-site6, FANCY-site4) (p < 0.01). In contrast, the C-to-G base editing efficiency of APOBEC-nCas9-Ung1 was significantly higher than that of APOBEC-nCas9-Ung at 4 out of 5 targeted sites (p < 0.01). Thus, to improve the C-to-G base editing efficiency of GBE, we replaced the original Ung in APOBEC-nCas9-Ung with Ung1 from S. cerevisiae to obtain APOBEC-nCas9-Ung1. To explore the editing window of APOBEC-nCas9-Ung1, we selected 12 loci (6 gRNA for non-C6 sites and 6 for C6 sites) for editing in HEK293T cells. The C-to-G base editing efficiencies of APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 are shown in Figures 1B and 1C. The experimental data confirmed that low C-to-G base editing was observed at the C5 and C7 positions with both editors, in addition to the high editing efficiency at the C6 positions. Since the average efficiency of APOBEC-nCas9-Ung1 at C6 was nearly 3.5- and 7.5-fold higher than that at the C5 and C7 positions, respectively, we considered C6 to be the major editing window.
Figure 1.
Base editing with different GBEs in HEK293T cells
(A) The C-to-G base editing efficiency of GBEs with different Ung homologs. (B) The average C-to-G base editing efficiency of APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 at 20 positions of protospacers from 12 loci (6 gRNA for non-C6 sites and 6 for C6 sites). (C) The C-to-G base editing efficiency of APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 at the C3–C8 position from 12 loci (6 gRNA for non-C6 sites and 6 for C6 sites). (D) The C-to-G base editing efficiency of APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 at the C6 position of 17 loci. (E) The average base editing (C-to-A, C-to-T, and C-to-G) efficiency of APOBEC-nCas9-Ung at 18 positions of protospacers from 17 loci (gRNA for C6 sites). (F) The average base editing (C-to-A, C-to-T, and C-to-G) efficiency of APOBEC-nCas9-Ung1 at 18 positions of protospacers from 17 loci (gRNA for C6 sites). ∗∗p < 0.01 was considered statistically significant. The bars represent the average editing efficiency, and error bars represent the SD of 3 independent biological replicates.
Next, base editing was performed with the parent editor, APOBEC-nCas9-Ung, and APOBEC-nCas9-Ung1 at the C6 sites from 17 chromosomal loci in HEK293T cells to compare their editing efficiency (Figures 1D and S1). GBE mainly edited the C6 position of the protospacers, and the C6 base editing efficiency of APOBEC-nCas9-Ung1 at 12 loci was significantly improved compared with that of APOBEC-nCas9-Ung. Specifically, the C-to-G editing efficiency of APOBEC-nCas9-Ung1 at the loci RP11-177B4-1, PSMB2-1, RP1-97D16, HIRA-1, RP11-177B4-3, RP11-399K21-2, RP11-582C12-2, FANCF-site4, EMX1-site7, DNMT3B-OTS1, PPP1R12C-site3, and RP11-399K21-1 was significantly higher than that of APOBEC-nCas9-Ung (p < 0.01), while the base editing efficiency of APOBEC-nCas9-Ung1 at 2 loci (VISTA hs267, HIRA-2) was significantly lower than that of APOBEC-nCas9-Ung (p < 0.01), and there was no significant difference at 3 loci (RNF2, VEGFA, FANCY).
Among the 17 tested loci, the editing efficiency of APOBEC-nCas9-Ung1 was generally 2–3 times that of APOBEC-nCas9-Ung at some loci, such as RP11-177B4-1, PSMB2-1, HIRA-1, and RP11-177B4-3 (Figure 1D). For instance, the C-to-G base editing efficiency of APOBEC-nCas9-Ung1 was nearly double that of APOBEC-nCas9-Ung at RP11-177B4-1 (an increase from 26.80% to 52.30%), PSMB2-1 (an increase from 25.73% to 43.90%), and RP11-177B4-3 (an increase from 14.00% to 24.90%). Moreover, the C-to-G base editing efficiency of APOBEC-nCas9-Ung1 at HIRA-1 was almost 3 times that of APOBEC-nCas9-Ung (increase from 10.00% to 29.90%). In addition, the C-to-G base editing efficiency of APOBEC-nCas9-Ung1 reached a satisfactory level of approximately 20% at 3 loci, FANCF-site4, EMX1-site7, and DNMT3B-OTS1, while that of APOBEC-nCas9-Ung was less than 4%.
To obtain a comprehensive overview of the editing efficiency of APOBEC-nCas9-Ung1, the average base editing efficiencies of APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 at 18 positions of protospacers (C2, C3, C4, C6, C7, C8, C9, C10, C11, C12, C13, C14, C15, C16, C17, C18, C19, and C20) from the 17 tested loci were calculated and analyzed. We found that APOBEC-nCas9-Ung1 mainly edited at the C6 position, similar to APOBEC-nCas9-Ung. Nevertheless, the average C-to-G base editing efficiency of APOBEC-nCas9-Ung1 at the C6 position was improved from 15.54% to 23.48%, with the change of Ung to Ung1, which was an increase of 51.14% (Figures 1E and 1F). The low editing efficiency at positions C4 and C8 by APOBEC-nCas9-Ung was also increased to 5.08% and 4.24%, respectively, with APOBEC-nCas9-Ung1.
One problem we observed with APOBEC-nCas9-Ung1 was that the average efficiency of the noncanonical base conversions was also increased along with the C-to-G conversion. Both C-to-A and C-to-T conversions at C3, C4, C6, and C8 were increased, which caused a decrease in C-to-G purity (Figures 1E and 1F). The data revealed that APOBEC-nCas9-Ung1 nonspecifically increased the incidence of base conversions of all types, which suggested that the reason for the increased editing efficiency of the APOBEC-nCas9-Ung1 editor could be the improved glycosylase activity of the complex. Our next aim was to improve the C-to-G base editing efficiency at the C6 position while also increasing the C-to-G purity.
APOBEC(R33A)-nCas9-Rad51-Ung1 achieved higher GBE editing efficiency
The R33A mutation of APOBEC1 was first reported by the Joung group to reduce the transcriptome-wide off-target RNA editing of CBE,19 and then they introduced it to CGBEs.11 The results indicated that the APOBEC1 R33A variant could alter the activity of cytidine deaminase, resulting in increased C-to-G conversion efficiency of CGBEs, but the mechanism was unclear. In 2020, the Li group20 enhanced the editing efficiency of CBE by fusing a single-stranded DNA-binding domain (ssDBD) from Rad51. Rad51 is a critical protein involved in DNA repair, which may increase the affinity between the nucleotide deaminase and single-strand DNA (ssDNA) substrate, resulting in the increased editing efficiency of CBE. We hypothesized that fusing Rad51 to GBE may increase the affinity of uracil DNA glycosylase for uracil and ultimately alter the C-to-G transversion outcomes of GBE.
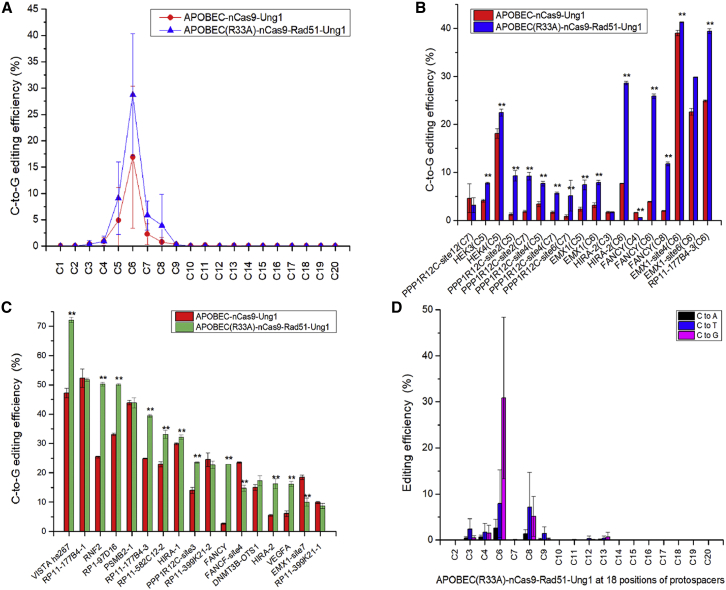
To further improve the C-to-G base editing efficiency and decrease the unexpected C-to-T and C-to-A conversions of APOBEC-nCas9-Ung1, we constructed the APOBEC(R33A)-nCas9-Rad51-Ung1 editor from APOBEC-nCas9-Ung1 by incorporating two reported beneficial modifications.19,20 To explore the editing window of APOBEC(R33A)-nCas9-Rad51-Ung1, we selected 12 loci (6 gRNA for non-C6 sites and 6 for C6 sites) for editing in HEK293T cells. The C-to-G base editing efficiency is shown in Figures 2A and 2B. The experimental data confirmed that similar to APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1, the C5 and C7 positions of protospacers were edited by APOBEC(R33A)-nCas9-Rad51-Ung1 with low efficiency, and the APOBEC(R33A)-nCas9-Rad51-Ung1 editor had even lower C-to-G base editing at the C8 position of protospacers. Since the average efficiency of APOBEC(R33A)-nCas9-Rad51-Ung1 at C6 was almost 3-, 5-, and 7-fold higher than that at the C5, C7, and C8 positions, respectively, we considered C6 to be the major editing window.
Figure 2.
Base editing with APOBEC-nCas9-Ung1 and APOBEC(R33A)-nCas9-Rad51-Ung1 in HEK293T cells
(A) The average C-to-G base editing efficiency of APOBEC-nCas9-Ung1 and APOBEC(R33A)-nCas9-Rad51-Ung1 at 20 positions of protospacers from 12 loci (6 gRNA for non-C6 sites and 6 for C6 sites). (B) The C-to-G base editing efficiency of APOBEC-nCas9-Ung1 and APOBEC(R33A)-nCas9-Rad51-Ung1 at the C3–C8 position from 12 loci (6 gRNA for non-C6 sites and 6 for C6 sites). (C) The C-to-G base editing efficiency of APOBEC-nCas9-Ung1 and APOBEC(R33A)-nCas9-Rad51-Ung1 at the C6 position of 17 loci (gRNA for C6 sites). (D) The average base editing (C-to-A, C-to-T, and C-to-G) efficiency of APOBEC(R33A)-nCas9-Rad51-Ung1 at 18 positions of protospacers from 17 loci (gRNA for C6 sites). ∗∗p < 0.01 was considered statistically significant. The bars represent the average editing efficiency, and error bars represent the SD of 3 independent biological replicates.
Next, base editing was performed with APOBEC-nCas9-Ung1 and APOBEC(R33A)-nCas9-Rad51-Ung1 at the C6 position of 17 chromosomal loci in HEK293T cells to compare their editing efficiency (Figures 2C and S2). The C-to-G editing efficiency of APOBEC(R33A)-nCas9-Rad51-Ung1 at 10 loci, VISTA hs267, RNF2, RP1-97D16, RP11-177B4-3, RP11-582C12-2, HIRA-1, PPP1R12C-site3, FANCY, HIRA-2, and VEGFA, was significantly higher than that of APOBEC-nCas9-Ung1 (p < 0.01), while the efficiency at 2 loci, FANCF-site4 and EMX1-site7, was significantly lower than that of APOBEC-nCas9-Ung1 (p < 0.01). There was no significant difference at 5 loci, RP11-177B4-1, PSMB2-1, RP11-399K21-2, DNMT3B-OTS1, and RP11-399K21-1. The C-to-G base editing efficiency of APOBEC(R33A)-nCas9-Rad51-Ung1 ranged from 8.70%–72.10%, while that of APOBEC-nCas9-Ung1 ranged from 2.63% to 52.3%. Among the 17 loci, the C-to-G base editing efficiency was approximately 1.5 times higher at the VISTA hs267 (from 47.23% to 72.10%), RP1-97D16 (from 33.07% to 50.07%), RP11-177B4-3 (from 24.90% to 39.43%), RP11-582C12-2 (from 22.87% to 33.07%) and PPP1R12C-site3 (from 14.00% to 23.53%) loci. Compared with that of APOBEC-nCas9-Ung1, the C-to-G base editing efficiency of APOBEC(R33A)-nCas9-Rad51-Ung1 was nearly twice that of RNF2 (increased from 25.40% to 50.23%) and triple that of HIRA-2 (increased from 5.53% to 16.27%) and VEGFA (increased from 6.07% to 16.17%). In addition, the C-to-G base editing efficiency of APOBEC(R33A)-nCas9-Rad51-Ung1 reached 22.90% at FANCY, while the C-to-G base editing efficiency of APOBEC-nCas9-Ung1 at this locus was only 2.63%. Thus, the editing efficiency was generally further increased with APOBEC(R33A)-nCas9-Rad51-Ung1, which was designated GBE2.0.
To obtain a comprehensive overview of the editing efficiency of GBE2.0, its average base editing efficiency at 18 positions of protospacers from the 17 tested loci was calculated and analyzed (Figure 2D). Similar to APOBEC-nCas9-Ung but unlike APOBEC-nCas9-Ung1, the average C-to-A conversion efficiency of GBE2.0 at the 18 positions of protospacers was very low (less than 2.6%), and the major C-to-T base editing at the positions of C6 and C8 was also limited to under 8.0%. GBE2.0 also mainly converted C-to-G at the C6 position, with an average editing efficiency of 30.88%, which was 31.50% and 98.75% higher than that of APOBEC-nCas9-Ung1 and APOBEC-nCas9-Ung, respectively. In addition, the C-to-G conversion of GBE2.0 at the C3, C4, and C13 positions decreased compared with that of APOBEC-nCas9-Ung1.
APOBEC(R33A)-nCas9-Rad51-Ung1 achieved high C-to-G purity
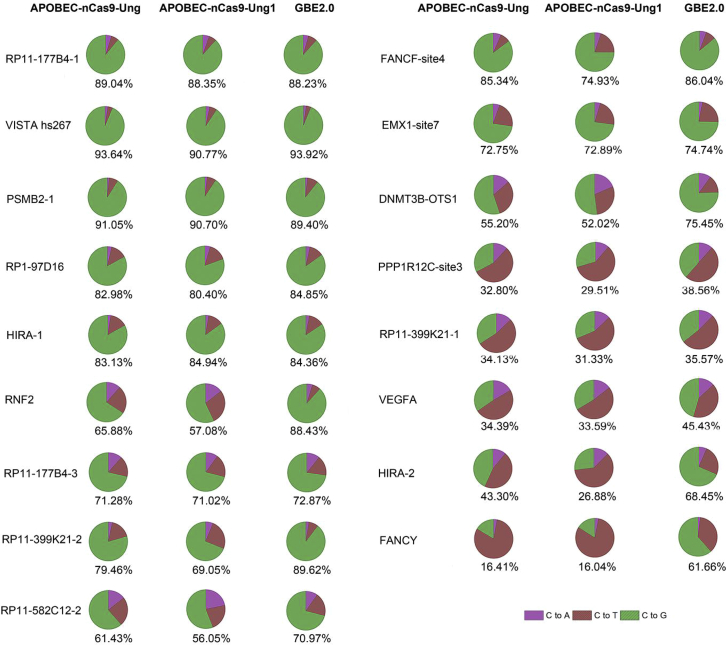
The average C-to-G conversion purity at the C6 position of protospacers from 17 loci using APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0 in HEK293T cells is illustrated in Figure 3. The average C-to-G purities of the 3 base editors APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0 were 64.25%, 60.33%, and 73.44%, respectively. In other words, replacing Ung in APOBEC-nCas9-Ung with Ung1 reduced the C-to-G purity of GBE, while the reconstructed glycosylase base editor GBE2.0 achieved high C-to-G conversion purity.
Figure 3.
The C-to-G purity with APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0 in HEK293T cells
As shown in Figure 3, the C-to-G conversion purity from 17 loci of APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0 ranged from 16.41% to 93.64%, 16.04% to 90.77%, and 35.57% to 93.92%, respectively. Among the 3 base editors, GBE2.0 edited the most loci, with a purity of more than 80%, followed by APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1. Specifically, GBE2.0 edited 8 loci, with a purity of more than 80%, while APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 edited only 6 and 5 loci, respectively, with a purity of more than 80%. Similarly, GBE2.0 edited the most loci, with a purity of 60–80%, followed by APOBEC-nCas9-Ung (5 loci) and APOBEC-nCas9-Ung1 (4 loci). In addition, the C-to-G purity of GBE2.0 was lower than 60% at only 3 sites (PPP1R12C-site3, RP11-399K21-1, and VEGFA), while that of APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 was lower than 60% at 6 and 8 loci, respectively. In summary, compared with APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1, the GBE2.0 editor achieved higher C-to-G purity.
Indel data were obtained by deep sequencing. Seventeen loci were tested, and the observed average indel frequencies of APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0 were 0.15%, 0.83%, and 0.26%, respectively (Figure S3). After replacing Ung in APOBEC-nCas9-Ung with Ung1, the indel frequencies of the APOBEC-nCas9-Ung1 editor were significantly increased (p < 0.01). Nevertheless, there was no significant difference in the indel frequencies between GBE2.0 and APOBEC-nCas9-Ung (p > 0.01). These observations indicated that GBE2.0 did not increase the indel frequencies of GBE.
To analyze the off-target editing frequencies of APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0, approximately 10 loci containing protospacer sequences most similar to RP11-177B4-1 and PSMB2-1 were amplified by PCR and subjected to deep-sequence analysis (Figure S4). The results showed that the editing rates of APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 at these potential off-target sites were lower than 0.4%, and the mutation rates of GBE2.0 were also in the same range. Therefore, the off-target effects of APOBEC-nCas9-Ung1 and GBE2.0 were comparable to those of the original APOBEC-nCas9-Ung.
Base editing in other cell types
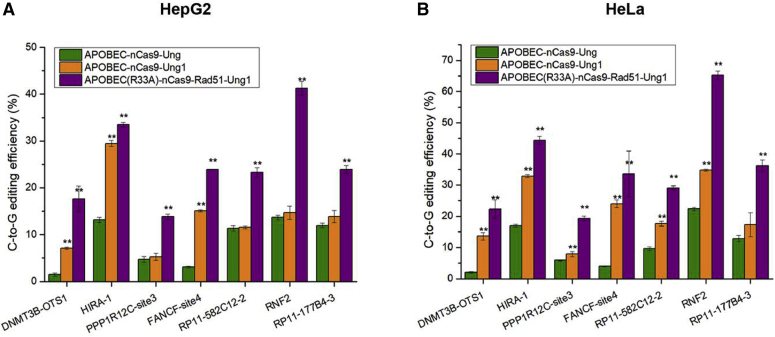
To test how APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0 performed in human cell lines other than HEK293T cells, we assayed the ability of these editors to edit 7 target loci (guide RNA [gRNA] for C6 sites) in HepG2 and HeLa cells (Figure 4). The target loci could be edited by APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1 and GBE2.0 in both HepG2 and HeLa cells. In addition, these 3 GBE editors showed different C-to-G editing efficiency in HepG2 and HeLa cells. Specifically, the C-to-G editing efficiency of APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0 in HepG2 cells ranged from 1.47% to 13.67%, with an average of 8.49%; from 5.23% to 29.47%, with an average of 13.85%; and from 13.83% to 41.23%, with an average of 25.33%. The C-to-G editing efficiency of APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0 in HeLa cells ranged from 2.07% to 22.35%, with an average of 10.54%; from 7.93% to 34.83%, with an average of 21.18%; and 19.33% to 65.23%, with an average of 35.74%, respectively.
Figure 4.
C-to-G base editing using APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0
(A) Use in HepG2 cells and (B) use in HeLa cells. ∗∗p < 0.01 was considered statistically significant. The bars represent the average editing efficiency, and error bars represent the SD of 3 independent biological replicates.
As illustrated in Figure 4, compared with APOBEC-nCas9-Ung, the APOBEC-nCas9-Ung1 editor led to a significant increase in C-to-G editing efficiency at 3 of 7 targeted sites (DNMT3B-OTS1, HIRA-1, FANCF-site4) in HepG2 cells (p < 0.01), and 6 of 7 targeted sites (DNMT3B-OTS1, HIRA-1, PPP1R12C-site3, FANCF-site4, RP11-582C12-2, RNF2) in HeLa cells (p < 0.01). Nevertheless, the GBE2.0 editor significantly improved the C-to-G editing efficiency of all 7 targeted sites in HepG2 and HeLa cells (p < 0.01) compared with APOBEC-nCas9-Ung. The above results indicated that the APOBEC-nCas9-Ung1 and GBE2.0 editors could enhance the C-to-G editing efficiency in both HepG2 and HeLa cells compared with APOBEC-nCas9-Ung. Although the cell types were different, the trend of efficiency increase was similar. The C-to-G editing efficiency of APOBEC-nCas9-Ung1 for each tested site was higher than that of APOBEC-nCas9-Ung, and GBE2.0 showed the highest C-to-G editing efficiency across different cell types.
Comparison of GBE2.0 with recently reported GBEs
In 2018, a patent by Liu and Koblan entitled “Cytosine to Guanine Base Editing” first demonstrated the possibility of C-to-G editing using uracil DNA glycosylase.21 In 2020 and 2021, 3 cytosine transversion base editors were reported, including GBE constructed by our group,10 CGBE1 constructed by the Joung group,11 and rAPOBEC1-nCas9-XRCC1 constructed by the Chew group.22 GBE and CGBE1 were designed using the same strategy, comprising APOBEC, nCas9, and Ung. The generation of AP sites by APOBEC and Ung, coupled with nicking at the nonedited strand by nCas9, initiates DNA repair and replication, and eventually leads to C-to-G base editing.23 In contrast to the Ung-mediated base excision initiation strategy, Chew et al. fused rAPOBEC1-nCas9 to XRCC1, a base excision repair protein, which could recruit other BER proteins to repair DNA damage, also resulting in cytosine transversion.22 The average editing efficiencies of GBE, CGBE1, and miniCGBE1 were reported to be 24.14%, 14.4%, and 13%, respectively,10,11 while the rAPOBEC1-nCas9-XRCC1 base editor exhibited a 15.4% mean editing efficiency.22
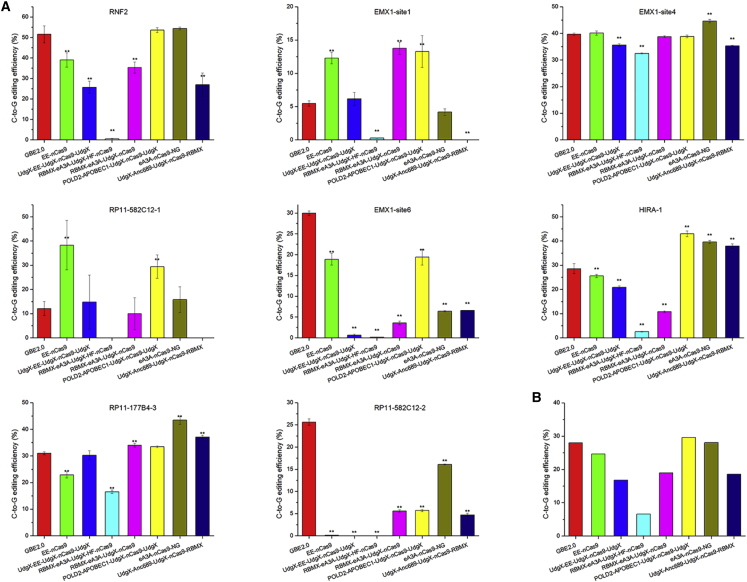
Very recently, the Liu group used various fusion proteins with functions related to DNA repair to construct multiple CGBEs with enhanced performance.17 To evaluate the performance of GBE2.0, we compared its editing performance with that of the 7 top CGBEs: EE-nCas9, UdgX-EE-UdgX-nCas9-UdgX, RBMX-eA3A-UdgX-HF-nCas9, RBMX-eA3A-UdgX-nCas9, POLD2-APOBEC1-UdgX-nCas9-UdgX, eA3A-nCas9-NG, and UdgX-Anc689-UdgX-nCas9-RBMX developed by Liu’s group17 from 8 loci in HEK293T cells. As illustrated in Figure 5A, the C-to-G editing efficiencies of GBE2.0, EE-nCas9, UdgX-EE-UdgX-nCas9-UdgX, RBMX-eA3A-UdgX-HF-nCas9, RBMX-eA3A-UdgX-nCas9, POLD2-APOBEC1-UdgX-nCas9-UdgX, eA3A-nCas9-NG, and UdgX-Anc689-UdgX-nCas9-RBMX ranged from 5.47% to 51.57%, 0.13% to 40.13%, 0% to 35.63%, 0% to 32.47%, 3.6% to 38.8%, 5.7% to 53.57%, 4.17% to 54.4%, and 0% to 37.93%, respectively.
Figure 5.
Comparison of GBE2.0 developed in this study with recently reported advanced GBEs
(A) The C-to-G base editing efficiency of GBE2.0 and CGBEs from 8 loci (gRNA for C6 sites). (B) The average C-to-G base editing efficiency of GBE2.0 and CGBEs from 8 loci (gRNA for C6 sites). ∗∗p < 0.01 was considered statistically significant. The bars represent the average editing efficiency, and error bars represent the SD of 3 independent biological replicates.
We found that different target loci were preferentially edited by different CGBEs. Specifically, at the EMX1-site6 and RP11-582C12-2 loci, the C-to-G editing efficiency of GBE2.0 was significantly higher than that of all of the other base editors (p < 0.01). At the RNF2 and loci, the C-to-G editing efficiency of GBE2.0 was significantly higher than that of EE-nCas9, UdgX-EE-UdgX-nCas9-UdgX, RBMX-eA3A-UdgX-HF-nCas9, RBMX-eA3A-UdgX-nCas9, and UdgX-Anc689-UdgX-nCas9-RBMX (p < 0.01), but there was no significant difference between GBE2.0, POLD2-APOBEC1-UdgX-nCas9-UdgX, and eA3A-nCas9-NG (p > 0.01). At the EMX1-site4 locus, the C-to-G editing efficiency of GBE2.0 was significantly higher than that of UdgX-EE-UdgX-nCas9-UdgX, RBMX-eA3A-UdgX-HF-nCas9 and UdgX-Anc689-UdgX-nCas9-RBMX and lower than that of eA3A-nCas9-NG (p < 0.01), but there was no significant difference between GBE2.0, EE-nCas9, RBMX-eA3A-UdgX-nCas9, and POLD2-APOBEC1-UdgX-nCas9-UdgX (p > 0.01). In addition, RBMX-eA3A-UdgX-nCas9, EE-nCas9, POLD2-APOBEC1-UdgX-nCas9-UdgX, and eA3A-nCas9-NG showed the highest C-to-G editing efficiency at EMX1-site1, RP11-582C12-1, HIRA-1, and RP11-177B4-3, respectively.
The above observations suggested that the enhanced CGBE editors and GBE2.0 exhibited different editing efficiencies across different loci, and the GBE2.0 editor had the highest editing efficiency at a portion of the loci (e.g., EMX1-site6, RP11-582C12-2) (Figure 5A). Among these base editors, GBE2.0, POLD2-APOBEC1-UdgX-nCas9-UdgX, and eA3A-nCas9-NG showed better editing efficiencies than other CGBEs, and their average editing efficiencies were 28.01%, 29.59%, and 28.06%, respectively (Figure 5B). The results suggested that the GBE2.0 editor showed high editing efficiency even among the new generation of C-to-G base editors. Moreover, since GBE2.0 was constructed using different strategies from the enhanced CGBEs from Liu’s group, the application of the preference of editing loci may be different from that of the enhanced CGBEs.
Correction of disease-associated SNVs using GBE2.0
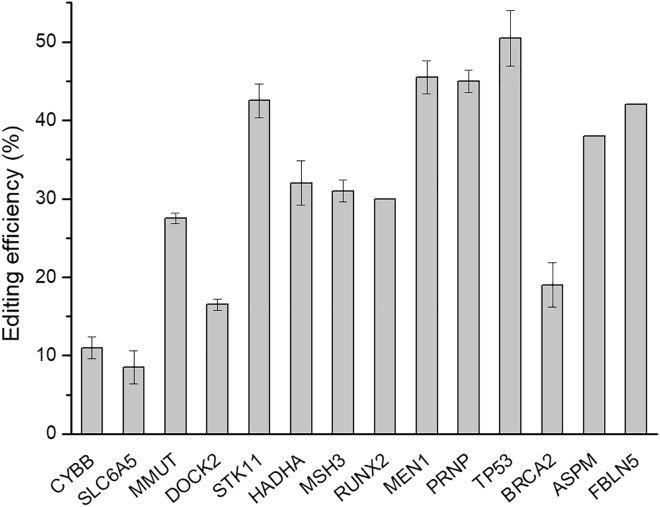
SNV data are retrieved from the ClinVar database. From the database, we selected 14 point mutations that can be corrected by GBEs, mainly the ones carrying C/G mutations at the C6 position of protospacer with NGG protospacer adjacent motifs (PAMs). The protospacer sequences of disease-associated SNVs are listed in Table S3, and the correction outcomes of C/G mutations related to human diseases using GBE2.0 are shown in Figure 6.
Figure 6.
Correction of SNVs using GBE2.0. The bars represent the average editing efficiency, and error bars represent the SD of 3 independent biological replicates.
As illustrated in Figure 6, GBE2.0 enabled C-to-G editing at disease-associated SNVs, and it corrected 5 SNVs to their wild-type coding sequence at >40% editing efficiency. For example, the C-to-G SNV in STK11 associated with Peutz-Jeghers syndrome (PJS),24 MEN1 associated with multiple endocrine neoplasia type 1,25,26 PRNP associated with cerebral amyloid angiopathy,27 TP53 associated with Li-Fraumeni syndrome,28,29 and FBLN5 associated with age-related macular degeneration 3.30 In addition, GBE2.0 could correct some recessive genetic diseases caused by C/G mutations, such as CYBB associated with chronic granulomatous disease,31 MMUT associated with methylmalonic acidemia,32 DOCK2 associated with immunodeficiency 40,33 and HADHA associated with long-chain 3-hydroxyacyl-coenzyme A (CoA) dehydrogenase deficiency.34 Collectively, these results revealed efficient correction of disease-related SNVs by GBE2.0, and the GBE2.0 editor could be used for a candidate gene therapy.
Discussion
The uracil DNA glycosylase Ung, which excises the uracils from the DNA single strand to create AP sites, is a key component of the GBE complex. The relatively specific C-to-G base conversion is obtained by cellular DNA repair of the AP sites.10,11 Previous studies have shown that Ung from different homologs could affect the C-to-G editing efficiency of GBEs.11,17,18 The Joung group11 replaced the hUNG present in BE4max(R33A)ΔUGI-hUNG with a homologous UNG from Escherichia coli (eUNG). The results revealed that the C-to-G editing frequencies induced by BE4max(R33A)ΔUGI-eUNG and eUNG-BE4max(R33A)ΔUGI were higher than that of BE4max(R33A)ΔUGI-hUNG for 6 of 7 gRNAs tested in HEK293T cells. Recently, the Liu17 group fused a variety of glycosylases (UNG, UgdX, SMUG1, MBD4, and TDG) to the AC scaffold (bpNLS-APOBEC1-Cas9 D10A-bpNLS, similar to other reported CGBEs),10,11,22 and evaluated the C-to-G editing efficiency at 5 genomic loci in HEK293T cells. The results showed that fusing UNG, SMUG1, MBD4 and TDG to the AC scaffold did not enhance the C-to-G transversion efficiency. Nevertheless, fusion of a UNG ortholog from Mycobacterium smegmatis (UdgX) was found to improve the editing yield and product purity of C-to-G base editing. The Zuo group18 developed 4 CGBEs (hUNG-CGBE, eUNG-CGBE, mUNG-CGBE, cUNG-CGBE) by substituting the UGI of BE3 with UNGs from humans, E. coli, mice, or Caenorhabditis elegans. They found that the C-to-G editing efficiencies of eUNG-CGBE and cUNG-CGBE were much higher than that of hUNG-CGBE for 34 endogenous sites in HEK293T cells. These studies indicated that the differences in C-to-G editing efficiency were related to the enzymatic activities of Ung orthologs from different species, and Ungs with higher enzymatic activity may be helpful to increase the performance of GBE.
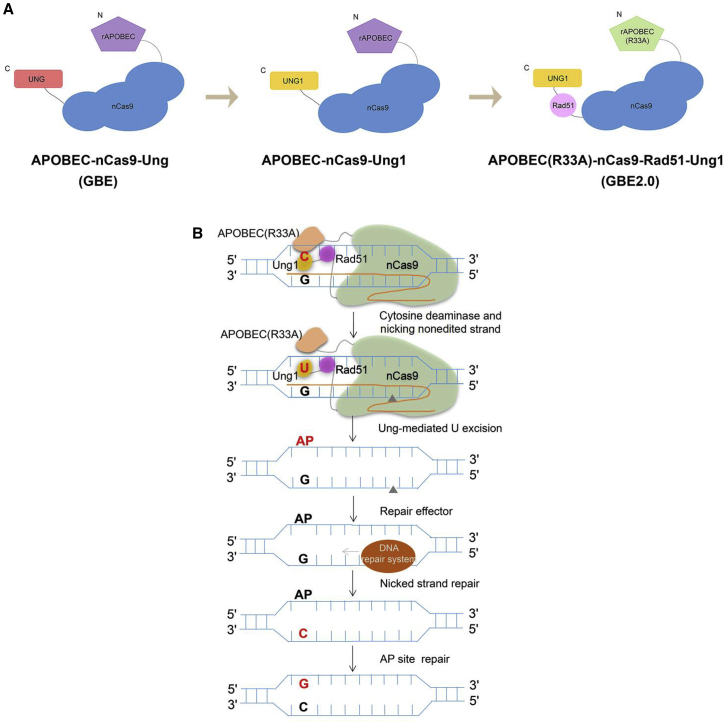
In our work, we replaced Ung in GBE with Ung orthologs from Corynebacterium glutamicum and S. cerevisiae and tested whether they could improve the C-to-G editing efficiency of GBE. As a result, APOBEC-nCas9-cgUng (cgUng from Corynebacterium glutamicum) did not increase the C-to-G editing efficiency, while APOBEC-nCas9-Ung1 (Ung1 from S. cerevisiae) led to a significant improvement in HEK293T cells. Thus, we replaced the original Ung in APOBEC-nCas9-Ung with Ung1 from S. cerevisiae to obtain APOBEC-nCas9-Ung1 for subsequent experiments (Figure 7A). The APOBEC-nCas9-Ung1 editor could improve the C-to-G editing efficiency, which may have been due to the improved uracil glycosylase activity of the GBE complex. The enzymatic activity of Ung1 was probably higher than that of Ung, so APOBEC-nCas9-Ung1 could remove more uracils and create more AP sites, ultimately resulting in an increase in C-to-G conversion efficiency. As a result, the APOBEC-nCas9-Ung1 editor also nonspecifically increased the frequency of base conversions of all types (C-to-T, C-to-A, and C-to-G) and decreased the C-to-G product purity of GBE. In addition, the editing efficiency at the base positions of the whole editing window was generally improved. These facts may also serve as indirect proof that the increased editing efficiency may be due to increased Ung1 activity.
Figure 7.
Schematics and potential mechanism of GBEs developed in this study
(A) Schematic of the APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0 complexes. (B) Potential functional mechanism of GBE2.0.
To further improve the C-to-G base editing efficiency while also increasing the C-to-G purity of APOBEC-nCas9-Ung1, we constructed APOBEC(R33A)-nCas9-Rad51-Ung1 with two reported beneficial modifications (Figure 7A). Previously, the Joung group indicated that the R33A mutation of APOBEC1 could reduce the transcriptome-wide off-target RNA editing of CBE,19 after which they introduced the R33A variant to CGBEs. The results showed that the APOBEC1 R33A variant may alter the activity of cytidine deaminase, resulting in increased C-to-G conversion efficiency, but the mechanism was unclear.11 Similarly, the introduction of APOBEC(R33A) increased the C-to-G editing efficiency of APOBEC(R33A)-nCas9-Rad51-Ung1 in our study. In 2020, the Li group20 reported that the single-stranded DNA-binding domain (ssDBD) from Rad51, a critical protein involved in DNA repair, could dramatically enhance the editing efficiency of CBE. The reason for the enhancement may be that Rad51 could increase the affinity between the nucleotide deaminase and the ssDNA substrate. Thus, we hypothesized that fusing Rad51 to GBE may increase the affinity of uracil-DNA glycosylase for uracil and ultimately alter the C-to-G transversion outcomes of GBE. In view of this, we combined all of the beneficial traits and constructed APOBEC(R33A)-nCas9-Rad51-Ung1, which was designated GBE2.0. The results showed that the GBE2.0 editor developed in this study could convert C-to-G with high efficiency and high product purity. In addition, APOBEC-nCas9-Ung, APOBEC-nCas9-Ung1, and GBE2.0 mainly edited the C6 position of protospacers. However, GBE2.0 could not only edit the C5 and C7 positions of protospacers with low efficiency but also the C8 position with even lower efficiency. This was probably due to Rad51 functioning as a long linker sequence in addition to ssDNA binding (Figure 7B).20
GBE2.0 provides high C-to-G editing efficiency and purity in mammalian cells, which makes it a powerful genetic tool for scientific research or potential genetic therapies for human diseases caused by G/C mutations.
Materials and methods
Plasmid construction
Plasmids APOBEC-nCas9-Ung1 and APOBEC(R33A)-nCas9-Rad51-Ung1 were assembled with the Golden Gate method.35 PCR primers for Golden Gate assembly were designed with the J5 Device Editor.36 The backbones of APOBEC-nCas9-Ung1 and APOBEC(R33A)-nCas9-Rad51-Ung1 were PCR amplified from APOBEC-nCas9-Ung. Ung1 used in S. cerevisiae was synthesized (GenScript, China). The R33A residue was reintroduced via PCR during its generation. Rad51 was PCR amplified from hyBE4max.20 The gRNA expression plasmids were assembled with the Golden Gate method, with the protospacer sequence embedded in the primers, and RNF2 sgRNA expression plasmids7 were used as the template. All of the DNA templates were PCR amplified with Phusion DNA polymerase (NEB, USA). PCR products were gel purified, digested with DpnI restriction enzyme (NEB, USA), and assembled with the Golden Gate assembly method. The protospacer sequences and associated primers used in this study are listed in Table S1.
Cell culture and transfection
HEK293T, HepG2, and HeLa cells (from ATCC) were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) at 37°C under 5% CO2. Cell transfection was performed according to the method used by Zhang et al.10 Briefly, cells were seeded in 24-well plates (Corning, USA) for approximately 24 h and transfected at approximately 40% confluency. Then, 600 ng Cas9 plasmid and 300 ng single-guide RNA (sgRNA)-expressing plasmid were cotransfected. Twenty-four hours after transfection, the medium was replaced with new medium containing puromycin (Merck, USA). Finally, genomic DNA was extracted from the cells using the QuickExtract DNA Extraction Solution (Epicentre, USA).
High-throughput sequencing of genomic DNA samples and data analysis
The high-throughput DNA sequencing of genomic DNA samples and data analysis were performed according to the method used by Zhang et al.10 Briefly, the next-generation sequencing library preparations were constructed according to the manufacturer’s protocol (VAHTS Universal DNA Library Prep Kit for Illumina). For each sample, the library was directly prepared with purified PCR fragments (>50 ng). The PCR reaction was performed at 95°C for 3 min, 30 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 10 s, and a final extension at 72°C for 5 min. Then, libraries with different indices were multiplexed and loaded on an Illumina HiSeq instrument according to the manufacturer’s instructions. Base substitution frequencies were calculated according to the method used by the Bae group.37 The C-to-G purity was calculated as C-to-G editing efficiency/(C-to-T editing efficiency + C-to-A editing efficiency + C-to-G editing efficiency). Indel frequencies were calculated according to the method used by Zhang et al,10 with some modifications. We required ≥1/1,000 of the total reads for calculating indel frequencies. Library members not meeting this condition were filtered.
Selection of DNA off-target sites
The DNA off-target sites of selected target loci were analyzed by Cas-OFFinder,38 and approximately 10 loci containing the most similar sequences of selected target loci were chosen as predicted off-target sites. The off-target sites and associated primers are listed in Table S2.
Generation of the SNV cell library
The generation of the SNV cell library was performed according to a previous method of our laboratory,39 with some minor modifications. First, the SNV target sequences was synthesized, amplified, and cloned into the lentiviral vector lentiGuide-Puro (Addgene cat. no. 52963) using the Golden Gate assembly.35 Second, lentivirus was produced by co-transfection of 3 transfection plasmids, including lentiGuide-Puro carrying SNV target sequences, psPAX2, and pMD2.G. Then HEK293T cells were infected with lentivirus to select cells with stably integrated lentivirus. Finally, the integrated cells were co-transfected with GBE2.0 plasmid and SNV sgRNA-expressing plasmid. The protospacer sequences and relevant information of SNVs are listed in Table S3.
Statistical analysis
All of the data were from 3 independent cell cultures.
Acknowledgments
This research was financially supported by the National Key Research and Development Program of China (2018YFA0903700), the National Natural Science Foundation of China (31861143019 and 31770105), a Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-001), the Tianjin Natural Science Foundation (20JCYBJC00310), and the China Postdoctoral Science Foundation (2021M693353).
Author contributions
X.Z. and C.B. designed the research, analyzed the data, and wrote the manuscript. N.S. designed the research, performed the experiments, analyzed the data, and wrote the manuscript. D.Z. designed the research. Z.Z. performed the experiments.
Declaration of interests
A provisional patent has been submitted, in part entailing the reported approach.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.03.023.
Contributor Information
Changhao Bi, Email: bi_ch@tib.cas.cn.
Xueli Zhang, Email: zhang_xl@tib.cas.cn.
Supplemental information
Data availability
The high-throughput sequencing data in this study are available in the NCBI database (PRJNA778911).
References
- 1.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanjurjo-Soriano C., Erkilic N., Mamaeva D., Kalatzis V. CRISPR/Cas9-mediated genome editing to generate clonal iPSC lines. Methods Mol. Biol. 2021:1–18. doi: 10.1007/7651_2021_362. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y., Zeng J., Roscoe B.P., Liu P., Yao Q., Lazzarotto C.R., Clement K., Cole M.A., Luk K., Baricordi C., et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med. 2019;25:776–783. doi: 10.1038/s41591-019-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frangoul H., Altshuler D., Cappellini M.D., Chen Y.S., Domm J., Eustace B.K., Foell J., de la Fuente J., Grupp S., Handgretinger R., et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 2021;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 5.Stadtmauer E.A., Fraietta J.A., Davis M.M., Cohen A.D., Weber K.L., Lancaster E., Mangan P.A., Kulikovskaya I., Gupta M., Chen F., et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367:eaba7635. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y., Xue J., Deng T., Zhou X., Yu K., Deng L., Huang M., Yi X., Liang M., Wang Y., et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020;26:732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 7.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y., et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 9.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao D., Li J., Li S., Xin X., Hu M., Price M.A., Rosser S.J., Bi C., Zhang X. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021;39:35–40. doi: 10.1038/s41587-020-0592-2. [DOI] [PubMed] [Google Scholar]
- 11.Kurt I.C., Zhou R., Iyer S., Garcia S.P., Miller B.R., Langner L.M., Grünewald J., Joung J.K. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021;39:41–46. doi: 10.1038/s41587-020-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z., Chen S., Shan H., Jia Y., Chen M., Song Y., Lai L., Li Z. Efficient base editing with high precision in rabbits using YFE-BE4max. Cell Death Dis. 2020;11:36. doi: 10.1038/s41419-020-2244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y., Leete T.C., Born D.A., Young L., Barrera L.A., Lee S.J., Rees H.A., Ciaramella G., Gaudelli N.M. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat. Commun. 2020;11:2052. doi: 10.1038/s41467-020-15887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter M.F., Zhao K.T., Eton E., Lapinaite A., Newby G.A., Thuronyi B.W., Wilson C., Koblan L.W., Zeng J., Bauer D.E., et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudelli N.M., Lam D.K., Rees H.A., Solá-Esteves N.M., Barrera L.A., Born D.A., Edwards A., Gehrke J.M., Lee S.J., Liquori A.J., et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 2020;38:892–900. doi: 10.1038/s41587-020-0491-6. [DOI] [PubMed] [Google Scholar]
- 17.Koblan L.W., Arbab M., Shen M.W., Hussmann J.A., Anzalone A.V., Doman J.L., Newby G.A., Yang D., Mok B., Replogle J.M., et al. Efficient C·G-to-G·C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat. Biotechnol. 2021;39:1414–1425. doi: 10.1038/s41587-021-00938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan T., Yan N., Fei T., Zheng J., Meng J., Li N., Liu J., Zhang H., Xie L., Ying W., et al. Optimization of C-to-G base editors with sequence context preference predictable by machine learning methods. Nat. Commun. 2021;12:4902. doi: 10.1038/s41467-021-25217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grünewald J., Zhou R., Garcia S.P., Iyer S., Lareau C.A., Aryee M.J., Joung J.K. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019;569:433–437. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X.H., Chen L., Zhu B., Wang L., Chen C., Hong M., Huang Y., Li H., Han H., Cai B., et al. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat. Cell Biol. 2020;22:740–750. doi: 10.1038/s41556-020-0518-8. [DOI] [PubMed] [Google Scholar]
- 21.Liu D.R., Koblan L.W. Liu, D.R., Koblan, L.W; 2018. Cytosine to Guanine Base Editor. WO2018165629A1. [Google Scholar]
- 22.Chen L., Park J.E., Paa P., Rajakumar P.D., Prekop H.T., Chew Y.T., Manivannan S.N., Chew W.L. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat. Commun. 2021;12:1384. doi: 10.1038/s41467-021-21559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molla K.A., Qi Y., Karmakar S., Baig M.J. Base editing landscape extends to perform transversion mutation. Trends Genet. 2020;36:899–901. doi: 10.1016/j.tig.2020.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Hernan I., Roig I., Martin B., Gamundi M.J., Martinez-Gimeno M., Carballo M. De novo germline mutation in the serine-threonine kinase STK11/LKB1 gene associated with Peutz-Jeghers syndrome. Clin. Genet. 2004;66:58–62. doi: 10.1111/j.0009-9163.2004.00266.x. [DOI] [PubMed] [Google Scholar]
- 25.Schaaf L., Pickel J., Zinner K., Hering U., Höfler M., Goretzki P.E., Spelsberg F., Raue F., von zur Mühlen A., Gerl H., et al. Developing effective screening strategies in multiple endocrine neoplasia type 1 (MEN 1) on the basis of clinical and sequencing data of German patients with MEN 1. Exp. Clin. Endocrinol. Diabetes. 2007;115:509–517. doi: 10.1055/s-2007-970160. [DOI] [PubMed] [Google Scholar]
- 26.Kövesdi A., Tóth M., Butz H., Szücs N., Sármán B., Pusztai P., Tőke J., Reismann P., Fáklya M., Tóth G., et al. True MEN1 or phenocopy? Evidence for geno-phenotypic correlations in MEN1 syndrome. Endocrine. 2019;65:451–459. doi: 10.1007/s12020-019-01932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Revesz T., Holton J.L., Lashley T., Plant G., Frangione B., Rostagno A., Ghiso J. Genetics and molecular pathogenesis of sporadic and hereditary cerebral amyloid angiopathies. Acta Neuropathol. 2009;118:115–130. doi: 10.1007/s00401-009-0501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X., Blanden A.R., Narayanan S., Jayakumar L., Lubin D., Augeri D., Kimball S.D., Loh S.N., Carpizo D.R. Small molecule restoration of wildtype structure and function of mutant p53 using a novel zinc-metallochaperone based mechanism. Oncotarget. 2014;5:8879–8892. doi: 10.18632/oncotarget.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandelker D., Zhang L., Kemel Y., Stadler Z.K., Joseph V., Zehir A., Pradhan N., Arnold A., Walsh M.F., Li Y., et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA. 2017;318:825–835. doi: 10.1001/jama.2017.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auer-Grumbach M., Weger M., Fink-Puches R., Papić L., Fröhlich E., Auer-Grumbach P., El Shabrawi-Caelen L., Schabhüttl M., Windpassinger C., Senderek J., et al. Fibulin-5 mutations link inherited neuropathies, age-related macular degeneration and hyperelastic skin. Brain. 2011;134:1839–1852. doi: 10.1093/brain/awr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reis B.C.S., Cunha D.P., Bueno A.P.S., Carvalho F.A.A., Dutra J., Mello F.V., Ribeiro M.C.M., Milito C.B., da Costa E.S., Vasconcelos Z. Chronic granulomatous disease and myelodysplastic syndrome in a patient with a novel mutation in CYBB. Genes. 2021;12:1476. doi: 10.3390/genes12101476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu J., Pupavac M., Watkins D., Tian X., Feng Y., Chen S., Fenter R., Zhang V.W., Wang J., Wong L.-J., Rosenblatt D.S. Next generation sequencing of patients with mut methylmalonic aciduria: validation of somatic cell studies and identification of 16 novel mutations. Mol. Genet. Metab. 2016;118:264–271. doi: 10.1016/j.ymgme.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Dobbs K., Domínguez Conde C., Zhang S.Y., Parolini S., Audry M., Chou J., Haapaniemi E., Keles S., Bilic I., Okada S., et al. Inherited DOCK2 deficiency in patients with early-onset invasive infections. N. Engl. J. Med. 2015;372:2409–2422. doi: 10.1056/NEJMoa1413462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sykut-Cegielska J., Gradowska W., Piekutowska-Abramczuk D., Andresen B.S., Olsen R.K., Ołtarzewski M., Pronicki M., Pajdowska M., Bogdańska A., Jabłońska E., et al. Urgent metabolic service improves survival in long-chain 3-hydroxyacyl-CoA dehydrogenase (LCHAD) deficiency detected by symptomatic identification and pilot newborn screening. J. Inherit. Metab. Dis. 2011;34:185–195. doi: 10.1007/s10545-010-9244-x. [DOI] [PubMed] [Google Scholar]
- 35.Engler C., Kandzia R., Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hillson N.J., Rosengarten R.D., Keasling J.D. j5 DNA assembly design automation software. ACS Synth. Biol. 2012;1:14–21. doi: 10.1021/sb2000116. [DOI] [PubMed] [Google Scholar]
- 37.Hwang G.H., Park J., Lim K., Kim S., Yu J., Yu E., Kim S.T., Eils R., Kim J.S., Bae S. Web-based design and analysis tools for CRISPR base editing. BMC Bioinformatics. 2018;19:542. doi: 10.1186/s12859-018-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B., Li Y.Q., Zhao D.D., Yang J., Ma Y.H., Bi C.H., Zhang X.L. Sequence motifs and prediction model of GBE editing outcomes based on target library analysis and machine learning. J. Genet. Genomics. 2021;49:254–257. doi: 10.1016/j.jgg.2021.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The high-throughput sequencing data in this study are available in the NCBI database (PRJNA778911).