Abstract
We are in an emerging era of gene-based therapeutics with significant promise for rare genetic disorders. The potential is particularly significant for genetic central nervous system disorders that have begun to achieve Food and Drug Administration approval for select patient populations. This review summarizes the discussions and presentations of the National Institute of Mental Health-sponsored workshop “Gene-Based Therapeutics for Rare Genetic Neurodevelopmental Psychiatric Disorders,” which was held in January 2021. Here, we distill the points raised regarding various precision medicine approaches related to neurodevelopmental and psychiatric disorders that may be amenable to gene-based therapies.
Keywords: neurodevelopment, psychiatric disorders, gene therapy
Graphical abstract

Davidson, Gao, Sahin, et al. summarize the National Institute of Mental Health-sponsored workshop “Gene-Based Therapeutics for Rare Genetic Neurodevelopmental Psychiatric Disorders,” held in January 2021. They distill the points raised regarding precision medicine approaches for neurodevelopmental and psychiatric disorders that may be amenable to gene-based therapies.
Introduction
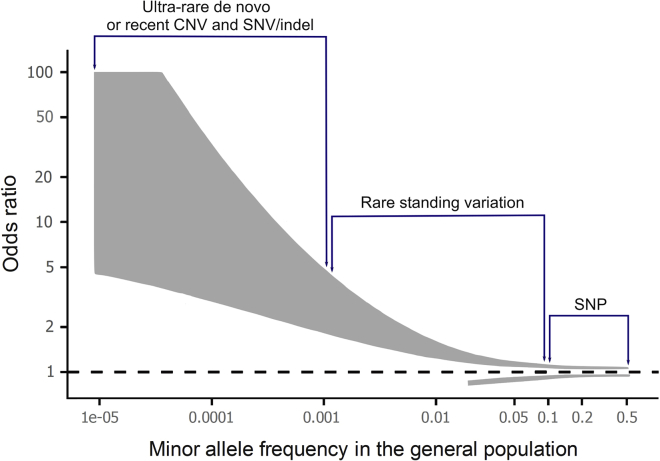
The heritability and therefore genetic basis of psychiatric disorders is substantial, ranging from 64% to 91% reported for autism spectrum disorder (ASD) to somewhere between 30% and 40% for symptoms of anxiety and depression.1, 2, 3 Heritability, as estimated from family and population studies, reflects genetic variation across a wide spectrum of both frequency in the population and effect size. Genetic variation that substantially increases risk for psychiatric disorders is likely to be rare, given purifying selection. For this reason, common genetic variation, most frequently in the form of single-nucleotide polymorphism (SNP), is associated with very small effect sizes (odds ratios of 1.1 or less).4, 5, 6, 7, 8, 9 In contrast, rare variation can be associated with substantial effect sizes (odds ratios of 10, 20, or more); such rare deleterious variants are likely recent, and frequently de novo, in the population, given their impact on neurodevelopment. Polygenic risk reflects the impact of a vast number of common variants thought to operate additively; more broadly, it is useful to consider risk as the additive impact of both common and rare variation.
Taking ASD as an example, most genetic risk for ASD resides with common variation from the population perspective; however, rare deleterious variation can be the primary determinant in specific individuals (Figure 1).10,11 To date, genome-wide sequencing studies have identified more than 100 genes that, when mutated, can confer high risk to ASD,12 with many more to be discovered (Figure 2). These studies are most powered to identify autosomal dominant loci, while more traditional clinical genetic studies have identified many autosomal and X-linked genes that can harbor dominant or recessive mutations that confer high risk for ASD.13, 14, 15 Studies to date confirm that common and rare variation appear to act additively in ASD.16,17 Importantly, the diagnostic yield of exome sequencing in identifying such variants is very high,18 leading to several recent papers recommending that exome sequencing be a first-line genetic test offered to those with ASD.19,20
Figure 1.
Effect sizes of genetic variation in neurodevelopmental disorders
Odds ratios, based on prior findings, are shown in gray, with the width of the line reflecting the range of odds ratios that have been reported for significant variants. Single-nucleotide polymorphisms (SNPs) have very small effect sizes, and some SNPs have been shown to be protective. Rare standing variation can show stronger effect sizes, but as effect sizes increase, such standing variation will be under stronger purifying selection. Ultra-rare variation showing significant association with one or more neurodevelopmental disorders includes copy number variation (CNV), single-nucleotide variation (SNV), and insertions/deletions (indel) and is often de novo or recent within a family, given strong purifying selection.
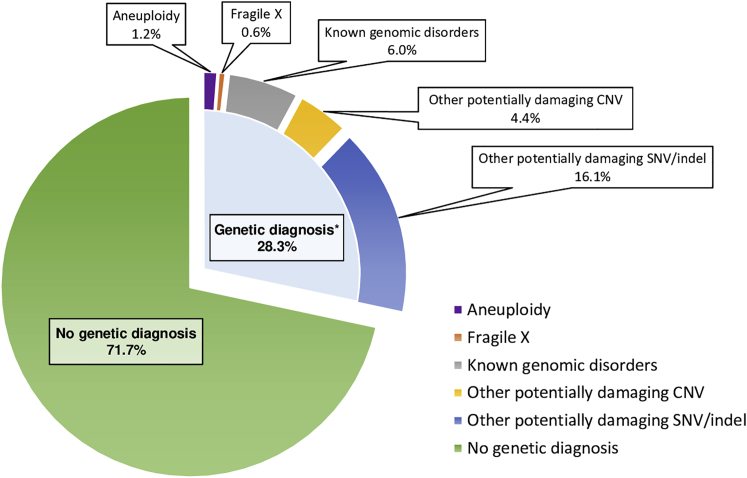
Figure 2.
Proportion of individuals with autism carrying a potentially damaging variant
The PAGES epidemiological sample of individuals with autistic disorder was analyzed for rare structural variation (aneuploidies and damaging CNV) and for point mutations (damaging SNV and indels). Potentially damaging variation was identified when structural variation was large or impacted a known genomic disorder region or when point mutations or indels impacted high-confidence autism or developmental delay genes. Cases with a diagnosis of Fragile X syndrome or Down syndrome were also tabulated. Potentially damaging point mutations were more common than potentially damaging structural variation, and the combined yield of potentially damaging variation was substantial at 28%. As more genes for autism are discovered, the yield will likely increase. Adapted from Mahjani et al.18
As sequencing continues in ASD, it is becoming possible to map the mechanisms associated with various gene mutations. For example, the vast majority of genes identified by genome-wide sequencing studies to date are acting through protein-truncating variants (PTVs), implying a loss-of-function (LoF) mechanism.10,12 This haploinsufficiency occurs in dosage-sensitive genes, where loss of one functional copy of the gene leads to reduced overall expression of the native protein. In such cases, increasing levels of the gene product should ameliorate the impact of the mutation. Other genes only show missense mutations, and in such cases, the mechanism is likely to be through gain of function (GoF), where the mutation confers altered function to the protein, leading to the ultimate phenotype. For example, KCNQ3 shows de novo missense variants modifying arginine residues in the voltage-sensing fourth transmembrane domain.10,12 For GoF mutations, it may be sufficient to block the formation of the mutant protein. Other psychiatric disorders with evidence of a significant role for rare deleterious variation include intellectual disability, attention deficit hyperactivity disorder, and obsessive-compulsive disorder;21, 22, 23 even later-onset disorders such as schizophrenia and bipolar disorder show evidence of a role for rare deleterious variation in some cases.24,25
With increased application of exome sequencing in clinical care, it is likely that rare genetic variants will be identified with increased frequency in clinical cohorts, leading to mechanistic insights about the etiology of psychiatric disorders affecting those individuals and paving the way for targeted gene-based therapeutic approaches to specific patient subsets. Given the promise of gene-based therapeutics to address deficits in the genetic architecture of neurodevelopmental and psychiatric conditions, it is necessary to consider how novel gene-based therapeutics could be evaluated clinically for efficacy in a safe and ethical way.
Current state of strategies and tools in gene-targeted therapies
Gene-based therapeutic strategies can be broadly categorized into two classes. The first strategy is in vivo gene therapy, in which therapeutic agents are given directly to humans to treat disease. The second category is ex vivo therapy, in which extracted human cells are genetically modified, expanded, and returned to the patient, where those modified cells function as a living treatment. Although ex vivo therapy has been used for some neurological disorders such as metachromatic leukodystrophy,26 its utility for treating ASD and other neurodevelopmental disorders remains to be explored. Five approaches for gene-based therapies are actively being pursued: (1) gene replacement—delivery of a functional gene copy to replace the faulty endogenous one, (2) gene addition—overexpression of exogenous or endogenous genes, (3) gene silencing—prevention of endogenous gene expression, (4) gene editing—gene modulation through technologies that alter a nucleotide sequence in the genome directly, and (5) gene activation or deactivation by using small-molecular drugs or CRISPR and other technologies.27,28
There are three key areas to consider for in vivo gene-based therapy approaches: (1) vector, (2) therapeutic payload, and (3) safe and efficient routing to target cells and tissues. The vector employed for delivery can include viral- and non-viral-based approaches. For ex vivo gene modulation, lentiviruses or non-viral agents are used to modify the cells prior to re-infusion. For in vivo gene delivery, specific serotypes of adeno-associated viruses (AAVs) are most often used for CNS disorders based on their tropism that includes specific CNS cell types. Considerations for any gene delivery vector include the transduction efficiency and expression level required for efficacy, long-term stability, low immunotoxicity, and low genotoxicity (reviewed in29). For in vivo gene delivery, recombinant adeno-associated virus vectors (rAAVs) fulfill most of these criteria and are therefore the most advanced and the most widely used. Currently, there are more than 250 rAAV gene therapy drugs under clinical development, representing approximately 28% of all clinical trials that use viral vectors for gene therapy.27,28 CNS-targeted applications account for approximately 20% of these trials. A rAAV is comprised of a single-stranded (ss) or double-stranded (ds, self-complementary and thus half-sized) DNA genome packaged in a viral capsid. The specific capsid is critical because it directs tissue tropism and intracellular trafficking, which may dictate many host immune responses. Therefore, significant effort is directed at capsid discovery during the vector development process. There are four main strategies for capsid discovery: (1) naturally occurring vectors from humans and other organisms,27,28 (2) directed evolution of extant serotypes,30, 31, 32, 33, 34 (3) rational design from evolved targeting ligands or known ligands,31,35,36 and (4) in silico design.37 Additionally, machine learning can be applied to each approach to generate further variations.38,39
The composition of the DNA payload in a rAAV confers the therapeutic benefit, determines long-term stability in transduced cells, and sometimes triggers innate and adaptive transgene-related immunities. There are numerous aspects of the DNA payload that must be carefully designed to minimize potential toxicity. Engineering the vector genome includes consideration of the inverted terminal repeats (ITRs), which are the only viral sequences remaining in rAAVs. ITRs guide vector genome replication and packaging. Selection of a modified ITR can mediate the generation of a self-complementary vector DNA that bypasses second-strand synthesis and achieves early-onset high gene expression.40,41 However, it also halves the DNA cargo size limit from 5.0 kb to 2.5 kb, imposing a limit on the size of potential therapeutic genes. Promoters and enhancers that regulate transcription can be engineered to meet specific therapeutic needs, such as sustained, regulated, tissue-specific, or cell-specific transgene expression.42, 43, 44 Alternatively, post-transcriptional modification can fine-tune transgene expression. For example, incorporating binding sites for tissue- or cell-type-specific endogenous micro-RNAs (miRNAs) in the transgene can de-target expression from a tissue and therefore reduce off-target transgene expression.45, 46, 47, 48, 49 Finally, complementary DNA (cDNA) can be modified for higher transgene expression, such as codon optimization. However, codon optimization is most effective when accompanied by a reduction of CpG content, which enhances transgene stability by reducing immunogenicity.50,51 Where possible, the availability of relevant pre-clinical models is key to study efficacy and safety. While safety can be measured by relatively standard outcomes, efficacy measurements need to be tailored to each disease and ideally should target clinically relevant phenotypes such as pathological changes, neurological functions, and quality and longevity of life.
Several recent reviews have addressed challenges common to many gene therapy programs, such as navigating the immune response after delivery, AAV manufacturing, and the necessity of collaborative networks for rare diseases.27, 28, 29,51,52 In this workshop, we focused on several challenges that are especially important for neurodevelopmental psychiatric diseases: gene dosage, biodistribution to the CNS, timing of treatment, and access to reliable natural history data and outcome measures (Table 1).
Table 1.
Recommendations to advance gene therapy in neurodevelopmental psychiatric disorders
Special challenge: Gene dosage
Special challenge: Delivery to the CNS
Special challenge: Timing of intervention and therapeutic windows of opportunity
Special challenge: How to measure impact and success
|
Special challenge: Gene dosage
Haploinsufficiency is a common genetic cause of neurodevelopmental and psychiatric disorders,53 and treatment can be challenging because of the Goldilocks effect: i.e., either too little or too much gene expression is detrimental,54 necessitating precise control of therapeutic-transgene expression. Potential therapeutic approaches include cDNA gene augmentation,55 direct mutation repair by DNA/RNA editing,56 boosting expression of the mutation-free allele by CRISPR activation (CRISPRa),57 or antisense oligonucleotides (ASOs),58 and readthrough therapy for nonsense mutations by small-molecule compounds or suppressor tRNAs.59,60
Gene therapy for MeCP2: An example for feedback regulation of dose-sensitive genes
A major confounding variable for gene replacement therapy of many neurodevelopmental psychiatric disorders is dose sensitivity to the target-gene product. One can see this effect with certain genetic disorders in humans where loss of function of a gene causes one disorder, whereas duplication of that same gene causes a different, but related disorder. A few examples of this are Rett syndrome and MeCP2 duplication syndrome (MECP2), Angelman syndrome and 15q11-q13 duplication syndrome (UBE3A), and Phelan-McDermid syndrome and 22q13 duplication syndrome (SHANK3). As mentioned above, these conditions reflect what is commonly referred to as a Goldilocks scenario, where either too little or too much expression produces a disease phenotype, and just the right level of gene expression is necessary for typical function.
Rett syndrome is an X-linked neurodevelopmental disorder primarily affecting girls. In classical Rett syndrome, children have apparently normal early psychomotor development followed by profound developmental stagnation and regression after the age of 6 months, with loss of fine motor skills and language, and acquisition of hand stereotypies and autistic behavior. Later in the disease, other clinical manifestations such as seizures, respiratory and autonomic dysfunction, gastrointestinal disease, anxiety, and sleep disorders become prominent.
Properly regulating transgene expression within a tight developmental time window for maximum therapeutic benefit is a daunting obstacle for any gene transfer approach. Rett syndrome provides a clear example of these challenges, and a new technical innovation was developed for MeCP2 gene therapy to begin to address this issue. Rett syndrome is caused by pathogenic mutations in MeCP2 (i.e., loss of function), whereas MeCP2 duplication syndrome results from partial chromosomal duplications that include the MeCP2 gene (NCT02738281). Mecp2 knockout (KO) mice as well as transgenic mice that overexpress Mecp2 each have severe disease phenotypes,61 demonstrating the clear dose sensitivity of MeCP2. Adding to the difficulty of any gene replacement strategy is the fact that MeCP2 is an X-linked gene, and the vast majority of Rett syndrome patients are heterozygous females. As such, they are mosaics for cells expressing MeCP2 due to random X chromosome inactivation, with approximately 50% of cells expressing wild-type (WT) MeCP2 and 50% expressing the mutant MeCP2. In a perfect scenario where one copy of MeCP2 is delivered to each cell and each transgene is expressed at WT levels, the cells of the patient expressing the mutant MeCP2 gene would be corrected, but the cells expressing the WT MeCP2 gene would be pushed to a MeCP2 duplication state, an undesirable clinical outcome. The scenario of Rett syndrome and MeCP2 dose sensitivity exemplifies a common problem with many neurodevelopmental psychiatric disorders that involve dose-sensitive genes and the need to regulate transgene expression on a cell-by-cell basis.
A potential solution to properly regulating a MeCP2 transgene includes general strategies described over a decade ago that take advantage of miRNA-based regulation of mRNA levels for gene therapy applications.49 For MeCP2, miRNA-mediated modulation was designed to be dose responsive by taking advantage of miRNAs whose expression levels are responsive to the transgene being expressed. The basic approach is the inclusion of miRNA-binding sites to miRNAs that fluctuate in response to MeCP2 levels to repress or permit MeCP2 transgene expression. For MeCP2 gene therapy, a panel of miRNA-binding sites for a set of MeCP2-responsive miRNAs (termed collectively as miRNA-responsive autoregulatory element, or miRARE) were engineered into the 3′ untranslated region (UTR) of the exogenously expressed MeCp2 mRNA. When an AAV/MeCP2-miRARE vector transduced a WT cell, expression would be suppressed because MeCP2 overexpression would elevate repressive miRNAs that would temper that overexpression. However, transduction of a MeCP2 KO cell would permit transgene expression until normal levels were reached. The miRARE element would also be expected to prevent overexpression of the MeCP2 transgene if too many vector genomes entered a single cell. In this manner, the element would be expected to normalize variable transfection efficiencies as well as endogenous gene activities. Supporting the utility of the miRARE regulation for MeCP2, Sinnett et al.62 found that intrathecal administration of an AAV9/miniMeCP2-miRARE vector at 4–5 weeks old could extend survival of MeCP2 KO mice. It is also encouraging that whereas a MeCP2 vector without miRARE induced adverse behavioral phenotypes and some premature death in WT mice, AAV9/miniMeCP2-miRARE was well tolerated in these animals.
One caveat to this approach is that the miRNA-binding sites incorporated in miRARE are over-represented in the 3′ UTRs of numerous dose-sensitive genes involved in neurodevelopmental disorders, including MeCP2, UBE3A, TCF4, ATRX, and MEF2C.62 Thus, if the miRNAs are sponged away from their normal targets, modest increases in these neurodevelopmental genes may occur, which could be consequential. Thus, appropriate titration is important.
Angelman syndrome: ASO-based gene reactivation as an example of harnessing intrinsic transcript regulation as a therapeutic target
Angelman syndrome (AS) is a neurodevelopmental disorder associated with severe intellectual disability and expressive language deficits. Most individuals with AS are non-verbal and experience very delayed motor skills, ataxia, tremor, laughing spells, seizures, sleep problems, excessive sociability, and behavior problems, including separation anxiety, hyperexcitability, and aggression.63 AS results from defects in the maternally derived UBE3A gene, a gene imprinted exclusively in the brain. Normally in brain tissue, the paternally derived copy of UBE3A is silenced, and only the maternal UBE3A gene is expressed. Defects in UBE3A can include maternal deletion of the 15q11–13 region (∼50%–60%), uniparental disomy (UPD) for the paternal chromosome 15 (∼20%), an imprinting center (IC) mutation on the maternal chromosome (<10%), or a mutation in the maternally derived copy of UBE3A, which renders it non-functional (20%). Individuals diagnosed with AS who have UPD, IC, and some UBE3A mutations tend to be higher functioning compared with those with maternal deletion of the 15q11–13 region, and they may be able to speak a few words, have fewer seizures, and develop better motor function.64
Typically, in the brain, an antisense transcript (UBE3A-ATS) induced by the imprinting center in 15q11–13 binds to UBE3A and blocks transcription of the paternal UBE3A sequence. Expression of UBE3A-ATS is repressed on the maternal chromosome, allowing the maternal copy of UBE3A to be transcribed. Thus, if a defect in the maternal UBE3A gene is present, there is insufficient production of UBE3A, resulting in AS. In a key study in the UBE3A-deficient mouse model, ASOs were designed to absorb the UBE3A-ATS transcript that is responsible for silencing the paternal allele and thereby decease its repressive activity. After ASO infusion, de-repression of the paternal allele occurred, resulting in increased UBE3A production in neurons throughout the brain.65 Strategies for genetic reversal of imprinted alleles for AS using ASOs are in development by GeneTx, Roche, and Biogen/Ionis (NCT04259281, NCT04428281). The initial ASO to be used in humans with AS (GTX-102) targets an exon of the anti-sense transcript that is identical across humans and non-human primates (NHPs), thus allowing pharmacodynamic studies in a NHP model, which showed that GTX-102 treatment could reduce UBE3A-ATS and increase UBE3A protein.
In the Phase 1/2 open-label trial with intrathecal administration of GTX-102, children and adolescents diagnosed with AS ages 4–17 years who carried a maternal UBE3A deletion were treated in multiple cohorts with ascending doses. Five patients were treated with cumulative doses ranging from 20 to 105.3 mg before a serious adverse event of acute inflammatory polyradiculopathy occurred, resulting in leg weakness in all patients and inability to stand in two of them, causing the study to be put on clinical hold. All patients recovered over several months when the drug was discontinued. These adverse events are uncommon for ASOs and probably reflect an adverse effect related to the sequence and/or chemistry of this specific ASO. Previous studies had documented sequence-specific pro-inflammatory effects of phosphorothioate-modified ASOs.66,67 However, the polyradiculopathy in the case of GTX-102 does not appear to be immune mediated and is likely due to direct local toxicity from the higher concentration near the injection site (E. Berry-Kravis, personal communication). Despite these adverse events, the treated patients showed surprising rapid improvements, even at the lowest doses. All five participants (ages 5–15 years) showed clinical improvement 4.5 months after the baseline dose, with a mean clinical global impression (CGI-AS) increase of +2.4 (between much and very much improved), and all patients were much improved or better in at least two domains (domains evaluated were sleep, behavior, communication, fine motor, and gross motor). The youngest patients (ages 5–6 years) were the most improved. Improvements were confirmed on multiple measures including parent diaries, Bayley expressive/receptive communication scales, Observer-Reported Communication Ability (ORCA) scale, the Vineland Adaptive Behavior scales, an Actimyo device and electroencephalogram (EEG) delta power, epileptiform discharges and notched delta, all characteristic findings on AS EEG which moved toward normal values in the study. Improvements included acquisition of vocabulary, signs, and gestures; better use of augmentative and alternative communication devices; better ability to respond, follow commands, and focus on tasks; and acquisition of capabilities such as self-feeding with a fork, learning to swim on their own, catching and throwing a ball, improved gait (narrower base, more stable), and posture, allowing the child to walk up hills, walk on sand without falling, step up on curbs, stop and turn better, and walk faster and farther. GTX-102 is planned to be further tested using lower doses that are intended to avoid the polyradiculopathy. The early pilot data suggest that the use of ASOs to activate the paternal UBE3A gene could be successful in improving development in patients with AS.
Special challenge: Delivery to the CNS
Biodistribution of therapeutics in the CNS
AAV9 has been used most often for CNS gene transfer, initially due to its ability to cross the blood-brain barrier (BBB) in various animal models as well as in NHPs.68,69 However, there are significant barriers to using systemic delivery of the vector to achieve CNS tropism in NHPs and eventually in humans, including reduced CNS and high peripheral tissue tropism in NHPs and humans,70,71 loss of transduction efficiency as well as a change in tropism with increased age of the host, and the high prevalence and titer of neutralizing antibodies (NAbs) against AAV9 in NHPs and potentially in adult humans.69,72 Therefore, identifying novel AAVs that have improved CNS cell tropism after direct or peripheral delivery is a major ongoing effort.27,30,31,73, 74, 75
As the field of gene therapy expands to include more complex diseases like neurodevelopmental psychiatric disorders, AAV gene therapy design and testing has, in turn, become more challenging. One major challenge of AAV gene therapy, not exclusive to neuropsychiatric disorders, is achieving appropriate expression in the correct cell types in the brain. In the ideal situation, one aims to express the endogenous levels of a gene/protein in every cell throughout different stages of development. Autopsy studies of children receiving AAV9 therapy via the intravenous (i.v.) route for spinal muscular atrophy (SMA) have shown broad CNS distribution of both viral genomes and transgene.70 While these data are very encouraging, the translation of AAVs to other CNS disorders is being met head on with newer capsids developed through various engineering approaches,71,76, 77, 78, 79, taking advantage of various routes of delivery. The gene therapy community is actively trying to improve the design of vectors for appropriate expression in specific cell types, discover miRNAs to de-target expression from unwanted cells,48,49,79,80 and engineer capsids with increased tropism for the brain and/or specific CNS cell types (reviewed in52,71,78,81,82). Although the AAV gene therapy field will likely have a toolkit of super capsids and highly selective promoters in the future, at present it is restricted to partial gene transfer in some brain regions but without broad transduction in cerebrum and cerebellum and often lacking gene transfer in deep brain areas such as the caudate and putamen. Fortunately, this level of expression has been sufficient to achieve transformative outcomes that led to the approval of Zolgensma for SMA83 with early results in first-in-human gene trials for giant axonal neuropathy (GAN), mucopolysaccharidosis type IIIA (MPS IIIA), and GM1 gangliosidosis (GM1-G) (Clinicaltrials.gov IDs NCT02362438 [l.i.t.], NCT04360265 [i.v.], and NCT03952637 [i.v.], respectively). The initial data from the GAN clinical trial indicate that the therapy is safe and well tolerated and that it shows promising efficacy indicating that disease progression may be slowed and it may improve regeneration of nerve fibers. Data from the MPS IIIA and GM1-G trials have not yet been released.
One of the largest challenges associated with i.v. CNS gene transfer is the non-uniform and generally sparse nature of gene transfer to CNS, especially in deep-brain structures.69,84 In pre-clinical models, AAV9 and AAVrh10 i.v. administration consistently provided only modest transduction of the forebrain compared with the hindbrain (cerebellum, brain stem) and spinal cord.85,86 For neurological disorders like SMA, where the disease features localize to motor neurons in the spinal cord, i.v. delivery with these capsids has the potential to drive transformative therapeutic outcomes. For disorders with CNS and systemic manifestations, such as GM1 and the MPS, i.v. delivery of AAV9 may be beneficial because it provides a therapy for central and peripheral disease. Given this, it is notable that clinical trials for MPS IIIA have been more modest, with efficacy reported at higher doses. However, higher doses have been associated with undesirable innate immune responses and dramatic adverse events.87,88 Additionally, pre-existing AAV NAbs may render i.v. AAV gene therapy ineffective in some patients (∼40%–50%), although methods to overcome this issue have been recently reported.89
Cerebrospinal fluid (CSF) delivery is an attractive alternative to i.v. delivery because CSF surrounds and penetrates the brain via the Virchow-Robin spaces of the glymphatic system. There are three major CSF delivery points: lumbar intrathecal (l.i.t.), cisterna magna (i.c.m.), and intracerebroventricular (i.c.v.) injection. L.i.t. delivery is the least invasive route and can be performed as an outpatient procedure without the need for anesthesia. The downside of l.i.t. delivery is the potential for reduced distribution to the brain due to physical distance;90,91 i.c.v. injection is more invasive than l.i.t. or i.c.m. but is considered safe because lateral ventricular catheterization is a routine procedure as the standard of care for hydrocephalus (reviewed in92). I.c.v. injection exposes the choroid plexus and ependyma to AAV, and with the correct capsid, these cells may be transduced and produce therapeutic proteins for the CNS.93 Additionally, the AAVs can penetrate throughout the CNS following CSF flow routes.94 Depending on the vector serotype or engineered capsid, this method can provide broad coverage or selective transduction of cells lining the ventricular system; i.c.m. injection can also achieve improved biodistribution to the brain in large animals, but requires technical expertise given its proximity to the brainstem. Notably, safe i.c.m. injections using a soft intravascular microcatheter have recently been trialed (NCT04669535).84,95
Although CSF delivery seems to be able to largely circumvent pre-existing Nabs,96 CSF delivery is not without deleterious immune responses, which limits usable dosages.97 Some recent mouse studies using direct parenchymal injection of AAV have reported neuronal toxicity,98,99 but in most models using CSF delivery, the major concern has been the dorsal root ganglia (DRG) toxicity, with reports of toxicity in pre-clinical studies (pigs, NHPs) and more recently in a patient.100, 101, 102 CSF turns over about five times per day, and the outflow is largely along cranial and spinal nerve roots. This combined with DRG exposure to general circulation (fenestrated capillaries and no BBB) illustrates that DRGs are inherently exposed to extraordinarily high vector doses.103,104 This likely contributes to DRG toxicity, which has been hypothesized to be caused by a combination of strong AAV tropism and supraphysiologic expression of the transgene encoded in the AAV.100,101 Therefore, appropriate dosing, cautious evaluation of DRGs in pre-clinical studies, and diligent evaluation of patients enrolled in clinical trials are warranted. Additional measures, such as engineering AAVs30 to avoid DRGs or reducing expressing within DRGs are under development.46
Special challenge: Timing of intervention and therapeutic windows of opportunity
In addition to distribution challenges, the timing of intervention in neurodevelopmental disorders is another critical consideration. Several animal models of genetic disorders have clearly demonstrated that there are likely to be critical periods for treatment initiation such that treating early in life can rescue more deficits that treating later in life whether the invention is behavioral, pharmacological, or by gene-induction.105, 106, 107 Similarly, pre-clinical and clinical data strongly suggest that earlier intervention with AAV treatment results in better outcomes. As was suggested in pre-clinical studies in a mouse model,108 high-dose, systemic delivery of a gene therapy in the AAV9 vector ABT-001 to MPS IIIA patients improved outcomes in young patients compared with advanced-stage patients.109 These findings may represent higher efficacy of intervention earlier in the disease and/or better vector biodistribution in younger patients. Intervention at an early, specific time point is likely to be a key factor for maximal outcome in other rapid-onset diseases. This hypothesis has been supported by observations of substantial benefits in survival, motor function, and developmental milestone achievements relative to natural history, which were particularly striking for several SMA patients treated at younger ages.83 In addition to being more efficacious, dosing younger patients may also increase the safety profile, particularly for i.v. administration of AAV. A recent study reported the deaths of 4 of 17 patients treated with systemic high doses of an AAV8 gene therapy for X-linked myotubular myopathy (XLMTM), AT132. The patients who died were the oldest and heaviest, which, combined with a more advanced disease state and XLMTM-related underlying cholestatic liver disease, proved to be a fatal combination (NCT03199469) and highlights the need for possible dose reduction and/or liver de-targeting by using more specific capsids. To maximize both safety and efficacy, determining the appropriate therapeutic window, route, and AAV dose will be critical.
Gene replacement in recessive neurological diseases: Spinal muscular atrophy and giant axonal neuropathy
SMA occurs due to mutations in SMN1, resulting in reduced SMN1 protein levels in spinal cord motor neurons, causing neuronal death and muscle weakness. Interestingly, humans have a duplicated paralog of SMN1, known as SMN2, that is appropriately spliced approximately 10% of the time to a full-length SMN2 protein that can complement SMN1 deficiency. Humans have variation in the number of copies of SMN2, which imparts a range in the level of protection, with more copies being more protective.110 Adrian Krainer and colleagues worked with Ionis Pharmaceutics to develop anti-sense oligonucleotides that promote exon 7 inclusion in SMN2 transcripts, which increases the levels of full-length SMN2 to compensate for SMN1 deficiency. Approved in 2016, this ASO, Spinraza (Biogen), is administered intrathecally and has been shown to increase survival and motor function of infants with SMA.111 Notably, this was among the first FDA-approved nucleic acid therapies for a neurological disease. Another approach takes advantage of gene replacement strategies. Jerry Mendell and colleagues at Nationwide Children’s Hospital, working with the company Avexis (now Novartis Gene Therapies), developed a gene transfer approach to treat SMA. SMN1 cDNAs driven by a ubiquitous promoter and packaged into AAV9 serotype vectors were administered intravenously to infants with SMA. Open-label trials showed the safety and efficacy of this approach.83,112 Similar to pre-clinical studies, the timing of delivery was essential, with early treatment of symptomatic infants before 6 months or treatment of asymptomatic infants showing the best neurological outcomes.83 This AAV9-mediated SMN1 gene therapy (known as Zolgensma or onasemnogene abeparvovec-xioi) was approved by the FDA in 2019. Finally, Roche has also developed a small molecule that enhances the inclusion of exon 7 in SMN2 transcripts, resulting in the 2020 FDA approval of the third therapeutic strategy to treat SMA.113
Giant axonal neuropathy (GAN) is a rare pediatric neurodegenerative, hereditary neuropathy affecting the central and peripheral nervous systems. Recessive GAN mutations cause dysfunction of gigaxonin, a cytoskeletal-regulatory protein, leading to progressive sensorimotor and optic neuropathy, CNS involvement and respiratory failure with death by the second to third decade. The first-in-human intrathecal AAV9-mediated gene transfer trial for GAN (NCT02362438) is a single site, phase I, non-randomized, open-label dose escalation study. Fourteen subjects have been dosed at four dose levels (i.e., 0.35, 1.2, 1.8, and 3.5 × 1014 vector genomes (vg)/patient) with scAAV9-JeT-GAN, with follow-up data as far as 36 months post-gene transfer. Interim safety and efficacy analysis at the 1.8 × 1014 vg/patient dose level supports the safety and therapeutic potential of an intrathecal gene-therapy approach using AAV9 to target the CNS as well as allow for its limitations be considered, such as to the optimal timing of intervention in a neurodegenerative disease. Analysis at the 3.5 × 1014 vg level is ongoing (C. Bonnemann, personal communication).
Although still ongoing and thus preliminary, conclusions relevant to CNS-directed gene therapy of scAAV90JeT-GAN include that i.t. gene transfer is feasible, that i.t. administration with Trendelenburg positioning (i.e., the participant supine on the table with their head angled down) during and after i.t. infusion helps distribution along the neuraxis and that peripheral anti-AAV neutralizing antibodies at baseline are not an exclusion criterion for i.t. dosing as there are no detectable i.t.-neutralizing antibodies. However, after i.t. transduction, a robust anti-capsid immune response as evidenced by neutralizing antibodies, is elicited including in the i.t. space by 3 months post-dosing and likely limiting possible redosing; i.t. gene transfer using AAV9 can be done safely at the doses explored thus far. A clinically asymptomatic dose-dependent CSF pleocytosis evident at 3 months after dosing can be suppressed effectively with extended steroid coverage. Using a T-cell -directed immune modulation protocol, both CRIM-positive and CRIM-negative patients can be dosed.
Similar to the SMA experience described earlier, treating a neurodegenerative disorder ultimately resulting in neuronal death means that there is a therapeutic window—which in a disorder like GAN means that the state of the disease is dependent on patient age relative to the age of onset and the length of the axon, with longer axons generally showing earlier manifestations. Thus, dosing as early as possible in the disease course would be expected to be the most beneficial.
Special challenge: How to measure impact and success
The importance of natural history and outcome measure studies
A major key to the development of gene therapy for rare diseases is the concept of a natural history study. A natural history study may be used exclusively for academic purposes to define and characterize the disease in question; it may also be used for therapeutic development to establish a comparator group to support clinical trials evaluating treatment in a rare disease. In some cases, natural history studies initiated by academicians are used later by sponsors as external comparators to support therapeutic approval, so the concepts are not mutually exclusive. Natural history studies are often conducted in the absence of a therapy under development to systematically collect data to understand the pathogenesis and/or progression of disease or to be the anticipated historical comparator for an open-label study for regulatory approval. It is important, however, to distinguish a natural history study from a simple registry. The former is generally a rigorous, longitudinal study with clearly defined endpoints, whereas the latter, in contrast, is often dependent exclusively on patient or caregiver input or collated medical records. Although both types of data sources have their place in understanding aspects of (particularly) rare disease, a simple registry is unlikely to provide the systematic details that would be useful for defining disease trajectory in the granular, longitudinal, and rigorous manner needed for either comprehensive understanding of disease course or serving as the basis for an external comparator group to support regulatory approval of novel therapeutics. A crucial element of natural history studies is the use and identification of assessable and clinically meaningful outcome measures that can be used to identify quantifiable endpoints for therapeutic clinical trials. Reliable outcome measures are needed to determine how many patients will be sufficient to power an interventional trial. There is a significant need for development of novel outcome measures for very young infants and cognitively impaired populations.
The purpose of a natural history study is to understand and quantify the key attributes of the disease, including but not limited to signs, symptoms, biological markers, and other parameters. Although the genetic abnormalities may have been identified and the biology of the disease partially studied, the normal course of the disease may not have been systematically studied and understood. Often, the knowledge of a disease as defined in the reported literature is subject to biases, including increased reporting of more severe forms of a disease, and prior reports often reflect antiquated standards of care. To evaluate therapeutic effects, particularly in a rare disease, it is essential to understand the details of the normal course of the disease for the entire range of the population captured by the study inclusion criteria and during the concurrent era reflecting modern standards of care. Once these have been established, meaningful, consistent, and predictable endpoints can be established for clinical trials. Natural histories should seek to understand psychosocial aspects of the disease to facilitate choosing a clinically significant endpoint that matters to the families and affected patients.114,115 For instance, while improvement of function and “cure” may be a satisfactory outcome for academics, stabilization of the disease may be considered a very desirable outcome for the patients and families. In considering outcomes such as reversal or stabilization of disease, the natural history study may—in the context of additional knowledge of disease pathophysiology—shed light on the critical issue of window for intervention (i.e., when is it possible to intervene to either improve or stabilize the disease course and when is it too late?). As a result, a natural history study can help to inform and define key inclusion and exclusion criteria that may be applied to a study to ensure enrollment of a population of patients most likely to benefit from the therapy; enrollment of such a population is critical for demonstrating the efficacy needed to allow for regulatory approval and potentially guide usage once approved.
Conducting natural history studies
Natural history studies can be conducted in a purely retrospective or purely prospective way, as well as in a combination of both ways. The most useful data are collected when the study is designed with systematic rigorous endpoints in a contemporaneous fashion to the interventional study. Practically, this means that a prospective natural history study should be commenced early in the treatment development process, ideally at the time that the investigational new drug (IND)-enabling work is done, if not before. This takes foresight and resources to execute; well-designed, prospective studies are expensive, and gene and other advanced therapies are often initially developed in academic institutions where the funding for such a study may not be available. Therefore, one of the key areas of focus for the gene therapy community, whether in academia or in industry, is how to initiate and fund these studies. For instance, if a therapy is licensed by a company at the point that it enters Phase I studies, it may be too late to feasibly collect the quantity and quality of prospective data that is needed to plan and inform the clinical study and endpoints. Patient advocacy groups and the National Institutes of Health (NIH) can play an essential role in supporting studies that generate high-quality natural history data.
For the reasons outlined above, it is important for those who are developing therapies to consider what is known about a disease’s natural history and what an appropriate comparator group would be for establishing efficacy very early in the clinical program. It is often helpful to engage experts in clinical drug development, ideally those who also have expertise and experience in planning and executing natural history studies. One of the key challenges facing the rare-disease community is that often several groups—be they academic investigators, patient advocacy groups, or biopharma sponsors—are independently and in parallel working to establish separate natural history cohorts. Aggregating and sharing natural history data would be a very powerful approach, saving precious time and resources. It is therefore incumbent upon all key stakeholders—the patient community, academic investigators, biopharma sponsors, and regulators—to put in place appropriate incentives to ensure aggregation and sharing of natural history data. Several for-profit companies, as well as some non-profit consortia, have emerged over the past few years in an attempt to fill this need; the ability of these various efforts to deliver on the goal of generating rigorous and comprehensive natural history data will be closely watched.
It is also important to recognize that not all prospective natural history data (e.g., data collected as part of academic studies) may be sufficient or appropriate to serve as an external comparator group for a treatment study. Specifically, the natural history dataset may not be easily matched to the population, outcome measures, symptoms, and characteristics that would ideally be collected in the clinical trial to establish efficacy and safety data appropriate for regulatory approval. Although the data might be invaluable, it might have several limitations in forming the basis for a regulatory approval. As mentioned above, one of the most critical features of natural history data that allows for use as an external comparator is that the data must be either contemporaneous with the data collected in the interventional study or collected in exactly the same way. Of note, standards of care change over time, and these changes may have an important effect on the course of the disease. For example, the routine use of gastrostomy tubes (g-tubes) for feeding may have changed the survival age for children with many diseases. The use of feeding tubes may then lead, for example, to achievement of different (additional) development milestones, and because milestone achievement is often used as a clinical endpoint in therapeutic studies, the changes in standards of care may affect patient outcome rather than the therapy. An additional limitation of these studies may be the sparsity of data at key time points or identifying patients that have more than one clinical visit. Population modeling of these data is therefore quite difficult, further complicating the ability to establish the usual course of the disease. Although it may produce some useful data, a retrospective literature search magnifies these effects: data are often not collected in a systematic way but rather could be limited to usual clinical observations, and some important data points may not be included at all. There are large prospective, longitudinal natural history studies currently ongoing in several genetic neurodevelopmental psychiatric disorders, and they will provide crucial unmet clinical trial readiness needs (e.g., NCT02461459, NCT02461420, and NCT02461446).
Key regulatory considerations
Rare and ultra-rare diseases provide some unique considerations from a regulatory standpoint. For any product to be approved, data need to be presented to the FDA, or other regulatory agencies, that clearly demonstrate both the safety and the efficacy of the product under consideration. Since the datasets are usually very small and it is not possible to conduct a rigorous randomized, controlled study in most cases, regulatory bodies including the FDA have expressed openness to the idea of performing open-label studies and using externally controlled data where natural history studies form the basis of this external comparator group. However, for clear conclusions to be drawn from this comparison, there are some specifics that should be met from the comparator data including:
-
•
The dataset should be robust enough to be able to clearly follow the normal course of the disease, based on a sufficient number of patients with systematically collected data;
-
•
The data should be contemporaneous with the clinical study to exclude changes in standard of care;
-
•
The primary endpoint that is chosen should be demonstrated as something obvious that without therapy these patients would not achieve, such as the attainment of a development milestone (e.g., sitting without assistance for an infant or breathing without a ventilator);
-
•
The safety profile of the product should be differentiated from a possible adverse event that is part of the normal course of the disease (e.g., the development of seizures).
Clearly, meeting all these criteria is very difficult using solely either a retrospective or a literature-based study. This does not mean that these data are not helpful, and in fact they may be all the data that are available, particularly in a rare disease. However, this does make it much more difficult to demonstrate specific effects that regulators would be looking for.
In summary, when one is considering the use of a natural history study as an external comparator group for therapeutic development, it is important to think about the need for such a study very early on in the development process. If a survey of the existing literature indicates that there is such a need, it is critical to plan a rigorous, longitudinal natural history study and, equally important, to involve the patient’s voice in this planning process to ensure that the endpoints chosen reflect issues and concepts that are meaningful to patients and caregivers. The limitations of the natural history as a comparator should be considered when one is designing a registrational program for a product, and an early review with the regulatory agencies should be considered an essential part of the program.
Acknowledgments
The workshop was organized by NIMH. The views expressed in this workshop review are those of the individual authors and do not represent the views of the NIH.
Author contributions
All the authors contributed to writing parts of the manuscript and edited the final version. G.G., M.S., B.L.D., and S.T.-W. were co-chairs of the workshop; B.D.L., T.R.F., C.T., and S.T-W. chaired the discussion panels. All authors contributed to the development and implementation of the workshop. A list of workshop and working group participants is detailed in Table S1.
Declaration of interests
B.L.D. serves an advisory role and/or receives sponsored research support from Homology Medicines, Saliogen Therapeutics, Patch Bio, Moment Bio, Panorama Medicines, Resilience, Spirovant Sciences, Novartis (NBIR), Roche, and Sanofi. G.G. is a scientific co-founder of Voyager Therapeutics and Aspa Therapeutics and holds equity in these companies. G.G. is an inventor on patents with potential royalties licensed to Voyager Therapeutics, Aspa Therapeutics, RegenxBio, and other biopharmaceutical companies. A.M.B. is a beneficiary of a licensing agreement with Axovant Gene Therapies (royalties) and is an inventor on a patent pending related to a GALC Vector: Optimized GALC Genes and Expression Cassettes and Their Use (PCT/US2019/067727). J.D.B. is a consultant for BridgeBio Pharma and received sponsored research support from Takeda California, Inc. G.R.C. is a previous employee of Sio Gene Therapies. S.J.G. received royalty income from Abeona Therapeutics and Taysha Gene Therapies. R.J.K. is an employee of Biogen. A.S. is an employee and shareholder of BridgeBio Pharma. H.Y.Z. is co-founder of Cajal Neuroscience, on the Board of Directors of Regeneron Pharmaceuticals, Inc., the Scientific Advisory Board of The Column Group, and the Institutional Advisory Board of VIB. T.R.F. is a paid consultant for Ferring Ventures, SA. S.T.-W. is an employee of Novartis. M.S. received sponsored research support from Novartis, Biogen, Astellas, Aeovian, Bridgebio, and Aucta and has served on scientific advisory boards for Novartis, Roche, Regenxbio, SpringWorks Therapeutics, Jaguar Therapeutics, and Alkermes.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.05.014.
Contributor Information
Beverly L. Davidson, Email: davidsonbl@chop.edu.
Guangping Gao, Email: guangping.gao@umassmed.edu.
Mustafa Sahin, Email: mustafa.sahin@childrens.harvard.edu.
Supplemental information
References
- 1.Bai D., Yip B.H.K., Windham G.C., Sourander A., Francis R., Yoffe R., Glasson E., Mahjani B., Suominen A., Leonard H., et al. Association of genetic and environmental factors with autism in a 5-country cohort. JAMA Psychiatry. 2019;76:1035–1043. doi: 10.1001/jamapsychiatry.2019.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tick B., Bolton P., Happe F., Rutter M., Rijsdijk F. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J. Child. Psychol. Psychiatry. 2016;57:585–595. doi: 10.1111/jcpp.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hebebrand J., Scherag A., Schimmelmann B.G., Hinney A. Child and adolescent psychiatric genetics. Eur. Child. Adolesc. Psychiatry. 2010;19:259–279. doi: 10.1007/s00787-010-0091-y. [DOI] [PubMed] [Google Scholar]
- 4.Visscher P.M., Yengo L., Cox N.J., Wray N.R. Discovery and implications of polygenicity of common diseases. Science. 2021;373:1468–1473. doi: 10.1126/science.abi8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J. 10 Years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vicente C.T., Revez J.A., Ferreira M.A.R. Lessons from ten years of genome-wide association studies of asthma. Clin. Transl Immunol. 2017;6:e165. doi: 10.1038/cti.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Ercument Cicek A., Kou Y., Liu L., Fromer M., Walker S., et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall C.R., Howrigan D.P., Merico D., Thiruvahindrapuram B., Wu W., Greer D.S., Antaki D., Shetty A., Holmans P.A., Pinto D., et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh T., Poterba T., Curtis D., Akil H., Al Eissa M., Barchas J.D., Bass N., Bigdeli T.B., Breen G., Bromet E.J., et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature. 2022;604:509–516. doi: 10.1038/s41586-022-04556-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaugler T., Klei L., Sanders S.J., Bodea C.A., Goldberg A.P., Lee A.B., Mahajan M., Manaa D., Pawitan Y., Reichert J., et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grove J., Ripke S., Als T.D., Mattheisen M., Walters R.K., Won H., Pallesen J., Agerbo E., Andreassen O.A., Anney R., et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y., Peng M., Collins R., Grove J., Klei L., et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. e523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neri G., Schwartz C.E., Lubs H.A., Stevenson R.E. X-linked intellectual disability update 2017. Am. J. Med. Genet. A. 2018;176:1375–1388. doi: 10.1002/ajmg.a.38710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 15.Doan R.N., Lim E.T., De Rubeis S., Betancur C., Cutler D.J., Chiocchetti A.G., Overman L.M., Soucy A., Goetze S., Autism Sequencing C., et al. Recessive gene disruptions in autism spectrum disorder. Nat. Genet. 2019;51:1092–1098. doi: 10.1038/s41588-019-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiner D.J., Wigdor E.M., Ripke S., Walters R.K., Kosmicki J.A., Grove J., Samocha K.E., Goldstein J.I., Okbay A., Bybjerg-Grauholm J., et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat. Genet. 2017;49:978–985. doi: 10.1038/ng.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klei L., McClain L.L., Mahjani B., Panayidou K., De Rubeis S., Grahnat A.C.S., Karlsson G., Lu Y., Melhem N., Xu X., et al. How rare and common risk variation jointly affect liability for autism spectrum disorder. Mol. Autism. 2021;12:66. doi: 10.1186/s13229-021-00466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahjani B., De Rubeis S., Gustavsson Mahjani C., Mulhern M., Xu X., Klei L., Satterstrom F.K., Fu J., Talkowski M.E., Reichenberg A., et al. Prevalence and phenotypic impact of rare potentially damaging variants in autism spectrum disorder. Mol. Autism. 2021;12:65. doi: 10.1186/s13229-021-00465-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srivastava S., Love-Nichols J.A., Dies K.A., Ledbetter D.H., Martin C.L., Chung W.K., Firth H.V., Frazier T., Hansen R.L., Prock L., et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 2019;21:2413–2421. doi: 10.1038/s41436-019-0554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manickam K., McClain M.R., Demmer L.A., Biswas S., Kearney H.M., Malinowski J., Massingham L.J., Miller D., Yu T.W., Hisama F.M., Directors A.B.o. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2021;23:2029–2037. doi: 10.1038/s41436-021-01242-6. [DOI] [PubMed] [Google Scholar]
- 21.Satterstrom F.K., Walters R.K., Singh T., Wigdor E.M., Lescai F., Demontis D., Kosmicki J.A., Grove J., Stevens C., Bybjerg-Grauholm J., et al. Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat. Neurosci. 2019;22:1961–1965. doi: 10.1038/s41593-019-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halvorsen M., Samuels J., Wang Y., Greenberg B.D., Fyer A.J., McCracken J.T., Geller D.A., Knowles J.A., Zoghbi A.W., Pottinger T.D., et al. Exome sequencing in obsessive-compulsive disorder reveals a burden of rare damaging coding variants. Nat. Neurosci. 2021;24:1071–1076. doi: 10.1038/s41593-021-00876-8. [DOI] [PubMed] [Google Scholar]
- 23.Cappi C., Oliphant M.E., Peter Z., Zai G., Conceicao do Rosario M., Sullivan C.A.W., Gupta A.R., Hoffman E.J., Virdee M., Olfson E., et al. De novo damaging DNA coding mutations are associated with obsessive-compulsive disorder and overlap with tourette's disorder and autism. Biol. Psychiatry. 2020;87:1035–1044. doi: 10.1016/j.biopsych.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganna A., Satterstrom F.K., Zekavat S.M., Das I., Kurki M.I., Churchhouse C., Alfoldi J., Martin A.R., Havulinna A.S., Byrnes A., et al. Quantifying the impact of rare and ultra-rare coding variation across the phenotypic spectrum. Am. J. Hum. Genet. 2018;102:1204–1211. doi: 10.1016/j.ajhg.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh T., Walters J.T.R., Johnstone M., Curtis D., Suvisaari J., Torniainen M., Rees E., Iyegbe C., Blackwood D., McIntosh A.M., et al. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat. Genet. 2017;49:1167–1173. doi: 10.1038/ng.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S., et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 27.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulcha J.T., Wang Y., Ma H., Tai P.W.L., Gao G. Viral vector platforms within the gene therapy landscape. Signal. Transduct Target. Ther. 2021;6:53. doi: 10.1038/s41392-021-00487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris J.A., Boshoff C.H., Schor N.F., Wong L.M., Gao G., Davidson B.L. Next-generation strategies for gene-targeted therapies of central nervous system disorders: a workshop summary. Mol. Ther. : J. Am. Soc. Gene Ther. 2021;29:3332–3344. doi: 10.1016/j.ymthe.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nonnenmacher M., Wang W., Child M.A., Ren X.Q., Huang C., Ren A.Z., Tocci J., Chen Q., Bittner K., Tyson K., et al. Rapid evolution of blood-brain-barrier-penetrating AAV capsids by RNA-driven biopanning. Mol. Ther. Methods Clin. Dev. 2021;20:366–378. doi: 10.1016/j.omtm.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidsson M., Wang G., Aldrin-Kirk P., Cardoso T., Nolbrant S., Hartnor M., Mudannayake J., Parmar M., Bjorklund T. A systematic capsid evolution approach performed in vivo for the design of AAV vectors with tailored properties and tropism. Proc. Natl. Acad. Sci. U S A. 2019;116:27053–27062. doi: 10.1073/pnas.1910061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabebordbar M., Lagerborg K.A., Stanton A., King E.M., Ye S., Tellez L., Krunnfusz A., Tavakoli S., Widrick J.J., Messemer K.A., et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell. 2021;184:4919–4938.e22. doi: 10.1016/j.cell.2021.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J., Chu K., Leborgne C., Dane A.P., Haft A., et al. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol. Ther. J. Am. Soc. Gene Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimm D., Buning H. Small but increasingly mighty: latest advances in AAV vector research, design, and evolution. Hum. Gene Ther. 2017;28:1075–1086. doi: 10.1089/hum.2017.172. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y.H., Chang M., Davidson B.L. Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat. Med. 2009;15:1215–1218. doi: 10.1038/nm.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan D., Buning H., Ling C. Rational design of gene therapy vectors. Mol. Ther. Methods Clin. Dev. 2019;12:246–247. doi: 10.1016/j.omtm.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landegger L.D., Pan B., Askew C., Wassmer S.J., Gluck S.D., Galvin A., Taylor R., Forge A., Stankovic K.M., Holt J.R., Vandenberghe L.H. A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat. Biotechnol. 2017;35:280–284. doi: 10.1038/nbt.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryant D.H., Bashir A., Sinai S., Jain N.K., Ogden P.J., Riley P.F., Church G.M., Colwell L.J., Kelsic E.D. Deep diversification of an AAV capsid protein by machine learning. Nat. Biotechnol. 2021;39:691–696. doi: 10.1038/s41587-020-00793-4. [DOI] [PubMed] [Google Scholar]
- 39.Ogden P.J., Kelsic E.D., Sinai S., Church G.M. Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science. 2019;366:1139–1143. doi: 10.1126/science.aaw2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarty D.M., Fu H., Monahan P.E., Toulson C.E., Naik P., Samulski R.J. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z., Ma H.I., Li J., Sun L., Zhang J., Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- 42.Rivera V.M., Gao G.P., Grant R.L., Schnell M.A., Zoltick P.W., Rozamus L.W., Clackson T., Wilson J.M. Long-term pharmacologically regulated expression of erythropoietin in primates following AAV-mediated gene transfer. Blood. 2005;105:1424–1430. doi: 10.1182/blood-2004-06-2501. [DOI] [PubMed] [Google Scholar]
- 43.Pacak C.A., Sakai Y., Thattaliyath B.D., Mah C.S., Byrne B.J. Tissue specific promoters improve specificity of AAV9 mediated transgene expression following intra-vascular gene delivery in neonatal mice. Genet. Vaccin. Ther. 2008;6:13. doi: 10.1186/1479-0556-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monteys A.M., Hundley A.A., Ranum P.T., Tecedor L., Muehlmatt A., Lim E., Lukashev D., Sivasankaran R., Davidson B.L. Regulated control of gene therapies by drug-induced splicing. Nature. 2021;596:291–295. doi: 10.1038/s41586-021-03770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie J., Xie Q., Zhang H., Ameres S.L., Hung J.H., Su Q., He R., Mu X., Seher Ahmed S., Park S., et al. MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Mol. Ther. : J. Am. Soc. Gene Ther. 2011;19:526–535. doi: 10.1038/mt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hordeaux J., Buza E.L., Jeffrey B., Song C., Jahan T., Yuan Y., Zhu Y., Bell P., Li M., Chichester J.A., et al. MicroRNA-mediated inhibition of transgene expression reduces dorsal root ganglion toxicity by AAV vectors in primates. Sci. Transl Med. 2020;12:eaba9188. doi: 10.1126/scitranslmed.aba9188. [DOI] [PubMed] [Google Scholar]
- 47.Xiao Y., Muhuri M., Li S., Qin W., Xu G., Luo L., Li J., Letizia A.J., Wang S.K., Chan Y.K., et al. Circumventing cellular immunity by miR142-mediated regulation sufficiently supports rAAV-delivered OVA expression without activating humoral immunity. JCI Insight. 2019;5:e99052. doi: 10.1172/jci.insight.99052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muhuri M., Zhan W., Maeda Y., Li J., Lotun A., Chen J., Sylvia K., Dasgupta I., Arjomandnejad M., Nixon T., et al. Novel combinatorial MicroRNA-binding sites in AAV vectors synergistically diminish antigen presentation and transgene immunity for efficient and stable transduction. Front. Immunol. 2021;12:674242. doi: 10.3389/fimmu.2021.674242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown B.D., Gentner B., Cantore A., Colleoni S., Amendola M., Zingale A., Baccarini A., Lazzari G., Galli C., Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 50.Wright J.F. Codon modification and PAMPs in clinical AAV vectors: the tortoise or the hare? Mol. Ther. : J. Am. Soc. Gene Ther. 2020;28:701–703. doi: 10.1016/j.ymthe.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muhuri M., Maeda Y., Ma H., Ram S., Fitzgerald K.A., Tai P.W., Gao G. Overcoming innate immune barriers that impede AAV gene therapy vectors. J. Clin. Invest. 2021;131:e143780. doi: 10.1172/JCI143780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 53.Parenti I., Rabaneda L.G., Schoen H., Novarino G. Neurodevelopmental disorders: from genetics to functional pathways. Trends Neurosci. 2020;43:608–621. doi: 10.1016/j.tins.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 54.Rice A.M., McLysaght A. Dosage sensitivity is a major determinant of human copy number variant pathogenicity. Nat. Commun. 2017;8:14366. doi: 10.1038/ncomms14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinnett S.E., Gray S.J. Recent endeavors in MECP2 gene transfer for gene therapy of Rett syndrome. Discov. Med. 2017;24:153–159. [PubMed] [Google Scholar]
- 56.Sinnamon J.R., Kim S.Y., Fisk J.R., Song Z., Nakai H., Jeng S., McWeeney S.K., Mandel G. In vivo repair of a protein underlying a neurological disorder by programmable RNA editing. Cell Rep. 2020;32:107878. doi: 10.1016/j.celrep.2020.107878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colasante G., Lignani G., Brusco S., Di Berardino C., Carpenter J., Giannelli S., Valassina N., Bido S., Ricci R., Castoldi V., et al. dCas9-Based Scn1a gene activation restores inhibitory interneuron excitability and attenuates seizures in dravet syndrome mice. Mol. Ther. J. Am. Soc. Gene Ther. 2020;28:235–253. doi: 10.1016/j.ymthe.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lim K.H., Han Z., Jeon H.Y., Kach J., Jing E., Weyn-Vanhentenryck S., Downs M., Corrionero A., Oh R., Scharner J., et al. Antisense oligonucleotide modulation of non-productive alternative splicing upregulates gene expression. Nat. Commun. 2020;11:3501. doi: 10.1038/s41467-020-17093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martins-Dias P., Romao L. Nonsense suppression therapies in human genetic diseases. Cell Mol Life Sci. 2021;78:4677–4701. doi: 10.1007/s00018-021-03809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J., Zhang Y., Mendonca C.A., Ren L., Liang J., Zhou C., Li J., Gao G., Wang D. In vivo delivery of suppressor tRNA overcomes a pathogenic nonsense mutation in mice. Mol. Ther. 2021;29:128. [Google Scholar]
- 61.Sandweiss A.J., Brandt V.L., Zoghbi H.Y. Advances in understanding of Rett syndrome and MECP2 duplication syndrome: prospects for future therapies. Lancet Neurol. 2020;19:689–698. doi: 10.1016/S1474-4422(20)30217-9. [DOI] [PubMed] [Google Scholar]
- 62.Sinnett S.E., Boyle E., Lyons C., Gray S.J. Engineered microRNA-based regulatory element permits safe high-dose miniMECP2 gene therapy in Rett mice. Brain. 2021;144:3005–3019. doi: 10.1093/brain/awab182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rotaru D.C., Mientjes E.J., Elgersma Y. Angelman syndrome: from mouse models to therapy. Neuroscience. 2020;445:172–189. doi: 10.1016/j.neuroscience.2020.02.017. [DOI] [PubMed] [Google Scholar]
- 64.Bird L.M. Angelman syndrome: review of clinical and molecular aspects. Appl. Clin. Genet. 2014;7:93–104. doi: 10.2147/TACG.S57386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meng L., Ward A.J., Chun S., Bennett C.F., Beaudet A.L., Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krieg A.M. Therapeutic potential of Toll-like receptor 9 activation. Nat. Rev. Drug Discov. 2006;5:471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 67.Bennett C.F., Baker B.F., Pham N., Swayze E., Geary R.S. Pharmacology of antisense drugs. Annu. Rev. Pharmacol. Toxicol. 2017;57:81–105. doi: 10.1146/annurev-pharmtox-010716-104846. [DOI] [PubMed] [Google Scholar]
- 68.Duque S., Joussemet B., Riviere C., Marais T., Dubreil L., Douar A.M., Fyfe J., Moullier P., Colle M.A., Barkats M. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol. Ther. : J. Am. Soc. Gene Ther. 2009;17:1187–1196. doi: 10.1038/mt.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Foust K.D., Nurre E., Montgomery C.L., Hernandez A., Chan C.M., Kaspar B.K. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomsen G., Burghes A.H.M., Hsieh C., Do J., Chu B.T.T., Perry S., Barkho B., Kaufmann P., Sproule D.M., Feltner D.E., et al. Biodistribution of onasemnogene abeparvovec DNA, mRNA and SMN protein in human tissue. Nat. Med. 2021;27:1701–1711. doi: 10.1038/s41591-021-01483-7. [DOI] [PubMed] [Google Scholar]
- 71.Meseck E.K., Guibinga G., Wang S., McElroy C., Hudry E., Mansfield K. Intrathecal sc-AAV9-CB-GFP: systemic distribution predominates following single-dose administration in cynomolgus macaques. bioRxiv. 2021 doi: 10.1101/2021.11.28.470258. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gray S.J., Matagne V., Bachaboina L., Yadav S., Ojeda S.R., Samulski R.J. Preclinical differences of intravascular AAV9 delivery to neurons and glia: a comparative study of adult mice and nonhuman primates. Mol. Ther. J. Am. Soc. Gene Ther. 2011;19:1058–1069. doi: 10.1038/mt.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin K., Zhong X., Li L., Ying M., Yang T., Zhang Z., He X., Xu F. AAV9-Retro mediates efficient transduction with axon terminal absorption and blood-brain barrier transportation. Mol. Brain. 2020;13:138. doi: 10.1186/s13041-020-00679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lukashchuk V., Lewis K.E., Coldicott I., Grierson A.J., Azzouz M. AAV9-mediated central nervous system-targeted gene delivery via cisterna magna route in mice. Mol. Ther. Methods Clin. Dev. 2016;3:15055. doi: 10.1038/mtm.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Macdonald J., Marx J., Buning H. Capsid-engineering for central nervous system-directed gene therapy with adeno-associated virus vectors. Hum. Gene Ther. 2021;32:1096–1119. doi: 10.1089/hum.2021.169. [DOI] [PubMed] [Google Scholar]
- 77.Kondratov O., Kondratova L., Mandel R.J., Coleman K., Savage M.A., Gray-Edwards H.L., Ness T.J., Rodriguez-Lebron E., Bell R.D., Rabinowitz J., et al. A comprehensive study of a 29-capsid AAV library in a non-human primate central nervous system. Mol. Ther. : J. Am. Soc. Gene Ther. 2021;29:2806–2820. doi: 10.1016/j.ymthe.2021.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goertsen D., Flytzanis N.C., Goeden N., Chuapoco M.R., Cummins A., Chen Y., Fan Y., Zhang Q., Sharma J., Duan Y., et al. AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset. Nat. Neurosci. 2021;25:106–115. doi: 10.1038/s41593-021-00969-4. [DOI] [PubMed] [Google Scholar]
- 79.Wang D., Li S., Gessler D.J., Xie J., Zhong L., Li J., Tran K., Van Vliet K., Ren L., Su Q., et al. A rationally engineered capsid variant of AAV9 for systemic CNS-directed and peripheral tissue-detargeted gene delivery in neonates. Mol. Ther. Methods Clin. Dev. 2018;9:234–246. doi: 10.1016/j.omtm.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dhungel B., Ramlogan-Steel C.A., Steel J.C. Synergistic and independent action of endogenous microRNAs 122a and 199a for post-transcriptional liver detargeting of gene vectors. Sci. Rep. 2018;8:15539. doi: 10.1038/s41598-018-33801-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu D., Zhu M., Zhang Y., Diao Y. Crossing the blood-brain barrier with AAV vectors. Metab. Brain Dis. 2021;36:45–52. doi: 10.1007/s11011-020-00630-2. [DOI] [PubMed] [Google Scholar]
- 82.Deverman B.E., Ravina B.M., Bankiewicz K.S., Paul S.M., Sah D.W.Y. Gene therapy for neurological disorders: progress and prospects. Nat. Rev. Drug Discov. 2018;17:641–659. doi: 10.1038/nrd.2018.110. [DOI] [PubMed] [Google Scholar]
- 83.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K., et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 84.Taghian T., Marosfoi M.G., Puri A.S., Cataltepe O.I., King R.M., Diffie E.B., Maguire A.S., Martin D.R., Fernau D., Batista A.R., et al. A safe and reliable technique for CNS delivery of AAV vectors in the cisterna magna. Mol. Ther. : J. Am. Soc. Gene Ther. 2020;28:411–421. doi: 10.1016/j.ymthe.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ballon D.J., Rosenberg J.B., Fung E.K., Nikolopoulou A., Kothari P., De B.P., He B., Chen A., Heier L.A., Sondhi D., et al. Quantitative whole-body imaging of I-124-Labeled adeno-associated viral vector biodistribution in nonhuman primates. Hum. Gene Ther. 2020;31:1237–1259. doi: 10.1089/hum.2020.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gurda B.L., De Guilhem De Lataillade A., Bell P., Zhu Y., Yu H., Wang P., Bagel J., Vite C.H., Sikora T., Hinderer C., et al. Evaluation of AAV-mediated gene therapy for central nervous system disease in canine Mucopolysaccharidosis VII. Mol. Ther. J. Am. Soc. Gene Ther. 2016;24:206–216. doi: 10.1038/mt.2015.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilson J.M., Flotte T.R. Moving forward after two deaths in a gene therapy trial of myotubular myopathy. Hum. Gene Ther. 2020;31:695–696. doi: 10.1089/hum.2020.182. [DOI] [PubMed] [Google Scholar]
- 88.Shieh P.B., Bonnemann C.G., Muller-Felber W., Blaschek A., Dowling J.J., Kuntz N.L., Seferian A.M. Re: "moving forward after two deaths in a gene therapy trial of myotubular myopathy" by wilson and flotte. Hum. Gene Ther. 2020;31:787. doi: 10.1089/hum.2020.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Monahan P.E., Negrier C., Tarantino M., Valentino L.A., Mingozzi F. Emerging immunogenicity and genotoxicity considerations of adeno-associated virus vector gene therapy for hemophilia. J. Clin. Med. 2021;10:2471. doi: 10.3390/jcm10112471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Edsbagge M., Tisell M., Jacobsson L., Wikkelso C. Spinal CSF absorption in healthy individuals. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;287:R1450–R1455. doi: 10.1152/ajpregu.00215.2004. [DOI] [PubMed] [Google Scholar]
- 91.Ringstad G., Vatnehol S.A.S., Eide P.K. Glymphatic MRI in idiopathic normal pressure hydrocephalus. Brain. 2017;140:2691–2705. doi: 10.1093/brain/awx191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cohen-Pfeffer J.L., Gururangan S., Lester T., Lim D.A., Shaywitz A.J., Westphal M., Slavc I. Intracerebroventricular delivery as a safe, long-term route of drug administration. Pediatr. Neurol. 2017;67:23–35. doi: 10.1016/j.pediatrneurol.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 93.Chen Y., Zheng S., Tecedor L., Davidson B.L. Overcoming limitations inherent in sulfamidase to improve Mucopolysaccharidosis IIIA gene therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2018;26:1118–1126. doi: 10.1016/j.ymthe.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murlidharan G., Samulski R.J., Asokan A. Biology of adeno-associated viral vectors in the central nervous system. Front. Mol. Neurosci. 2014;7:76. doi: 10.3389/fnmol.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Katz N., Goode T., Hinderer C., Hordeaux J., Wilson J.M. Standardized method for intra-cisterna magna delivery under fluoroscopic guidance in nonhuman primates. Hum. Gene Ther. Methods. 2018;29:212–219. doi: 10.1089/hgtb.2018.041. [DOI] [PubMed] [Google Scholar]
- 96.Gray S.J., Nagabhushan Kalburgi S., McCown T.J., Jude Samulski R. Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther. 2013;20:450–459. doi: 10.1038/gt.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Perez B.A., Shutterly A., Chan Y.K., Byrne B.J., Corti M. Management of neuroinflammatory responses to AAV-mediated gene therapies for neurodegenerative diseases. Brain Sci. 2020;10:119. doi: 10.3390/brainsci10020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Suriano C.M., Verpeut J.L., Kumar N., Ma J., Jung C., Boulanger L.M. Adeno-associated virus (AAV) reduces cortical dendritic complexity in a TLR9-dependent manner. bioRxiv. 2021 doi: 10.1101/2021.09.28.462148. Preprint at. [DOI] [Google Scholar]
- 99.Johnston S., Parylak S.L., Kim S., Mac N., Lim C., Gallina I., Bloyd C., Newberry A., Saavedra C.D., Novak O., et al. AAV ablates neurogenesis in the adult murine hippocampus. Elife. 2021;10:e59291. doi: 10.7554/eLife.59291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hinderer C., Katz N., Buza E.L., Dyer C., Goode T., Bell P., Richman L.K., Wilson J.M. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 2018;29:285–298. doi: 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hordeaux J., Hinderer C., Goode T., Katz N., Buza E.L., Bell P., Calcedo R., Richman L.K., Wilson J.M. Toxicology study of intra-cisterna magna adeno-associated virus 9 expressing human alpha-L-iduronidase in rhesus macaques. Mol. Ther. Methods Clin. Dev. 2018;10:79–88. doi: 10.1016/j.omtm.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mueller C., Berry J.D., McKenna-Yasek D.M., Gernoux G., Owegi M.A., Pothier L.M., Douthwright C.L., Gelevski D., Luppino S.D., Blackwood M., et al. SOD1 suppression with adeno-associated virus and MicroRNA in familial ALS. N. Engl. J. Med. 2020;383:151–158. doi: 10.1056/NEJMoa2005056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cserr H.F., Harling-Berg C.J., Knopf P.M. Drainage of brain extracellular fluid into blood and deep cervical lymph and its immunological significance. Brain Pathol. 1992;2:269–276. doi: 10.1111/j.1750-3639.1992.tb00703.x. [DOI] [PubMed] [Google Scholar]
- 104.Pardridge W.M. CSF, blood-brain barrier, and brain drug delivery. Expert Opin. Drug Deliv. 2016;13:963–975. doi: 10.1517/17425247.2016.1171315. [DOI] [PubMed] [Google Scholar]
- 105.Achilly N.P., Wang W., Zoghbi H.Y. Presymptomatic training mitigates functional deficits in a mouse model of Rett syndrome. Nature. 2021;592:596–600. doi: 10.1038/s41586-021-03369-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tsai P.T., Rudolph S., Guo C., Ellegood J., Gibson J.M., Schaeffer S.M., Mogavero J., Lerch J.P., Regehr W., Sahin M. Sensitive periods for cerebellar-mediated autistic-like behaviors. Cell Rep. 2018;25:357–367.e4. doi: 10.1016/j.celrep.2018.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Silva-Santos S., van Woerden G.M., Bruinsma C.F., Mientjes E., Jolfaei M.A., Distel B., Kushner S.A., Elgersma Y. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J. Clin. Invest. 2015;125:2069–2076. doi: 10.1172/JCI80554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fu H., Cataldi M.P., Ware T.A., Zaraspe K., Meadows A.S., Murrey D.A., McCarty D.M. Functional correction of neurological and somatic disorders at later stages of disease in MPS IIIA mice by systemic scAAV9-hSGSH gene delivery. Mol. Ther. Methods Clin. Dev. 2016;3:16036. doi: 10.1038/mtm.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Flanigan K.M., Smith N.J.C., Couce M.L., Escolar M., Truxal K.V., McBride K.L., et al. Updated results of Transpher A, a multicenter, single-dose, Phase 1/2 clinical trial of ABO-102 investigational gene therapy for Sanfilippo syndrome type A (mucopolysaccharidosis IIIA) (S39.008) Neurology. 2022;98:213. [Google Scholar]
- 110.Feldkotter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Finkel R.S., Mercuri E., Darras B.T., Connolly A.M., Kuntz N.L., Kirschner J., Chiriboga C.A., Saito K., Servais L., Tizzano E., et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 112.Day J.W., Finkel R.S., Chiriboga C.A., Connolly A.M., Crawford T.O., Darras B.T., Iannaccone S.T., Kuntz N.L., Pena L.D.M., Shieh P.B., et al. Onasemnogene abeparvovec gene therapy for symptomatic infantile-onset spinal muscular atrophy in patients with two copies of SMN2 (STR1VE): an open-label, single-arm, multicentre, phase 3 trial. Lancet Neurol. 2021;20:284–293. doi: 10.1016/S1474-4422(21)00001-6. [DOI] [PubMed] [Google Scholar]
- 113.Baranello G., Darras B.T., Day J.W., Deconinck N., Klein A., Masson R., Mercuri E., Rose K., El-Khairi M., Gerber M., et al. Risdiplam in type 1 spinal muscular atrophy. N. Engl. J. Med. 2021;384:915–923. doi: 10.1056/NEJMoa2009965. [DOI] [PubMed] [Google Scholar]
- 114.McDougall F., Willgoss T., Hwang S., Bolognani F., Murtagh L., Anagnostou E., Rofail D. Development of a patient-centered conceptual model of the impact of living with autism spectrum disorder. Autism. 2018;22:953–969. doi: 10.1177/1362361317718987. [DOI] [PubMed] [Google Scholar]
- 115.Willgoss T., Cassater D., Connor S., Krishnan M.L., Miller M.T., Dias-Barbosa C., Phillips D., McCormack J., Bird L.M., Burdine R.D., et al. Measuring what matters to individuals with angelman syndrome and their families: development of a patient-centered disease concept model. Child. Psychiatry Hum. Dev. 2021;52:654–668. doi: 10.1007/s10578-020-01051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.