Abstract
Proneural (PN) to mesenchymal (MES) transition (PMT) is a crucial phenotypic shift in glioblastoma stem cells (GSCs). However, the mechanisms driving this process remain poorly understood. Here, we report that Fos-like antigen 1 (FOSL1), a component of AP1 transcription factor complexes, is a key player in regulating PMT. FOSL1 is predominantly expressed in the MES subtype, but not PN subtype, of GSCs. Knocking down FOSL1 expression in MES GSCs leads to the loss of MES features and tumor-initiating ability, whereas ectopic expression of FOSL1 in PN GSCs is able to induce PMT and maintain MES features. Moreover, FOSL1 facilitates ionizing radiation (IR)-induced PMT and radioresistance of PN GSCs. Inhibition of FOSL1 enhances the anti-tumor effects of IR by preventing IR-induced PMT. Mechanistically, we find that FOSL1 promotes UBC9-dependent CYLD SUMOylation, thereby inducing K63-linked polyubiquitination of major nuclear factor κB (NF-κB) intermediaries and subsequent NF-κB activation, which results in PMT induction in GSCs. Our study underscores the importance of FOSL1 in the regulation of PMT and suggests that therapeutic targeting of FOSL1 holds promise to attenuate molecular subtype switching in patients with glioblastomas.
Keywords: glioblastoma stem cells, proneural-to-mesenchymal transition, FOSL1, UBC9/CYLD/NF-κB axis
Graphical abstract
Here, we unveil the crucial role of FOSL1 in driving PMT in GSCs and provide an important rationale for targeting FOSL1. Our findings not only provide a better understanding of the molecular mechanisms underlying PMT in GSCs but also have important implications in the development of FOSL1-specific inhibitors for glioblastoma.
Introduction
Glioblastoma (GBM) is the most common and aggressive primary brain malignancy in adults.1,2 The standard treatment consists of maximal feasible surgical resection followed by radiation and temozolomide-based chemotherapy. Unfortunately, this therapeutic option shows many limitations in its efficacy, and almost all patients suffer from recurrence after the initial treatment.3,4 GBM is highly heterogeneous and can be subdivided into at least three subtypes with distinct genetic alterations, namely proneural (PN), mesenchymal (MES), and classical (CL), and these subtypes are associated with varying responses to intensive therapy and distinct clinical outcomes.5, 6, 7
GBM contains a heterogeneous subpopulation of cancer cells with stem cell characteristics, termed GBM stem cells (GSCs), which can propagate tumors and are thought to be responsible for the source of tumor recurrence and therapeutic resistance.8, 9, 10 Similar to bulk GBM tumors, gene expression profiling has classified GSCs into three subtypes: PN, MES, and CL.11,12 Of these, the MES GSCs are the most biologically aggressive and highly resistant to radiotherapy. Accumulating evidence has documented that PN GSCs may acquire more aggressive potential and radioresistance by shifting their phenotypic and transcriptomic signatures toward MES GSCs, a process called PN-to-MES transition (PMT).12, 13, 14, 15, 16, 17, 18, 19 Thus, unraveling the underlying mechanisms regulating PMT is of great importance in identifying novel and effective molecular targets for GBM.
FOS-like antigen 1 (FOSL1) is a member of the activator protein 1 (AP1) complex that heterodimerizes with members of the JUN family for proficient transcriptional activity. FOSL1 plays essential roles in various biological processes, including cell proliferation, differentiation, survival, and embryonic development. Numerous studies have demonstrated that FOSL1 is overexpressed in the majority of human cancers including GBM and exerts its oncogenic role by transcriptionally activating a subset of genes involved in cancer initiation and progression.20,21 For instance, FOSL1 has been shown to be a major inducer of epithelial-to-MES transition (EMT), a crucial step during tumor growth and invasiveness.22, 23, 24, 25, 26, 27 Moreover, FOSL1 serves as a key reprogramming factor for normal cells with potent tumor transformation potential.28 Furthermore, FOSL1 is sufficient to reprogram differentiated tumor cells into tumor-propagating stem-like cells by regulating stemness-related transcription factors.29 Additionally, directly targeting FOSL1 by using small-molecule inhibitors or knocking down its expression was reported by several groups, suggesting FOSL1 inhibition as a potential therapeutic strategy.30, 31, 32
Notably, a recent genome-wide expression profiling study has revealed that FOSL1 is one of the top upregulated transcription factors in MES GSCs when compared with PN GSCs and neural progenitors.33 Thus far, however, the specific roles of FOSL1 in different subtypes of GSCs remain unclear. Here, we identify FOSL1 as a key player in inducing the phenotypic transition from PN to MES subtype, either spontaneously or in response to ionizing radiation (IR). Mechanistically, we find that FOSL1 transcriptionally induces the expression of E2 SUMO-conjugating enzyme UBC9 and greatly enhances UBC9-mediated SUMOylation of CYLD. This leads to K63-linked polyubiquitination of multiple nuclear factor κB (NF-κB) signaling intermediaries, thereby unleashing NF-κB signaling, which eventually promotes PMT in GSCs.
Results
FOSL1 maintains the MES phenotype in GSCs
First, we examined the expression of FOSL1 in two distinct subtypes of GSCs (PN 35, PN 182, MES 21, and MES 505) from patient-derived xenografts as we previously described.34 The results showed that FOSL1 was highly expressed in MES 21 and MES 505 GSCs but was almost undetectable in PN 35 and PN 182 GSCs (Figure 1A). We next analyzed FOSL1 expression in ALDH1-positive (ALDH+) and ALDH1-negative (ALDH−) subpopulations of MES 21 and MES 505 GSCs and found that ALDH+ cells expressed higher levels of FOSL1 compared with ALDH− cells (Figures 1B and 1C). Using immunofluorescence staining of MES GSC- or PN GSC-derived tumorspheres, we observed that FOSL1 co-expressed with the MES marker CD44 but not with the PN markers OLIG2 and SOX2 (Figures 1D, 1E, and S1A). Furthermore, we assessed FOSL1 expression between undifferentiated MEC GSCs and their differentiated derivatives. After incubation with osteogenic differentiation medium for the indicated days, the expression of FOSL1 declined rapidly with a sharp contrast in the marked induction of an osteogenic differentiation marker RUNX2 (Figure S1B). Consistent with these in vitro results, elevated levels of FOSL1 were also found in mice bearing MES GSC-derived tumors (Figures S1C and S1D). These data suggest that FOSL1 is predominantly expressed in the MES subtype of GSCs.
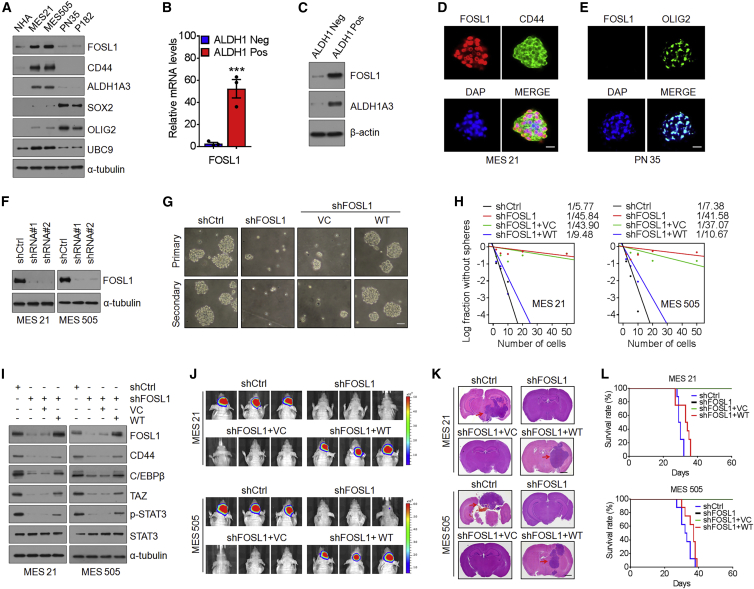
Figure 1.
FOSL1 maintains the MES phenotype in GSCs
(A) Immunoblot (IB) analysis of FOSL1, CD44, ALDH1A3, SOX2, OLIG2, and UBC9 in NHAs, two MES GSCs, and two PN GSCs. α-Tubulin was used as internal control. (B) qRT-PCR analysis of FOSL1 mRNA expression in ALDH1-positive and ALDH1-negative subpopulations of MES 21 glioma spheres. (C) IB analysis of FOSL1 protein expression in ALDH1-positive and ALDH1-negative subpopulations of MES 21 glioma spheres. α-Tubulin was used as internal control. (D) Representative immunofluorescence (IF) images of FOSL1 and CD44 expression in MES 21 GSCs. FOSL1 is in red and CD44 in green. Nuclei were counterstained with DAPI (blue). Scale bar, 25 μm. (E) Representative IF images of FOSL1 and OLIG2 expression in PN 35 GSCs. FOSL1 is in red and OLIG2 in green. Nuclei were counterstained with DAPI (blue). Scale bar, 25 μm. (F) IB analysis of FOSL1 in MES 21 and MES 505 GSCs after transduction with two independent lentiviral shRNA constructs targeting FOSL1 (FOSL1 shRNA1 and shRNA2; one targeting the open reading frame and one targeting the 3′ UTR). α-Tubulin was used as internal control. (G) Representative images of primary or secondary neurosphere formation of MES 21 GSCs with indicated modifications. Scale bar, 25 μm. (H) Limiting dilution neurosphere-forming assay in MES 21 and MES 505 GSCs transduced with shCtrl or shFOSL1 (targeting the 3′ UTR), reconstituted with vector control or wild-type (WT) FOSL1. Stem cell frequencies were estimated as the ratio 1/x with the upper and lower 95% confidence intervals, where 1 = stem cell and x = all cells. (I) IB analysis of FOSL1, CD44, C/EBPβ, TAZ, p-STAT3 (Tyr705), and STAT3 in MES 21 and 505 GSCs transduced with shCtrl or shFOSL1 (targeting the 3′ UTR), with or without re-expression of an shRNA-resistant FOSL1. (J) Representative bioluminescent (BLI) images of intracranial GBM xenografts derived from luciferase-expressing MES 21 and MES 505 GSCs with indicated modifications. Colored scale bars represent photons/s/cm2/steradian. (K) Representative H&E-stained brain sections from mice intracranially implanted with MES 21 and MES 505 GSCs with indicated modifications. Red arrows indicate tumors. Scale bar, 1 mm. (L) Kaplan-Meier survival curves of mice intracranially injected with MES 21 and MES 505 GSCs with indicated modifications (n = 8). Data are presented as means ± SD of three independent experiments. ∗∗∗p < 0.001, two-tailed Student’s t test.
To examine whether FOSL1 plays a role in the maintenance of GSCs, we used two distinct short hairpin RNAs (shRNAs) to knock down FOSL1 expression in luciferase-labeled MES 21 and MES 505 GSCs (Figure 1F). An in vitro limiting dilution assay illustrated that depletion of FOSL1 dramatically decreased the tumorsphere-forming capability of MES 21 and MES 505 GSCs (Figure 1G). Moreover, immunoblotting of dissociated tumorspheres showed that knockdown of FOSL1 reduced the expression of CD44 as well as C/EBPβ, TAZ, and p-STAT3, the core transcription factors (TFs) that regulate the MES features of GSCs (Figure 1H). Furthermore, we performed in vivo xenograft experiments whereby our data demonstrated that depletion of FOSL1 remarkably abrogated the in vivo tumorigenicity of MES GSCs (Figures 1I–1K).
To rule out the potential off-target effects of the FOSL1 shRNA, we expressed an shRNA-resistant FOSL1 in MES 21 and MES 505 GSCs that had been depleted of endogenous FOSL1. Reconstitution of FOSL1, but not an empty vector control, was able to restore sphere-forming frequency in vitro (Figures 1G and 1H) and tumorigenicity in the orthotopic xenografts (Figures 1I–1K), thus confirming that the impairment of MES GSC self-renewal and tumorigenicity is specifically due to the knockdown of FOSL1. Together, these results suggest that FOSL1 is required for the maintenance of MES features and tumorigenic potential of GSCs.
FOSL1 promotes transformation of PN GSCs into an MES state
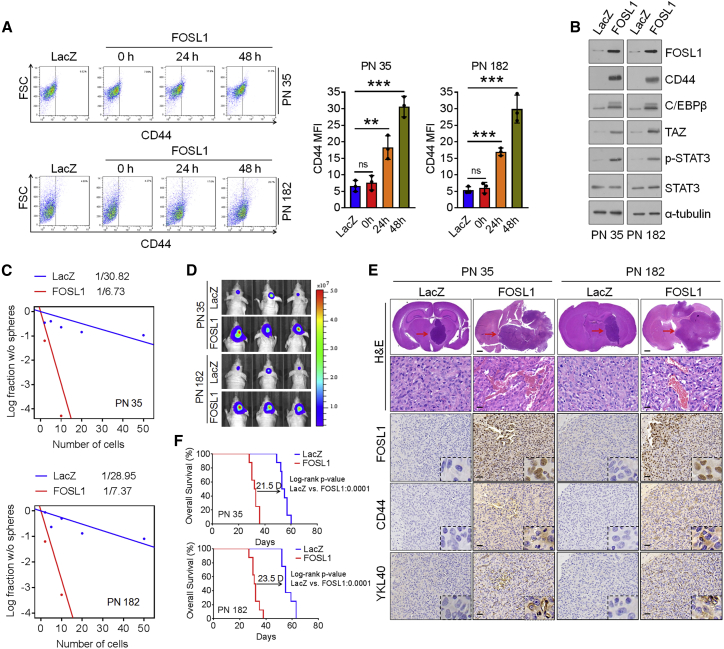
Recent evidence has documented that PMT could be prompted by certain cell-intrinsic signals.12,13,35 Given the key role of FOSL1 in sustaining MES features of GSCs, coupling with the differential expression levels of FOSL1 between PN GSCs and MES GSCs, we asked whether FOSL1 is involved in the regulation of PMT. To that end, we ectopically expressed lenti-LacZ or lenti-FOSL1 in PN 35 and PN 182 GSCs, which exhibited a very low level of endogenous FOSL1 protein. Flow-cytometric and immunoblot analysis showed that ectopic expression of FOSL1 in two PN GSCs led to an increase in MES marker CD44 expression (Figure 2A). Moreover, increased expression of the master MES TFs, including C/EBPβ, TAZ, and p-STAT3, were observed in FOSL1-tranduced PN GSCs (Figure 2B). The results of in vitro and in vivo models demonstrated that, compared with untransduced or LacZ-transduced PN GSCs, FOSL1-transduced PN CSCs could increase sphere-forming abilities and form hypervascular and invasive tumors (Figures 2C–2E). The elevation of the MES markers (CD44, YKL40, and vimentin) was also validated in FOSL1-transduced tumors (Figure 2E). Correspondingly, mice implanted with FOSL1-transduced PN GSCs exhibited considerably shortened survival relative to those implanted with LacZ-transduced PN GSCs (Figure 2F). Together, these results indicate that FOSL1 enables PN GSCs to acquire an MES phenotype with enhanced ability to promote tumorigenesis.
Figure 2.
FOSL1 promotes transformation of PN GSCs into an MES state
(A) Representative FACS plots of CD44+ subpopulation in PN 35 and PN 182 GSCs transduced with vector control or FOSL1. Median fluorescence intensity of CD44 is shown on the right. (B) IB analysis of FOSL1, CD44, C/EBPβ, TAZ, p-STAT3 (Tyr705), and STAT3 in PN 35 and PN 182 GSCs ectopically expressing FOSL1 or vector control. α-Tubulin was used as internal control. (C) Limiting dilution neurosphere-forming assay in PN 35 and PN 182 GSCs transduced with FOSL1 or vector control. (D) Representative BLI images of mice bearing xenografts derived from luciferase-expressing PN 35 and PN 182 GSCs transduced with FOSL1 or vector control. Colored scale bars represent photons/s/cm2/steradian. (E) Representative H&E-stained brain sections and IHC-staining images of FOSL1, CD44, and YKL40 in mice bearing xenografts derived from PN 35 and PN 182 GSCs with indicated modifications. Red arrows indicate tumors. Scale bars, 1 mm (H&E staining) and 25 μm (IHC staining). (F) Kaplan-Meier survival curves of mice intracranially implanted with PN 35 and PN 182 GSCs with indicated modifications (n = 8).
FOSL1 facilitates IR-induced PMT and radioresistance of PN GSCs
It is becoming increasingly clear that the extrinsic factors that can induce PMT are IR and tumor necrosis factor α (TNF-α).11,12,16,17,36 Thus, we wondered whether FOSL1 is involved in IR- or TNF-α-induced PMT. As shown in Figures 3A and S2A, treatment of PN 35 and PN 182 GSCs with either IR (5 Gy) or TNF-α (10 ng/mL) markedly induced the expression of FOSL1, which preceded the induction of the MES marker CD44 and the master MES TFs (C/EBPβ, TAZ, and p-STAT3), indicating that elevation of FOSL1 is an early event linked to extrinsic stimuli-induced PMT. We next established a tetracycline (doxycycline [Dox])-inducible lentiviral shRNA system to knock down FOSL1. As expected, IR- or TNF-α-induced MES transdifferentiation was largely abrogated in two PN GSCs when FOSL1 expression was depleted by the addition of Dox (Figures 3B and S2B).
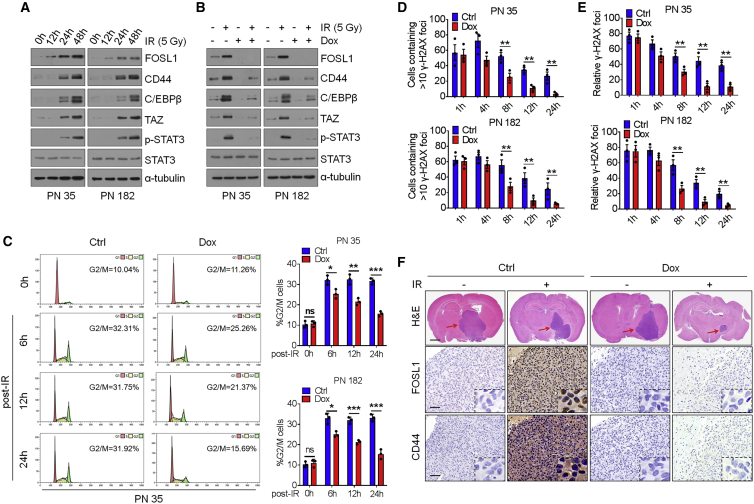
Figure 3.
FOSL1 facilitates IR-induced PMT and radioresistance of PN GSCs
(A) IB analysis of FOSL1, CD44, C/EBPβ, TAZ, p-STAT3, and STAT3 in PN 35 and PN 182 GSCs at the indicated time points following treatment with 5 Gy IR. α-Tubulin was used as internal control. (B) IB analysis of indicated antibodies in PN 35 and PN 182 GSCs transduced with Dox-inducible lentiviral vectors expressing FOSL1 shRNA or Ctrl shRNA in the presence or absence of 5 Gy IR. (C) Cell-cycle analysis of PN 35 and PN 182 GSCs transduced with Dox-inducible FOSL1 shRNA or Ctrl shRNA. Left: cell-cycle plots. The percentage of cells in G2/M phase is indicated within each plot. Right: quantification of percentage of cells in G2/M phase in FOSL1 shRNA or Ctrl shRNA-transduced PN 35 and PN 182 GSCs. (D) Quantitation of cells containing >10 γ-H2AX foci at the indicated time points after IR (5 Gy) treatment. Percentage of cells containing >10 γ-H2AX foci in ten random microscopic fields was calculated. (E) The average of mean number of γ-H2AX foci per nucleus in FOSL1- and LacZ-transduced PN 35 and PN 182 GSCs at the indicated time points after IR treatment. (F) Representative H&E-stained brain sections and IHC-staining images of FOSL1 and CD44 in mice bearing xenografts derived from PN 35 and PN 182 GSCs with indicated modifications. Red arrows indicate tumors. Scale bars, 1 mm (H&E staining) and 25 μm (IHC staining). Data are presented as means ± SD of three independent experiments. ∗∗p < 0.01, two-tailed Student’s t test.
Since PMT is closely associated with IR resistance, we next determined whether FOSL1 contributes to GSC radioresistance. We analyzed the survival of mice inoculated with FOSL1- or LacZ-transduced PN 35 and PN 182 GSCs following treatment with IR (2.5 Gy × 4). The results showed that IR treatment prolonged the survival of mice bearing tumors derived from LacZ-transduced PN GSCs by 46 days (PN 35) and 47.5 days (PN 182). By contrast, in mice bearing FOSL1-transduced tumors, IR extended mouse survival for only 20 days (PN 35) and 18 days (PN 182), thus suggesting that FOSL1 promotes the radioresistance of PN GSCs in vivo (Figure S2C). Given that activation of the DNA damage checkpoint response is critical for the radioresistance of cancer cells, we therefore evaluated cell-cycle distribution and found that LacZ-transduced PN GSCs exhibited a profound G2/M phase arrest at 24 h after IR, whereas FOSL1-transduced PN GSCs only display a modest arrest in G2/M upon exposure to IR (Figure 3C). We further measured the kinetics of nuclear foci formation of γ-H2AX, a well-known sensor protein of DNA damage, using immunofluorescence (IF) staining. One hour after IR treatment, FOSL1- and LacZ-transduced PN GSCs displayed comparable γ-H2AX foci formation. However, fewer γ-H2AX-positive cells were observed at 8, 12, or 24 h after IR in FOSL1-transduced PN GSCs than in LacZ-transduced PN GSCs (Figures 3D and 3E). These results suggest that FOSL1 promotes the radioresistance of PN GSCs by preferential activation of the DNA damage response.
Next, we examined the combined effect of FOSL1 depletion with IR on PN GSC-derived xenograft tumors. Mice were intracranially inoculated with PN 35 and PN 182 GSCs transduced with Dox-inducible FOSL1 shRNA and then treated with four cycles of 2.5 Gy IR on consecutive days, in the absence or presence of Dox. Compared with mice receiving IR treatment alone, those receiving the combined treatment (Dox plus IR) showed a remarkable delay in tumor growth and prolonged survival (Figures 3F and S2D). Immunohistochemistry (IHC) analysis of PN GSC-derived tumor tissues revealed that the MES marker CD44 was strongly induced by IR, which was substantially attenuated by Dox-induced FOSL1 knockdown (Figure 3F). These results suggest that inhibition of FOSL1 could enhance the anti-tumor effects of IR at least in part by preventing IR-induced PMT.
FOSL1 induces activation of NF-κB signaling
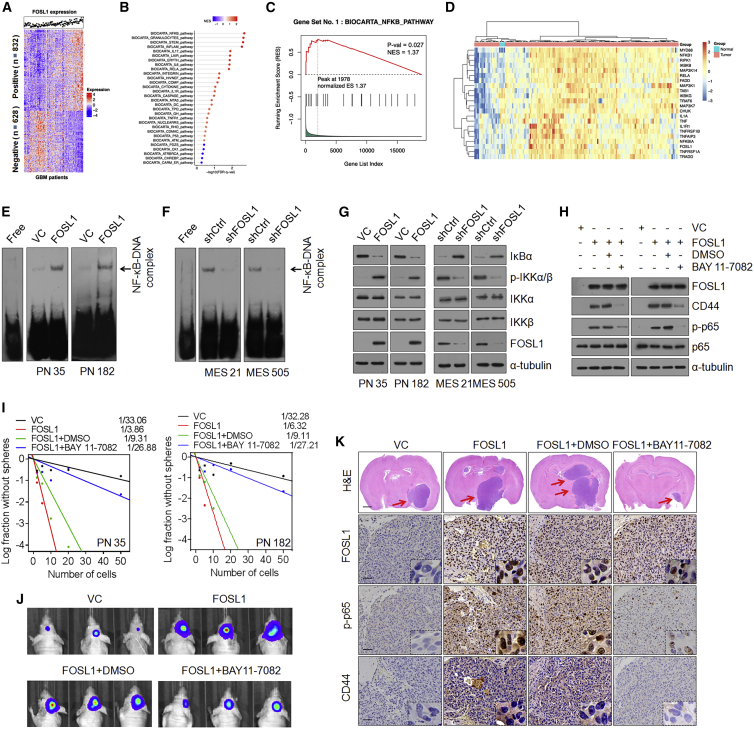
To gain insight into the mechanisms that underpin FOSL1-mediated PMT, we sought to explore the possible downstream pathways that FOSL1 could potentially regulate. We assessed the correlation between 1,460 FOSL1-associated genes and FOSL1 expression across 168 GBM tumors from The Ca (Figure 4A). Gene set enrichment analysis (GSEA) showed that NF-κB, a master regulator that mediates PMT in GSCs,12 most significantly correlated with FOSL1 (Figures 4B, 4C, and S3A). We next analyzed the differentially expressed genes in 5 normal brain tissues and 168 GBM specimens from The Cancer Genome Atlas (TCGA) database. We found that 137 tumors expressing high levels of FOSL1 display high NF-κB expression, whereas 21 FOSL1 low-expressing tumors exhibit low NF-κB expression (Figure 4D), suggesting that FOSL1 strongly correlated with NF-κB expression.
Figure 4.
FOSL1 induces activation of NF-κB signaling
(A) Cluster heatmap showing 832 FOSL1 positively correlated and 628 FOSL1 negatively correlated genes in TCGA database. (B) Gene set enrichment analysis (GSEA) showing signaling pathways related to FOSL1 in human primary GBM. The color gradation and location of the dot respectively indicate the GSEA score and p value. (C) Correlation between the enrichment of FOSL1 and NF-κB pathway gene expression in GBM by GSEA analysis. (D) Heatmap depicting genes differentially expressed between FOSL1 and NF-κB pathways/targets in normal brain tissues (n = 5) and GBM (n = 168). Genes are labeled and clustered using hierarchical clustering. (E and F) EMSA analysis of NF-κB DNA-binding activity in PN 35 and PN 182 (E), and MES 21 and MES 505 (F) GSCs with indicated modifications. (G) IB analysis of IκBα, p-IKKα/β (Ser180/181), IKKα, IKKβ, and FOSL1 in PN 35, PN 182, MES 21, and MES 505 GSCs with indicated modifications. α-Tubulin was used as internal control. (H) IB analysis of FOSL1, CD44, C/EBPβ, TAZ, p-STAT3 (Tyr705), STAT3, p-p65 (Ser536), and p65 in PN 35 and PN 182 GSCs transduced with vector control or FOSL1 in the presence or absence of BAY 11-7082. α-Tubulin was used as internal control. (I) Limiting dilution neurosphere-forming assay in PN 35 and PN 182 GSCs expressing FOSL1 or vector control, treated with dimethyl sulfoxide (DMSO) or BAY 11-7082. (J) Representative BLI images of mice bearing xenografts derived from luciferase-expressing PN 35 and PN 182 GSCs transduced with FOSL1 or vector control, with or without BAY 11-7082 treatment. (K) Representative H&E-stained brain sections and IHC-staining images of FOSL1, p-p65, and CD44 in indicated PN GSC-derived xenografts treated with DMSO or BAY 11-7082. Red arrows indicate tumors. Scale bars, 1 mm (H&E staining) and 25 μm (IHC staining).
To examine whether FOSL1 might interfere with NF-κB signaling, we performed a dual-luciferase reporter assay and an electrophoretic mobility shift assay (EMSA). The results demonstrated that ectopic expression of FOSL1 in PN 35 and PN 182 GSCs resulted in a marked increase in NF-κB transcriptional and DNA-binding activities (Figures 4E and S3B), whereas depletion of FOSL1 in MES 21 and MES 505 GSCs had the opposite effects (Figures 4F and S3C). The canonical NF-κB signaling includes the activation of catalytic IκB kinases IKKα and IKKβ, which promotes the assembly of ubiquitination and proteasomal degradation of IκBα, thereby triggering translocation of NF-κB to the nucleus and activation of target genes.37 Therefore, we analyzed the NF-κB signaling components IκBα and IKKα/β. As shown in Figure 4G, an increase in IKKα/β phosphorylation and a concomitant decrease in IκBα protein levels were detected in FOSL1-transduced PN 35 and PN 182 GSCs, while the opposite effect was observed in MES 21 and MES 505 GSCs depleted of FOSL1. Correspondingly, a key set of NF-κB target genes was substantially upregulated in two FOSL1-transduced PN GSCs (Figure S3D) and was decreased in two FOSL1-depleted MES GSCs (Figure S3E). Furthermore, we used a specific NF-κB inhibitor, BAY 11-7082, to treat FOSL1-transduced PN 35 and PN 182 GSCs. BAY 11-7082 treatment effectively reduced sphere-forming capacity of FOSL1-transduced PN 35 and PN 182 GSCs and suppressed intracranial tumor growth in athymic nude mice (Figures 4I–4K and S3F). Immunoblotting and IHC analysis revealed that FOSL1-induced upregulation of NF-κB p-p65 and CD44 were largely abrogated by BAY 11-7082 (Figures 4H and 4K). Consistent with these data, BAY 11-7082 treatment also markedly inhibited tumor growth of GBM xenografts derived from MES 21 and 505 GSCs expressing high endogenous levels of FOSL1 (Figure S3G). Together, these results indicate that FOSL1 might be an upstream regulator of NF-κB signaling.
FOSL1 promotes CYLD SUMOylation to impair deubiquitination of NF-κB signaling intermediaries
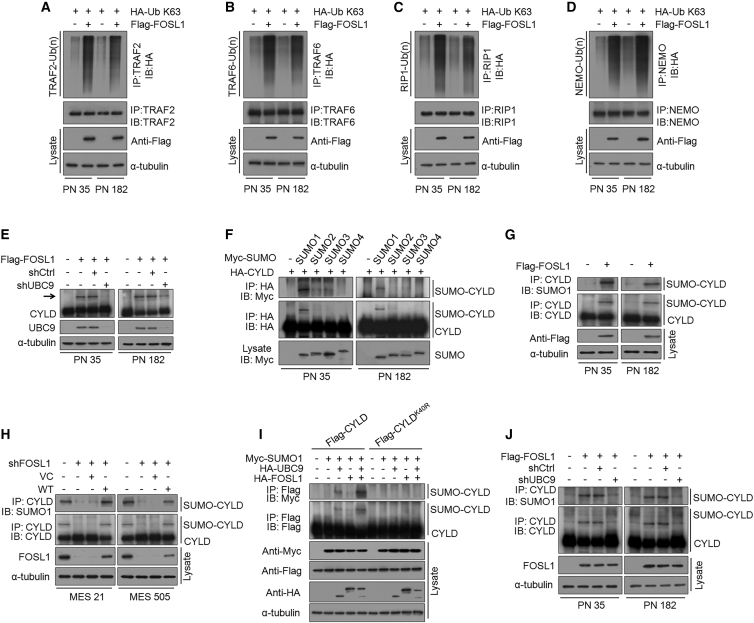
It is well established that NF-κB signaling is tightly controlled by multiple positive and negative regulators. In particular, ubiquitination and deubiquitination (DUB) of the core NF-κB signaling components have been identified as crucial steps in the control of NF-κB signaling pathways.38 Accordingly, we assessed the effect of FOSL1 on the polyubiquitin levels of a series of NF-κB signaling intermediaries, including TRAF2, TRAF6, RIP1, TAK1, NEMO, and BCL3. Intriguingly, we found that ectopic expression of FOSL1 in PN 35 and PN 182 GSCs induced K63-linked polyubiquitin levels of TRAF2, TRAF6, RIP1, and NEMO, but not TAK1 and BCL3 (Figures 5A–5D). Moreover, we examined the expression of several key DUBs, including CYLD, A20, Cezanne, OTUB1/2, and OTULIN, which could switch off NF-κB signaling through specifically dismantling polyubiquitin chains from NF-κB signaling intermediaries. However, no significant change in the protein levels of CYLD, A20, Cezanne, OTUB1/2, and OTULIN were observed after FOSL1 overexpression. Remarkably, we observed two distinct immunoreactive CYLD bands (a predominant form at 107 kDa and an upper slow-migrating form at 125 kDa) in FOSL1-transduced PN 35 and PN 182 GSCs (Figure 5E). Recent evidence suggests that CYLD is regulated by SUMOylation, a post-translational modification involving the conjugation of a small ubiquitin-like modifier (SUMO) to protein, which could impair its deubiquitinating activity toward NF-κB signaling intermediaries.39,40 Since CYLD was detected as two bands in two PN GSCs expressing FOSL1, we asked whether FOSL1 could affect CYLD SUMOylation in PN GSCs. To that end, we co-transfected hemagglutinin (HA)-CYLD together with each one of the four 5× Myc-tagged SUMO isoforms (SUMO1–4) in PN 35 and PN 182 GSCs. As shown in Figure 5F, a slower-migrating form of HA-CYLD was detected only when co-transfected with Myc-SUMO1, but not with other SUMO isoforms (SUMO2, 3, 4), indicating that SUMO1 modification of CYLD existed in two PN GSCs. Indeed, a co-immunoprecipitation assay showed that ectopic expression of FOSL1 in PN 35 and PN 182 GSCs led to an increase in levels of SUMO1-conjugated CYLD (Figure 5G). By contrast, knocking down FOSL1 expression in MES 21 and MES 505 GSCs largely abrogated the formation of SUMO1-CYLD conjugates, and this effect could be restored by re-expression of RNAi-resistant wild-type (WT) FOSL1 (Figure 5H). Furthermore, we transduced FLAG-tagged WT or K40R (the unSUMOylatable mutant) CYLD, together with HA-tagged FOSL1, Myc-tagged SUMO1, and HA-tagged UBC9 into HEK293T cells. We noted that WT, but not K40R, CYLD was moderately SUMOylated by SUMO1 and UBC9, which could be strongly strengthened by ectopic expression of FOSL1 (Figure 5I). These findings support a critical role of FOSL1 in the promotion of CYLD SUMOylation in GSCs.
Figure 5.
FOSL1 promotes CYLD SUMOylation to impair deubiquitination of NF-κB signaling intermediaries
(A–D) K63-linked polyubiquitin chains of TRAF2 (A), TRAF6 (B), RIP1 (C), and NEMO (D) were analyzed in PN 35 and PN 182 GSCs with indicated modifications. (E) IB analysis of CYLD protein expression in PN 35 and PN 182 GSCs expressing exogenous FOSL1 or vector control, with or without FOSL1 depletion. Arrow indicates additional slow-migrating band in FOSL1-transduced PN GSCs. α-Tubulin was used as internal control. (F) PN 35 and PN 182 GSCs were co-transfected with HA-CYLD and one of the Myc-tagged SUMO isoforms (SUMO1–4). Immunoprecipitated HA-CYLD was either incubated with anti-HA antibody (middle panel) or was probed for SUMOylation using anti-Myc antibody (top panel). The expression level of SUMO1–4 in the cell lysates is also shown (bottom panel). (G) PN 35 and PN 182 GSCs were transduced with FLAG-tagged FOSL1 or vector control. CYLD was immunoprecipitated and then incubated with anti-SUMO1 antibody or anti-CYLD antibody, respectively. (H) MES 21 and MES 505 GSCs were transduced with shCtrl or shFOSL1 (targeting the 3′ UTR), reconstituted with WT FOSL1 or vector control. CYLD was immunoprecipitated and then incubated with anti-SUMO1 antibody or anti-CYLD antibody, respectively. (I) HEK293T cells were transduced with FLAG-tagged WT CYLD or K40R CYLD (an unSUMOylatable mutant), together with HA-tagged FOSL1, Myc-tagged SUMO1, and HA-tagged UBC9. FLAG-tagged CYLD was immunoprecipitated and then incubated with anti-Myc antibody or anti-FLAG antibody, respectively. (J) PN 35 and PN 182 GSCs were transduced with FLAG-tagged FOSL1 or vector control, with or without UBC9 knockdown. CYLD was immunoprecipitated and then incubated with anti-SUMO1 antibody or anti-CYLD antibody, respectively.
FOSL1 facilitates CYLD SUMOylation, NF-κB activation, and PMT by transcriptionally activating UBC9
Protein SUMOylation is catalyzed by a set of enzymes: E1-activating enzyme (SAE1/SAE2), E2-conjugating enzyme (UBC9), and E3 ligases. Intriguingly we noted that, without co-transfection of UBC9, FOSL1 alone could not strengthen CYLD SUMOylation in HEK293T cells (Figure 5I), even in the presence of SUMO1. Moreover, when UBC9 was depleted in FOSL1-transduced PN 35 and PN 182 GSCs, FOSL1-mediated SUMOylation of CYLD was markedly attenuated (Figures 5E and 5J), indicating that FOSL1 might possibly facilitate CYLD SUMOylation in a manner that depends on UBC9.
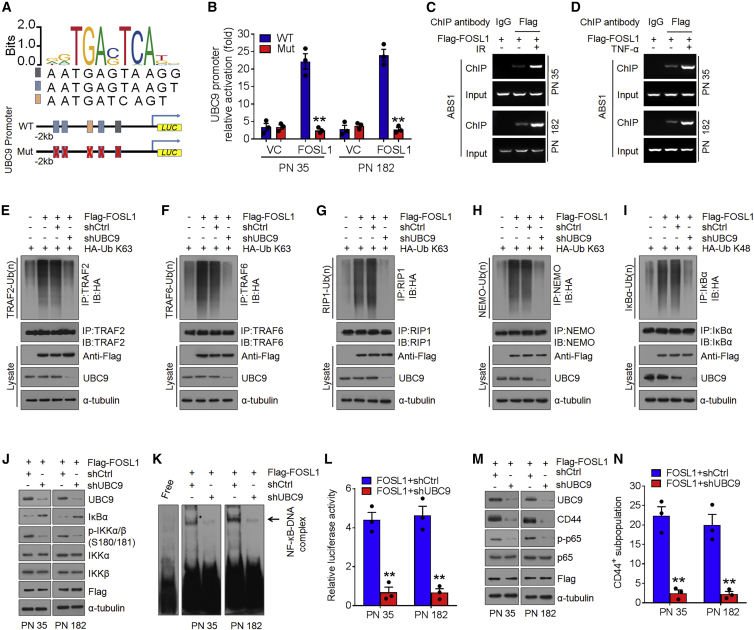
To examine the relationship between FOSL1 and UBC9, we analyzed the expression of UBC9 in PN and MES GSCs as well as their corresponding tumor xenografts. Similar to the expression pattern of FOSL1, elevated levels of UBC9 were observed in two MES GSCs and their xenografts (Figures 1A and S1D). Intriguingly, we noticed that UBC9 promoter encompassed five putative FOSL1-binding sites (A1–A5) by using the sequence analysis of the UBC9 promoter region (http://jaspar.genereg.net/), implicating that FOSL1 possibly regulates UBC9 expression at transcriptional levels (Figure 6A). Consistent with this notion, we found that ectopic expression of FOSL1 increased, whereas knockdown of FOSL1 reduced UBC9 mRNA and protein levels (Figures S4A–S4D). We next cloned a firefly luciferase reporter construct harboring approximately 2 kb of the UBC9 promoter or mutations in all five putative AP1-binding sites (ABS), located at −838 bp (ABS1), −1,181 bp (ABS2-1), −1,317 bp (ABS3), −1,615 bp (ABS2-2), and −1,659 bp (ABS2-3). Ectopic expression of FOSL1 in PN 35 and PN 182 GSCs increased the activity of the WT UBC9 promoter but not the mutated UBC9 promoter (Figure 6B). To ascertain which ABS was responsive to FOSL1-mediated transcriptional activation of UBC9 promoter, we performed an ABS-directed mutagenesis assay and observed that only the constructs containing ABS1 mutation could abolish the activation of UBC9 promoter activity (Figures S4E and S4F). Chromatin immunoprecipitation assays confirmed that FOSL1 directly bound to ABS1 rather than other ABSs (Figure S4G) in the UBC9 promoter, and this effect could be further enhanced by IR or TNF-α (Figures 6C and 6D).
Figure 6.
FOSL1 facilitates CYLD SUMOylation, NF-κB activation, and PMT via transcriptionally activating UBC9
(A and B) The five AP1-binding sites of the UBC9 promoter were mutated to generate the mutant UBC9 promoter (A). The relative luciferase activity of WT or mutant UBC9 promoters was determined in PN 35 and PN 182 GSCs transfected with vector control or FOSL1 (B). (C and D) Chromatin immunoprecipitation assays on AP1-binding site 1 of UBC9 promoter were performed in FLAG-FOSL1-transduced PN 35 and PN 182 GSCs treated with DMSO, 5 Gy IR (C), or 10 ng/mL TNF-α (D). (E–H) K63-linked polyubiquitin chains of TRAF2 (E), TRAF6 (F), RIP1 (G), and NEMO (H) were detected in PN 35 GSCs expressing exogenous FOSL1 or vector control, with or without UBC9 knockdown. (I) K48-linked polyubiquitin chains of IκBα were analyzed in PN 35 GSCs with indicated modifications. (J) IB analysis of UBC9, IκBα, p-IKKα/β (Ser180/181), IKKα, IKKβ, and FLAG-FOSL1 in PN 35 and PN 182 GSCs with indicated modifications. α-Tubulin was used as internal control. (K) EMSA analysis of NF-κB DNA-binding activity in PN 35 and PN 182 GSCs with indicated modifications. (L) Relative luciferase reporter activity of NF-κB in PN 35 and PN 182 GSCs with indicated modifications. (M) IB analysis of UBC9, CD44, p-p65 (Ser536), p65, and FLAG-FOSL1 in PN 35 and PN 182 GSCs with indicated modifications. α-Tubulin was used as internal control. (N) FACS analysis of CD44+ subpopulation in PN 35 and PN 182 GSCs with indicated modifications. Data are presented as means ± SD of three independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001, two-tailed Student’s t test.
To assess the role of UBC9 in FOSL1-mediated NF-κB activation and PMT, we introduced the shRNA against UBC9 into FOSL1-tranduced PN 35 and PN 182 GSCs. Silencing of UBC9 severely compromised FOSL1-induced K63-linked polyubiquitin levels of TRAF2, TRAF6, RIP1, and NEMO (Figures 6E–6H), and K48-linked polyubiquitin levels of IκBα (Figure 6I), which leads to upregulation of IκBα and downregulation of p-IKKα/β (Figure 6J) as well as increased NF-κB DNA-binding and transcriptional activities (Figures 6K and 6L). Moreover, following UBC9 knockdown, upregulation of CD44, p-p65, BCL-XL, CCND1, MMP-9, TWIST1, and VEGFC induced by FOSL1 were drastically inhibited (Figures 6M and S4H). Furthermore, depletion of UBC9 substantially abrogated the promoting effects of FOSL1 on sphere-forming frequency in vitro and tumorigenicity of orthotopic GBM xenografts (Figures 6N and S4I–S4L). Additionally, we overexpressed WT UBC9 or its SUMOylation defective mutant UBC9 (C93S) in MES 21 and MES 505 GSCs in which endogenous FOSL1 had been depleted. We found that exogenous WT, but not the C93S mutant UBC9, could largely rescue the inhibitory effect of FOSL1 depletion on the accumulation of SUMO1-CYLD conjugates (Figure S5A), the DNA-binding and transcriptional activities of NF-κB (Figures S5B and S5C), and the ubiquitination status of TRAF2, TRAF6, RIP1, NEMO, and IκBα (Figures S5E–S5I). Consistent with this, WT, but not C93S, UBC9 was sufficient to restore the loss of MES marker CD44 and master MES TFs (Figure S5D), sphere formation efficiency (Figure S5J), and tumorigenicity (Figure S5K) caused by FOSL1 knockdown. Together, these results indicate that FOSL1-mediated transcriptional activation of UBC9 is crucial for CYLD SUMOylation, NF-κB activation, and PMT in GSCs.
Clinical relevance of FOSL1/NF-κB-driven PMT in human GBM
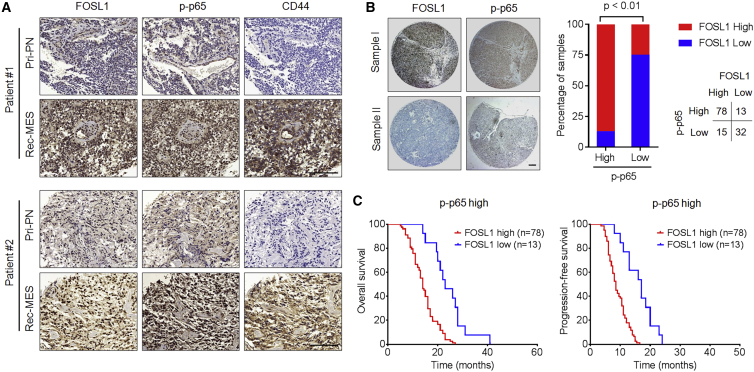
To examine the potential clinical relevance of our findings, we assessed the expression of FOSL1, NF-κB (p-p65), CD44, and UBC9 in six primary GBM specimens (PN subtype) and corresponding recurrent GBM specimens (MES subtype). We found that FOSL1, NF-κB (p-p65), CD44, and UBC9 were significantly upregulated in recurrent MES tumors compared with those in primary PN tumors (Figures 7A and S6). Moreover, we analyzed the expression of FOSL1 and p-p65 in a tissue microarray containing 138 primary GBM specimens. As shown in Figure 7B, FOSL1 strongly correlated with p-p65 expression. Specifically, about 83.9% of the high-FOSL1 samples exhibited high p-p65 expression, whereas 71.1% of samples with low FOSL1 exhibited low p-p65 expression. Furthermore, Kaplan-Meier survival analysis showed that high-FOSL1 patients displayed significantly shorter overall survival and progression-free survival in the high p-p65 group (Figure 7C), indicating that FOSL1 has a prognostic value. Collectively, these results strongly support our experimental findings that FOSL1 induces NF-κB activation, thereby promoting PMT in GBM.
Figure 7.
Clinical relevance of FOSL1/NF-κB-driven PMT in human GBMs
(A) Representative FOSL1, CD44, and p-p65 expression levels are shown in consecutive sections of two matched pairs of primary PN tumors and corresponding relapsed MES tumors. Scale bars, 50 μm. (B) Representative IHC-staining images of FOSL1 and p-p65 in tissue microarray containing 138 primary GBM samples (left). Correlations of IHC data for high or low FOSL1 expression relative to level of p-p65 are shown (right). Scale bar, 1 mm. (C) Kaplan-Meier curves showing overall survival (left) and progression-free survival (right) of GBM patients divided on the basis of FOSL1 expression in p-p65high tumors.
Discussion
PMT can endow GSCs with more aggressive phenotypes, which are closely associated with therapeutic resistance, tumor recurrence, and unfavorable prognosis. Therefore, unraveling the mechanisms that drive PMT is of great significance to the design of better therapeutic strategies for treating GBM. Thus far, there are several possible explanations for the initiation of PMT: (1) cell-intrinsic factors (e.g., NF1 mutations, together with NF-κB transcriptional programs); (2) the pro-inflammatory factors present in the GBM microenvironment; (3) exposure to cytotoxic treatments, such as radio-/chemotherapy and anti-angiogenic therapy. Recent investigations have identified several molecular effectors involved in PMT;12,13,15,41, 42, 43 however, the molecular mechanisms responsible for modulating this process remain to be determined.
In this study, we investigated the role of FOSL1 in patient-derived PN and MES GSCs. Our results revealed that FOSL1 is a critical inducer of the transition from PN GSCs to MES differentiation. Specifically, we found that FOSL1 is preferentially expressed in the MES subtype of GSCs, but not the PN subtypes. Ectopic expression of FOSL1 in PN GSCs results in the acquisition of the MES phenotype, whereas knockdown of FOSL1 in MES GSCs leads to the loss of MES features and tumor-initiating ability. Moreover, we uncovered FOSL1 as a potential link between PMT and tumor radioresistance in GSCs. IR induced upregulation of FOSL1, thereby activating a set of PMT-associated regulators, which led to radioresistance of PN GSCs. Importantly, depletion of FOSL1 severely attenuated IR-induced PMT, sensitized GSCs to IR, and conferred survival benefit in mice bearing GSC-derived tumors. Our findings clearly demonstrated that targeting FOSL1 could be considered as a new option for treating GBM. Consistent with this notion, several recent studies have suggested that a gene therapy approach targeting FOSL1 expression through RNAi, CRISPR-Cas9, or PROTAC may represent a promising alternative option for the treatment of human cancers.30 Although TFs have historically proven to be difficult to target for therapeutic purposes, in part due to their high activity, targeting FOSL1 by two novel small-molecule inhibitors, SR11302 and LY-1816, has recently been shown to display potent efficacy in pre-clinical models, which provide groundwork for attempted FOSL1 pharmacological inhibition.31,32
NF-κB is a pleiotropic TF that controls the expression of many genes related to inflammation and immune responses as well as biological processes central to the development of malignancies. Deregulated hyperactivation of NF-κB has been frequently observed in diverse types of cancers, including GBM. Accumulating evidence has suggested that NF-κB signaling plays crucial roles in maintaining proliferation, self-renewal, and tumorigenicity of GSCs. More recently, NF-κB was found to be a master regulator that mediates PMT in GSCs. However, the molecular mechanisms underpinning sustained NF-κB activation during PMT remain undetermined. Here, we demonstrated that FOSL1 is able to induce PMT by modulating the IKKβ-IκBα-NF-κB signaling pathway. We found that ectopic expression of FOSL1 in PN GSCs constitutively induces multiple processes, including phosphorylation of IKKβ on Ser181, degradation of IκBα, and targeting of NF-κB p65 to the nucleus. Thus, our current work reveals a novel mechanism whereby FOSL1 might activate the canonical NF-κB signaling, which could be a key node in driving PMT and sustaining MES identity in GSCs.
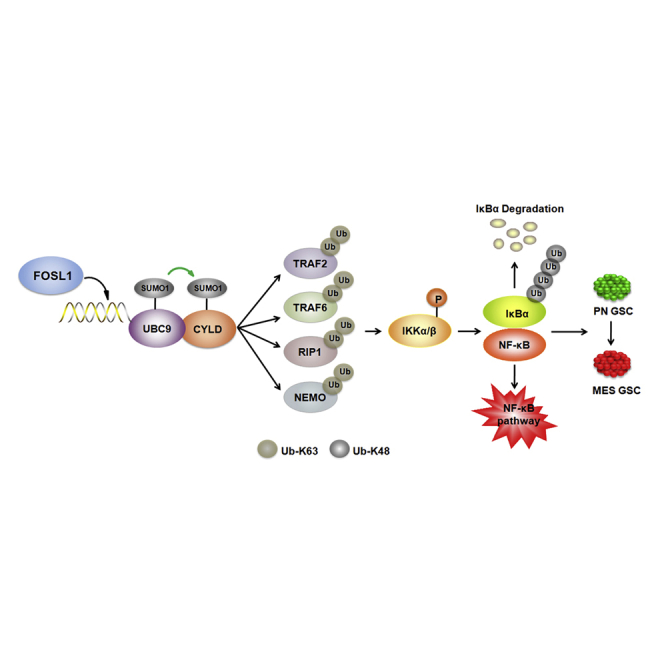
CYLD, a K63-specific deubiquitinating enzyme, has been shown to switch off NF-κB signaling through removing K63-linked polyubiquitin chains from multiple NF-κB signaling intermediaries, including RIP1, NEMO, TRAF2, TRAF6, TAK1, and BCL3. It has been reported that CYLD can be transcriptionally regulated by multiple transcriptional repressors, such as Snail and Hes1. Nevertheless, the results of our current study showed that expression of CYLD mRNA is not altered after manipulation of FOSL1 expression. Instead we found that FOSL1 increases the SUMOylation of CYLD, impairing deubiquitination of master NF-κB signaling intermediaries including TRAF2/6, RIP1, and NEMO, which ultimately leads to NF-κB activation (Figure 7D). This effect is dependent on the FOSL1-mediated transcriptional activation of UBC9, a unique SUMO E2 enzyme known to conjugate SUMO to target substrates. When UBC9 was depleted, FOSL1-induced CYLD SUMOylation, polyubiquitination of NF-κB intermediaries, and subsequent NF-κB activation were severely compromised. On the basis of the above findings, we establish a mechanistic link between FOSL1 and UBC9/CYLD/NF-κB axis and clearly demonstrate that sustained NF-κB activation is driven by a FOSL1-mediated signaling cascade. Given the abundant expression of NF-κB in a variety of cancers, future studies will need to further explore the role of FOSL1 in other cancer types that are dependent on NF-κB for tumorigenesis.
In conclusion, our study unveils the crucial role of FOSL1 in driving the process of PMT in GSCs and provides an important rationale for targeting FOSL1 alone or in combination with IR to treat GBM patients. Our findings not only provide a better understanding of the molecular mechanisms underlying PMT in GSCs but also have important implications in the development of FOSL1-specific inhibitors for cancers including GBM.
Materials and methods
Human subjects
Four freshly resected GBM specimens were obtained from the Department of Neurosurgery, the First Affiliated Hospital of Nanjing Medical University (Table S1). Tissue microarray consisting of 138 formalin-fixed paraffin-embedded GBM specimens was obtained from the Department of Neurosurgery, the Second and Fourth Affiliated Hospitals of Harbin Medical University, as described previously.44,45 All tumor collections and analyses were approved by the Institutional Review Board and the Ethics Committee of Nanjing Medical University and Harbin Medical University. Informed consent was obtained from all individual participants.
Cell lines, primary cell cultures, and GSCs
Human embryonic kidney HEK293T cell lines were obtained from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS). Normal human astrocytes (NHAs) were obtained from ScienCell Research Laboratories and cultured in the astrocyte growth media supplemented with recombinant human epidermal growth factor (rhEGF), insulin, ascorbic acid, GA-1000, L-glutamine, and 5% FBS. GSCs were isolated from primary GBM tumors or patient-derived GBM xenografts as previously described.46,47 In brief, GBM cells were dissociated from the freshly resected surgical specimens using the Papain Dissociation system (Worthington Biochemical). The isolated GBM cells were recovered in DMEM/F12 medium supplemented with B27 (1:50), basic fibroblast growth factor, and EGF (20 ng/mL each). Cells were then labeled with CD133 antibody or CD44 antibody at 4°C for 1 h followed by fluorescence-activated cell sorting (FACS) to isolate PN GSCs (PN 35 and PN 182) and MES GSCs (MES 21 and MES 505). The sorted GSCs were validated by GSC enrichment markers and functionally characterized by self-renewal potential (in vitro limiting dilution assay), multilineage differentiation potency (serum-induced cell differentiation assay), and tumorigenic capacity (in vivo limiting dilution tumor formation assay). GSCs were constantly maintained as GBM xenografts and were only dissociated, sorted, and cultured in the neurobasal medium for the functional experiments. Only early-passage GSCs were used for the study. The unique identity of all patient-derived GSCs was authenticated by short tandem repeat analysis as described in Table S2. All cells were routinely tested for mycoplasma contamination bimonthly using MycoAlert PLUS kits (Lonza).
Mice and animal housing
Female athymic nude mice at 4–6 weeks were purchased from the Experimental Animal Center of Nanjing Medical University. Mice were housed in groups of five animals in large plastic cages and maintained under pathogen-free conditions. All animal experiments were conducted with the approval of Institutional Animal Care and Use Committee of Nanjing Medical University and in conformity with National Guide for the Care and Use of Laboratory Animals.
Intracranial xenograft tumor models and treatments
Mice were randomly assigned to experimental groups for all of the experiments. For orthotopic tumor models, luciferase-expressing PN or MES GSCs were established by infection with pLenti-CMV-Puro-LUC (Addgene) in the presence of puromycin (0.5 μg/mL). The GSCs were then transduced with the indicated lentiviral vectors. Forty-eight hours after the lentiviral transduction, 1 × 104 PN or MES GSCs were stereotactically injected into the right striatum of nude mice. Kinetics of tumor growth was monitored via bioluminescent imaging using the IVIS 200 Spectrum system and quantified by Living Image software. Four to five weeks after GSC implantation, the mice were humanely killed and their brains were harvested, paraffin embedded, stained with hematoxylin and eosin (H&E) to confirm the presence of tumors, and subjected to immunohistochemical staining. For the animal survival analysis, mice were maintained until manifestation of pathological symptoms (i.e., hunched back, loss of body weight, reduced food consumption, and inactivity) from tumor burden developed or 80 days after injection.
Fractionated whole-brain irradiation (2.5 Gy daily for 4 consecutive days) of tumor-burdened mice was performed using the X-RAD 225Cx image-guided small animal stereotactic irradiator (Precision X-Ray), which mimics the clinical regimen of radiation therapy for GBM.
Bioinformatics analysis of FOSL1 expression
TCGA RNA-sequencing data of GBM samples were downloaded from the TCGA website (https://tcga-data.nci.nih.gov/docs/publications/gbm_exp/), including 57 PN, 33 neural, 54 CL, and 58 MES tumors.
Plasmid construction, lentiviral production, and transfection
The full-length cDNA of FOSL1 was amplified by RT-PCR and then cloned into the lentiviral vector pLenti6.2/V5-DEST vector to generate pLenti6.2-FOSL1 vector. Lentiviral vectors expressing FLAG- or HA-tagged FOSL1, FLAG-tagged WT or mutant (K40R) CYLD, HA-tagged UBC9, and HA-tagged CYLD were generated by cloning their open reading frame with the N-terminal FLAG or HA sequence into the pCDH-CMV-MCS-EF1α-Puro vector. SUMO1, SUMO2, SUMO3, or SUMO4, respectively was cloned into the pcDNA3.1 vector harboring an N-terminal Myc tag. Plasmids coding for HA-tagged ubiquitin-K63 (pRK5-HA-ubiquitin-K63) and HA-tagged ubiquitin-K48 (pRK5-HA-ubiquitin-K48) were obtained from Addgene. Site-directed mutagenesis in CYLD was performed with a QuikChange mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. The authenticity of all constructs was confirmed by DNA sequencing.
Lentiviral vectors expressing non-target control shRNA or specific shRNA constructs (FOSL1 and UBC9) were obtained from Dharmacon. For the FOSL1 rescue experiment, shRNA targeting 3′ UTR of FOSL1 was used for knockdown. RNAi oligonucleotide sequences are listed in Table S3.
All transfections were performed using Lipofectamine 3000 (Invitrogen) or X-tremeGENE HP DNA Transfection Reagent (Roche) following the manufacturer’s instructions.
EdU incorporation
5-Ethynyl-2′-deoxyuridine (EdU) staining was performed using the Click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen). In brief, dissociated GSCs were plated into wells of laminin pre-coated 8-well chamber slides. EdU was added to the culture medium in a final concentration of 10 μM for 3 h. Cells were then fixed with 4% paraformaldehyde in PBS for 15 min and penetrated with 0.5% Triton X-100 for 30 min. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Five fields of view per slide were examined for EdU-positive cells.
Extreme limiting dilution assay and neurosphere formation
For the in vitro limiting dilution assay, GSCs with indicated modification or treatment were dissociated to single cells and then plated in 96-well plates at a density of 1, 5, 10, 20, or 50 cells per well. After 7 days for MES 21 and MES 505 GSCs and 14 days for PN 35 and PN 182 GSCs, each well was examined for formation of tumor spheres. Stem cell frequency was measured and analyzed by Extreme Limiting Dilution Analysis software (http://bioinf.wehi.edu.au/software/elda). For the primary neurosphere assay, dissociated single cells were plated at a density of 1 cell/μL, and the spheres that formed after 7 days were counted. For the secondary neurosphere assay, established tumorspheres were dissociated into single cells and seeded at a density of 1 cell/μL, and the spheres that formed after 7 days were counted.
BrdU comet assay
Bromodeoxyuridine (BrdU) comet assays were performed as described in our previous study.44, 45 In brief, cells were pulse-labeled in the presence of BrdU for 20 min and irradiated using a GammaCell 1000 Elite Tissue Irradiator (dose rate 2.94 Gy/min, 102 s to deliver a dose of 5 Gy to cells). BrdU-labeled DNA was chased in the presence of 4× deoxynucleoside triphosphates for 0, 2, 4, 6, and 12 h. The cells were lysed and subjected to electrophoresis. Analysis of DNA migration was done by staining DNA with a specific anti-BrdU antibody. Data analysis was performed using TriTek Comet Score Freeware v1.5 software.
FACS analysis
GSCs with indicated modification were dissociated into single cells with Accutase (Sigma) for 5 min and stained with fluorescein isothiocyanate-conjugated CD44 antibody. Cells without primary antibody were used for negative control. Data were analyzed using FlowJo software.
Cell-cycle analysis
GSCs treated with or without 5 Gy IR were fixed in ice-cold 70% ethanol, incubated with propidium iodide and RNase A, and analyzed by FACS analysis.
γ-H2AX foci formation assay
For analysis of γ-H2AX foci formation, 1 × 104 cells were plated onto 16-well chambered coverglass coated with poly-L-lysine and incubated overnight in 5% FBS medium. Cells were then treated with or without 5 Gy IR and recovered for 6 h. Cells were fixed with 4% formaldehyde for 20 min at 4°C and immunostained with primary anti-phosphohistone H2AX (Ser139) and secondary Alexa Fluor 488-conjugated immunoglobulin G (IgG). Cells were finally counterstained with DAPI and visualized by an inverted fluorescence microscope (Leica DMI3000B).
RNA isolation and quantitative PCR
Total RNA was extracted from GSCs using the RNeasy Mini Kit (Qiagen) and reverse transcribed with an iScript cDNA Synthesis Kit (Bio-Rad). Expression levels of target genes were determined using the 2−ΔΔCt method and normalized to the housekeeping gene GAPDH. The oligonucleotide primers are shown in Table S3.
Immunoblotting, immunohistochemistry, and immunofluorescence
For immunoblot (IB) analysis, cells were harvested and lysed in RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM EDTA) containing protease inhibitor and phosphatase inhibitor cocktail (Sigma-Aldrich). Cell lysates were subjected to SDS-PAGE on 4%–12% gels and transferred onto polyvinylidene difluoride membranes (Roche Diagnostics). The membranes were incubated with the indicated primary antibodies, washed, and probed with horseradish peroxidase (HRP)-conjugated secondary antibodies. Signals were detected using a SuperSignal West Femto Maximum Sensitivity Substrate Trial Kit (Thermo Fisher Scientific). For IHC staining, GBM xenografts or surgical specimen tissue slides were deparaffinized and rehydrated through a descending alcohol series, followed by antigen retrieval with sodium citrate buffer. Tumor sections were blocked with 1% bovine serum albumin (BSA) with 0.25% Triton X-100 and 3% H2O2 in PBS for 1 h at room temperature. The slides were then incubated with the indicated primary antibodies overnight at 4°C, followed by incubation with an HRP-conjugated secondary antibody for 1 h at room temperature. Signals were detected by using a 3,3′-diaminobenzidine substrate kit. The slides were captured using a Pannoramic MIDI digital slide scanner, and the images were analyzed by Pannoramic Viewer software 1.15.2 (3D-Histech). For immunofluorescence (IF) staining, patient-derived GSCs were fixed with 4% formaldehyde, permeabilized with 0.25% Triton X-100, and blocked with 1% BSA for 1 h at room temperature. Cells were probed with the indicated primary antibodies overnight at 4°C. After being washed with PBS-T, cells were incubated with appropriate Alexa Fluor 488 or Alexa Fluor 594 secondary antibodies and DAPI-containing Vectashield mounting solution (Vector Laboratories), then visualized by a confocal laser scanning microscope (LSM5 PASCAL; Carl Zeiss).
Immunoprecipitation
GSCs transfected with the indicated constructs were collected and lysed in immunoprecipitation (IP) lysis buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA, protease, and phosphatase inhibitors). Cell extracts were pre-cleared with protein A/G beads and incubated with the indicated antibody overnight at 4°C. The beads were washed three times with IP buffer and the bound proteins eluted, followed by IB analysis.
Electrophoretic mobility shift assay
EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific). In brief, equal amounts of nuclear extracts prepared from GSCs was incubated with biotin-end-labeled NF-κB binding probe (Table S3). The nuclear protein-oligonucleotide complexes were then resolved by electrophoresis on a 5% non-denaturing polyacrylamide gel in 1 × Tris-borate-EDTA buffer at 120 V for 2 h at 4°C. The gels were dried and exposed to Kodak XAR-5 film for approximately 1–3 days.
SUMOylation and ubiquitination assays
For detection of CYLD-SUMO1 in vivo, pre-cleared cell lysates containing 20 mM N-ethylmaleimide (NEM), a potent inhibitor of deSUMOylation enzymes, were rotated with anti-CYLD or anti-SUMO1 antibody at 4°C for 4 h. Immunoprecipitates, collected on protein A/G beads, were washed three times with IP buffer and subjected to SDS-PAGE and IB analysis. For SUMOylated-CYLD detection in HEK293T cells that were transduced with FLAG-CYLD, HA-FOSL1, HA-UBC9, and Myc-SUMO1, cell lysates containing NEM were incubated with anti-FLAG antibody, precipitated with protein A/G beads, washed three times with IP buffer, and subjected to IB analysis. For ubiquitination assay, cell lysates were incubated with TRAF2, TRAF6, RIP1, NEMO, and IκBα antibodies for 4 h and protein A/G agarose beads for a further 8 h at 4°C. The precipitated proteins were then released from the beads by boiling for 10 min in 1% SDS and diluted 10× in IP buffer. HA-tagged ubiquitin-conjugated proteins were detected by IB analysis.
NF-κB luciferase assay
GSCs were co-transfected with 100 ng of NF-κB-derived luciferase (pGL4.32[luc2P/NF-κB-RE/Hygro]) and 1 ng of CMV-Renilla (pGL4.75[hRluc/CMV]) vectors (Promega). The activities of firefly luciferase and Renilla luciferase were measured 24 h after transfection using the Dual Luciferase Reporter Assay Kit (Promega). The luminescent signal was measured using a FLUOstar Optima Microplate Reader (BMG Labtech).
Chromatin immunoprecipitation assay
GSCs were crosslinked by adding formaldehyde directly to neurobasal medium to a final concentration of 1% and incubated for 10 min at 37°C. The cell lysates were sonicated to shear crosslinked chromatin into a size range of 100–300 bases followed by centrifugation at 16,000 × g for 10 min. Equal aliquots of chromatin supernatants were separated and incubated with 5 μg of anti-FLAG antibodies or an anti-IgG antibody overnight at 4°C with rotation. Precipitated protein-DNA complexes were eluted and reverse crosslinked. DNA was recovered by phenol-chloroform extraction and analyzed by real-time PCR using primers against UBC9 promoters.
Magnetic resonance imaging
Magnetic resonance imaging studies were performed on a Bruker 7.0T scanner (Bruker BioSpin) with a 16 cm bore. T2-weighted coronal images were acquired by rapid acquisition with relaxation enhancement (RARE) sequence with the following parameters: repetition time 3,000 ms, echo time 60 ms, RARE factor 12, average 4, field of view 40 × 30 mm, in-plane resolution 156 × 156 μm2, slice thickness 0.75 mm, and slice gap 0.25 mm. Tumor volume was assessed by contouring the lesions in the T2-weighted images using ImageJ software (https://imagej.nih.gov/ij/download.html).
Statistical analysis
Data are presented as the mean ± standard deviation. Significance was calculated by a two-tailed Student’s t test using GraphPad Prism 5.0 software or one-way ANOVA for multiple comparisons. Log-rank analysis was used to determine statistical significance of Kaplan-Meier survival curves.
Acknowledgments
We would like to thank the Core Facility of Jiangsu Provincial People’s Hospital for its help in the detection of experimental samples; NJMU Animal Core Facility for excellent mouse husbandry and care; NJMU Pathology Facility for assistance with paraffin processing, embedding, and histopathological analysis; and NJMU Flow Facility for expert cell sorting. This work was supported by grants from the National Natural Science Foundation of China (82072783, 81772681, and 81201978 to H.W., 81670153 and 81200362 to S.W.), the Natural Science Foundation of Jiangsu Province (BK20160098 and BK2012483 to H.W.), the Program for Advanced Talents within Six Industries of Jiangsu Province (WSN-019 to H.W.), the National High Technology Research, Development Program of China (863 Program) (2012AA02A508) and the National Key Research and Development Plan (2016YFC0902500), the Program for Development of Innovative Research Team in the First Affiliated Hospital of NJMU, and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author contributions
Z.C. and H.W. conceived the project and designed the study. Z.C., S.W., H.-L.L., H.L., X.W., J.L. and H.-W.W. performed the experiments. Y.C., D.C., W.-T.W., S.Z., Q.H. and D.L. provided intellectual contribution and technical assistance. N.L., Y.Y., W.W., and H.W. analyzed and interpreted the data. H.W. wrote the manuscript and provided study supervision.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.10.028.
Contributor Information
Wei Wu, Email: weiwu1974@126.com.
Huibo Wang, Email: hbwang@njmu.edu.cn.
Supplemental information
References
- 1.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frattini V., Trifonov V., Chan J.M., Castano A., Lia M., Abate F., Keir S.T., Ji A.X., Zoppoli P., Niola F., et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat. Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu B., Wang Q., Wang Y.A., Hua S., Sauve C.G., Ong D., Lan Z.D., Chang Q., Ho Y.W., Monasterio M.M., et al. Epigenetic activation of WNT5A drives glioblastoma stem cell differentiation and invasive growth. Cell. 2016;167:1281–1295.e18. doi: 10.1016/j.cell.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen P.Y., Kesari S. Malignant gliomas in adults. N. Engl. J. Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 5.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L., et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 7.Brennan C., Momota H., Hambardzumyan D., Ozawa T., Tandon A., Pedraza A., Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemmati H.D., Nakano I., Lazareff J.A., Masterman-Smith M., Geschwind D.H., Bronner-Fraser M., Kornblum H.I. Cancerous stem cells can arise from pediatric brain tumors. Proc. Natl. Acad. Sci. U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh S.K., Clarke I.D., Terasaki M., Bonn V.E., Hawkins C., Squire J., Dirks P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 10.Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 11.Mao P., Joshi K., Li J., Kim S.H., Li P., Santana-Santos L., Luthra S., Chandran U.R., Benos P.V., Smith L., et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl. Acad. Sci. U S A. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhat K.P.L., Balasubramaniyan V., Vaillant B., Ezhilarasan R., Hummelink K., Hollingsworth F., Wani K., Heathcock L., James J.D., Goodman L.D., et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S.H., Ezhilarasan R., Phillips E., Gallego-Perez D., Sparks A., Taylor D., Ladner K., Furuta T., Sabit H., Chhipa R., et al. Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-kappaB-dependent manner. Cancer Cell. 2016;29:201–213. doi: 10.1016/j.ccell.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minata M., Audia A., Shi J., Lu S., Bernstock J., Pavlyukov M.S., Das A., Kim S.H., Shin Y.J., Lee Y., et al. Phenotypic plasticity of invasive edge glioma stem-like cells in response to ionizing radiation. Cell Rep. 2019;26:1893–1905.e7. doi: 10.1016/j.celrep.2019.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narayanan A., Gagliardi F., Gallotti A.L., Mazzoleni S., Cominelli M., Fagnocchi L., Pala M., Piras I.S., Zordan P., Moretta N., et al. The proneural gene ASCL1 governs the transcriptional subgroup affiliation in glioblastoma stem cells by directly repressing the mesenchymal gene NDRG1. Cell Death Differ. 2018;26:1813–1831. doi: 10.1038/s41418-018-0248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno M., Pedrosa L., Pare L., Pineda E., Bejarano L., Martinez J., Balasubramaniyan V., Ezhilarasan R., Kallarackal N., Kim S.H., et al. GPR56/ADGRG1 inhibits mesenchymal differentiation and radioresistance in glioblastoma. Cell Rep. 2017;21:2183–2197. doi: 10.1016/j.celrep.2017.10.083. [DOI] [PubMed] [Google Scholar]
- 17.Segerman A., Niklasson M., Haglund C., Bergstrom T., Jarvius M., Xie Y., Westermark A., Sonmez D., Hermansson A., Kastemar M., et al. Clonal variation in drug and radiation response among glioma-initiating cells is linked to proneural-mesenchymal transition. Cell Rep. 2016;17:2994–3009. doi: 10.1016/j.celrep.2016.11.056. [DOI] [PubMed] [Google Scholar]
- 18.Behnan J., Finocchiaro G., Hanna G. The landscape of the mesenchymal signature in brain tumours. Brain. 2019;142:847–866. doi: 10.1093/brain/awz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozawa T., Riester M., Cheng Y.K., Huse J.T., Squatrito M., Helmy K., Charles N., Michor F., Holland E.C. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26:288–300. doi: 10.1016/j.ccr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakumoto K., Sasai K., Sukezane T., Oneyama C., Ishimaru S., Shibutani K., Mizushima H., Mekada E., Hanafusa H., Akagi T. FRA1 is a determinant for the difference in RAS-induced transformation between human and rat fibroblasts. Proc. Natl. Acad. Sci. U S A. 2006;103:5490–5495. doi: 10.1073/pnas.0601222103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollock C.B., Shirasawa S., Sasazuki T., Kolch W., Dhillon A.S. Oncogenic K-RAS is required to maintain changes in cytoskeletal organization, adhesion, and motility in colon cancer cells. Cancer Res. 2005;65:1244–1250. doi: 10.1158/0008-5472.CAN-04-1911. [DOI] [PubMed] [Google Scholar]
- 22.Diesch J., Sanij E., Gilan O., Love C., Tran H., Fleming N.I., Ellul J., Amalia M., Haviv I., Pearson R.B., et al. Widespread FRA1-dependent control of mesenchymal transdifferentiation programs in colorectal cancer cells. PLoS One. 2014;9:e88950. doi: 10.1371/journal.pone.0088950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakiri L., Macho-Maschler S., Custic I., Niemiec J., Guio-Carrion A., Hasenfuss S.C., Eger A., Muller M., Beug H., Wagner E.F. Fra-1/AP-1 induces EMT in mammary epithelial cells by modulating Zeb1/2 and TGFbeta expression. Cell Death Differ. 2015;22:336–350. doi: 10.1038/cdd.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H., Ren G., Wang T., Chen Y., Gong C., Bai Y., Wang B., Qi H., Shen J., Zhu L., et al. Aberrantly expressed Fra-1 by IL-6/STAT3 transactivation promotes colorectal cancer aggressiveness through epithelial-mesenchymal transition. Carcinogenesis. 2015;36:459–468. doi: 10.1093/carcin/bgv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreolas C., Kalogeropoulou M., Voulgari A., Pintzas A. Fra-1 regulates vimentin during Ha-RAS-induced epithelial mesenchymal transition in human colon carcinoma cells. Int. J. Cancer. 2008;122:1745–1756. doi: 10.1002/ijc.23309. [DOI] [PubMed] [Google Scholar]
- 26.Desmet C.J., Gallenne T., Prieur A., Reyal F., Visser N.L., Wittner B.S., Smit M.A., Geiger T.R., Laoukili J., Iskit S., et al. Identification of a pharmacologically tractable Fra-1/ADORA2B axis promoting breast cancer metastasis. Proc. Natl. Acad. Sci. U S A. 2013;110:5139–5144. doi: 10.1073/pnas.1222085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sayan A.E., Stanford R., Vickery R., Grigorenko E., Diesch J., Kulbicki K., Edwards R., Pal R., Greaves P., Jariel-Encontre I., et al. Fra-1 controls motility of bladder cancer cells via transcriptional upregulation of the receptor tyrosine kinase AXL. Oncogene. 2012;31:1493–1503. doi: 10.1038/onc.2011.336. [DOI] [PubMed] [Google Scholar]
- 28.Maurus K., Hufnagel A., Geiger F., Graf S., Berking C., Heinemann A., Paschen A., Kneitz S., Stigloher C., Geissinger E., et al. The AP-1 transcription factor FOSL1 causes melanocyte reprogramming and transformation. Oncogene. 2017;36:5110–5121. doi: 10.1038/onc.2017.135. [DOI] [PubMed] [Google Scholar]
- 29.Pecce V., Verrienti A., Fiscon G., Sponziello M., Conte F., Abballe L., Durante C., Farina L., Filetti S., Paci P. The role of FOSL1 in stem-like cell reprogramming processes. Sci. Rep. 2021;11:14677. doi: 10.1038/s41598-021-94072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallejo A., Erice O., Entrialgo-Cadierno R., Feliu I., Guruceaga E., Perugorria M.J., Olaizola P., Muggli A., Macaya I., O'Dell M., et al. FOSL1 promotes cholangiocarcinoma via transcriptional effectors that could be therapeutically targeted. J. Hepatol. 2021;75:363–376. doi: 10.1016/j.jhep.2021.03.028. [DOI] [PubMed] [Google Scholar]
- 31.Zhang M., Hoyle R.G., Ma Z., Sun B., Cai W., Cai H., Xie N., Zhang Y., Hou J., Liu X., et al. FOSL1 promotes metastasis of head and neck squamous cell carcinoma through super-enhancer-driven transcription program. Mol. Ther. J. Am. Soc. Gene Ther. 2021;29:2583–2600. doi: 10.1016/j.ymthe.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W., Meng L., Chen K., Tian C., Peng B., Zhong L., Zhang C., Yang X., Zou J., Yang S., Li L. Preclinical pharmacodynamic evaluation of a new Src/FOSL1 inhibitor, LY-1816, in pancreatic ductal adenocarcinoma. Cancer Sci. 2019;110:1408–1419. doi: 10.1111/cas.13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng P., Wang J., Waghmare I., Sartini S., Coviello V., Zhang Z., Kim S.H., Mohyeldin A., Pavlyukov M.S., Minata M., et al. FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res. 2016;76:7219–7230. doi: 10.1158/0008-5472.CAN-15-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z., Wang H.W., Wang S., Fan L., Feng S., Cai X., Peng C., Wu X., Lu J., Chen D., et al. USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells. J. Clin. Invest. 2019;129:2043–2055. doi: 10.1172/JCI126414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fedele M., Cerchia L., Pegoraro S., Sgarra R., Manfioletti G. Proneural-mesenchymal transition: phenotypic plasticity to acquire multitherapy resistance in glioblastoma. Int. J. Mol. Sci. 2019;20:2746. doi: 10.3390/ijms20112746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q., Hu B., Hu X., Kim H., Squatrito M., Scarpace L., deCarvalho A.C., Lyu S., Li P., Li Y., et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32:42–56.e6. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedmann-Morvinski D., Narasimamurthy R., Xia Y., Myskiw C., Soda Y., Verma I.M. Targeting NF-kappaB in glioblastoma: a therapeutic approach. Sci. Adv. 2016;2:e1501292. doi: 10.1126/sciadv.1501292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harhaj E.W., Dixit V.M. Deubiquitinases in the regulation of NF-kappaB signaling. Cell Res. 2011;21:22–39. doi: 10.1038/cr.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi T., Masoumi K.C., Massoumi R. Deubiquitinating activity of CYLD is impaired by SUMOylation in neuroblastoma cells. Oncogene. 2015;34:2251–2260. doi: 10.1038/onc.2014.159. [DOI] [PubMed] [Google Scholar]
- 40.Masoumi K.C., Marfany G., Wu Y., Massoumi R. Putative role of SUMOylation in controlling the activity of deubiquitinating enzymes in cancer. Future Oncol. 2016;12:565–574. doi: 10.2217/fon.15.320. [DOI] [PubMed] [Google Scholar]
- 41.Yin J., Oh Y.T., Kim J.Y., Kim S.S., Choi E., Kim T.H., Hong J.H., Chang N., Cho H.J., Sa J.K., et al. Transglutaminase 2 inhibition reverses mesenchymal transdifferentiation of glioma stem cells by regulating C/EBPbeta signaling. Cancer Res. 2017;77:4973–4984. doi: 10.1158/0008-5472.CAN-17-0388. [DOI] [PubMed] [Google Scholar]
- 42.Carro M.S., Lim W.K., Alvarez M.J., Bollo R.J., Zhao X., Snyder E.Y., Sulman E.P., Anne S.L., Doetsch F., Colman H., et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhat K.P., Salazar K.L., Balasubramaniyan V., Wani K., Heathcock L., Hollingsworth F., James J.D., Gumin J., Diefes K.L., Kim S.H., et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo H., Chen Z., Wang S., Zhang R., Qiu W., Zhao L., Peng C., Xu R., Chen W., Wang H.W., et al. c-Myc-miR-29c-REV3L signalling pathway drives the acquisition of temozolomide resistance in glioblastoma. Brain. 2015;138:3654–3672. doi: 10.1093/brain/awv287. [DOI] [PubMed] [Google Scholar]
- 45.Peng C., Chen Z., Wang S., Wang H.W., Qiu W., Zhao L., Xu R., Luo H., Chen Y., Chen D., et al. The error-prone DNA polymerase kappa promotes temozolomide resistance in glioblastoma through rad17-dependent activation of ATR-chk1 signaling. Cancer Res. 2016;76:2340–2353. doi: 10.1158/0008-5472.CAN-15-1884. [DOI] [PubMed] [Google Scholar]
- 46.Hu J., Sun T., Wang H., Chen Z., Wang S., Yuan L., Liu T., Li H.R., Wang P., Feng Y., et al. MiR-215 is induced post-transcriptionally via HIF-drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell. 2016;29:49–60. doi: 10.1016/j.ccell.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., Sun T., Hu J., Zhang R., Rao Y., Wang S., Chen R., McLendon R.E., Friedman A.H., Keir S.T., et al. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J. Clin. Invest. 2014;124:4489–4502. doi: 10.1172/JCI75284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.