Abstract
The development of new therapeutic avenues that target the early stages of Alzheimer’s disease (AD) is urgently necessary. A disintegrin and metalloproteinase domain 10 (ADAM10) is a sheddase that is involved in dendritic spine shaping and limits the generation of amyloid-β. ADAM10 endocytosis increases in the hippocampus of AD patients, resulting in the decreased postsynaptic localization of the enzyme. To restore this altered pathway, we developed a cell-permeable peptide (PEP3) with a strong safety profile that is able to interfere with ADAM10 endocytosis, upregulating the postsynaptic localization and activity of ADAM10. After extensive validation, experiments in a relevant animal model clarified the optimal timing of the treatment window. PEP3 administration was effective for the rescue of cognitive defects in APP/PS1 mice only if administered at an early disease stage. Increased ADAM10 activity promoted synaptic plasticity, as revealed by changes in the molecular compositions of synapses and the spine morphology. Even though further studies are required to evaluate efficacy and safety issues of long-term administration of PEP3, these results provide preclinical evidence to support the therapeutic potential of PEP3 in AD.
Keywords: synapse, ADAM10, cell-permeable peptide, endocytosis, Alzheimer’s disease, synaptic dysfunction, cognitive deficits, sAPPα
Graphical abstract

Musardo and colleagues demonstrated that restoring ADAM10 synaptic availability during early, but not late, stages of Alzheimer’s disease is a successful approach to counteract both synaptic failure and cognitive deficits in animal models, showcasing the importance of targeting early synaptic dysfunction as an effective Alzheimer’s therapeutic strategy.
Introduction
Synapse dysfunction and loss are crucial to the pathophysiology of Alzheimer’s disease (AD) and strongly correlate to cognitive decline.1,2
It is widely accepted that synapses are an early target of amyloid-β (Aβ), which has been described as a central component of AD pathogenesis.3 Aβ is generated from amyloid precursor protein (APP) by the concerted actions of β-site APP cleaving enzyme 1 (BACE1) and γ-secretase.4 In an alternative, non-amyloidogenic pathway, a disintegrin and metalloproteinase domain 10 (ADAM10) cleaves APP within the Aβ domain5 in a process known as ectodomain shedding that precludes Aβ generation and results in the secretion of the neuroprotective soluble APPα (sAPPα).6
Aβ targeting has been at the heart of therapeutic developments in AD research for more than 25 years.7 In the cascade of AD pathogenesis, synaptic failure, triggered by Aβ and tau, has been identified as a key step and an early event in AD, preceding the occurrence of neurodegeneration.8
We previously identified ADAM10 as an enzyme that is active at the crossroads between the amyloid cascade and synaptic loss. ADAM10 is a component of the excitatory postsynaptic density (PSD),9 and its postsynaptic abundance and activity are controlled by the interaction between its cytoplasmic tail and specific protein partners: the synapse-associated protein 97 (SAP97) and the clathrin adaptor hetero-tetramer AP2.9,10 SAP97 mediates local ADAM10 trafficking from dendritic Golgi outposts to the PSD,11 whereas the interaction with AP2 triggers the removal of ADAM10 from the synaptic membrane.10 ADAM10 cleaves neuronal adhesion molecules,12,13 and thereby plays a pivotal role in the sequence of events that regulate dendritic spine formation and stabilization.14 Therefore, ADAM10 synaptic localization and activity are controlled by activity-dependent synaptic plasticity phenomena,10 further strengthening the relevance of ADAM10 at the synapse. Long-term depression (LTD) enhances ADAM10 membrane insertion by fostering the SAP97-mediated forward trafficking of ADAM10 to the synaptic membrane, an event required for the LTD-induced spine shrinkage.10 On the other hand, long-term potentiation (LTP) reduces the membrane levels of ADAM10 by inducing its endocytosis in order to stabilize synaptic contacts.10 Remarkably, the short-term exposure to Aβ oligomers has been shown to impair ADAM10 endocytosis and to increase ADAM10 synaptic levels,15 in line with a disorder in LTP mechanism in AD. Interestingly, these mechanisms that regulate ADAM10 synaptic localization are impaired in the hippocampus of AD patients. A reduction in the ADAM10/SAP97 interaction because of decreased protein kinase C (PKC)-dependent phosphorylation of SAP97 and the concomitant increased association of ADAM10 with AP2 have been observed in AD patients compared with healthy control subjects.10,11,16 These defects in ADAM10 local trafficking lead to a significant reduction of synaptic ADAM10 levels in AD patients,16 eventually contributing to AD pathogenesis through Aβ generation and impaired synaptic plasticity.
Considering the crucial role played by ADAM10 in synaptic function and in the amyloid cascade, we propose that the pharmacological modulation of ADAM10 synaptic localization and activity may serve as a therapeutic approach for AD.
To upregulate the synaptic availability of ADAM10, we developed cell-permeable peptides (CPPs) that are capable of interfering with the ADAM10/AP2 interaction. The results indicated that administration of a CPP capable of inhibiting ADAM10 endocytosis rescued cognitive and synaptic function when administered during the early disease stage in AD model mice, with no evidence of associated safety issues, suggesting that this therapeutic strategy warrants further attention.
Results
Identification and validation of CPPs targeting the ADAM10/AP2 interaction
AP2 exists as a heterotetrameric assembly of two large subunits (α and β2), a medium subunit termed μ2, and a small subunit, σ.17 We have previously reported that the AP2-binding region is located in the C terminus of ADAM10 and contains two positively charged residues, R735 and R737, and a hydrophilic amino acid, Q736 (Figure 1A).10 The two arginine residues are crucial for the binding between these two proteins, because the double mutation of both arginines to alanine residues completely abolished the interaction.10 To better characterize the interaction between ADAM10 and the AP2 complex, we performed a computational analysis and found that the residues P733, P734, Q736, and R737 of ADAM10 are likely to interact with the residues F469, G472, and E471 in the β2 subunit of AP2. Moreover, the residue D437 of the β2 subunit is properly positioned to establish an electrostatic interaction with the R735 residue in ADAM10.10 As previously reported,10 a blast search of the Protein Data Bank (PDB) revealed a peptide with a high degree of identity with the C-terminal region of ADAM10 (Figure 1A). Based on this result, we designed four active CPPs (PEP1, PEP2, PEP3, and PEP4) to interfere with ADAM10/AP2 interaction to increase ADAM10 synaptic localization and activity. Figure S1A shows the structural organization of the four CPPs, in which each peptide harbors the TAT sequence (YGRKKRRQRR) and a short linker (composed of four amino acids: three glycines and one serine, GGSG). Considering that the CPPs were designed to compete with endogenous ADAM10 for the association to AP2, the different amino acid compositions of the four CPPs were determined to make the key residues (i.e., R735 and R737) more prone to interact with AP2 by improving their solvent accessibility and the general stability of the peptides in solution. Because ADAM10/β2 subunit association implies an electrostatic interaction, four corresponding inactive CPPs (inPEP1, inPEP2, inPEP3, inPEP4) were designed substituting the key arginine residues R735 and R737 with a negatively charged amino acid (i.e., glutamate) and were used as controls in each experiment.
Figure 1.
PEP2 and PEP3 affect ADAM10 endocytosis, and thereby its synaptic localization
(A) Alignment between the C terminus of ADAM10 and the peptide identified in the Protein Data Bank (PDB). The two arginines of ADAM10 (i.e., 735 and 737) are crucial for the binding with AP2. The ribbon representation of the NMR structure of the peptide in complex with the c-Src-SH3 domain (PDB: 1QWE) is also reported. The residues of the peptide are represented as stick, while the backbone is in red ribbon. (B) Antibody uptake assays were performed on COS7 cells transfected with TacADAM10-RAR; internalization was analyzed after treatment with either PEP2 or PEP3 active or inactive peptides (1 μM, 30 min). Representative images of cells returned to 37°C to allow endocytosis. The quantification shows a significant decrease in the internalization index for both PEP2 and PEP3 (inPEP2: 100.0% ± 1.78%, n = 74 cells; PEP2: 92.39% ± 2.43%, n = 54 cells; ∗∗Mann-Whitney U test = 1,338, p = 0.013; inPEP3: 100.0% ± 1.55%, n = 59 cells; PEP3: 85.91% ± 1.29%, n = 60 cells; ∗∗∗unpaired t test: t = 6.983, degrees of freedom [df] = 117, p < 0.0001). Scale bars, 20 μm. (C) Experimental paradigm: primary hippocampal neurons were treated with either PEP2, PEP3, or the corresponding inactive peptides at 1 μM for 30 min. (D) Homogenates were immunoprecipitated with anti-ADAM10 antibody, and α-Adaptin, μ2, and ADAM10 were evaluated. Both CPPs significantly decrease ADAM10/AP2 complex co-precipitation (n = 3, α-Adaptin, one-way ANOVA F(3,8) = 14.37, p = 0.0014, ∗∗p = 0.003, ∗p = 0.0232; μ2, one-way ANOVA F(3,8) = 11.79, p = 0.0026, ∗∗p = 0.006). (E) WB analysis of ADAM10 in the synaptic fraction of CPPs-treated neurons. Both active peptides increase ADAM10 synaptic localization (n = 3, one-way ANOVA F(3,8) = 32.36, p < 0.0001; Bonferroni’s test inPEP2 versus PEP2, ∗∗p = 0.0082; inPEP3 versus PEP3, ∗∗p = 0.0003; PEP2 versus PEP3, ∗p = 0.0264). (F–H) qRT-PCR analysis of ADAM10 (F, one-way ANOVA F(3,16) = 0.022, p = 0.995, n = 5), BACE1 (G, one-way ANOVA F(3,16) = 0.063, p = 0.978, n = 5), and APP (H, one-way ANOVA F(3,16) = 0.166, p = 0.917, n = 5) expression in hippocampal neurons treated with either PEP2, PEP3, or corresponding inactive peptides. No significant changes are detected. The treated samples are normalized on control condition. Data are reported as mean ± standard error (SE).
To test the efficacy of the CPPs for uncoupling the ADAM10/AP2 complex, we performed pilot experiments in in vitro and in vivo models. We treated acute rat hippocampal slices with either the active CPPs or corresponding inactive CPPs at 10 μM for 30 min (Figure S1B). Co-immunoprecipitation (coIP) experiments showed that PEP2 and PEP3 were able to significantly reduce the interaction between ADAM10 and AP2 (Figure S1C). To examine the effects of PEP2 and PEP3 on the levels of ADAM10 at the membrane, we performed a bis(sulfosuccinimidyl)suberate (BS3) cross-linking assay.10,18 BS3 is a membrane-impermeable, irreversible, amine-reactive cross-linking reagent, and BS3 treatment results in the cross-linking of membrane-inserted ADAM10, which forms high-molecular-weight aggregates that barely enter the polyacrylamide gel. Therefore, the ADAM10 surface pool was not detectable, and the observed modifications to the ADAM10 intracellular pool reflected changes in ADAM10 expression at the surface. The western blot (WB) analysis showed a substantial reduction in the intracellular ADAM10 levels when the slices are exposed to either PEP2 (Figure S1D) or PEP3 (Figure S1E), indicating an increase in the level of ADAM10 membrane insertion.
To verify that PEP2 and PEP3 treatment affected ADAM10 endocytosis, we performed a fluorescence-based “antibody uptake” assay to assess ADAM10 internalization in COS7 cells transfected with TacADAM10-RAR, a chimera between the surface reporter protein Tac (a human interleukin [IL]-2 receptor α subunit)19 and a modified version of the ADAM10 C-terminal tail, in which the endoplasmic reticulum (ER)-retention motif was mutated, allowing for its exit from the ER and delivery to the plasma membrane surface.20 COS7 cells were treated with either PEP2 or PEP3 and corresponding control peptides, using a lower peptide concentration than previous experiments (1 μM for 30 min) to further test CPPs efficacy. As shown in Figure 1B, the internalization index of TacADAM10-RAR, corresponding to the internalized/total fluorescence ratio, was significantly reduced on both PEP2 and PEP3 treatment, indicating that both peptides were able to impair the endocytosis of ADAM10.
Because ADAM10 is an integral component of the glutamatergic PSD,9 we examined whether PEP2 or PEP3 modulated the postsynaptic levels of ADAM10 in primary hippocampal neurons (Figure 1C). After confirming that both active peptides reduced the interaction between ADAM10 and the AP2 complex (Figure 1D), the postsynaptic Triton-insoluble fraction (TIF, which is highly enriched in all categories of PSD proteins)21,22 was purified from both control and treated neurons. A quantitative WB analysis showed that both CPPs were able to increase the postsynaptic availability of ADAM10 (Figure 1E). Importantly, PEP2 and PEP3 treatment does not affect ADAM10 mRNA level (Figure 1F) and does not modify the expression of the other players of the amyloid cascade, i.e., BACE1 and APP (Figures 1G and 1H). To strengthen our results, we performed a biotinylation assay that further confirmed the efficacy of PEP2 and PEP3 in increasing ADAM10 membrane levels in primary neuronal cultures (Figures S1F–S1H).
In light of these results, we tested the efficacy of PEP2 and PEP3 in vivo. Using a previously described experimental design,9 we administered PEP2, PEP3, the corresponding inactive peptides, or saline solution to 5-month-old wild-type mice via intraperitoneal injections at a dose of 3 nmol/g (Figure 2A). After 24 h, we confirmed the capability of the peptides to cross the blood-brain barrier (Figure S2) and to disrupt the interaction between ADAM10 and the AP2 complex (Figure 2B). To assess the effects of peptide treatment on ADAM10 localization and activity, we performed WB analysis to measure the levels of ADAM10 in the total homogenate and the postsynaptic fraction and of sAPPα in the soluble fraction. Although neither PEP2 nor PEP3 had any effects on ADAM10 total protein levels (Figure 2C), both active peptides increased ADAM10 postsynaptic levels (Figure 2D) and stimulated sAPPα release (Figure 2E). Our analyses showed that both PEP2 and PEP3 were able to positively affect ADAM10 localization and activity. However, because PEP3 was more effective, we focused on PEP3 for further experiments.
Figure 2.
PEP2 and PEP3 affect ADAM10 synaptic localization and activity in vivo
(A) Experimental paradigm: 5-month-old wild-type mice were treated with either PEP2 or PEP3 (3 nmol/g) via intraperitoneal injection, and 24 h later biochemical analyses were carried out. (B) Forebrain total homogenates were immunoprecipitated with anti-ADAM10 antibody, and AP2 subunit co-precipitation was evaluated. PEP2 and PEP3 significantly reduced ADAM10/AP2 co-precipitation (n = 3; α-Adaptin: one-way ANOVA F(3,8) = 16.73, p = 0.0008, PEP2 ∗∗p = 0.0042, PEP3 ∗∗p = 0.0051; β2-Adaptin: one-way ANOVA F(3,8) = 104.6, p < 0.0001, PEP2 ∗∗∗p < 0.0001, PEP3 ∗∗∗p < 0.0001; μ2: one-way ANOVA F(3,8) = 13.80, p = 0.0016, PEP2 ∗p = 0.0119, PEP3 ∗∗p = 0.0058). (C and D) WB analysis of ADAM10 levels in homogenate (C) and synaptic fraction (D) of CPP-treated mouse forebrain. Both peptides promote ADAM10 synaptic localization without affecting total protein level (C, homogenate, n = 3, one-way ANOVA F(3,8) = 0.08472, p = 0.9665; D, TIF, n = 3, one-way ANOVA F(3,8) = 10.55, p = 0.0037; Bonferroni’s multiple comparisons test inPEP2 versus PEP2, ∗p = 0.0237; inPEP3 versus PEP3, ∗p = 0.026). (E) WB analysis of sAPPα (detected with the ab2072 antibody recognizing the N-terminal region of Aβ) and sAPPtot (revealed with the 22c11 antibody directed against the N-terminal domain of APP) levels in soluble fraction purified from the forebrain. sAPPα/sAPPtot ratio is increased in mice treated with PEP2 and PEP3 (Brown-Forsythe and Welch’s ANOVA test F(3,8.096) = 10.34, p = 0.0038, W(3,7.832) = 13.35, p = 0.0019; Dunnett’s T3 test inPEP2 versus PEP2, ∗p = 0.0336; inPEP3 versus PEP3, ∗∗∗p = 0.0021). Data are reported as mean ± SE.
PEP3 upregulates ADAM10 activity without associated safety issues
To design a PEP3 sub-chronic treatment regimen, we tested the efficacy of two different doses (1 or 3 nmol/g) administered through two different routes (subcutaneous or intraperitoneal injections to 5-month-old wild-type mice), by assessing three parameters, i.e., ADAM10/AP2 association, ADAM10 postsynaptic levels, and sAPPα release. PEP3 displayed efficacy only when administered at the higher tested dose (3 nmol/g) by intraperitoneal, but not subcutaneous, injections (Figure S3), and its effect could be detected 24 h after the injection and not at various time points after the injection (Figure S4).
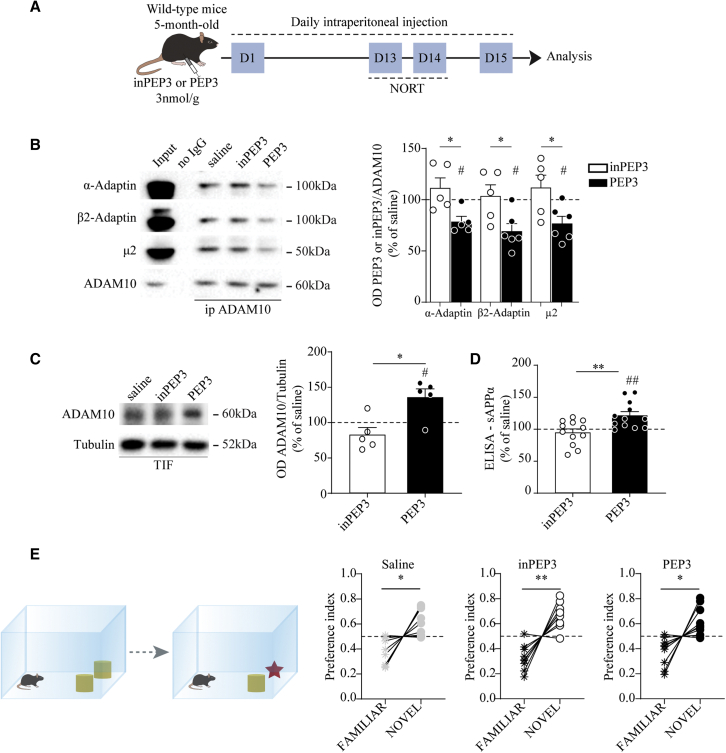
Considering these results, 5-month-old wild-type mice were treated for 14 days, and the safety and efficacy of PEP3 were assessed using a battery of tests (Figure 3A). During the treatment, no changes in body weights and food intake were observed (Figures S5A and S5B). The results of the erythrogram, platelet count, leukogram, and clinical chemistry analyses suggested that the PEP3 treatment did not affect the main systems or organs and did not induce significant systemic dysmetabolism (Tables S1–S3). After 14 days of treatment, we confirmed target engagement by assessing the interaction between ADAM10 and the AP2 complex (Figure 3B), ADAM10 postsynaptic availability (Figure 3C), and ADAM10 activity (Figure 3D). We measured no changes in APP and BACE1 at mRNA and protein levels (Figures S5C and S5D), as well as no modifications in ADAM10 gene expression (Figure S5D). To test the specificity of PEP3 for the ADAM10/AP2 interaction, we verified that PEP3 treatment did not affect the association between AP2 and the β3 subunit of the γ-aminobutyric acid type A receptor (GABA-A β3 subunit), whose endocytic sequence is similar to the AP2 binding motif in the ADAM10 cytoplasmic tail (Figure S5F). PEP3 administration did not modify the ADAM10/SAP97 interaction (Figure S5E) and the association of AP2 to APP (Figure S5G) and BACE1 (Figure S5H), confirming that the increase in ADAM10 synaptic levels and activity toward APP cannot be ascribed to modifications in forward trafficking or in alterations of APP or BACE1 internalization or expression. In addition, we performed a novel object recognition test (NORT), which is a behavioral test that measures an animals’ preference for exploring novelty and can serve as a robust indicator of potential cognitive impairment. The analysis showed that all tested mice preferred the novel object over the familiar one, indicating that PEP3 treatment did not affect the cognitive function of wild-type mice (Figure 3E). Furthermore, the molecular composition of the PSD is not changed in PEP3-treated mice (Figure S5I).
Figure 3.
Sub-chronic treatment with PEP3 promotes ADAM10 activity without affecting cognitive function
(A) Experimental paradigm: 5-month-old wild-type C57BL/6j mice were treated with PEP3 or inPEP3 at 3 nmol/g via intraperitoneal daily injection for 14 days. From day 12 to 14, NORT was performed, and at 15 day, biochemical analyses were carried out. (B) Representative immunoblot of α-Adaptin, β2-Adaptin, μ2, and ADAM10 from forebrain homogenates immunoprecipitated with anti-ADAM10 antibody. OD quantification shows that PEP3 significantly reduces ADAM10 interaction with AP2 subunits (∗unpaired t test: α-Adaptin: t = 2.950, df = 8, p = 0.0184; β2-Adaptin: t = 2.720, df = 9, p = 0.0236; μ2: t = 2.603, df = 9, p = 0.0286; #PEP3 one-sample t test against 100%: α-Adaptin: t = 4.129, df = 4, p = 0.0145; β2-Adaptin: t = 4.192, df = 5, p = 0.0086; μ2: t = 3.323, df = 5, p = 0.0209). (C) Representative immunoblot of ADAM10 WB analysis in TIF fraction purified from the forebrain. ADAM10/tubulin OD ratio revealed a significant increase in ADAM10 synaptic levels in animals treated with PEP3 (Mann-Whitney test U = 1, ∗p = 0.0159, #PEP3 one-sample t test against 100%: t = 3.004, df = 4, p = 0.0398). (D) ELISA analysis on forebrain samples shows an increase in sAPPα levels in PEP3-treated mice (∗∗unpaired t test: t = 3.233, df = 22, p = 0.0038, ##one-sample t test against 100%: t = 3.587, df = 11, p = 0.0043). (E) NORT was performed on animal treated with either PEP3 or inPEP3. Preference index was calculated as the ratio between the familiar (or novel) object exploration time divided by total exploration time (saline: familiar 0.387 ± 0.036, novel 0.613 ± 0.036, paired t test t = 3.154, df = 8, p = 0.0135; inPEP3: familiar 0.329 ± 0.035, novel 0.671 ± 0.035, paired t test t = 4.952, df = 8, p = 0.0011; PEP3: familiar 0.377 ± 0.039, novel 0.623 ± 0.039, paired t test t = 3.183, df = 8, p = 0.0129). Data are reported as mean ± SE.
PEP3 treatment administered at the full-blown AD disease fails to rescue cognitive impairments in APP/PS1 mice
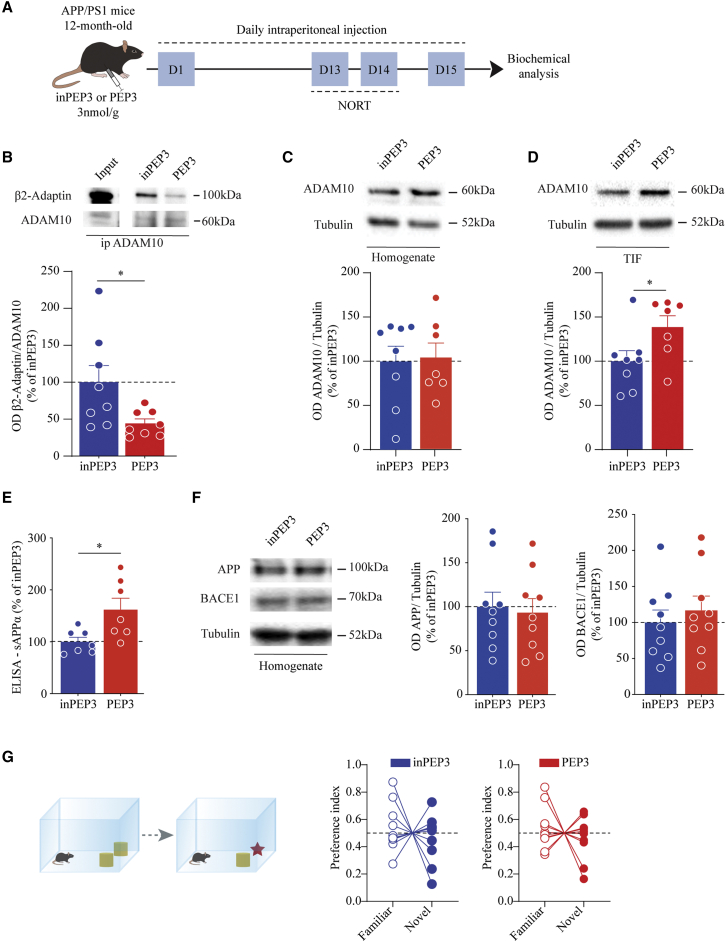
Considering the positive results obtained in wild-type mice, we performed a sub-chronic treatment in an animal model of AD to assess the therapeutic efficacy of PEP3. We administered either PEP3 or the inactive control peptide inPEP3 to APP/PS1 mice at the age of 12 months, when mice show cognitive deficits (Figure 4A).23 First, we evaluated target engagement, and we confirmed the efficacy of PEP3 treatment in reducing ADAM10/β2-Adaptin association (Figure 4B), in increasing ADAM10 postsynaptic availability without modifying the total enzyme levels (Figures 4C and 4D), and in promoting sAPPα release (Figure 4E) in APP/PS1 mice. The increase in sAPPα levels could not be ascribed to modifications in APP or BACE1 levels (Figure 4F). To assess the functional effects of PEP3 treatment on cognitive processes, we performed NORT. The results showed that APP/PS1 mice treated with PEP3 were not able to discriminate between the novel and the familiar object, similar to the inPEP3-treated APP/PS1 mice, revealing that both groups displayed cognitive deficits (Figure 4G).
Figure 4.
PEP3 does not restore the cognitive deficits of 12-month-old APP/PS1 mice
(A) Experimental paradigm: 12-month-old APP/PS1 mice were treated with PEP3 or inPEP3 at 3 nmol/g via daily intraperitoneal injections. After performing NORT (days 12–14), at day 15 biochemical analyses were carried out. (B) Representative immunoblot of β2-Adaptin immunoprecipitated from hippocampal homogenate with anti-ADAM10 antibody. OD quantification shows a reduction in ADAM10/AP2 interaction in APP/PS1 mice treated with PEP3 (unpaired t test: t = 2.385, df = 14, ∗p = 0.031). (C and D) Representative immunoblot of ADAM10 in homogenate (C) and TIF fraction purified from the forebrain. OD quantification of TIF fraction revealed a significant increase in ADAM10 synaptic availability in PEP3-treated APP/PS1 mice, without changes in the total homogenate (C, homogenate, unpaired t test: t = 0.1516, df = 13, p = 0.882; D, TIF, unpaired t test: t = 2.201, df = 13, ∗p = 0.0464). (E) ELISA analysis on the soluble fraction purified from the cortex shows an increase in sAPPα release after treatment with active peptide (unpaired t test, t = 2.738, df = 12, ∗p = 0.018). (F) Representative immunoblot and quantification of APP and BACE1 in forebrain homogenate show no changes in total levels of the proteins (APP, unpaired t test, t = 0.273, df = 16, p = 0.788; BACE1, unpaired t test, t = 0.667, df = 16, p = 0.514). (G) Novel object recognition was performed on animal treated with either active or inactive PEP3. Preference index shows no preference for novel object for both groups of mice (inPEP3: familiar 0.537 ± 0.055, novel 0.463 ± 0.055, paired t test, t = 0.6672, df = 9, p = 0.5214; PEP3: familiar 0.535 ± 0.050, novel 0.465 ± 0.050, paired t test, t = 0.6962, df = 9, p = 0.5039). Data are reported as mean ± SE.
PEP3 administration during the early disease stage restores cognitive defects and synaptic failure in APP/PS1 mice
In light of these results, we focused on earlier pathological stages. We tested the interaction between ADAM10 and the AP2 complex and examined the ADAM10 synaptic levels in APP/PS1 mice at the ages of 3 and 6 months. We found a significant increase in the association between ADAM10 and β2-Adaptin and a concomitant significant reduction in ADAM10 synaptic levels in APP/PS1 mice at 6 months compared with wild-type mice, whereas no changes were detected at 3 months of age (Figures S6A and S6B). No modifications in ADAM10 mRNA levels were detected at 3 and 6 months of age (Figure S6C).
Considering these results, we opted to target the ADAM10/AP2 association in 6-month-old APP/PS1 mice (Figure 5A). As expected, PEP3 administration reduced the interaction between ADAM10 and the AP2 complex (Figure 5B), and increased the ADAM10 postsynaptic availability without changing the total enzyme protein levels (Figures 5C and 5D). PEP3 promoted the generation of sAPPα (Figure 5E), without modifying APP and BACE1 levels (Figure 5F), also in 6-month-old APP/PS1 mice.
Figure 5.
PEP3 treatment promotes ADAM10 activity in APP/PS1 mice at the age of 6 months
(A) Experimental paradigm: 6-month-old APP/PS1 mice were treated with PEP3 or inPEP3 at 3 nmol/g via intraperitoneal daily injections and, after performing Y-maze test and NORT, at day 15 analyses were carried out. (B) Representative immunoblot of β2-Adaptin from hippocampal homogenate immunoprecipitated with anti-ADAM10 antibody. OD quantification shows a significant reduction in ADAM10/AP2 interaction in the PEP3-treated transgenic mice (unpaired t test, t = 2.457, df = 15, ∗p = 0.0267). (C and D) Representative immunoblot of ADAM10 in homogenate and TIF fraction purified from the forebrain. OD quantification of TIF fraction revealed significant increase in ADAM10 synaptic levels in APP/PS1 mice treated with PEP3 without changes in the total level of the enzyme (C, homogenate, unpaired t test: t = 1.695, df = 17, p = 0.108; D, TIF, Kolmogorov-Smirnov test: D = 0.6667, ∗p = 0.0336). (E) ELISA analysis of hippocampal samples shows an increase in sAPPα levels in PEP3-treated mice (unpaired t test: t = 2.506, df = 15, ∗p = 0.0242). (F) Representative immunoblot and quantification of APP and BACE1 in forebrain homogenate. No changes in the total protein levels are detected (APP, unpaired t test: t = 1.731, df = 13, p = 0.107; BACE1, unpaired t test: t = 0.54, df = 11, p = 0.6). Data are reported as mean ± SE. (G) Scatterplot of Aβ plaques/mm2 analyzed with primary antibodies recognizing different Aβ epitopes. Each antibody was able to detect a specific pattern of plaque deposits, which were then categorized according to the diameter into small (<30 μm) and large plaques (>30 μm). The three-way ANOVA showed that, after considering the antibody effect, the larger plaques were significantly different from the smaller plaques. The number of larger plaques was 2.65-fold lower than that of smaller plaques, although plaques with diameters >30 μm occupied a larger area of the hippocampus compared with the area occupied by smaller ones. When we adjusted for the diameter effect, the mean number of plaques stained with the D9A3A antibody was 1.9-fold lower than the mean number of plaques stained with the D54D2 antibody, with no significant interaction between diameter and antibodies. The mean number of plaques stained with the OC antibody was not significantly different from those of D54D2-positive plaques (Table S4). (H) Scatterplot of the percentage of hippocampal area occupied by larger or smaller plaques. When the plaques were stained with the Aβ C-terminal epitope D9A3A antibody, the effect on the percentage of area occupied by plaques was attenuated compared with the staining performed using the Aβ N-terminal domain D54D2 antibody (p = 0.019 for the interaction between diameter and antibody; see Table S4). The percentage of area occupied by plaques stained with the OC antibody was not significantly different from those of D54D2-positive plaques (Table S4). Least-squares means and the corresponding standard errors are superimposed over the plots. p values from tests on single parameters and ANOVA are reported in Table S4.
To investigate changes in the Aβ burden in the hippocampus of 6-month-old APP/PS1 mice, we performed confocal microscopy analysis of brain slices immunolabeled with three independent antibodies raised against different Aβ epitopes: the OC antibody against fibrillar epitopes,24 the D54D2 antibody against the Aβ N-terminal (human-specific) epitope, and the D9A3A antibody that selectively recognizes the Aβ42 C-terminal neo-epitope. Each antibody was able to detect a specific pattern of plaque deposits, which were then categorized according to the diameter into small and large plaques (Figure S7). We performed a comparative analysis of immunoreactivity across all available antibodies using linear mixed-effect models (LMMs) to analyze the density and percentage area occupied by the plaques, considering the hierarchical structure of the data. The three-way ANOVA analysis showed that plaques recognized by the D9A3A antibody, which are primarily composed of Aβ42, represented a subset of the overall Aβ plaque burden generically recognized by the OC and D54D2 antibodies. Remarkably, this multifaceted characterization of Aβ plaques did not show significant differences between APP/PS1 mice treated with PEP3 and those treated with inPEP3 for both the percentage area occupied by plaques and the number of plaques in the hippocampal area, after adjusting for the previous variables (Figures 5G and 5H; Table S4). This finding suggested that PEP3 treatment had no effect on Aβ plaque numbers, extension, or composition.
To assess the cognitive functions of PEP3-treated APP/PS1 mice, we performed two different behavioral tasks, the NORT and the Y-maze test. The NORT results showed that APP/PS1 mice treated with the inactive control inPEP3 peptide were unable to discriminate between the novel and familiar objects, which is indicative of cognitive impairment. Interestingly, APP/PS1 mice treated with PEP3 displayed a clear preference for the novel object compared with the familiar one, indicating that PEP3 treatment reversed the cognitive impairments of APP/PS1 mice (Figure 6A). To strengthen these results, we performed a Y-maze test, which can be used to evaluate the hippocampus in terms of spatial recognition and memory processes. As expected, inPEP3-treated APP/PS1 mice did not show any preferences, whereas the APP/PS1 mice treated with PEP3 spent more time exploring the novel arm compared with the familiar one (Figure 6B). Importantly, we analyzed NORT and Y-maze data considering separately males and females, and the results revealed no gender differences (Figure S8), demonstrating that PEP3 is effective in restoring cognitive function in a sex-independent manner. These results further confirmed that PEP3 could reverse early cognitive deficits in an AD model.
Figure 6.
PEP3 ameliorates cognitive performances and synaptic dysfunction in APP/PS1 mice
(A) NORT was performed on an animal treated with either active or inactive PEP3. Preference index shows preference for novel object for mice treated with PEP3 (inPEP3: familiar 0.470 ± 0.026, novel 0.530 ± 0.026, paired t test t = 1.144, df = 16, p = 0.2695; PEP3: familiar 0.400 ± 0.020, novel 0.600 ± 0.0204, paired t test t = 4.918, df = 14, ∗∗∗p = 0.0002). (B) Y-maze test was performed on animal treated with either active or inactive PEP3. Preference index shows a preference for the novel arm in mice treated with PEP3 (inPEP3: familiar 0.515 ± 0.040, novel 0.485 ± 0.040, paired t test t = 0.368, df = 15, p = 0.7179; PEP3: familiar 0.387 ± 0.022, novel 0.613 ± 0.022, paired t test t = 5.099, df = 15, ∗∗∗p = 0.0001). (C) Representative immunoblots of synaptic proteins in hippocampal homogenate and TIF fraction. OD quantification reveals a significant increase in GluN2A synaptic levels in PEP3-treated mice (GluN2A, unpaired t test: t = 2.360, df = 17, ∗p = 0.0305). (D) Representative confocal images of spine morphology from hippocampal slices. The stacked bar chart shows the percentage of the different types of spines in inPEP3- and PEP3-treated APP/PS1 mice. The joint distribution of dendritic spine type and treatment is represented in the table together with the p value of the chi-square test of independence. The odds ratios of becoming mushroom starting either stubby or thin when mice were treated with PEP3, as compared with inPEP3 control peptide, were presented in the table on the right together with the corresponding confidence interval. No changes in spines density were detected (inPEP3, 9.41 ± 0.72 spines/10 μm; PEP3, 9.9 ± 0.60 spines/10 μm, p > 0.05). Scale bar, 5 μm. Data are reported as mean ± SE.
To investigate the molecular modifications triggered by PEP3 treatment at the synaptic level, we analyzed the synaptic localization of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) and N-methyl-D-aspartate receptor (NMDAR) subunits (Figure 6C). PEP3 administration led to a significant increase in the postsynaptic level of the NMDAR subunit GluN2A, without changing the levels of GluN1 and GluN2B and without affecting AMPAR (Figure 6C). We also assessed whether the PEP3 treatment could affect the dendritic spine morphology in the hippocampus (Figure 6D). The evaluation of spine length and width did not provide any significant evidence (Table S4), while different spine categories (mushroom, stubby, thin, and filopodia) were associated with PEP3 treatment (Figure 6D, chi-square test, p = 0.00017). In addition, results from the two generalized LMMs (GLMMs) showed that the stubby spines of PEP3-treated mice had a significant 3-fold odds ratio (OR) of becoming a mushroom spine compared with the stubby spines of inPEP3-treated mice. A similar OR was found for the thin spines, although the OR failed to reach significance (Figure 6D).
We next examined the effects of PEP3 on human neurons. We differentiated human induced pluripotent stem cells (hiPSCs) into cortical neurons. After 40 days of differentiation, we treated hiPSC-derived neurons with either PEP3 or inPEP3 (10 μM, 30 min; Figure 7A). PEP3 exposure promoted ADAM10 postsynaptic localization (Figure 7B) and increased spine density (Figure 7C) in human neurons. Interestingly, the augment of spine density was maintained also after longer treatments with PEP3 (10 μM, 24 h) (Figures 7D and 7E).
Figure 7.
PEP3 increases ADAM10 synaptic localization and spine density in human neurons
(A) Experimental paradigm: hiPSC-derived human neurons were treated with either inPEP3 or PEP3 (10μM) for 30 min. (B) Representative immunoblot of ADAM10 in the synaptic fraction. OD quantification shows an increase of ADAM10 on PEP3 treatment (paired t test, t = 10.95, df = 3, ∗∗p = 0.0016). (C) Representative confocal images of dendrites and spines. Spine density analysis shows an increase after 30 min of PEP3 treatment (one-way ANOVA F(2,59) = 3.252, p = 0.0457, Fisher’s LSD test inPEP3 versus control [Ctrl], p = 0.689; inPEP3 versus PEP3, ∗p = 0.022; PEP3 versus Ctrl, ∗p = 0.047). Scale bar, 2 μm. (D) Experimental paradigm: hiPSC-derived human neurons were treated with either inPEP3 or PEP3 (10μM) for 24 h. (E) Representative confocal images of spines along dendrites. Spine density analysis shows an increase after 24 h of PEP3 treatment (one-way ANOVA F(2,39) = 3.565, p = 0.0378, Fisher’s LSD test inPEP3 versus Ctrl, p = 0.868; inPEP3 versus PEP3, ∗p = 0.0263; PEP3 versus Ctrl, ∗p = 0.0263). Scale bar, 6 μm. Data are reported as mean ± SE.
Discussion
Drug discovery in AD requires an unconventional approach that explores the therapeutic potential of molecular pathways at the crossroads between the multiple alterations that occur during the early phase of the disease.
Here, we demonstrated that targeting ADAM10 endocytosis to restore early synaptic dysfunction represents a powerful approach for rescuing cognitive performance in APP/PS1 mice. Furthermore, our evidence suggested that PEP3, the CPP we developed to target ADAM10 endocytosis, represents a safe and specific brain-penetrating peptide capable of restoring ADAM10 activity at the synapse. Therefore, PEP3 is not just a regulator of ADAM10 but also targets a specific feature associated with synaptic dysfunction in AD.
In the last 25 years, drug development strategies in the AD field have primarily focused on reducing Aβ levels, which is believed to be the primary trigger of the pathogenesis cascade.3 However, the development of disease-modifying treatments for AD has been challenging, and recently, the US Food and Drug Administration has granted a controversial accelerated approval to the anti-amyloid antibody aducanumab,25 targeting the pathological oligomeric forms of Aβ.26 Aducanumab approval marks a turning point in the drug development landscape because it highlights the importance of focusing on early phases of the disease progression. In particular, soluble Aβ aggregates have been implicated in synaptic dysfunction, a step that precedes full-blown neurodegeneration. Normal synapse numbers and function are necessary for normal cognitive performance, and the synaptic attack by Aβ oligomers impairs neuronal communication, resulting in cognitive impairment. Synaptic dysfunction and loss correlate strongly with the pathological cognitive decline observed in AD.27 Importantly, in mouse models of AD, the effects of Aβ on synapses28,29 are reversible, indicating that compounds capable of reducing synapse damage might be useful as AD treatments.2
Considering the synaptic framework, we targeted ADAM10, an enzyme that is associated with both the amyloid cascade and synaptic function. The ADAM10-mediated cleavage of adhesion molecules is required for changes in spine morphology triggered by activity-dependent synaptic plasticity.10 ADAM10 is also strictly associated with AD because it acts as the α-secretase for the neuronal cleavage of APP5 and has been identified as a risk locus for AD in genome-wide association studies (GWASs).30,31 Platelet levels of ADAM10 are reduced in AD,32, 33, 34 and plasma and cerebrospinal fluid (CSF) levels of a soluble and inactive ADAM10 form were significantly increased in mild AD,35 thus suggesting that alterations in ADAM10 are linked to the disease. ADAM10 activity not only limits Aβ generation but also liberates the neuroprotective sAPPα.36 sAPPα levels are reduced in the CSF of AD patients,32 suggesting that ADAM10 upregulation might have great potential as a therapeutic strategy in AD.37,38 Supporting this hypothesis, the neuronal overexpression of ADAM10 in AD transgenic mice was found to alleviate cognitive deficits.39
ADAM10 synaptic activity is spatially and temporally controlled by trafficking mechanisms that are altered in AD.10,16 In the synaptic context, endocytosis is a critical trafficking pathway in AD, and GWASs have identified several genes associated with endocytosis and synaptic function as risk loci for AD.40 In addition, ADAM10 endocytosis is an early target of Aβ oligomers,15 and the increase in ADAM10 binding to AP2 could be a synaptic signature of AD.
To target such synaptic features of AD and restore physiological levels of ADAM10 endocytosis, we developed CPPs capable of interfering with ADAM10/AP2 association. The use of CPPs has previously been exploited in clinical studies for acute neurological disorders,41 and the efficacy of CPP-mediated ADAM10 inhibition for the treatment of animal models of neurodevelopmental and neurodegenerative disorders has also been demonstrated.42,43
We used a biocomputational study to design four CPPs that could interfere with ADAM10/AP2 association. Using a variety of approaches, we identified and fully characterized PEP3, which was able to target ADAM10 endocytosis without affecting the associations with other ADAM10 or AP2 protein partners.
The sub-chronic administration of PEP3 to wild-type mice resulted in increased ADAM10 synaptic localization and activity and demonstrated the good safety profile of PEP3 treatment. Although ADAM10 cleaves several substrates,44 the moderate activation of ADAM10 has been reported to be a safe approach. ADAM10 transgenic mice have a normal phenotype,39 and the administration of the synthetic retinoid acitretin, which enhances ADAM10 expression, has been shown to be safe in AD patients, indicating that the upregulation of ADAM10 in humans is well tolerated.45
When we treated APP/PS1 mice with PEP3 at the full-blown pathological stage, the treatment was unable to alleviate the well-established cognitive deficits, despite the target engagement. The lack of efficacy for rescuing cognitive defects could be ascribed to the advanced stage of the pathology. To challenge this hypothesis, we administered PEP3 at an earlier stage, when ADAM10/AP2 association and ADAM10 synaptic availability were unbalanced in the hippocampus. The treatment of 6-month-old APP/PS1 mice effectively increased ADAM10 synaptic levels and promoted sAPPα generation. The virus-mediated gene transfer of sAPPα was shown to mitigate synaptic failure and ameliorate plaque pathology in APP/PS1 mice.46 In our experimental conditions, neither plaque density nor the area of Aβ deposition was affected by PEP3 treatment. However, PEP3 administration consistently promoted the maturation of stubby spines into mature mushroom spines. At the molecular level, this effect on spine shape was accompanied by a significant increase in the synaptic levels of the NMDAR GluN2A subunit, suggesting that PEP3 promotes synaptic function. Interestingly, two behavioral tests showed that PEP3 treatment restored cognitive deficits in APP/PS1 mice. Previously, sAPPα has been shown to contribute to spatial memory, and the upregulation of NMDAR function is a downstream effect of sAPPα action.47 It is therefore legitimate to hypothesize that the PEP3-triggered rescue of AD synaptic dysfunction could be mediated by sAPPα. However, we are unable to exclude the possibility that another ADAM10-dependent pathway might promote synaptic plasticity. Consistent with our hypothesis and as a proof of concept for the translational potential of PEP3, hiPSC-derived human neurons exposed to PEP3 showed an increase in spine density.
However, further studies are required to determine the effect of PEP3 in AD hiPSC-derived human neurons, to assess the therapeutic efficacy and the safety of long-term administration of PEP3, and to investigate whether a chronic treatment is able to affect Aβ deposition and neuroinflammation.
Overall, our findings indicated that targeting ADAM10 endocytosis may represent a powerful strategy for treating synaptic dysfunction and cognitive deficits in AD if applied during an optimal treatment window. These findings suggest that the synapse is highly likely to represent a critical substrate that links Aβ pathology with AD cognitive symptoms, and the development of synaptic therapy approaches could provide great efficacy for the rescue of cognitive deficits in AD patients.
Materials and methods
CPPs
The peptides were designed combining the structural results reported by Marcello et al.10 and different computational techniques.48 Basically, starting from the sequence QQPPRQRPRES (Figure 1A), we have designed four peptides changing the residues flanking the conserved portion (i.e., PPRQRPR) (Figure 1A). The CCPs were developed by coupling the active peptide sequence with HIV-1 TAT (Trans-activating transcriptional activator)-derived peptide, which is able to successfully deliver a large variety of cargoes such as peptides, proteins, and nucleic acids, by overcoming the lipophilic barrier of the cellular membranes. Subsequently, we have selected a linker sequence, with the aim to keep the TAT sequence and the peptides active sequence well spatially separated. To this end, we have carried out short molecular dynamics (MD) simulations (i.e., 20 ns for each peptide) that were used to analyze the structural and dynamical features of peptides in solutions. The MD simulations were performed as described by Di Marino et al.;49 we have performed analyses with the protocol described in Di Marino et al.,50 aimed at detecting the most frequently accessed conformations of the peptides in solution and the distribution of the surface electrostatic potential (data not shown). The linker was selected based on its ability to maintain the TAT sequence and the active portion of the peptide separated. The choice fell on the GGSG sequence, as shown in Figure S1A. As far as concern the inactive peptides (inPEP1, inPEP2, inPEP3, and inPEP4), the two arginines of the sequence RQR, which are the key residues for the activity of the peptide, were substituted with glutamate (EQE).
The sequences of the peptides are covered by patent application no. PCT/IB2018/060,511. Covalab (France) manufactured each CPP according to our designed sequences. Lyophilized CPPs were resuspended in sterile deionized water to a stock concentration of 1 mM and stored at −20°C.
Derivation of hiPSCs and neuronal differentiation
hiPSCs were obtained as described by Culotta et al.51 Three to four weeks after transduction, hiPSC colonies were manually picked for further expansion or analysis. For neural stem cells (NSCs) derivation, hiPSCs were detached with UltraPure EDTA (Thermo Fisher Scientific) and plated on Matrigel-coated six-well plates in Essential 8 medium. After 24 h, the medium was replaced with PSC Neural Induction Medium (Thermo Fisher Scientific) and subsequently was changed every other day following the manufacturer’s instructions. On day 7 of neuronal induction, NSCs (P0) were harvested with StemPro Accutase (Thermo Fisher Scientific) and plated on Matrigel-coated plates for further expansion.
NSCs were converted into glutamatergic cortical neurons by lentiviral transduction of tetracycline-inducible expression of the neuralizing transcription factor Neurogenin-2 (NGN2). To obtain mature glutamatergic neurons, we cultured transduced cells, for 4–6 weeks, in neuron culture medium (Neurobasal containing B27, 10 ng/mL NT-3, 20 ng/mL Brain-Derived Neurotrophic Factor (BDNF), 10 ng/mL glial cell line-derived neurotrophic factor [GDNF], 1 μM retinoic acid, and 100 μM dibutyryl cyclic adenosine monophosphate) in the presence of doxycycline (Dox) (4 μg/mL). The medium was replaced every 4 days. Cells were treated with either PEP3 or inPEP3 at the concentration of 10 μM for 30 min or 24 h.
Animal treatment with the CPPs
Adult C57BL/6j male mice were purchased from Charles River Laboratories, while the transgenic line B6; C3-Tg(APPswe, PSEN1dE9)85Dbo (APP/PS1) was bred in-house, and male/female transgenic mice at the age of 6 and 12 months were used for testing the CPP effect. APP/PS1 are double-transgenic mice expressing a chimeric mouse/human APP (Mo/HuAPP695swe) and a mutant human presenilin 1 (PS1-dE9).23 G Power Software52 was used to calculate the adequate sample size to study the effects of CPP administration.
Adult C57BL/6j mice were treated with either sterile saline solution or active peptides, or inactive peptides (3 nmol/g, diluted in sterile solution) by intraperitoneal injection. For biochemical and histological analyses, animals were euthanized 24 h after the injection (see Figures 2 and S2).
To assess the optimal sub-chronic administration protocol of PEP3, we treated adult C57BL/6j mice with either sterile saline solution or PEP3 or inPEP3 at different doses (1 or 3 nmol/g, diluted in sterile solution) by intraperitoneal or subcutaneous injections. At different time points (Figures S3 and S4), animals were euthanized, and the brain was rapidly dissected for biochemical analyses.
To evaluate efficacy and safety of a sub-chronic treatment, we administered adult C57BL/6j mice with either PEP3 or inPEP3 (3 nmol/g) by daily intraperitoneal injections for 2 weeks. For biochemical analyses, animals were euthanized 24 h after the last injection, and the brains were rapidly removed for the analysis (see Figure 3).
APP/PS1 mice received daily intraperitoneal injections of either inPEP3 or PEP3 for 15 days. After 24 h from the last injection, mice were euthanized for the following analyses (Figures 4 and 5).
CoIP assay
Aliquots of 40–50 μg of proteins from brain or neuronal culture homogenates were incubated with the antibody overnight at 4°C in radioimmunoassay (RIA) buffer containing 50 mM Tris HCl (pH 7.2), 150 mM NaCl, 1% Nonidet P-40 (NP-40), 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate (SDS), in a final volume of 150 μL. As a control, one sample was incubated in the absence of the antibody (no IgG lane). The day after, protein A/G-Sepharose beads (Santa Cruz) were added, and incubation was continued for 2 h at room temperature (RT). Beads were collected by centrifugation and washed three times with RIA buffer before adding 30 μL of Laemmli buffer for SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and the mixture was heated for 5 min before loading onto SDS-PAGE gels.
Antibody internalization assay
Tac-ADAM10-RAR-transfected COS7 cells were treated with either inactive or active peptides (1μM, 30 min). To label surface TacADAM10-RAR, we incubated COS7 cells with anti-Tac antibody for 45 min in medium at 4°C, in the presence of the peptides. After brief washing in DMEM, COS7 cells were returned to 37°C for 10 min. Cells were then fixed with 4% PFA and 4% sucrose in PBS (pH 7.4) and blocked with normal serum; remaining surface Tac chimeras were labeled with Alexa 488-conjugated secondary antibody (nonpermeabilized), and internalized receptors were labeled with Alexa 555-conjugated secondary antibody after Triton permeabilization. Wide-field fluorescence images were acquired with a Zeiss ×40 objective and a CoolSnap charge-coupled device (CCD) camera.
Dendritic spines labeling
Carbocyanine dye DiI (Invitrogen) was used to label neurons.53 DiI crystals were applied using a thin needle by delicately touching the region of interest on both sides of 2-mm coronal sections prepared from previously cardiacally fixed brains with 1.5% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB). DiI was left to diffuse for 1 day in the dark at RT; then sections were fixed again with 4% PFA in PB 0.1 M for 45 min at 4°C. 150-μm coronal sections were then obtained using a vibratome, and the first section was discarded. Sections were mounted on glass slides with Fluoromount mounting medium (Sigma-Aldrich) for confocal imaging. Fluorescence images from slides were then obtained using a Confocal microscope Zeiss LSM510 Meta system with an objective 63× performing sequential acquisition at a resolution of 1,024 × 1,024 pixels. z stack acquisitions were analyzed using ImageJ free software, and the length of spine, head width, and neck width for each spine were measured with straight line function.
Human neurons derived from hiPSCs were fixed with 4% PFA-4% sucrose in PBS solution for 5 min at 4°C and washed several times with PBS. Cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature (RT) and then blocked with 5% BSA in PBS for 1 h at RT. Cells were then labeled with anti-GFP primary antibody at 4°C overnight. Cells were washed and then incubated with secondary antibody for 1 h at RT. After, the cells were washed in PBS and mounted on glass slides with Fluoromount mounting medium (Sigma-Aldrich).
Images for the analysis of dendritic spines density of hiPSC-derived human neurons were acquired using Confocal microscope Zeiss with 63× oil objective, Plan Apochromat, NA 1.42 (1,024 × 1,024, 0.5-μm z steps).
Plaques staining
Coronal sections of 40 μm were obtained from fixed brains using a sliding microtome (Leica SM 2000R). Double immunofluorescences were performed on free-floating sections by sequential steps of blocking and permeabilization (20% normal donkey serum [NDS] in 0.3% Triton X-100/Tris-buffered saline [TBS]), incubation with primary antibodies (in 10% NDS/TBS) overnight at 4°C, and incubation with secondary antibodies (in 5% NDS/TBS). Plaques were immunodetected by D54D2 (rabbit monoclonal; CST), D9A3A (rabbit monoclonal; CST), OC (rabbit polyclonal; Millipore), primary antibodies, and anti-rabbit Alexa 488 secondary antibody (Alexa Fluor; Thermo Fisher Scientific). Neurons were stained with the neural marker NeuN (mouse monoclonal; Millipore) and anti-mouse Alexa 647 (Alexa Fluor; Thermo Fisher Scientific) secondary antibody. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI). Sequential images of hippocampus were captured at 20× magnification under an X-Light V2 spinning disk confocal microscope (Crisel Instrument) and processed by Metamorph software.
Experimental design and quantification of data
To minimize the possibility of bias in experimental results, we used randomization and blinding in the experimental design, imaging acquisitions, and analyses. Acquisition and quantification of WB was performed by means of computer-assisted imaging (ChemiDoc system and Image lab 4.0 software; Bio-Rad). The different protein levels were quantified as optical density (OD) readouts and were further normalized on tubulin protein OD. The OD of phosphorylated pS845-GluA1 subunit was normalized to GluA1 levels. In order to quantify the coIP of ADAM10/AP2 complex, we normalized the different subunits of AP2 on the OD of immunoprecipitated ADAM10. For quantification of internalization, cells were chosen randomly for quantification from four to eight different coverslips (two to three independent experiments), images were acquired using the same settings/exposure times, and at least 10 cells for each condition were analyzed. All images were analyzed using ImageJ, the average fluorescence intensities in the green (total) and red (internalized) channels were measured, background was subtracted, and the internalized/total fluorescence ratio was calculated for each cell. Morphological analysis of dendritic spines was performed on the total length of the dendrites using ImageJ software to measure spine length and head and neck width with straight line function, in order to classify spines in mushroom, stubby, thin, and filopodia. For plaques analysis, the reconstruction of the hippocampal area and the measurement of number, diameter, and area of the plaques were done using the Fiji software.
Statistics
We accounted for the hierarchical structure of the data using LMMs for the analysis of the following continuous variables: length and width of spines (Figure 6D; Table S4) and percentage and density of amyloid plaques (Figures 5G and 5H; Table S4). We specified one mixed-effects model54 for each experiment under investigation and outcome variable of interest within the experiment.55 GLMMs with a logit link function and binomial family were fitted for the type outcome variable in the experiments presented in Figure 6D; in these two analyses, the outcome variable was restricted to assume either categories mushroom/stubby (reference category) or mushroom/thin (reference category). From GLMMs estimates, we derived the ORs and the corresponding 95% confidence interval (CI) of a change in spine type going from either stubby or thin to the mushroom type when the treatment was PEP3 as compared with inPEP3.
In the spine morphology experiment, treatment was the only independent variable; in the plaques analysis, we considered the effect of treatment, as well as diameter of plaques (≤30 μm and >30 μm), antibodies used for plaques staining (D54D2, D9A3A, and OC), and two-way and three-way interactions between treatment, diameter, and antibody. In the absence of evidence supporting a three-way interaction term, all two-way interactions were evaluated. In the analysis of plaques density, the main-effects model was finally selected in the absence of evidence supporting two-way interactions. In the analysis of plaques percentage, the two-way interaction of diameter and antibody was statistically significant, and the overall interaction term was borderline significant; therefore, the final model selected included all three two-way interaction terms.
For each mixed-effects model, one or two random intercepts were specified depending on the available levels of the hierarchy. The mouse random intercept was possibly present in any model, whereas either the neuron or the antibody level of the hierarchy was present when the experimental unit of analysis was spine or slice, respectively. Correlations between random effects were allowed to be present and fitted within the model to better capture the hierarchical structure of the experiment. We reported information on which dependent and independent variables, as well as random effects, were used within each mixed-effects model in Table S4 (columns on the left). For model fitting, restricted maximum likelihood (REML) estimates were used, because they provided better estimates of variance components. To test model assumptions, we plotted residuals versus fitted values for each model; when outliers were detected, they were checked at the experimental level. Spine length and plaque analysis (density and percentage of amyloid plaques) showed model convergence issues related to null standard deviations of the mouse random effects; therefore, we fitted equivalent models showing a fixed-effect term for mouse instead of the corresponding mixed-effect one.
Hypothesis testing on the significance of single parameters was performed within the LMMs or GLMMs with a Student’s t distribution or a Gaussian distribution, respectively. The F statistics from the ANOVA analysis provided information on the overall significance of the independent variables entered into each model. Finally, the chi-square test of independence was used to assess the presence of an association between spine type and treatment in experiments presented in Figure 6D.
For all the other experiments, there was no hierarchical structure to take into account, and data followed a normal distribution; therefore, either a Student's t test or ANOVA, followed by a post hoc adjustment, was carried out.
Throughout the manuscript, when continuous variables were considered, values were reported as mean ± SE, and when qualitative variables were analyzed, absolute frequencies (raw numbers) or percentages were indicated. The type of parametric or non-parametric test used for each experiment and the corresponding p values, as well the type of adjustment for multiple comparisons (if any), were provided in the figure legends. All statistical tests were two-sided, and significance was assumed if p < 0.05. Calculations were carried out using the open-source statistical computing environment R56 with its libraries lme4,57 lmerTest58 and emmeans59 for the experiments in Figures 5G, 5H, and 6D and with Prism 6 and 9 (GraphPad, La Jolla, CA, USA) for all the other quantitative evaluations.
Ethics approval
All protocols involving animals were carried out in accordance with institutional guidelines in compliance with Italian law (D. Lgs no. 2014/26, implementation of the 2010/63/UE). The Ethics Committee of the University of Milano and the Italian Ministry of Health approved studies in mice (Italian Ministry of Health permit 124/2003 and 497/2015). Animals for in vivo and in vitro experiments were maintained in a controlled environment with a 12/12-h light/dark cycle in a temperature-controlled room (22°C) in cages with free access to food and water at the Department of Pharmacological and Biomolecular Sciences (University of Milan) animal facility. Housing in the animal facility is performed in conformity with local and European Community regulations under the control of veterinarians with the assistance of trained personnel. Animal handling and procedures were carried out with care taken to minimize discomfort and pain.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplemental information files. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
We acknowledge the donors of the Alzheimer Disease Research, a program of the BrightFocus Foundation, for supporting E.M. We thank Elisa Zianni and Annalisa Longhi for technical assistance, and Maria Daffara, Silvia Pedrini, Marta Corrà, Alberto Catanese, Gregorio P. Milani, and Andrea Salvatori for excellent practical work. Part of this work was carried out at NOLIMITS, an advanced imaging facility established by the Università degli Studi di Milano. This project has received support from the Alzheimer Disease Research program research fellowship of BrightFocus Foundation (A2014314F to E.M.), the Alzheimer’s Association (NIRP-14-304969 to E.M.), the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement 676144 (Synaptic Dysfunction in Alzheimer Disease [SyDAD]) to M.D.L., the Italian Ministry of University and Research (PRIN 2017MYJ5TH to M.D.L., PRIN 2017B9NCSX to E.M., MIUR Progetto Eccellenza), the Veronesi Foundation Young Investigator Research Programme 2013 (to E.M.), Fondazione Cariplo (Grant 2018-0511 to E.M.; Grant 2019-3396 to C.S.), an intramural grant of the University of Milan to E.M. (PSR2019_EMARC), and the Fondation Jerome Lejeune (project 1638 to C.S.; project 1938 to C.V.). This work was supported by MIUR - PON “Ricerca e Innovazione” PerMedNet ID project ARS01_01226.
Author contributions
S.M. designed and conducted the experiments and wrote and revised the manuscript; S.T. designed and conducted the experiments and wrote and revised the manuscript; S.P. designed and conducted the experiments; L.D.A. designed and conducted the experiments; R.S. designed and conducted the experiments; A.R. designed and conducted the experiments; A.M. designed and conducted the experiments; C.B. designed experiments and revised the manuscript; J.P. conducted the experiments; C.S. revised the manuscript; C.V. designed experiments and revised the manuscript; V.G. designed experiments; V.E. conducted statistical analysis (mixed-effects models) and revised the manuscript; G.F. revised the manuscript; F.G. revised the manuscript; G.M. designed experiments and revised the manuscript; D.D.M. designed the CPPs, conducted MD simulations, and revised the manuscript; M.D.L. performed conceptualization, funding acquisition, project administration, and supervision, and wrote and revised the manuscript; E.M. performed conceptualization, funding acquisition, project administration, and supervision, and wrote and revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.03.024.
Contributor Information
Monica Di Luca, Email: monica.diluca@unimi.it.
Elena Marcello, Email: elena.marcello@unimi.it.
Supplemental information
References
- 1.Selkoe D.J. Alzheimer’s disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 2.Colom-Cadena M., Spires-Jones T., Zetterberg H., Blennow K., Caggiano A., Dekosky S.T., Fillit H., Harrison J.E., Schneider L.S., Scheltens P., et al. The clinical promise of biomarkers of synapse damage or loss in Alzheimer’s disease. Alzheimers Res. Ther. 2020;12:21. doi: 10.1186/s13195-020-00588-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy J.A., Higgins G.A. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 4.Epis R., Marcello E., Gardoni F., di Luca M. Alpha, beta-and gamma-secretases in Alzheimer’s disease. Front. Biosci. (Schol. Ed.) 2012;4:1126–1150. doi: 10.2741/s322. [DOI] [PubMed] [Google Scholar]
- 5.Kuhn P.H., Wang H., Dislich B., Colombo A., Zeitschel U., Ellwart J.W., Kremmer E., Rossner S., Lichtenthaler S.F. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29:3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter M.C., Ludewig S., Winschel A., Abel T., Bold C., Salzburger L.R., Klein S., Han K., Weyer S.W., Fritz A.K., et al. Distinct in vivo roles of secreted APP ectodomain variants APPsα and APPsβ in regulation of spine density, synaptic plasticity, and cognition. EMBO J. 2018;37:e98335. doi: 10.15252/embj.201798335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tolar M., Abushakra S., Hey J.A., Porsteinsson A., Sabbagh M. Aducanumab, gantenerumab, BAN2401, and ALZ-801-the first wave of amyloid-targeting drugs for Alzheimer’s disease with potential for near term approval. Alzheimers Res. Ther. 2020;12:95. doi: 10.1186/s13195-020-00663-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mecca A.P., Chen M.K., O'Dell R.S., Naganawa M., Toyonaga T., Godek T.A., Harris J.E., Bartlett H.H., Zhao W., Nabulsi N.B., et al. In vivo measurement of widespread synaptic loss in Alzheimer’s disease with SV2A PET. Alzheimers Dement. 2020;16:974–982. doi: 10.1002/alz.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcello E., Gardoni F., Mauceri D., Romorini S., Jeromin A., Epis R., Borroni B., Cattabeni F., Sala C., Padovani A., Di Luca M. Synapse-associated protein-97 mediates alpha-secretase ADAM10 trafficking and promotes its activity. J. Neurosci. 2007;27:1682–1691. doi: 10.1523/JNEUROSCI.3439-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcello E., Saraceno C., Musardo S., Vara H., de La Fuente A.G., Pelucchi S., Di Marino D., Borroni B., Tramontano A., Pérez-Otaño I., et al. Endocytosis of synaptic ADAM10 in neuronal plasticity and Alzheimer’s disease. J. Clin. Invest. 2013;123:2523–2538. doi: 10.1172/JCI65401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saraceno C., Marcello E., di Marino D., Borroni B., Claeysen S., Perroy J., Padovani A., Tramontano A., Gardoni F., Di Luca M. SAP97-mediated ADAM10 trafficking from Golgi outposts depends on PKC phosphorylation. Cell Death Dis. 2014;5:e1547. doi: 10.1038/cddis.2014.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malinverno M., Carta M., Epis R., Marcello E., Verpelli C., Cattabeni F., Sala C., Mulle C., Di Luca M., Gardoni F. Synaptic localization and activity of ADAM10 regulate excitatory synapses through N-cadherin cleavage. J. Neurosci. 2010;30:16343–16355. doi: 10.1523/JNEUROSCI.1984-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardoni F., Saraceno C., Malinverno M., Marcello E., Verpelli C., Sala C., Di Luca M. The neuropeptide PACAP38 induces dendritic spine remodeling through ADAM10-N-cadherin signaling pathway. J. Cell Sci. 2012;125:1401–1406. doi: 10.1242/jcs.097576. [DOI] [PubMed] [Google Scholar]
- 14.Saftig P., Lichtenthaler S.F. The alpha secretase ADAM10: a metalloprotease with multiple functions in the brain. Prog. Neurobiol. 2015;135:1–20. doi: 10.1016/j.pneurobio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Marcello E., Musardo S., Vandermeulen L., Pelucchi S., Gardoni F., Santo N., Antonucci F., Di Luca M. Amyloid-β oligomers regulate ADAM10 synaptic localization through aberrant plasticity phenomena. Mol. Neurobiol. 2019;56:7136–7143. doi: 10.1007/s12035-019-1583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcello E., Epis R., Saraceno C., Gardoni F., Borroni B., Cattabeni F., Padovani A., Di Luca M. SAP97-mediated local trafficking is altered in Alzheimer disease patients’ hippocampus. Neurobiol. Aging. 2012;33:422.e1–422.e10. doi: 10.1016/j.neurobiolaging.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Bonifacino J.S., Traub L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 18.Hall R.A., Soderling T.R. Quantitation of AMPA receptor surface expression in cultured hippocampal neurons. Neuroscience. 1997;78:361–371. doi: 10.1016/s0306-4522(96)00525-8. [DOI] [PubMed] [Google Scholar]
- 19.Bonifacino J.S., Cosson P., Klausner R.D. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
- 20.Marcello E., Gardoni F., di Luca M., Pérez-Otano I. An arginine stretch limits ADAM10 exit from the endoplasmic reticulum. J. Biol. Chem. 2010;285:10376–10384. doi: 10.1074/jbc.M109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardoni F., Bellone C., Cattabeni F., di Luca M. Protein kinase C activation modulates alpha-calmodulin kinase II binding to NR2A subunit of N-methyl-D-aspartate receptor complex. J. Biol. Chem. 2001;276:7609–7613. doi: 10.1074/jbc.M009922200. [DOI] [PubMed] [Google Scholar]
- 22.Pelucchi S., Vandermeulen L., Pizzamiglio L., Aksan B., Yan J., Konietzny A., Bonomi E., Borroni B., Padovani A., Rust M.B., et al. Cyclase-associated protein 2 dimerization regulates cofilin in synaptic plasticity and Alzheimer’s disease. Brain Commun. 2020;2:fcaa086. doi: 10.1093/braincomms/fcaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jankowsky J.L., Fadale D.J., Anderson J., Xu G.M., Gonzales V., Jenkins N.A., Copeland N.G., Lee M.K., Younkin L.H., Wagner S.L., et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum. Mol. Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 24.Kayed R., Head E., Sarsoza F., Saing T., Cotman C.W., Necula M., Margol L., Wu J., Breydo L., Thompson J.L., et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cummings J., Aisen P., Lemere C., Atri A., Sabbagh M., Salloway S. Aducanumab produced a clinically meaningful benefit in association with amyloid lowering. Alzheimers Res. Ther. 2021;13:98. doi: 10.1186/s13195-021-00838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arndt J.W., Qian F., Smith B.A., Quan C., Kilambi K.P., Bush M.W., Walz T., Pepinsky R.B., Bussière T., Hamann S., et al. Structural and kinetic basis for the selectivity of aducanumab for aggregated forms of amyloid-β. Sci. Rep. 2018;8:6412. doi: 10.1038/s41598-018-24501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spires-Jones T.L., Hyman B.T. The intersection of amyloid beta and tau at synapses in Alzheimer’s disease. Neuron. 2014;82:756–771. doi: 10.1016/j.neuron.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spires-Jones T.L., Mielke M.L., Rozkalne A., Meyer-Luehmann M., de Calignon A., Bacskai B.J., Schenk D., Hyman B.T. Passive immunotherapy rapidly increases structural plasticity in a mouse model of Alzheimer disease. Neurobiol. Dis. 2009;33:213–220. doi: 10.1016/j.nbd.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balducci C., Beeg M., Stravalaci M., Bastone A., Sclip A., Biasini E., Tapella L., Colombo L., Manzoni C., Borsello T., et al. Synthetic amyloid-beta oligomers impair long-term memory independently of cellular prion protein. Proc. Natl. Acad. Sci. U S A. 2010;107:2295–2300. doi: 10.1073/pnas.0911829107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marioni R.E., Harris S.E., Zhang Q., McRae A.F., Hagenaars S.P., Hill W.D., Davies G., Ritchie C.W., Gale C.R., Starr J.M., et al. GWAS on family history of Alzheimer’s disease. Transl. Psychiatry. 2018;8:99. doi: 10.1038/s41398-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkle B.W., Grenier-Boley B., Sims R., Bis J.C., Damotte V., Naj A.C., Boland A., Vronskaya M., van der Lee S.J., Amlie-Wolf A., et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019;51:414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colciaghi F., Borroni B., Pastorino L., Marcello E., Zimmermann M., Cattabeni F., Padovani A., Di Luca M. [alpha]-Secretase ADAM10 as well as [alpha]APPs is reduced in platelets and CSF of Alzheimer disease patients. Mol. Med. 2002;8:67. [PMC free article] [PubMed] [Google Scholar]
- 33.Colciaghi F., Marcello E., Borroni B., Zimmermann M., Caltagirone C., Cattabeni F., Padovani A., Di Luca M. Platelet APP, ADAM 10 and BACE alterations in the early stages of Alzheimer disease. Neurology. 2004;62:498–501. doi: 10.1212/01.wnl.0000106953.49802.9c. [DOI] [PubMed] [Google Scholar]
- 34.Manzine P.R., de França Bram J.M., Barham E.J., de Assis Carvalho Do Vale F., Selistre-De-Araújo H.S., Cominetti M.R., Iost Pavarini S.C. ADAM10 as a biomarker for Alzheimer’s disease: a study with Brazilian elderly. Demen. Geriatr. Cogn. Disord. 2013;35:58–66. doi: 10.1159/000345983. [DOI] [PubMed] [Google Scholar]
- 35.Vanatabe I.P., Peron R., Grigoli M.M., Pelucchi S., de Cesare G., Magalhães T., Manzine P.R., Balthazar M.L.F., Luca M.D., Marcello E., et al. ADAM10 plasma and CSF levels are increased in mild Alzheimer’s disease. Int. J. Mol. Sci. 2021;22:1–13. doi: 10.3390/ijms22052416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mockett B.G., Richter M., Abraham W.C., Mülle U.C. Therapeutic potential of secreted amyloid precursor protein APPsα. Front. Mol. Neurosci. 2017;10:30. doi: 10.3389/fnmol.2017.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musardo S., Marcello E. Synaptic dysfunction in Alzheimer’s disease: from the role of amyloid β-peptide to the α-secretase ADAM10. Eur. J. Pharmacol. 2017;817:30–37. doi: 10.1016/j.ejphar.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 38.Marcello E., Borroni B., Pelucchi S., Gardoni F., di Luca M. ADAM10 as a therapeutic target for brain diseases: from developmental disorders to Alzheimer’s disease. Expert Opin. Ther. Targets. 2017;21:1017–1026. doi: 10.1080/14728222.2017.1386176. [DOI] [PubMed] [Google Scholar]
- 39.Postina R., Schroeder A., Dewachter I., Bohl J., Schmitt U., Kojro E., Prinzen C., Endres K., Hiemke C., Blessing M., et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J. Clin. Invest. 2004;113:1456–1464. doi: 10.1172/JCI20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karch C.M., Goate A.M. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry. 2015;77:43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill M.D., Martin R.H., Mikulis D., Wong J.H., Silver F.L., terBrugge K.G., Milot G., Clark W.M., Macdonald R.L., Kelly M.E., et al. Safety and efficacy of NA-1 in patients with iatrogenic stroke after endovascular aneurysm repair (ENACT): a phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2012;11:942–950. doi: 10.1016/S1474-4422(12)70225-9. [DOI] [PubMed] [Google Scholar]
- 42.Pasciuto E., Ahmed T., Wahle T., Gardoni F., D'Andrea L., Pacini L., Jacquemont S., Tassone F., Balschun D., Dotti C.G., et al. Dysregulated ADAM10-mediated processing of APP during a critical time window leads to synaptic deficits in fragile X syndrome. Neuron. 2015;87:382–398. doi: 10.1016/j.neuron.2015.06.032. [DOI] [PubMed] [Google Scholar]
- 43.Vezzoli E., Caron I., Talpo F., Besusso D., Conforti P., Battaglia E., Sogne E., Falqui A., Petricca L., Verani M., et al. Inhibiting pathologically active ADAM10 rescues synaptic and cognitive decline in Huntington’s disease. J. Clin. Invest. 2019;129:2390–2403. doi: 10.1172/JCI120616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhn P.H., Colombo A.V., Schusser B., Dreymueller D., Wetzel S., Schepers U., Herber J., Ludwig A., Kremmer E., Montag D., et al. Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. Elife. 2016;5:e12748. doi: 10.7554/eLife.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endres K., Fahrenholz F., Lotz J., Hiemke C., Teipel S., Lieb K., Tüscher O., Fellgiebel A. Increased CSF APPs-α levels in patients with Alzheimer disease treated with acitretin. Neurology. 2014;83:1930–1935. doi: 10.1212/WNL.0000000000001017. [DOI] [PubMed] [Google Scholar]
- 46.Fol R., Braudeau J., Ludewig S., Abel T., Weyer S.W., Roederer J.P., Brod F., Audrain M., Bemelmans A.P., Buchholz C.J., et al. Viral gene transfer of APPsα rescues synaptic failure in an Alzheimer’s disease mouse model. Acta Neuropathol. 2016;131:247–266. doi: 10.1007/s00401-015-1498-9. [DOI] [PubMed] [Google Scholar]
- 47.Taylor C.J., Ireland D.R., Ballagh I., Bourne K., Marechal N.M., Turner P.R., Bilkey D.K., Tate W.P., Abraham W.C. Endogenous secreted amyloid precursor protein-alpha regulates hippocampal NMDA receptor function, long-term potentiation and spatial memory. Neurobiol. Dis. 2008;31:250–260. doi: 10.1016/j.nbd.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 48.D’Annessa I., di Leva F.S., la Teana A., Novellino E., Limongelli V., di Marino D. Bioinformatics and biosimulations as toolbox for peptides and peptidomimetics design: where are we? Front. Mol. Biosci. 2020;7:66. doi: 10.3389/fmolb.2020.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.di Marino D., Chillemi G., de Rubeis S., Tramontano A., Achsel T., Bagni C. MD and docking studies reveal that the functional switch of CYFIP1 is mediated by a butterfly-like motion. J. Chem. Theor. Comput. 2015;11:3401–3410. doi: 10.1021/ct500431h. [DOI] [PubMed] [Google Scholar]
- 50.di Marino D., D’Annessa I., Tancredi H., Bagni C., Gallicchio E. A unique binding mode of the eukaryotic translation initiation factor 4E for guiding the design of novel peptide inhibitors. Protein Sci. 2015;24:1370–1382. doi: 10.1002/pro.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Culotta L., Scalmani P., Vinci E., Terragni B., Sessa A., Broccoli V., Mantegazza M., Boeckers T., Verpelli C. SULT4A1 modulates synaptic development and function by promoting the formation of PSD-95/NMDAR complex. J. Neurosci. 2020;40:7013–7026. doi: 10.1523/JNEUROSCI.2194-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Faul F., Erdfelder E., Lang A.G., Buchner A. G∗Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 53.Kim B.G., Dai H.N., McAtee M., Vicini S., Bregman B.S. Labeling of dendritic spines with the carbocyanine dye DiI for confocal microscopic imaging in lightly fixed cortical slices. J. Neurosci. Methods. 2007;162:237–243. doi: 10.1016/j.jneumeth.2007.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinheiro J., Bates D. Springer Nature; 2009. Mixed-Effects Models in S and S-Plus. [Google Scholar]
- 55.Paternoster V., Rajkumar A.P., Nyengaard J.R., Børglum A.D., Grove J., Christensen J.H. The importance of data structure in statistical analysis of dendritic spine morphology. J. Neurosci. Methods. 2018;296:93–98. doi: 10.1016/j.jneumeth.2017.12.022. [DOI] [PubMed] [Google Scholar]
- 56.R Core Team . R Foundation for Statistical Computing; 2019. R: A Language and Environment for Statistical Computing.https://CRAN.Rproject.org/package¼emmeans [Google Scholar]
- 57.Bates D., Mächler M., Bolker B.M., Walker S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015;67:1–48. [Google Scholar]
- 58.Kuznetsova A., Brockhoff P.B., Christensen R.H.B. lmerTest package: tests in linear mixed effects models. J. Stat. Softw. 2017;82:1–26. [Google Scholar]
- 59.Lenth R., Singmann H., Love J., Buerkner P., Herve M. 2021. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. R package version 1.6.2-1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplemental information files. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.