Abstract
Predictable DNA off-target effect is one of the major safety concerns for the application of cytosine base editors (CBEs). To eliminate Cas9-dependent DNA off-target effects, we designed a novel effective CBE system with dual guiders by combining CRISPR with transcription activator-like effector (TALE). In this system, Cas9 nickase (nCas9) and cytosine deaminase are guided to the same target site to conduct base editing by single-guide RNA (sgRNA) and TALE, respectively. However, if nCas9 is guided to a wrong site by sgRNA, it will not generate base editing due to the absence of deaminase. Similarly, when deaminase is guided to a wrong site by TALE, base editing will not occur due to the absence of single-stranded DNA. In this way, Cas9- and TALE-dependent DNA off-target effects could be completely eliminated. Furthermore, by fusing TALE with YE1, a cytidine deaminase with minimal Cas9-independent off-target effect, we established a novel CBE that could induce efficient C-to-T conversion without detectable Cas9- or TALE-dependent DNA off-target mutations.
Keywords: cytosine base editor, CRISPR-Cas9, TALE, predictable DNA off-target effects
Graphical abstract

Zhou et al. develop a novel safe and effective CBE system with dual guiders by combining CRISPR with transcription activator-like effector (TALE) that can induce efficient C-to-T conversion without detectable Cas9- or TALE-dependent DNA off-target effects.
Introduction
CRISPR/Cas-based cytosine base editors (CBEs) can efficiently and accurately mediate the conversion of a nucleotide from C to T without generating double-stranded DNA (dsDNA) breaks.1,2 However, CBEs often generate undesired mutations including Cas9-dependent off-target DNA editing3 and Cas9-independent off-target DNA and RNA single-nucleotide variations,4, 5, 6, 7 which give rise to safety concerns during their applications in gene therapy and production of genetically modified organisms. Cas9-independent off-target effect, which is caused by random deamination without the participation of single-guide RNA (sgRNA), has already been successfully minimized through the application of YE1, a BE3 variant with the deaminase domain mutation, through reducing DNA- and RNA-binding activity.8,9 Cas9-dependent off-target effect, which is caused by mismatch tolerance between sgRNA and targeting sequence, has been mitigated by using high-fidelity Cas domain variants,10 anti-CRISPR proteins,11 or ribonucleoprotein delivery12 to some extent, but it cannot be completely eliminated. Moreover, the reduction of Cas9-dependent off-target effects is achieved by the methods above often at the cost of on-target base editing efficiency. Therefore, Cas9-dependent off-target effect is still a major hurdle for the practical application of CBE.
In engineered nuclease-mediated gene editing systems, nucleases need to be carried to target sites by guiders, which have the capacity to specifically combine desired DNA sequences, to conduct precise gene editing. A CBE comprises four components including Cas9 nickase (nCas9), cytosine deaminase, uracil DNA glycosylase inhibitor (UGI), and sgRNA, in which cytosine deaminase, nCas9, and UGI are fused into a single cassette and guide to target sites by single guider, sgRNAs, to carry out C-to-T conversion. In the TALEN editing system, truncated transcription activator-like effector (TALE) protein variant, a programmable DNA-binding domain, is used as a guider to carry FokI or VP64 to target sites to edit DNA or regulate gene expression.13,14 Previously, a TALE has been used to guide SpCas9 with attenuated DNA-binding affinity together with sgRNA to increase sequence targeting range and improve precision, by taking advantage of TALE’s inherent modularity with specificity and affinity.15 However, theoretically, the fusion of SpCas9 with TALE can only reduce Cas9-dependent DNA off-target effects to some extent but cannot eliminate them completely. Here, to completely eliminate Cas9-dependent DNA off-target effects in CBEs, we fused cytosine deaminase from BE3 to TALE, resulting in a new base editor (named TaC9-CBE) with dual guiders, namely, sgRNA and TALE. In this new base editor, nCas9 is still guided to target site by sgRNA, while deaminase catalytic domain is separately guided to the target sites by TALE. Due to the inability of unwinding the double helix, deaminase-TALE alone has no base editing activity at the target sites.16 The deaminase can convert C to T on the single strand only when nCas9 is guided to the target site of TALE by sgRNA to open the DNA duplex and form single-stranded DNA (ssDNA). With this system, if nCas9 is guided to a wrong site (mismatched off-target site) by sgRNA, it will not generate base editing due to the absence of deaminase. Similarly, when deaminase is guided to a wrong site by TALE (targeted dsDNA only), base editing will not occur due to the absence of ssDNA. Base editing will only happen when nCas9 and deaminase guided by sgRNA and TALE, respectively, are present in the same site (desired on-target site). In this way, Cas9- and TALE-dependent DNA off-target effects could be completely eliminated using our base editing system (Figure 1).
Figure 1.
Schematic of on-target effects and predictable DNA off-target effects of BE3 and TaC9-CBE
Results
Design and optimization of CBEs with dual guiders
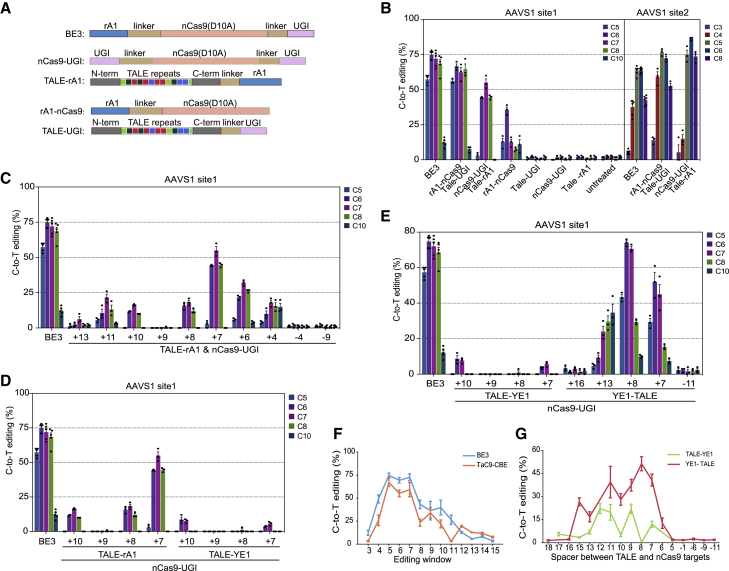
The most commonly used base editor was made of a cassette with three components, in which a rat cytosine deaminase APOBEC1 (rA1) was fused at the N-terminal, while UGI was located at the C-terminal of nCas9 (D10A). To explore the possibility of generating a dual-guider CBE possessing comparable base editing efficiency with ordinary CBEs, we constructed two fusion proteins, namely, TALE-UGI and TALE-rA1, by combining a corresponding CBE component with a TALE. BE3, nCas9-2×UGI, and rA1-nCas9 were also constructed and used for controls (Figure 2A). Two human AAVS1 loci (AAVS1 site1 and AAVS1 site2) were chosen to define the optimal combination with high editing efficiency. TALE repeats targeting AAVS1 loci were assembled by a simplified method based on degenerated codon, named dcTA, developed by our group.17 HEK293T cells were transiently transfected with sgRNAs together with different vectors that expressed BE3, TALE-UGI, TALE-rA1, nCas9-2×UGI, and rA1-nCas9. As shown in Figure 2B, the expression of an individual component, such as TALE-UGI, TALE-rA1, and nCas9-2×UGI, had no detectable gene mutation on the target site, similar to the untreated cells, while rA1-nCas9 had 15%−35% C-to-T conversion. However, rA-nCas9/TALE-UGI and TALE-rA1/nCas9-2×UGI groups, which both have two components binding targets with 7 bp distance, were transfected together with sgRNA into HEK293T cells. The efficiency of C-to-T conversion within the effective editing window C5–C8 was higher than 50%, close to that of the BE3 positive control. Although rA1-nCas9/TALE-UGI combination showed strong base editing capability, it does not provide any advantage of minimizing off-target effects relative to BE3. The TALE-rA1/nCas9-2×UGI group, named TaC9-CBE, not only had strong base-editing capability, but it also might eliminate Cas9-dependent DNA off-target effects given that rA1 and nCas9 would be guided to the target sites by TALE and sgRNA separately.
Figure 2.
Optimization of the TaC9-CBE system
(A) Schematic of BE3, TALE-UGI, rA1-nCas9, TALE-rA1, and nCas9-UGI constructs. (B) Comparison of the base-editing efficiencies of BE3 and different combinations of TALE-UGI, rA1-nCas9, TALE-rA1, and nCas9-UGI at the AAVS1 site1 and site2 loci in HEK293T cells. (C) Base-editing efficiency of the TaC9-CBE system with different target distances between TALEs and sgRNAs at AAVS1 site1 locus. (D) Comparison of the base-editing efficiencies of nCas9-UGI combined with either TALE-rA1 or its variant TALE-YE1 at the AAVS1 site1 locus. (E) Comparison of the base-editing efficiencies of TaC9-CBE with YE1 fusing at the N or C terminus of TALE and different spacers at AAVS1 site1 locus. (F) Base-editing efficiency of BE3 and TaC9-CBEYE1 across the entire protospacer of eight genomic loci. (G) Summary of the influence of spacer on editing efficiency across eight genomic loci. Data and error bars show the mean ± SEM of three or five independent experiments (n = 3 or 5). WT, wild-type.
The distance between TALE-rA1 and nCas9-2×UGI targets might affect the gene-editing efficiency. As shown in Figure S1A, the targeting sequence of TALE-rA1s located at distal (+) or proximal (−) sites of adjacent motif (PAM) with different distances resulted in different C-to-T conversion efficiencies on the AAVS1 locus. Effective base editing was achieved when both components bound targets with 4–13 bp distance, and the best effect was observed at a distance of 6–11 bp (Figures 2C and S1B). The main editing window of TaC9-CBE (edit efficiency >30%) was narrowed to C5 to C8 (∼4 nt) from the original C4 to C8 (∼5 nt) of BE3 with the same or slightly lower efficiency.
The fusion of YE1, an rA1 variant, with nCas9 has been reported to substantially reduce the levels of Cas9-independent DNA and RNA off-target editing.8,9 Therefore, to achieve a dual-guider CBE that not only eliminates Cas9- or TALE-dependent DNA off-target mutations, but also has minimized Cas9-independent DNA and RNA off-target editing, we next replaced rA1 with YE1 in our new base editor. We initially fused YE1 on the C-terminal of TALE as that of TALE-rA1, but we found that this replacement resulted in far less efficient base editing on site1 and site2 of the AAVS1 loci (Figures 2D and S1C). Thus, we further conducted a comparison test of editing efficiency by fusing YE1 to either the N-terminal or C-terminal of TALE and placing the target sites at either the distal or proximal sites of PAM (Figure S1D). We found that a high efficiency of base editing was achieved when YE1 was fused to the N-terminal of TALE (YE1-TALE) (Figures 2E and S1E). The effective TALE target sites were 6–15 bp distant to sgRNA targets at distal sites of PAM, while no base editing was detected when TALE was combined at proximal sites of PAM (Figures 2C, 2E, S1B, and S1E).
To further determine the effective universality of YE1 TaC9-CBE in different genes, six more loci located at AAVS1 (site1 and site2), EMX1, HEK, FANCF, and VEGFA were selected for editing (Figures S1F−S1K). A total of 39 TALEs and eight sgRNAs were designed to test the ability of TaC9-CBE in inducing C-to-T conversion (Tables S1–S4). Compared with BE3, TaC9-CBE showed lower efficiency when YE1 was on the C-terminal of TALE (Figures S1F, S1H, S1I, and S1K) in four sites, while only one site showed high efficiency when the distance between sgRNA and TALE was 11 bp. On the contrary, the efficiency was high and comparable to that of BE3 with mean editing frequencies ranging from 30% to 85% when YE1 was fused to the N-terminal of TALE (Figures S1G–S1K). We then analyzed the distribution of editing positions across all eight sgRNA targets. The results showed that the high frequency of C-to-T editing could be achieved at positions 5–7 bp of the protospacer of sgRNA, while moderate editing efficiency was also detected at positions 4 and 8–10 bp (Figure 2F). The editing efficiency proved that TaC9-CBE with YE1-TALE had significantly higher base editing efficiency than using TALE-YE1 (Figure 2G). It also showed that the effective distance between TALE and sgRNA targets was 6–15 bp. Therefore, YE1-TALE combining nCas9-2×UGI with a 6–15 bp spacer between sgRNA and TALE was defined as a new base editing system (named TaC9-CBEYE1) with a high base-editing efficiency and used for the following experiments. For TaC9-CBE with the highest efficiency, its corresponding TALE obeyed these TALE design rules at most sites (Figures S1F–S1J).
To further validate the universality of TaC9-CBEYE1 on different types of cells, we tested the base-editing efficiency of eight genome sites described above in two more cell types (HeLa and U2OS). The editing window and efficiency of TaC9-CBEYE1 were comparable to those of BE3 in all eight sites (Figure S2). No significant differences in base-editing efficiency were observed among three types of cells, including HEK293T, HeLa, and U2OS.
Efficient base editing of TaC9-CBE in mammalian embryos
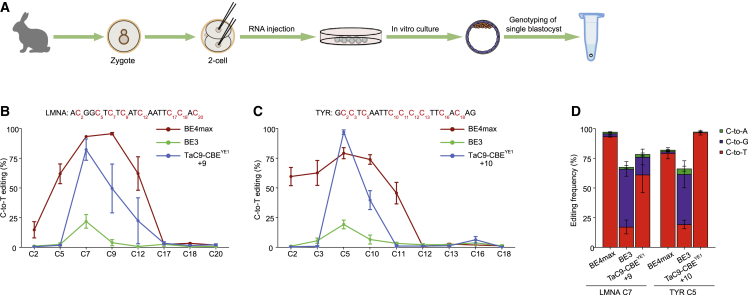
Previous studies have been proved that the CBE could be used to generate base editing animals efficiently by direct injection of the editing vectors into the one-cell stage of embryos.18 Therefore, we also tested whether TaC9-CBEYE1 could convert C to T efficiently in mammalian embryos. Rabbit LMNA and TYR genes were chosen as target sites. sgRNAs with one of the mRNAs expressing BE4max, BE3, or TaC9-CBEYE1 were co-injected into two-cell rabbit embryos (Figure 3A). The injected embryos developed to blastocysts at a rate of 60%–90% (Table S2). The rate of edited embryos reached 62.5% and 91.7% at LMNA and TYR sites, respectively, which was similar to or higher than those of BE3 and BE4max. For the editing window, TaC9-CBEYE1 and BE3 had narrowed editing windows (C7 and C9 at the LMNA locus and C5 and C10 at the TYR locus), while BE4max had larger editing windows (C5, C7, C9, and C12 at the LMNA locus and C2, C3, C5, C10, and C11 at the TYR locus). For the editing efficiency, TaC9-CBEYE1 induced more than 75% C-to-T conversion, similar to BE4max and much higher than BE3 (<25%) (Figures 3B and 3C). Further analysis demonstrated that BE3 also caused 50% C-to-G conversion, while TaC9-CBEYE1 and BE4max created very little C-to-G conversion on both targets (Figure 3D).
Figure 3.
Base editing of TaC9-CBEYE1 and BE3 in rabbit embryos
(A) Schematic of CBE mRNA and sgRNA that were co-injected into two-cell embryos. (B and C) Base editing of BE4max, BE3, and TaC9-CBEYE1 at rabbit LMNA and TYR genomic loci. (D) Comparison of C-to-A, C-to-G, and C-to-T base editing of three editors across LMNA or TYR genomic targets. Data and error bars show the mean ± SEM of three independent experiments (n = 3). WT, wild-type.
Detection of Cas9- and TALE-dependent DNA off-targeting of TaC9-CBE in human cells
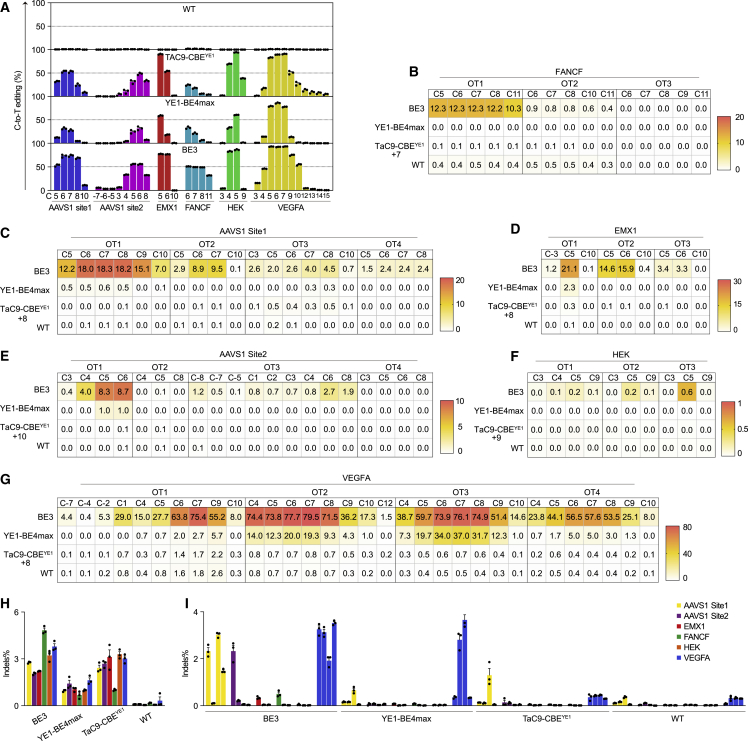
Next, we examined whether TaC9-CBEYE1 could eliminate predictable DNA off-target effect. Six of the eight genomic loci described above were used to test the on-target and Cas9- and TALE-dependent off-target editing activity in HEK293T cells using high-throughput sequencing (HTS). First, on-target analysis showed that the on-target editing frequencies of TaC9-CBEYE1 ranged from 24.5% to 94.2% at all six sites, which was comparable to those of BE3 and significantly higher than those of YE1-BE4max at four out of six targets (Figure 4A). Three to four potential Cas9-dependent off-target sites for each sgRNA were amplified for analysis (Table S6). Sequencing showed that 18 for BE3 (4 of 4 for AAVS1 site1, 2 of 4 for AAVS1 site2, 3 of 3 for EMX1, 3 of 3 for HEK, 2 of 3 for FANCF, and 4 of 4 for VEGFA) and six for YE1-BE4max (1 of 4 for AAVS1 site1, 1 of 4 for AAVS1 site2, 1 of 3 for EMX1, and 3 of 4 for VEGFA) out of 21 potential off-target sites had C-to-T base editing. Among them, 12 out of 18 for BE3 and four out of six for YE1-BE4max displayed higher than 2% off-target editing, indicating that BE3 and YE1-BE4max had extremely high activity at predictable DNA off-target sites. By contrast, TaC9-CBEYE1 exhibited no base editing in all of the 21 off-target sites (Figures 4B−G). Additionally, to verify the TALE-dependent off-target effects, we tested 20 predictable TALE off-target sites of TaC9-CBEYE1 and also did not observe any off-target mutagenesis (Tables S7 and S8). We found that all three base editors caused 1%–5% unwanted indels on all six target sites (Figure 4H). However, the unwanted indels in the 21 Cas9-dependent off-target sites were found in almost half (11/21) of the off-target sites by BE3 and three off-target sites by YE1-BE4max, while only in one off-target site by TaC9-CBEYE1 (Figure 4I).
Figure 4.
Efficiency of cytidine base editing and indel at Cas9 on-target sites and off-target sites
(A) On-target editing efficiency of BE3, YE1-BE4max, TaC9-CBEYE1, and WT across six genomic loci. (B−G) Heat maps showing the average base-editing efficiency at 21 Cas9-dependent off-target sites of six BE3 and TaC9-CBEYE1 editors. (B) Three off-target effects of FANCF targeting sgRNA. (C) Four off-target effects of AAVS1 site1 targeting sgRNA. (D) Three off-target effects of EMX1 targeting sgRNA. (E) Four off-target effects of AAVS1 site2 targeting sgRNA. (F) Three off-target effects of HEK targeting sgRNA. (G) Four off-target effects of VEGFA targeting sgRNA. (H) On-target indel frequencies of BE3, YE1-BE4max, TaC9-CBEYE1, and WT across six genomic loci. (I) Indel frequencies of BE3, TaC9-CBEYE1, and WT at 21 Cas9-dependent off-target sites of six sgRNAs. Data are mean ± SEM for n = 3 independent experiments. WT, wild-type.
Discussion
In this study, we developed a novel CBE TaC9-CBE by combining CBE and TALE. After several rounds of optimization, we achieved the effective base editing when cytosine deaminase YE1 was fused to the N-terminal of TALE and the spacer length between TALE and gRNA targets ranged from 7 bp to 11 bp. The efficient editing window was narrowed to the range between C5 and C7 compared with BE3 (C4 to C8). More importantly, TaC9-CBEYE1 completely eliminated Cas9- and TALE-dependent off-target DNA effects. Considering that the YE1 variant has been proved to have minimized Cas9-independent DNA and RNA off-target effects, TaC9-CBEYE1 should be an ideal base editing tool for gene therapy and generation of genetically modified organisms with minimized safety concern.
We found that the optimal spacer length between TALE and gRNA targets was 7–11 bp. Inefficient base editing occurred occasionally in some sites, although the spacers were in the optimal range (Figures 2C–2E, S1E and S1K). We speculated that the inefficient targeting of an individual TALEs should be the reason for the low efficiency of TaC9-CBE. A previous report showed that the nucleotide in the “−1 position” of the targeting sequence was important when designing the custom TAL effectors with high targeting efficiency.19 In most cases, the effective sequences of TALE targets were preceded by a T or C at the 5′-end. Therefore, except for spacer length, the TALE design rules should also be obeyed to achieve a TaC9-CBE with high efficiency.
A previous report has shown that compared with BE4, YE1-BE4max was able to dramatically reduce Cas9-dependent OT editing, but at the cost of on-target base-editing efficiency.8,9 In our study, YE1-BE4max truly resulted in less Cas9-dependent off-target effects than BE3, but it was not able to eliminate it completely since base edit effects occurred on six out of 21 Cas9-dependent off-target sites. Furthermore, YE1-BE4max gave rise to lower on-target editing efficiency than both BE3 and TaC9-CBEYE1.
A previous study showed that except for C-to-T conversion, CBE could cause unwanted indels, another safety concern for its application, in both on-targeting and off-targeting sites because a double-strand break could be randomly created by nCas9 at some rate.2 In our study, we found that TaC9-CBE caused extremely low indels at off-target sites compared with BE3, indicating that TaC9-CBE is more specific than BE3. Therefore, our TaC9-CBE could not only eliminate Cas9-dependent DNA off-targeting, but it could also reduce the possible Cas9-dependent indels in off-targeting sites, overcoming another safety concern for clinical application.
Recently, a transformer CBE (tBE) with a cleavable deoxycytidine deaminase inhibitor (dCDI) domain was developed to eliminate genome and transcriptome-wide off-target mutations. In the tBE system, the deaminase remained inert at both sgRNA-dependent and -independent OT sites. The dCDI domain will be removed only when tBE binds at on-target sites, leading to the activation of deaminase.20 However, the active deaminase may be released to the nucleus and become the potential off-target factor on both Cas9-dependent and independent sites. The tBE editor is made up of six independent components including nCas9, cytosine deaminase and its inhibitor fusing protein, two-halves of TEV enzymes, and two sgRNAs, and it is more complex to make the constructs than our TaC9-CBE containing only three components (i.e., nCas9, sgRNA, and TALE-cytosine deaminase). Moreover, our system can also be applied to ABE optimization if TALE fuses with adenine deaminase, which cannot be applied in the tBE system. Splitting of cytidine deaminases was also used for reducing the off-target effects. A dsDNA deaminase, DddAtox, was split into two parts and fused with a TALE to form DdCBE. However, compared with the BE system, DdCBE had lower editing efficiency and a wider editing window; thus, it is mainly used for mtDNA editing.21 Other split-engineered BEs allow two inactive fragments to be reassembled into a functional enzyme by inducing protein dimerization (CID), rapamycin, for controlling the activity of the base editor22 to reduce the chance of off-target effects. Off-target effects would not happen in the absence of rapamycin but are still inevitable in the presence of rapamycin. Thus, previously split BEs could not solve the off-target problem completely.
Here, in rabbit embryos, we found that TaC9-CBEYE1 and BE4max caused higher C-to-T conversion efficiency than BE3 on the same sites, while BE3 showed higher unwanted C-to-G conversion efficiency. Previous studies also showed that BE3 could give rise to C-to-G conversion at some DNA positions in both rabbit and porcine embryos with a high efficiency ranging from 10% to 50%.18,23 When the plasmid DNA of BEs was transfected into somatic cells in vitro, it would produce large amounts of RNPs with UGI transiently, which was sufficient to inhibit endogenous UDG, the key factor for C-to-G conversion,24,25 resulting in no obvious difference of C-to-T conversion efficiency between 1×UGI (BE3) and 2×UGI (BE4max or TaC9-CBEYE1). By contrast, when BE mRNA and sgRNA are injected into embryos, less RNP complex will form. It was reported that the expression level of UDG in oocytes and early embryos was higher than in most somatic cells.26,27 The amount of UDG could be inhibited more thoroughly by 2×UGI (BE4max or TaC9-CBEYE1) than by 1×UGI (BE3), which would result in higher C-to-T conversion efficiency. Thus, the additional UGI in both TaC9-CBEYE1 and BE4max may enhance the UDG-inhibiting ability and lead to higher C-to-T conversion efficiency in embryos than in somatic cells.
For a long time, the construction of the TALE vector was a huge challenge due to its highly repetitive sequence. Recently, we designed a simple, rapid, and efficient method (named dcTA) based on degenerated codon to assemble TALE repeats, in which TALE trimers with nonrepetitive DNA sequences were amplified by PCR and sequentially assembled via Gibson assembly.17 Therefore, by using dcTA, the construction of TALE was very simple and easy to master, which facilitated the application of TaC9-CBE.
The major obstacle for the in vivo applications of current base editors comes from the vector size, in which deaminase, nCas9, and UGI are fused together, forming a vector with a size of ∼5.5 kb, larger than the package limitation of AAV (4.8 kb). One possible way to deliver such base editors with AAV is to split the base editors into two parts.20,28, 29, 30 Our TaC9-CBE system can also deliver with dual AAV by constructing two vectors, one harboring nCas9-UGI cassette (4.6 kb) and the other harboring TALE-deaminase plus sgRNA cassettes (3.7 kb). In the future, all components can be packaged into a single AAV particle by replacing spCas9 with small CRISPR (e.g., Cas12F1, 1.4–1.6 kp31,32) and reducing TALE repeats, which is expected to increase the delivery efficiency of base editors both in vitro and in vivo.
In summary, we developed an efficient and precise CBE by combining CRISPR with TALE to minimize predictable DNA off-target effects, which would provide a safe tool for gene therapy and generation of genetically modified organisms.
Materials and methods
Vector construction
For the construction of sgRNAs, two complementary oligonucleotides were synthesized by Guangzhou IGE Biotechnology, annealed by PCR (Bio-Rad T100 thermal cycler), and cloned into the BbsI site of the sgRNA expression vector using T4 DNA Ligase (New England Biolabs, #M0202). pCMV-BE3 (#73021), pCMV-BE4max (#112093), and U6-sgRNA cloning vector (#48962) were purchased from Addgene. The mCherry gene was synthesized by Guangzhou IGE Biotechnology and cloned into pCMV-BE3 as pCMV-BE3-mcherry. pCMV-rAPOBEC1-nCas9-mcherry and pCMV-UGI-nCas9-UGI-mcherry were constructed on the basis of pCMV-BE3-mcherry. The rA1 gene was amplified from pCMV-BE3 and cloned into the TALEN expression vector. YE1 variant was mutated from rA1 gene. PCR was performed using PrimeSTAR Max DNA Polymerase (Takara, R045), and plasmids were constructed using the ClonExpress II One Step Cloning Kit (Vazyme, C112) in accordance with the manufacturer’s instructions. All TALE architectures were generated by using dcTA assembly as recently described.17 All restriction endonucleases were purchased from Thermo Fisher Scientific, and the sgRNA sequences are listed in Table S4. All subcloning experiments were conducted using TOP10 competent cells, and plasmid extraction was performed using the EndoFree Plasmid Mini Kit (Cwbio).
Cell culture and transfection
HEK293T, HeLa, and U2OS cells were cultured in Dulbecco’s modified Eagle’s medium (HyClone) supplemented with 10% fetal bovine serum (Gibco) at 37°C under 5% CO2. Then, they were seeded on 24-well plates (Greiner Bio-One) and transfected at approximately 90% density. After 24 h, 250 ng of sgRNA, 500 ng of plasmids with nCas9, and 300 ng of plasmids with TALE were transfected using 1 μL of Lipo8000 Transfection Reagent (Beyotime, C0533) per well. After 3 days of transfection, cells were collected and filtered through a 40-μm strainer (BD Falcon), and the transfected cells were collected on a Beckman Coulter MoFlo Astrios Cell Sorter (laser option: blue 488 nm and red 561 nm). About 10 μL of NP40 Lysis Buffer was added to the collected cells, and the reaction was incubated at 56°C for 1 h. The products were used as a template for amplification.
Sanger sequencing and analysis using EditR
PCR primers were synthesized by Guangzhou IGE Biotechnology. PCR was performed using PrimeSTAR Max DNA Polymerase (Takara, R045), and Sanger sequencing was performed by Guangzhou IGE Biotechnology. The results were quantified using EditR.33 All PCR primers are listed in Table S4.
HTS and data analysis
Cas9-dependent off-target sites were predicted by Cas-OFFinder.34 TALE-dependent off-target sites were selected with mismatches of less than 3 nt across the human genome. The PCR primers of HTS were synthesized by Guangzhou IGE Biotechnology. The targeted amplicons were amplified using Q5 High-Fidelity DNA Polymerase (New England Biolabs). Then, the targeted amplicon was used as a template to amplify two terminal index sequences using TruePrep Index Kit V4 for Illumina (Vazyme) according to the manufacturer’s instructions. The final PCR products were purified as HTS samples using HiPure Gel DNA Mini Kit (Magen). The Illumina HiseqX platform at Beijing Annoroad Gene Technology was used for sequencing of HTS samples. All HTS primers are listed in Table S5.
The FASTQ files of HTS results were analyzed using CRISPResso2 (CRISPResso2 provides accurate and rapid genome editing sequence analysis). All raw FASTQ files generated are available from the NCBI database.
mRNA preparation and microinjection
sgRNAs with T7 promoter were linearized by PCR amplification, and PCR primers were synthesized by Guangzhou IGE Biotechnology. Linearized sgRNA fragments were in vitro transcribed using the HiScribe T7 High Yield RNA Synthesis Kit (NEB) according to the manufacturer’s protocol. BE4max, BE3, and UGI-nCas9-UGI vectors were linearized by MssI (Thermo Fisher Scientific) through in vitro transcription using the HiScribe T7 ARCA mRNA Kit (NEB). YE1-TALE vectors were linearized by NotI (Thermo Fisher Scientific) through in vitro transcription using the High Yield Capped RNA Transcription Kit (Ambion, AM1340). mRNAs were purified by using the Mini Kit (Qiagen). The sgRNA primers used in in vitro transcription are listed in Table S3.
Rabbits were bred in standard cages at the Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences. All animal experiment protocols were approved by the Institutional Animal Care and Use Committees at the Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences. Experiments using donor rabbits were performed following previously published procedures.35 Rabbit zygotes were collected from donor rabbits, and 50 ng/μL of sgRNA; 100 ng/μL of BE4max, BE3, or nCas9-2×UGI mRNA; and 75 ng/μL of YE1-TALE mRNA were co-microinjected into the cytoplasm of zygotes. The injected zygotes were cultured in EmbryoMax Advanced KSOM Embryo Medium (Sigma-Aldrich) at 37°C under 5% CO2. After 3 days, morula or blastocyst stage embryos were individually collected and lysed with NP40 solution. Subsequently, PCR amplification of the sequence around the target site was performed with Sanger sequencing.
Statistical analysis
All graphic data were presented as mean ± SEM, and statistical analysis was performed with GraphPad Prism.
Acknowledgments
This work was supported by National Key Research and Development Program of China Stem Cell and Translational Research (2017YFA0105103), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16030503), the Science Foundation for Young Teachers of Wuyi University (2019TD05), the Natural Science Foundation of Guangdong Province of China (2019A1515110283), the Science and Technology Planning Project of Guangdong Province, China (2019A030317010, 2017B020231001, 2021B1212040016), the Science and Technology Program of Guangzhou, China (202007030003), the National Natural Science Foundation of China (82001974), and the Key Research & Development Program of Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory) (2018GZR110104004).
Author contributions
J.Z. and Y.Liu designed and performed most experiments and wrote the manuscript. Y.Wei and S.Z. performed experiments on plasmid construction and cell culture. S.G. performed the informatics analyses. Y.Y., T.L., M.C., Y.L., Q.Zhang, C.T., Y.Liu, Y.Wu, X.P., M.G., and J.W. provided technical assistance. L.L., Q.Zou, and K.Z. conceived and supervised the project. Q.Zou and L.L. reviewed and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.04.010.
Contributor Information
Kun Zhang, Email: kzhang@gdut.edu.cn.
Liangxue Lai, Email: lai_liangxue@gibh.ac.cn.
Qingjian Zou, Email: zouqj@wyu.edu.cn.
Supplemental information
References
- 1.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A∗T to G∗C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grunewald J., Zhou R., Garcia S.P., Iyer S., Lareau C.A., Aryee M.J., Joung J.K. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019;569:433–437. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R., et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019;571:275–278. doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 6.Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 8.Zuo E., Sun Y., Yuan T., He B., Zhou C., Ying W., Liu J., Wei W., Zeng R., Li Y., Yang H. A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects. Nat. Methods. 2020;17:600–604. doi: 10.1038/s41592-020-0832-x. [DOI] [PubMed] [Google Scholar]
- 9.Doman J.L., Raguram A., Newby G.A., Liu D.R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 2020;38:620–628. doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L., Sun N., Zeina C.M., Gao X., Rees H.A., Lin Z., Liu D.R. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang M., Sui T., Liu Z., Chen M., Liu H., Shan H., Lai L., Li Z. AcrIIA5 suppresses base editors and reduces their off-target effects. Cells. 2020;9:1786. doi: 10.3390/cells9081786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rees H.A., Komor A.C., Yeh W.H., Caetano-Lopes J., Warman M., Edge A.S.B., Liu D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017;8:15790. doi: 10.1038/ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J., et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 14.Maeder M.L., Linder S.J., Reyon D., Angstman J.F., Fu Y., Sander J.D., Joung J.K. Robust, synergistic regulation of human gene expression using TALE activators. Nat. Methods. 2013;10:243–245. doi: 10.1038/nmeth.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolukbasi M.F., Gupta A., Oikemus S., Derr A.G., Garber M., Brodsky M.H., Zhu L.J., Wolfe S.A. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat. Methods. 2015;12:1150–1156. doi: 10.1038/nmeth.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L., Briggs A.W., Chew W.L., Mali P., Guell M., Aach J., Goodman D.B., Cox D., Kan Y., Lesha E., et al. Engineering and optimising deaminase fusions for genome editing. Nat. Commun. 2016;7:13330. doi: 10.1038/ncomms13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng L., Zhou X., Zheng Y., Tang C., Liu Y., Zheng S., Liu Y., Zhou J., Li C., Chen M., et al. Simple and rapid assembly of TALE modules based on the degeneracy of the codons and trimer repeats. Genes (Basel) 2021;12:1761. doi: 10.3390/genes12111761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z., Chen M., Chen S., Deng J., Song Y., Lai L., Li Z. Highly efficient RNA-guided base editing in rabbit. Nat. Commun. 2018;9:2717. doi: 10.1038/s41467-018-05232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doyle E.L., Booher N.J., Standage D.S., Voytas D.F., Brendel V.P., Vandyk J.K., Bogdanove A.J. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L., Xue W., Zhang H., Gao R., Qiu H., Wei J., Zhou L., Lei Y.N., Wu X., Li X., et al. Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations. Nat. Cell Biol. 2021;23:552–563. doi: 10.1038/s41556-021-00671-4. [DOI] [PubMed] [Google Scholar]
- 21.Mok B.Y., de Moraes M.H., Zeng J., Bosch D.E., Kotrys A.V., Raguram A., Hsu F., Radey M.C., Peterson S.B., Mootha V.K., et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583:631–637. doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berrios K.N., Evitt N.H., DeWeerd R.A., Ren D., Luo M., Barka A., Wang T., Bartman C.R., Lan Y., Green A.M., et al. Controllable genome editing with split-engineered base editors. Nat. Chem. Biol. 2021;17:1262–1270. doi: 10.1038/s41589-021-00880-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie J., Ge W., Li N., Liu Q., Chen F., Yang X., Huang X., Ouyang Z., Zhang Q., Zhao Y., et al. Efficient base editing for multiple genes and loci in pigs using base editors. Nat. Commun. 2019;10:2852. doi: 10.1038/s41467-019-10421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao D., Li J., Li S., Xin X., Hu M., Price M.A., Rosser S.J., Bi C., Zhang X. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021;39:35–40. doi: 10.1038/s41587-020-0592-2. [DOI] [PubMed] [Google Scholar]
- 25.Kurt I.C., Zhou R., Iyer S., Garcia S.P., Miller B.R., Langner L.M., Grunewald J., Joung J.K. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021;39:41–46. doi: 10.1038/s41587-020-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J.C., Jang H.K., Kim J., Han J.H., Jung Y., Kim K., Bae S., Cha H.J. High expression of uracil DNA glycosylase determines C to T substitution in human pluripotent stem cells. Mol. Ther. Nucleic Acids. 2022;27:175–183. doi: 10.1016/j.omtn.2021.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu D., Chen L., Sun Q., Wu X., Jia S., Meng A. Uracil-DNA glycosylase is involved in DNA demethylation and required for embryonic development in the zebrafish embryo. J. Biol. Chem. 2014;289:15463–15473. doi: 10.1074/jbc.M114.561019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim C.K.W., Gapinske M., Brooks A.K., Woods W.S., Powell J.E., Zeballos C M.A., Zeballos C.M., Winter J., Perez-Pinera P., Gaj T. Treatment of a mouse model of ALS by in vivo base editing. Mol. Ther. 2020;28:1177–1189. doi: 10.1016/j.ymthe.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy J.M., Yeh W.H., Pendse N., Davis J.R., Hennessey E., Butcher R., Koblan L.W., Comander J., Liu Q., Liu D.R. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 2020;4:97–110. doi: 10.1038/s41551-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Zhou L., Tao R., Liu N., Long J., Qin F., Tang W., Yang Y., Chen Q., Yao S. sgBE: a structure-guided design of sgRNA architecture specifies base editing window and enables simultaneous conversion of cytosine and adenosine. Genome Biol. 2020;21:222. doi: 10.1186/s13059-020-02137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D.Y., Lee J.M., Moon S.B., Chin H.J., Park S., Lim Y., Kim D., Koo T., Ko J.H., Kim Y.S. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 2022;40:94–102. doi: 10.1038/s41587-021-01009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z., Zhang Y., Yu H., Pan D., Wang Y., Wang Y., Li F., Liu C., Nan H., Chen W., Ji Q. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat. Chem. Biol. 2021;17:1132–1138. doi: 10.1038/s41589-021-00868-6. [DOI] [PubMed] [Google Scholar]
- 33.Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J. 2018;1:239–250. doi: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song J., Zhong J., Guo X., Chen Y., Zou Q., Huang J., Li X., Zhang Q., Jiang Z., Tang C., et al. Generation of RAG 1- and 2-deficient rabbits by embryo microinjection of TALENs. Cell Res. 2013;23:1059–1062. doi: 10.1038/cr.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.