Abstract
The potential effect of UV radiation on the composition of coastal marine bacterioplankton communities was investigated. Dilution cultures with seawater collected from the surface mixed layer of the coastal North Sea were exposed to different ranges of natural or artificial solar radiation for up to two diurnal cycles. The composition of the bacterioplankton community prior to exposure was compared to that after exposure to the different radiation regimes using denaturing gradient gel electrophoresis (DGGE) of 16S rRNA and 16S ribosomal DNA. Only minor changes in the composition of the bacterial community in the different radiation regimes were detectable. Sequencing of selected bands obtained by DGGE revealed that some species of the Flexibacter-Cytophaga-Bacteroides (FCB) group were sensitive to UV radiation while other species of the FCB group were resistant. Overall, only up to ≈10% of the operational taxonomic units present in the dilution cultures appeared to be affected by UV radiation. Thus, we conclude that UV radiation has little effect on the composition of coastal marine bacterioplankton communities in the North Sea.
Research on the impact of UV (280- to 400-nm-wavelength) radiation on aquatic food webs has been stimulated by the notion that increasing levels of UVB (280- to 320-nm-wavelength) radiation are reaching the Earth's surface (9). Since bacterioplankton play a central role in the carbon and energy flux through marine food webs (4, 15), knowledge of the potential impact of UV radiation on bacterioplankton composition and activity is essential for understanding the biogeochemical cycling of elements in marine surface layers.
Early studies of the attenuation of UV radiation (22) indicated that the UV range of the solar spectrum is attenuated within the top few meters of the oceanic water column. More recent surveys using more sensitive instruments showed, however, that UV radiation penetrates to considerable depth (5, 16, 37).
UV radiation induces DNA damage via the formation of cyclobutane and pyrimidine-pyrimidone dimers (21, 27). No UV-protective compounds have been found in bacterioplankton (25). Furthermore, bacteria are considered to be too small to develop protective pigmentation against UV radiation (17). Therefore, bacterioplankton are more susceptible to the detrimental effects of UV radiation than other planktonic organisms (20). However, the effects of UV radiation on bacterioplankton are not only detrimental: long-wavelength UVA (360 to 400 nm) and short-wavelength photosynthetic active radiation (PAR; 400 to 430 nm) play crucial roles in the repair of DNA damage by activating the photoenzymatic repair mechanism PER (24).
In coastal marine and freshwater systems, exposure of dissolved organic matter (DOM) to UV radiation has been shown to result in subsequent elevated bacterial growth due to the enhanced availability of photochemically produced low-molecular-weight DOM (24, 26, 34). This is not a universal response, however, since in open oceanic waters, exposure of surface water DOM might also lead to reduced bacterial activity (7, 32). Whether UV exposure of DOM leads to postexposure enhanced or reduced bacterial activity depends on the original bioavailability of the DOM prior to exposure to solar radiation (33).
Recently, evidence has been presented that even under open-ocean conditions diurnal stratification of surface layers is a common phenomenon (14). Microorganisms and DOM confined to these diurnally stratified layers are therefore subjected to high UV radiation levels for almost the entire period of solar radiation. Thus, it might be reasonable to assume that microorganisms adapted to such high UV radiation levels dominate the bacterioplankton community in these layers. On one hand, Herndl et al. (18) found that surface water bacterioplankton are as sensitive to UV radiation as subpycnocline (>20-m depth) bacterioplankton. On the other hand, large interspecific differences in sensitivity to UV radiation and recovery from previous UV stress have been reported among marine bacterial isolates (3, 23). Hence, while measurements of the activity of the bulk bacterioplankton community indicate clear relationships between UV dose and inhibition in bacterioplankton activity (1, 18, 24, 38), interspecific differences in the response of selected bacterial isolates to UV radiation obviously do occur (3).
The aim of this study was therefore to determine possible alterations in the community composition of coastal marine bacterioplankton mediated by UV radiation. We hypothesized that interspecific differences in sensitivity to UV radiation and/or in efficiency of recovery from previous UV stress result in shifts in the composition of the bacterioplankton community. Using denaturing gradient gel electrophoresis (DGGE), the community structure was determined on the DNA and RNA levels. Due to the high number of ribosomes in active cells, rRNA is an indicator of metabolically active cells, whereas DNA reflects the general presence of a phylogenetic unit (39). Analysis of both DNA and RNA should therefore lead to a higher resolution of possible UV-induced alterations in the composition of the bacterioplankton community.
(This work is in partial fulfillment of the requirements for a M.S. degree from the University of Vienna [C.W.].)
MATERIALS AND METHODS
Sampling site and sample collection.
Ten to 15 liters of near-surface (0.5-m depth) seawater was collected from the Netherlands Institute for Sea Research (NIOZ) pier (53°00′ N, 4°45′ E) during high tide, when North Sea water enters the Wadden Sea. The seawater was collected in acid-cleaned polypropylene containers, brought back to the laboratory, and immediately processed as described below (Fig. 1). Additionally, 4-ml subsamples from this water were fixed with 4% formaldehyde (final concentration) to determine the abundance of the bacterioplankton as described below.
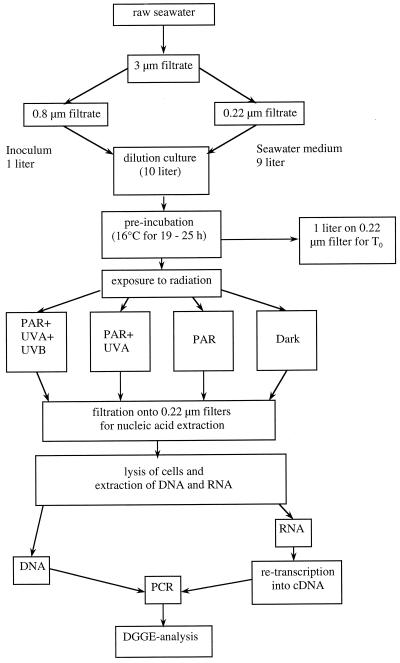
FIG. 1.
Flow chart of experimental protocol. For details and abbreviations see Materials and Methods.
Experimental protocol.
In total, nine experiments were performed between April and September 1998, two under natural surface solar radiation and seven under simulated solar radiation, as outlined in Fig. 1. Briefly, the collected water was first filtered through a 3-μm-pore-size polycarbonate membrane (Isopore TSTP; diameter, 142 mm; Millipore) to remove larger plankton and sediment particles. Seawater dilution cultures were established by filtering 1 liter of water through 0.8-μm-pore-size polycarbonate filters (Isopore ATTP; diameter, 47 mm; Millipore) and inoculating the filtrate into 9 liters of 0.22-μm-pore-size (Isopore GVWP; diameter, 142 mm; Millipore)-filtered water. The dilution cultures were established within 1.5 to 2 h after sampling of the original water.
The dilution culture was held in the dark at 16°C until the bacteria reached early exponential growth after 19 to 25 h, as determined by frequent bacterial enumeration (at 2- to 3-h intervals) (referred to below as preincubation in the dark). Subsequently, 1 liter of the dilution culture was filtered onto a 0.22-μm-pore-size filter (Isopore GVWP; diameter, 142 mm). This filter was cut into small pieces with ethanol-flamed scissors, transferred into 50-ml polypropylene tubes (Greiner), and immediately frozen in liquid nitrogen and stored at −80°C until extraction for later DGGE analysis of the bacterial community of the preexposure incubation. The remaining water of the dilution culture was split and used to fill acid-cleaned quartz glass tubes (volume per tube, ∼1 liter; inner diameter, ∼5 cm), and the tubes were closed with acid-rinsed silicon stoppers coated with Teflon and exposed in duplicate to natural or simulated solar radiation of different wavelength ranges (Fig. 1). The preincubation in the dark was performed because previous experiments revealed shifts in the species composition of the bacterioplankton community when confirmed in containers (data not shown). Such shifts unrelated to the different radiation regimes would have masked the possible effect of UV radiation on the composition of the bacterioplankton community. Through the introduction of a preincubation in the dark, those members of the bacterioplankton community sensitive to confinement are excluded.
Four different radiation regimes were established: (i) the full range of solar radiation (PAR plus UVA plus UVB) by exposing the quartz tubes directly to natural or simulated solar radiation, (ii) PAR plus UVA by wrapping a Mylar foil (Mylar-D; 50% transmittance at 325 nm; Dupont) around the quartz tubes, (iii) the PAR treatment, by excluding the entire UV range with acrylic glass (Plexiglass XT colorless 21570 AR; 3 mm thick; 50% transmittance at 390 nm; Thun-Hohenstein GmbH), and (iv) a dark treatment established by wrapping the incubation tubes in aluminum foil. For exposure to natural solar radiation, a flowthrough seawater bath was used to maintain in situ temperature. The incubation tubes were overlaid by a 1- to 2-cm-thick water layer. The experiments under simulated solar radiation were performed in a water bath (the temperature was controlled by an RC 6CS laboratory cooler [Lauda]) at in situ temperature. Underneath the water bath of the solar simulator, a second flowthrough bath with tap water (height of the cooling jacket, 1 cm) was placed to reduce the amount of heat originating from the PAR source which irradiated the incubation tubes from below. The bottoms of both water baths consisted of high-quality borosilicate glass. The PAR source of the solar simulator consisted of two HQI-T Powerstar lamps (each 250 W; Osram) in LEO/S 252-N-CR housings (catalog no. COD 05750013; SBP Company). The sources of UV radiation were mounted on top of the temperature-controlled water bath. UVA (320- to 400-nm-wavelength) irradiation was provided by a Philips TL100W/10R fluorescent tube; UVB irradiation was provided by two UVA-340 fluorescent lamps (Q-Panel Company, Bolton, United Kingdom). The radiation intensity was adjusted by varying the distance between the radiation sources and the incubation tubes. Details of the radiation regimes used in the different experiments are given in Table 1.
TABLE 1.
Summary of environmental conditions for the nine experiments performeda
| Parameter | Value
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Expt 4 | Expt 5 | Expt 6 | Expt 7 | Expt 8 | Expt 9 | |
| Date | 22 Jun | 25 Jun | 21 Jul | 25 Jul | 7 Aug | 17 Aug | 24 Aug | 31 Aug | 4 Sep |
| Temp (°C) | 16 | 16 | 18 | 18–21 | 18–21 | 18 | 18 | 16 | 17 |
| Preincubation periodb | 22 | 23 | 25 | 19 | 20 | 23 | 20 | 21 | 22 |
| Mean PARc on day 1 (day 2) | 650 | 650 | 650 | 958 | 880 (1,040) | 880 | 880 | 880 | 880 |
| Mean dose 305 nmd on day 1 (day 2) | 1.1 | 1.1 | 1.1 | 1.06 | 1.18 (1.45) | 1.48 | 1.48 | 1.48 | 1.48 |
| Mean dose 320 nmd on day 1 (day 2) | 7.68 | 7.68 | 7.68 | 14.26 | 13.37 (16.2) | 10.37 | 10.37 | 10.37 | 10.37 |
| Mean dose 340 nmd on day 1 (day 2) | 14.5 | 14.5 | 14.5 | 26.58 | 24.1 (28.9) | 19.56 | 19.56 | 19.56 | 19.56 |
| Mean dose 380 nmd on day 1 (day 2) | 30.16 | 30.16 | 30.16 | 35.46 | 32.04 (38) | 37.88 | 37.88 | 37.88 | 37.88 |
| Total dose PARe | 3,042 | 3,042 | 3,042 | 4,140 | 9,270 | 8,554 | 3,802 | 7,603 | 7,603 |
| Total dose 305 nmf | 0.05 | 0.05 | 0.05 | 0.05 | 0.12 | 0.14 | 0.06 | 0.13 | 0.13 |
| Total dose 320 nmf | 0.36 | 0.36 | 0.36 | 0.62 | 3.42 | 1.01 | 0.45 | 0.9 | 0.9 |
| Total dose 340 nmf | 0.68 | 0.68 | 0.68 | 1.15 | 2.55 | 1.9 | 0.85 | 1.69 | 1.69 |
| Total dose 380 nmf | 1.41 | 1.41 | 1.41 | 1.53 | 3.37 | 3.68 | 1.64 | 3.27 | 3.27 |
| Exposure period day 1b | 13 | 13 | 13 | 12 | 15 | 15 | 12 | 15 | 15 |
| Dark periodb | 0 | 11 | 11 | 0 | 9 | 9 | 0 | 9 | 9 |
| Exposure period day 2b | 12 | 12 | |||||||
The date, water temperature, duration of the preexposure incubation, and radiation dose for the PAR and for the different wavelengths in the UV range are given. Also given are the duration of the exposure periods for day 1, the dark period, and the subsequent exposure period (day 2). Experiments 4 and 5 were carried out under natural solar radiation; all the other experiments were performed under the solar simulator.
In hours.
In microeinsteins m−2 s−1.
In μW cm−2 nm−1.
In microeinsteins cm−2.
In W cm−2 nm−1.
After the seawater dilution cultures were exposed to the different radiation regimes, the bacterial communities from the different treatments were collected on 0.22-μm-pore-size filters (Isopore) within 1 h. Additionally, each tube was subsampled to determine bacterial abundance (described below).
Solar radiation measurements.
During the exposure of the incubation tubes to natural solar radiation, irradiance was monitored with a PUV 500 surface sensor (Biospherical Instruments) measuring PAR (400 to 700 nm) and also UV radiation at four distinct UV wavelengths (305, 320, 340, and 380 nm). The recording interval was 1 min, and the measurements were corrected for the zero offset determined for 20 min before and after the actual measurement. The irradiation intensity measured under a clear, cloudless sky (22 June 1998) was used to adjust the radiation intensities of the various light sources in the solar simulator as described above.
Bacterial abundance.
During the course of the preexposure incubation of the dilution culture in the dark and at the end of the exposure experiments, 4 ml of water was withdrawn from each tube, and the bacteria were stained with acridine orange and enumerated under a Zeiss Axioplan epifluorescence microscope (19). At least 30 fields or 350 bacteria were counted per sample.
Characterization of the composition of the bacterioplankton community. (i) Nucleic acid extraction.
The protocol used for nucleic acid extraction is a combination of two previously described methods consisting of four freeze-thaw cycles (−196 to +37°C) and subsequent treatment with lysozyme and proteinase K in 1% sodium dodecyl sulfate (6, 36). In detail, the filters were thawed on ice, and then 2 ml of 1× lysis buffer (50 mM Tris, 20 mM Na2-EDTA [HCl, pH 8.0]) was added. The four freeze-thaw cycles were performed by placing the tubes in liquid nitrogen and subsequently thawing the filters in a water bath at 37°C. Thereafter, 2 ml of lysis buffer was added again, and the filters were treated with lysozyme (catalog no. L7651; Sigma), at a final concentration of 1.25 mg ml−1 at 37°C for 30 min. Lysis of the cells was completed by adding sodium dodecyl sulfate and proteinase K (catalog no. 82456; Fluka) at final concentrations of 1% (wt/vol) and 100 μg ml−1, respectively, and incubating the vials at 55°C for 2 h. The lysis efficiency was checked by epifluorescence microscopy. No intact cells were found after the final incubation step, indicating complete lysis of the cells.
The liquid phase was extracted once with an equal volume of 1× TE (10 mM Tris-HCl [pH 8.0], 1 mM Na2-EDTA)-buffered phenol, a mixture of phenol, chloroform, and isoamyl alcohol (ratio, 25:24:1) buffered with 1× TE and with chloroform-isoamyl alcohol (24:1). Standard ethanol precipitation overnight was used to concentrate and clean the extracted nucleic acids. The pellet was washed twice with 200 μl of ice-cold 70% ethanol and redissolved in 100 μl of diethylpyrocarbonate (DEPC)-treated Milli-Q water (36).
(ii) DNA preparation and PCR conditions.
Five microliters of crude nucleic acid extract from each sample was loaded onto a 1% agarose gel (Bio-Rad), run at 5 V/cm in 1× TBE (89 mM Tris-HCl [pH 8.3], 89 mM boric acid, 2 mM Na2-EDTA) for 3.5 h, and poststained with ethidium bromide (0.1 μg ml−1) to check the integrity of the DNA (data not shown). To determine the optimal template concentration, dilutions were made from two subsamples per experiment. RNA was digested by adding 4.5 μl of a 1-mg ml−1 RNase I, A solution (catalog no. 27-0323 [Pharmacia Biotech]; made DNase-free by boiling at 100°C for 20 min) to 45 μl of nucleic acid extract and incubating the mixture in a heating block at 55°C for 30 min. DNA was cleaned with a QIAEX II gel extraction kit (Qiagen) as recommended by the manufacturer for fragments larger than 10 kbp (checked by agarose gel electrophoresis as described above). The DNA was dissolved in 20 μl of elution buffer (Qiagen).
The highest and lowest concentrations of the samples were serially diluted 1:10, 1:20, 1:50, 1:100, and 1:200 in elution buffer (Qiagen), and 1 μl of each dilution was used as a template for PCR amplification (35) with the eubacterial specific primer pair GM5f and 907r (31). The PCR conditions were similar to those described by Moeseneder et al. (29) except that we performed another 20 cycles after the initial 10 cycles of touchdown PCR (13). The PCR products had a length of 626 bp and corresponded to Escherichia coli 16S ribosomal DNA (rDNA) numbering positions 341 to 927. Every product was checked for by-products and proper size by applying 5 μl of 50-μl reaction mixtures on a 1.5% agarose gel run at 5 V/cm in 1× TBE for 50 min and poststained with ethidium bromide. The first dilution lacking unspecific by-products was chosen as the optimal template concentration and used for further analysis.
(iii) RNA preparation and cDNA synthesis.
Crude nucleic acid extracts of selected samples were diluted as described above. DNA was digested by adding 15.5 μl of DEPC-treated Milli-Q water, 4 μl of 10× assay buffer (40 mM Tris-HCl [pH 7.5], 6 mM MgCl2), and 0.5 μl of DNase I (catalog no. 27-0514 [Pharmacia Biotech]; 10 U/μl) to 20 μl of extract. The reaction mixtures were incubated at 37°C for 30 min and subsequently extracted once with an equal volume of 1× TE-buffered phenol–chloroform–isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1). Nucleic acids were ethanol-precipitated overnight and redissolved in 31 μl of DEPC-treated Milli-Q water. One microliter of each dilution was subjected to PCR as described above. The dilution with no visible product was chosen as the maximum concentration of DNA still digestible by the amount of DNase used. After all samples had been adjusted to the proper concentration, RNA was reverse transcribed into first-strand cDNA using Ready To Go You-Prime First-Strand Beads (catalog no. 27-9264-01; Pharmacia Biotech) as recommended by the manufacturer. The primers used were pd(N)6 (catalog no. 27-2166; Pharmacia Biotech) random hexamers at a final concentration of 200 ng per reaction. Immediately after the reactions were completed, RNA was digested by adding 2 μl of RNase I A (stock 200 ng μl−1) to each reaction mixture and incubating the mixtures at 55°C for 30 min. First-strand cDNA was cleaned using QIAquick spin columns (Qiagen). The final volume was 20 μl in elution buffer (Qiagen). We used the same procedures described above to determine the optimal template concentration for the PCRs. The PCR conditions were the same as for DNA.
(iv) DGGE and image analysis.
DGGE was performed on a DCode Universal Mutation detection system (Bio-Rad) as described by Moeseneder et al. (29). Per sample, four PCR products were pooled and precipitated with ethanol overnight, and the resulting pellet was redissolved in 9 μl of 1× TE buffer. One microliter of this solution and the Precision molecular mass ruler (catalog no. 170-8207 [Bio-Rad]; undiluted and in 1:2, 1:5, and 1:10 dilutions) were applied to a 1.5% agarose gel. Applying a mass ruler allowed us to load equal amounts of PCR products per lane and experiment. The total amount of PCR products loaded onto the DGGE gels was 500 to 800 ng per lane. The gels were poststained with GelStar (FMC) as recommended by the manufacturer for polyacrylamide gel applications.
The analysis of the resulting DGGE banding patterns and image acquisition was performed with a Fluor-S MultiImager (Bio-Rad) and the Multi-Analyst software (version 1.0.2 for Apple Power PC) as described by Moeseneder et al. (29). The banding patterns of the individual lanes were transformed into a density profile by using the Extract Profiles feature of the Multi-Analyst software. The profiles were aligned with the corresponding lanes to allow better visualization.
(v) Phylogenetic affiliation of selected DGGE bands.
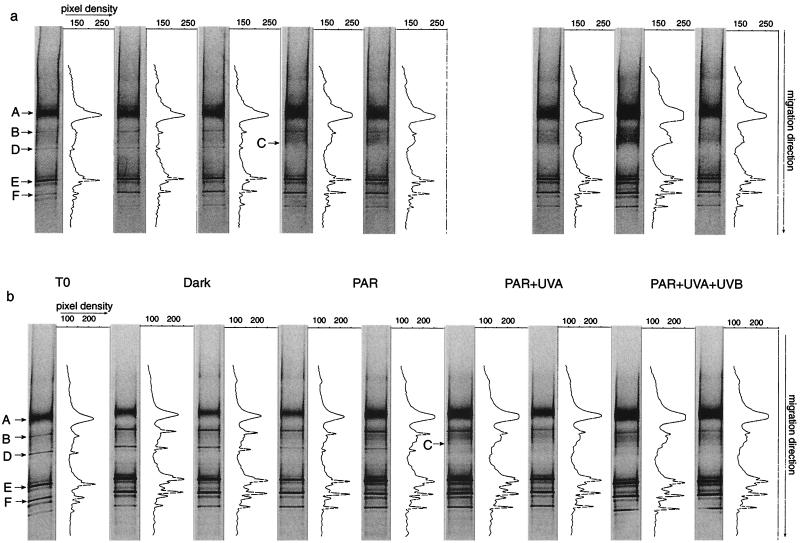
In total, six different bands were excised from the DNA and RNA DGGE gels of experiment 5 (see Fig. 4) and experiment 8 to confirm the alignment among DGGE gels of different experiments. The gel slices were overlaid with 200 μl of autoclaved Milli-Q water for 3 h. Thereafter, the Milli-Q water was replaced by 20 μl of autoclaved Milli-Q water and allowed to stand for 24 h. Between 1 and 5 μl of this water was then subjected to PCR and DGGE as described above to ensure the purity of the excised bands (data not shown). Sequencing of the reamplified bands was performed with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (catalog no. 4303152; Perkin-Elmer) in a PCR consisting of 25 cycles (denaturation at 96°C for 10 s, annealing at 45°C for 5 s, and elongation at 60°C for 4 min). We sequenced from both ends of the fragments by using the forward primer GM5f without the 40-bp GC clamp and, in another reaction, the reverse primer 907r as described above. The amplicons were subsequently purified by isopropanol precipitation, and sequences were obtained with an automated sequencer (ABI Prism 310; Perkin-Elmer–Applied Biosystems). Electrophoresis, data acquisition, and analysis were performed using the settings recommended by the manufacturer. First, the sequences were evaluated with the program Chimera Check of the Ribosomal Database Project II at Michigan State University (28) to exclude possible chimeric artifacts. Further analysis involved comparison to the 16S rDNA sequences at the GenBank nucleotide library by BLAST searching (2). The six DGGE bands excised (subsequently referred to as operational taxonomic units [OTUs] [31]) were chosen because they were present in all the experiments and in all radiation treatments (OTUs A, E, and F), were sensitive to UV radiation (OTUs B and D), or appeared in UV radiation but not in the dark treatments (OTU C).
FIG. 4.
OTU patterns and corresponding intensity profiles obtained for the bacterioplankton community of the preexposure incubations (T0) and after exposure to the different radiation regimes under natural surface solar radiation for experiment 5 at the DNA (a) and RNA (b) levels as revealed by DGGE. The OTUs marked by arrows were sequenced. In panel a, one of the duplicates for the PAR-UVA treatment was lost due to a mistake in the preparation for loading the sample onto the DGGE gel.
Nucleotide sequence accession numbers.
The 16S rDNA sequences obtained in this study were submitted to GenBank and are available under various accession numbers (see Table 3).
TABLE 3.
Summary of analysis of sequences obtained for OTUs
| OTU | Length of sequence (bp) | 16S rDNA identification (closest neighbor) | % Sequence similarity | Phylogenetic groupa | GenBank no. |
|---|---|---|---|---|---|
| A | 551 | Flexibacter maritimus (M64629) | 96 | FCB | AF207850 |
| B | 524 | P. irgensii (M61002) | 95 | FCB | AF207851 |
| C | 555 | Unidentified bacterium (UBZ88574) | 99 | FCB | AF207852 |
| D | 536 | Unidentified bacterium (AJ224942) | 91 | FCB | AF207849 |
| E | 534 | Uncultured marine bacterium D049 (AF177568) | 96 | α Proteobacteria | AF207853 |
| F | 559 | Alteromonas macleodii strain CH-516 (AMY18234) | 97 | γ Proteobacteria | AF207848 |
FCB, Flexibacter-Cytophaga-Bacteroides.
RESULTS
Initial bacterial abundance and bacterioplankton community pattern.
The bacterial abundance in the original seawater ranged from 8.0 × 106 to 16.3 × 106 cells ml−1 (Fig. 2a). Experiments 4 and 5 were performed during a massive bloom of Phaeocystis sp. For experiments 5 to 9, the loss of bacteria due to the initial filtration steps was determined. On average, (27 ± 4)% (n = 5) of the original bacterial abundance was lost by filtering through 3-μm-pore-size filters and (11 ± 3)% (n = 5) was lost when the 3-μm filtrate was filtered through 0.8-μm-pore-size filters. Due to the relatively high particle load of the water, we assume that mainly particle-associated bacteria were retained by the filtration steps. The 0.8-μm filtrate (i.e., the bacterioplankton community) was used in the subsequent exposure experiments.
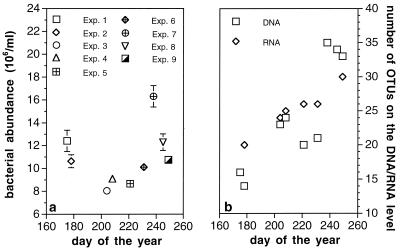
FIG. 2.
(a) Bacterial abundance in original, unfiltered seawater on different sampling dates. The vertical bars represent standard errors in duplicate samples. (b) Number of OTUs detected by DGGE at the DNA and RNA levels after preexposure incubation and in the dark treatment.
Over the sampling period, the number of OTUs per lane obtained by DGGE increased in the initial preexposure dilution cultures (Fig. 2b). For experiments 1 to 8, the number of OTUs at the RNA level was equal to or slightly higher than that at the DNA level. The variable incubation period of the subsequent exposure to different radiation regimes did not influence the number of OTUs detectable at the end of the exposure (Table 2). Thus, the preexposure incubation period was long enough to allow the growth of a diverse bacterial consortia, excluding, however, those bacterial species sensitive to confinement.
TABLE 2.
Summary of number of OTUs detected at DNA and RNA levels by DGGE
| Expt no. | No. of OTUsa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T0
|
Dark
|
PAR
|
PAR-UVA
|
PAR-UVA-UVB
|
||||||
| DNA | RNA | DNA | RNA | DNA | RNA | DNA | RNA | DNA | RNA | |
| 1 | 16 | 16 | 16 | 16 | 15 | 16 | 15 | 16 | 14 | 16 |
| 2 | 14 | 20 | 14 | 20 | 14 | 20 | 14 | 20 | 14 | 20 |
| 3 | 23 | 24 | 23 | 24 | 23 | 24 | 23 | 24 | 23 | 24 |
| 4 | 24 | 25 | 24 | 25 | 24 | 25 | 24 | 25 | 24 | 25 |
| 5 | 20 | 26 | 20 | 26 | 20 | 26 | 19 | 25 | 19 | 25 |
| 6 | 21 | 26 | 21 | 26 | 21 | 26 | 21 | 26 | 21 | 26 |
| 7 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 |
| 8 | 34 | 34 | 34 | 34 | 34 | 34 | 33 | 33 | 33 | 33 |
| 9 | 33 | 30 | 33 | 30 | 33 | 30 | 32 | 28 | 32 | 28 |
The number of OTUs is given for the end of the preexposure incubation held in the dark (T0) and after exposure to the different radiation regimes. The corresponding exposure periods and the radiation intensities are given in Table 1.
Radiation conditions.
The exposure conditions for all the experiments performed are shown in Table 1. The organisms in experiments 1 to 3, exposed to simulated solar irradiation, received much lower doses than those in all the other experiments. Experiment 5 was conducted under natural solar radiation over a total period of 36 h (Table 1). The radiation intensity measured during the first day of experiment 2 was used to adjust the radiation levels in the subsequent experiments (experiments 6 to 9) under the solar simulator. For the reference day, we obtained a radiation dose amounting to about 70% of the maximum daily dose measured during the investigation period.
Influence of different radiation regimes on bacterioplankton community composition.
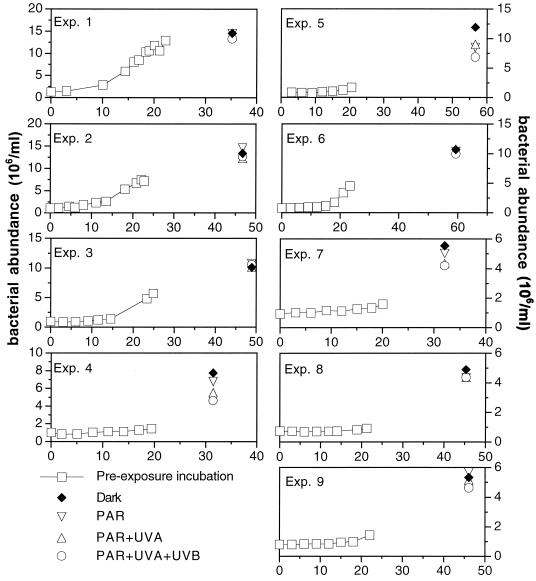
The development of bacterial abundance for all nine experiments in the preexposure incubation and subsequently, at the end of the exposure to different radiation ranges using simulated or natural solar radiation, is shown in Fig. 3. The differences in bacterial abundance among the different radiation regimes in experiments 1 to 3, 6, 8, and 9 at the end of the exposure were rather small (Fig. 3). Pooling all the experiments, significantly lower bacterial abundance in the treatment receiving full solar radiation (natural or simulated) was obtained compared to that in the PAR and the dark treatments (sign test, P = 0.039; n = 9).
FIG. 3.
Time course of bacterial abundance in the preexposure incubation and at the end of the exposure to different radiation regimes for all the experiments performed. Error bars are not visible because the standard error is smaller than the symbol.
A representative OTU pattern of the bacterial community resulting from the exposure to different radiation conditions is depicted in Fig. 4. The OTUs sensitive to UV radiation or present in all experiments and used for between-gel comparisons were sequenced (Fig. 4). The percent similarities to the closest related sequences and the corresponding phylogenetic groups are presented in Table 3. Experiment 5 covered two diurnal cycles of comparable natural solar radiation with one nocturnal cycle in between (Table 1). The OTU pattern revealed differences among the different radiation regimes. On the DNA level (Fig. 4a), OTU B was present in much lower intensities in the PAR-UVA and the PAR-UVA-UVB treatments, while OTU D was completely missing in these light treatments. These two OTUs (B and D), however, were clearly visible after the preexposure incubation and in the PAR and the dark treatments (Fig. 4a). After the preexposure incubation and in the dark treatment, OTU C was at the lower limit of detection, but it became clearly visible under PAR, PAR-UVA, and PAR-UVA-UVB radiation conditions. At the RNA level, the differences among the various radiation conditions were more pronounced (Fig. 4b). OTU B was detectable in all the radiation treatments, while OTUs C and D revealed the same pattern as at the DNA level (Fig. 4b).
OTU D, although obviously sensitive to UV stress, was detectable throughout the entire period of the study at the end of the preexposure incubation and in the dark treatment (Fig. 4). It was not detectable, however, at the RNA level in the treatments receiving full radiation in four out of the nine experiments and in three experiments in the PAR-UVA treatment. In experiments 8 and 9 (covering 15 h of the diurnal and 9 h of the nocturnal cycle), several OTUs were absent in the PAR-UVA and PAR-UVA-UVB treatments (Table 2). In experiments 2 and 3, no changes in the banding patterns of the different treatments were detected, even after a nocturnal period following exposure (Table 1). Exposure over two diurnal cycles and one nocturnal cycle (experiments 5 and 6) did not reveal a coherent OTU pattern reflecting the influence of UV radiation (Table 2). In experiment 5, carried out under natural solar radiation, some OTUs were absent, especially at the DNA level in the PAR-UVA and PAR-UVA-UVB treatments. The OTU pattern of experiment 6, conducted under radiation intensities similar to those of experiment 5 but under the solar simulator, was not affected by the presence of UV radiation (Table 2).
DISCUSSION
We used seawater dilution cultures in combination with DGGE to determine the possible influence of solar radiation on the composition of the bacterioplankton community. The filtration steps required to establish the dilution cultures resulted in a loss of ≈35% of the bacteria present in the original seawater. Because ≈27% of this overall loss can be assigned to the initial filtration step through 3-μm-pore-size filters, we attribute this loss primarily to the removal of particle-associated bacteria, which are abundant in the coastal North Sea.
In previous experiments with water from the northern Adriatic, we consistently obtained remarkable differences between the OTU patterns of the original bacterioplankton community and that after confinement for 24 to 48 h (unpublished data). Therefore, we established preexposure dilution cultures in the dark for 20 to 30 h before splitting the volume equally among the different radiation treatments. This procedure resulted in identical OTU patterns in the duplicate incubations in all the experiments and in the preexposure incubation and the dark treatment (Fig. 1 and 2). Thus, the preexposure incubation effectively reduced the noise in the data introduced by bacterial species sensitive to confinement.
Since bacterial growth was essential for the interpretation of our results, exposure experiments were started when the bacteria entered exponential growth in the preexposure incubation (except in experiment 1). With the exception of experiment 9, the number of OTUs after the preincubation and in the dark treatment was equal or higher on the RNA level than on the DNA level (Fig. 2b), indicating growth of bacteria in the different radiation treatments. Only actively growing bacterial cells produce ribosomes and therefore 16S rRNA for cellular metabolism. Bacterial growth in the different treatments allowed us to detect even OTUs at the RNA level which were below the detection limit at the DNA level (Table 2). This suggests that their contribution to the community in terms of cell numbers was too low for detection, but due to their high metabolic activity and the accompanying 16S rRNA synthesis, these OTUs were readily detectable at the RNA level (39). As mentioned in Materials and Methods, we checked every sample for DNA contamination by PCR amplifying the DNase digest before transcribing the remaining RNA into first-strand cDNA. Therefore, we are confident that bacterial cells were growing during the light incubation period.
The results presented in Table 2 show that the OTU patterns became more complex towards the end of the sampling period (September). This probably reflects increased species richness within the bacterial community as the massive Phaeocystis bloom at the initial phase of the study was replaced by a more diverse phytoplankton community towards the end of the sampling period, leading also to a larger diversity of potentially utilizable substrate (11). In each experiment, the OTU patterns at the end of the preexposure incubation and in the subsequent dark treatment were identical at both the DNA and RNA levels, indicating highly reproducible experimental conditions.
Exposing the dilution cultures to different radiation regimes resulted in significantly lower bacterial abundance in the treatment receiving full solar radiation than in the PAR and the dark treatments (Fig. 3). During exposure to solar radiation, photochemical degradation of DOM takes place, increasing the level of low-molecular-weight compounds utilizable by bacterioplankton (8, 12, 40). UV radiation, therefore, might indirectly counteract its direct negative effects exerted on bacterioplankton activity (18) by rendering a part of the DOM pool more labile for bacterioplankton (30).
At most three OTUs per experiment (out of 26) were affected by UV radiation (Fig. 4 and Table 2). Whenever the OTU pattern indicated differences between the UV and the non-UV treatments, OTU D was among the affected OTUs, indicating that this member of the Flexibacter-Cytophaga-Bacteroides group (Table 3) is highly sensitive to UV radiation. Whether this unidentified bacterium (OTU D) is an important member of the bacterioplankton community in terms of carbon and energy flux remains to be elucidated. OTU B also appears to be sensitive to UV radiation (Fig. 4) and is closely related to Polaribacter irgensii (Table 3) (10). Nevertheless, throughout the sampling period (June to early September 1998), OTUs B and D were readily detectable in the preexposure incubations of water collected from the surface layer of the North Sea, indicating that they are present in the water column throughout the summer despite their sensitivity to UV radiation. The persistence and survival of UV-sensitive bacteria in the North Sea is facilitated by the relatively high turbidity and wind-induced mixing. This might be in contrast to oligotrophic waters, with their lower attenuation of UV radiation and the establishment of diurnal stratification of the upper layers of the water column (14). Under such conditions, the microorganisms are trapped in a layer with high radiation levels for almost the entire period of solar radiation (14), possibly resulting in an efficient suppression of UV-sensitive bacterioplankton species in the euphotic zone.
In summary, we have shown that UVB and, to a lesser extent, also UVA radiation lead to only minor alterations in the species composition of the bacterioplankton community in dilution cultures established with coastal North Sea water. Only up to ≈10% (at most 3 out of 26 OTUs) of the bacterioplankton species are sensitive to UV radiation. The sensitive species were, however, readily detectable in the upper layers of the water column of the North Sea throughout the summer. Therefore, these species are never exposed to a dose of UV radiation high enough to lead to their complete disappearance under the conditions prevailing in the North Sea (such as high attenuation of UV radiation in the water column and wind-induced mixing). However, under subtropical or tropical open-ocean conditions with low attenuation of UV radiation, penetrating about half of the photic zone in a biologically affective dose, UV radiation might be an important factor influencing the species composition of the bacterioplankton community.
ACKNOWLEDGMENTS
We are grateful to D. Slezak for her help in planning the solar simulator and the discussions during experimental work and to the staff of the workshop at the NIOZ for constructing the solar simulator. We thank J. M. Arrieta, who was prepared to answer any question at any time. The continuous support of our colleagues at the NIOZ during the course of this study is gratefully acknowledged.
Funding was provided by the Austrian Students Exchange Program of the Ministry of Science and Transport, the MAST-MTP II MATER project (MAS3-CT96-0051), and the Environment and Climate Program of the European Union (MICOR; project EV5V-CT94-0512).
Footnotes
Publication no. 3548 of the Netherlands Institute for Sea Research.
REFERENCES
- 1.Aas P, Lyons M M, Pledger R, Mitchell D L, Jeffrey W H. Inhibition of bacterial activities by solar radiation in nearshore waters and the Gulf of Mexico. Aquat Microb Ecol. 1996;11:229–238. [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Arrieta J M, Weinbauer M G, Herndl G J. Interspecific variability in sensitivity to UV radiation and subsequent recovery in selected isolates of marine bacteria. Appl Environ Microbiol. 2000;66:1468–1473. doi: 10.1128/aem.66.4.1468-1473.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azam F, Fenchel T, Field J G, Gray J S, Meyer-Reil L A, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 5.Baker K S, Smith R C. Spectral irradiance penetration in natural waters. In: Calkins J, editor. The role of solar ultraviolet radiation in marine ecosystems. New York, N.Y: Plenum Press; 1982. pp. 233–246. [Google Scholar]
- 6.Bej A K, Mahbubani M H, Dicesare J L, Atlas R M. Polymerase chain reaction-gene probe detection of microorganisms by using filter-concentrated samples. Appl Environ Microbiol. 1991;57:3529–3534. doi: 10.1128/aem.57.12.3529-3534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benner R, Biddanda B. Photochemical transformations of surface and deep marine dissolved organic matter: effects on bacterial growth. Limnol Oceanogr. 1998;43:1373–1378. [Google Scholar]
- 8.Bertilsson S, Tranvik L J. Photochemically produced carboxylic acids as substrates for freshwater bacterioplankton. Limnol Oceanogr. 1998;43:885–895. [Google Scholar]
- 9.Blumthaler M, Ambach W. Indication of increasing solar ultraviolet-B radiation flux in Alpine regions. Science. 1990;248:206–208. doi: 10.1126/science.2326634. [DOI] [PubMed] [Google Scholar]
- 10.Bowman J P, McCammon S A, Brown M V, Nichols D S, McMeekin T A. Diversity and association of psychrophilic bacteria in Antarctic Sea ice. Appl Environ Microbiol. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brussaard C P D, Riegman R, Noordeloos A A M, Cadée G C, Witte H, Kop A J, Nieuwland G, Duyl F C V, Bak R P M. Effects of grazing, sedimentation and phytoplankton cell lysis on the structure of a coastal pelagic food web. Mar Ecol Prog Ser. 1995;123:259–271. [Google Scholar]
- 12.Bushaw K L, Zepp R G, Tarr M A, Schulz-Jander D, Bourbonniere R A, Hodson R E, Miller W L, Bronk D A, Moran M A. Photochemical release of biologically available nitrogen from aquatic dissolved organic nitrogen. Nature. 1996;381:404–407. [Google Scholar]
- 13.Don R H, Cox P T, Wainwright B, Baker K, Mattick J S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doney S C, Najjar R G, Stewart S. Photochemistry, mixing and diurnal cycles in the upper ocean. J Mar Res. 1995;53:341–369. [Google Scholar]
- 15.Ducklow H W, Carlson C A. Oceanic bacterial production. In: Marshall K C, editor. Advances in microbial ecology. New York, N.Y: Plenum Press; 1992. pp. 113–181. [Google Scholar]
- 16.Fleischmann E M. The measurement and penetration of ultraviolet radiation into tropical marine water. Limnol Oceanogr. 1989;34:1623–1629. [Google Scholar]
- 17.Garcia-Pichel F. A model for the internal self-shading in planktonic organisms and its implications for the usefulness of ultraviolet sunscreens. Limnol Oceanogr. 1994;39:1704–1717. [Google Scholar]
- 18.Herndl G J, Mueller-Niklas G, Frick J. Major role of ultraviolet-B in controlling bacterioplankton growth in the surface layer of the ocean. Nature. 1993;361:717–719. [Google Scholar]
- 19.Hobbie J E, Daley R J, Jasper S. Use of nucleopore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffrey W H, Haven R V, Hoch M P, Coffin R B. Bacterioplankton RNA, DNA, protein content and relationships to rates of thymidine and leucine incorporation. Aquat Microb Ecol. 1996;10:87–95. [Google Scholar]
- 21.Jeffrey W H, Pledger R J, Aas P, Hager S, Coffin R B, Haven R V, Mitchell D L. Diel and depth profiles of DNA photodamage in bacterioplankton exposed to ambient solar ultraviolet radiation. Mar Ecol Prog Ser. 1996;137:283–291. [Google Scholar]
- 22.Jerlov N. Ultra-violet radiation in the sea. Nature. 1950;166:111–112. doi: 10.1038/166111a0. [DOI] [PubMed] [Google Scholar]
- 23.Joux F, Jeffrey W H, LeBaron P, Mitchell D L. Marine bacterial isolates display diverse responses to UV-B radiation. Appl Environ Microbiol. 1999;65:3820–3827. doi: 10.1128/aem.65.9.3820-3827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaiser E, Herndl G J. Rapid recovery of marine bacterioplankton activity after inhibition by radiation in coastal waters. Appl Environ Microbiol. 1997;63:4026–4031. doi: 10.1128/aem.63.10.4026-4031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karentz D, Bothwell M L, Coffin R B, Hanson A, Herndl G J, Kilham S S, Lesser M P, Lindell M, Moeller R E, Morris D P, Neale P J, Sanders R W, Weiler C S, Wetzel R G. Impact of UV-B radiation on pelagic freshwater ecosystems: report of the working group on bacteria and phytoplankton. Arch Hydrobiol. 1994;43:31–69. [Google Scholar]
- 26.Lindell M, Granéli W, Tranvik L J. Enhanced bacterial growth in response to photochemical transformation of dissolved organic matter. Limnol Oceanogr. 1995;40:195–199. [Google Scholar]
- 27.Lyons M M, Aas P, Pakulski J D, Waasbergen L V, Miller R V, Mitchell D L, Jeffrey W H. DNA damage induced by ultraviolet radiation in coral-reef microbial communities. Mar Biol. 1998;130:537–543. [Google Scholar]
- 28.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moeseneder M M, Arrieta J M, Muyzer G, Winter C, Herndl G J. Optimization of terminal restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:3518–3525. doi: 10.1128/aem.65.8.3518-3525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran M A, Zepp R G. Role of photoreactions in the formation of biologically labile compounds from dissolved organic matter. Limnol Oceanogr. 1997;42:1307–1316. [Google Scholar]
- 31.Muyzer G, Teske A, Wirsen C O. Phylogenetic relationships of Thiomicrospira species and their identification in deep sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 32.Obernosterer I, Kraay G, Ranitz E D, Herndl G J. Dynamics of low molecular weight carboxylic acids and carbonyl compounds in the Aegean Sea (Eastern Mediterranean) and the turnover of pyruvic acid. Aquat Microb Ecol. 1999;20:147–156. [Google Scholar]
- 33.Obernosterer I, Reitner B, Herndl G J. Contrasting effects of solar radiation on dissolved organic matter and its bioavailability to marine bacterioplankton. Limnol Oceanogr. 1999;44:1645–1654. [Google Scholar]
- 34.Reitner B, Herndl G J, Herzig A. Role of ultraviolet-B radiation on photochemical and microbial oxygen consumption in a humic-rich shallow lake. Limnol Oceanogr. 1997;42:950–960. [Google Scholar]
- 35.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Smith R C, Baker K S. Optical properties of the clearest natural water (200–800 nm) Appl Opt. 1981;20:177–184. doi: 10.1364/AO.20.000177. [DOI] [PubMed] [Google Scholar]
- 38.Sommaruga R, Obernosterer I, Herndl G J, Psenner R. Inhibitory effect of solar radiation on thymidine and leucine incorporation by freshwater and marine bacterioplankton. Appl Environ Microbiol. 1997;63:4178–4184. doi: 10.1128/aem.63.11.4178-4184.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wawer C, Jetten M S M, Muyzer G. Genetic diversity and expression of the [NiFe] hydrogenase large-subunit gene of Desulfovibrio spp. in environmental samples. Appl Environ Microbiol. 1997;63:4360–4369. doi: 10.1128/aem.63.11.4360-4369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wetzel R G, Hatcher P G, Bianchi T S. Natural photolysis by ultraviolet irradiance of recalcitrant dissolved organic matter to simple substrates for rapid bacterial metabolism. Limnol Oceanogr. 1995;40:1369–1380. [Google Scholar]