Summary
LDLR, as the uptake receptor of low-density lipoprotein, plays a crucial role in lipid metabolism. However, the detailed mechanism by which LDLR affects hepatic triglyceride (TG) accumulation has rarely been reported. Here, we found that knockdown of LDLR effectively mitigated PA-induced TG accumulation. Further analysis revealed that the expression of LDLR was controlled by SREBP2 directly and indirectly. On one hand, transcription factor SREBP2 activated the transcription of LDLR directly. On the other hand, SREBP2 indirectly regulated LDLR by increasing the transcription of lncRNA LDLR-AS in fish. Mechanism analysis found that LDLR-AS functioned as an RNA scaffold to recruit heterogeneous nuclear ribonucleoprotein R (hnRNPR) to the 5′ UTR region of LDLR mRNA, which stabilized LDLR mRNA at the post-transcription level. In conclusion, our study demonstrates that increased LDLR transcription and mRNA stability is regulated by SREBP2 directly or indirectly, and promotes hepatic TG accumulation by endocytosing LDL in fish.
Subject areas: Biological sciences, Zoology, Cell biology, Functional aspects of cell biology
Graphical abstract

Highlights
-
•
PA-mediated LDLR increases triglyceride accumulation via the uptake of LDL in fish
-
•
SREBP2 activated by TNFα promotes LDLR transcription in fish
-
•
LncRNA LDLR-AS increases LDLR mRNA stability by recruiting hnRNPR in fish
Biological sciences; Zoology; Cell biology; Functional aspects of cell biology
Introduction
LDLR is responsible for the endocytosis of LDL from plasma to the liver and plays a critical role in lipid metabolism. The LDL-LDLR complex internalized via the clathrin-coated pits is delivered to endosomes, where LDL released from LDLR is transported to lysosomes for degradation, and LDLR is recycled to the cell surface (Xia et al., 2021). However, an excessive increase in LDLR on the cell membrane can disrupt lipid homeostasis in the liver by ingesting large amounts of LDL. Although a previous study has reported that LDLR is associated with hepatic TG accumulation (Minahk et al., 2008), it is poorly understood the detailed mechanism by which LDLR affects hepatic TG levels.
To our knowledge, LDLR expression is regulated by multiple regulators, such as inducible degrader of LDLR (IDOL), proprotein convertase subtilisin/kexin type 9 (PCSK9), and SREBP2 (Lin et al., 2021; Yu et al., 2021). IDOL can bind the intracellular tail of LDLR to induce its degradation via E3 ubiquitylation (Zelcer et al., 2009), while PCSK9 can bind to the EGF-A domain of LDLR, which promotes LDLR degradation by targeting this receptor to the lysosome (Barale et al., 2021). In contrast to PCSK9 and IDOL degrading LDLR, SREBP2 can increase LDLR expression to some extent (Yang et al., 2020a; 2020b). Furthermore, in recent years, non-coding RNAs as new regulators also play a regulatory role in LDLR function. In mice, hsa-miR-335 and hsa-miR-6825 can increase LDLR mRNA expression by reducing PCSK9/SREBP2 interaction (Lebeau et al., 2022), suggesting that LDLR function may be synergistically affected by various factors. Similar to microRNAs (miRNAs), long non-coding RNAs (lncRNAs), as a class of non-coding RNAs, also have attracted increasing attention. Although sixty lncRNAs have been demonstrated to be novel participants in lipid metabolism-related biological processes in birds and mammals, including chickens, humans, mice, rats, and pigs (Ananthanarayanan 2016; Chen, 2015; Muret et al., 2019), lncRNA related to LDLR function has not been found.
Fish are the largest group of vertebrates in the world. Although fish are evolutionarily inferior to mammals, the nutrient-sensing system and metabolic process are evolutionarily conserved. Our previous studies have found that large yellow croaker (Larimichthys crocea) shares a similar regulation in lipid metabolism with that of mammals (Chen et al., 2021; Du et al., 2020; Li et al., 2022; Xu et al., 2020; Yang et al., 2020a, 2020b), suggesting that this species is an appropriate model to investigate the mechanisms of abnormal lipid deposition. Large yellow croaker is sensitive to vegetable oils, which can lead to abnormal lipid accumulation in the liver (Li et al., 2019a; Zhu et al., 2018). Therefore, this study aims to investigate the role and regulatory mechanism of LDLR in PA-induced triglycerides accumulation in fish. Our findings may be used to develop therapeutic strategies for alleviating hepatic steatosis.
Results
Increased LDLR expression induced by PA promotes TG accumulation via the uptake of LDL
At present, the inclusion of n-3 LC-PUFA-rich fish oil in aquaculture has decreased with the increased utilization of vegetable oil. Moreover, unbalance of fatty acid profile in vegetable oil disrupted lipid metabolism in the liver. In the present study, to understand the effect of vegetable oil on the lipid accumulation in the liver, fish were randomly fed with fish oil (FO) diets (as the control), soybean oil (SO) diets, olive oil (OO) diets, or palm oil (PO) diets for ten weeks. Transmission electron microscope (TEM) observations revealed that hepatocytes in fish fed with FO diets showed a clear structure in which lipid droplets were smaller and fewer, while numerous big lipid droplets were found in fish fed with vegetable oil diets, especially in OO and PO diets (Figure 1A). These results suggested that OO and PO diets induced obvious hepatic steatosis in fish. To explore the reasons for the difference between fish oil and vegetable oil diets, fatty acid profiles of four diets were analyzed using an HP6890 gas chromatograph. The obtained results showed that FO diets, SO diets, OO diets, and PO diets were rich in docosahexaenoic acid (DHA), linoleic acid (LA), oleic acid (OA), and palmitic acid (PA), respectively (Table S1). Next, fish hepatocytes were stained using Oil Red O after treatment with DHA, LA, OA, and PA, respectively. An obvious lipid accumulation was observed in fish hepatocytes treated with PA followed by OA and LA compared to DHA (Figure 1B).
Figure 1.
LDLR induced by PA promotes TG accumulation via the uptake of LDL
(A) Lipid droplets in the liver were identified by transmission electron microscope (TEM) in fish fed with FO, SO, OO, and PO diets (Scale bars, 5 μm). The red arrow indicates lipid droplets. N, nucleus; LD, lipid droplets.
(B) Lipid accumulation in fish hepatocytes was observed by Oil Red O staining after treatment with DHA, LA, OA, or PA for 12 h (Scale bars, 100 μm).
(C and D) RT-qPCR and Western blot assays were used to evaluate the expressions of LDLR in the liver of fish fed with different diets. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(E and F) RT-qPCR and Western blot assays were used to evaluate the expressions of LDLR in the liver of fish fed with PO diets after injection with control siRNA and si-LDLR for 48 h. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗p < 0.05, ∗∗p < 0.01.
(G and H) Plasma and hepatic TG contents were tested in fish fed with PO diets after injection with control siRNA and si-LDLR for 48 h. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗∗p < 0.01.
(I) Oil Red O staining was used to evaluate lipid accumulation in the liver from fish fed with PO diets after injection with control siRNA and si-LDLR for 48 h (Scale bars, 100 μm).
(J) Fluorescent microscopy images were used to evaluate lipid droplets accumulation in fish hepatocytes treated with PA alone, Dil-LDL alone, or a combination of PA and Dil-LDL for 6 h (Scale bars, 25 μm). DAPI displays blue; BODIPY displays green; Dil-LDL displays red. FO, fish oil; SO, soybean oil; OO, olive oil, PO, palm oil; DHA, docosahexaenoic acid; LA, linoleic acid; OA, oleic acid; PA, palmitic acid. See also Figure S1.
LDLR-mediated endocytosis is essential for lipoprotein uptake and lipid homeostasis. To investigate whether LDLR was involved in vegetable oil-mediated lipid accumulation, we analyzed the effect of vegetable oils on LDLR expression. The results revealed that liver LDLR mRNA level was significantly increased in the SO, OO, and PO diets compared to the FO diets (Figure 1C), while LDLR protein level was significantly increased only in the OO and PO diets (Figure 1D). In vitro, we examined the effect of different fatty acids on LDLR expression after exposure of fish hepatocytes to DHA, LA, OA, and PA, respectively. The highest expression of LDLR in fish hepatocytes was observed in the PA group followed by the OA group compared to the DHA group (Figures S1A and S1B). The trend of LDLR expression in fish hepatocytes was consistent with the result from lipid accumulation by Oil Red O staining. In vivo and in vitro experiments illustrated that PO or PA was more prone to induce lipid accumulation and LDLR expression in fish.
Given the above findings, we conjectured that the LDLR expression was positively correlated with the TG content. To further test whether LDLR affected the TG metabolism of fish fed with PO diets, LDLR was successfully knocked down in fish after injections of small interfering RNA (si-LDLR) (Figures 1E and 1F). As expected, LDLR knockdown inhibited the TG accumulation in the plasma and liver induced by PO diets (Figures 1G and 1H). Meanwhile, the Oil Red O staining assay showed a visible lipid reduction in the liver after LDLR was knocked down (Figure 1I). The result from fluorescent microscopy found that LDL was colocalized with lipid droplets, which indicated that LDL endocytosed by LDLR might be involved in the esterification of TG (Figure 1J). Also, the number of lipid droplets was more in fish hepatocytes exposed to PA and LDL in combination than those exposed to PA alone or LDL alone (Figure 1J). These observations suggested that LDLR expression upregulated by PA resulted in TG accumulation via the uptake of LDL.
LDLR is regulated by the transcription factor of SREBP2 under PA stimulation
To elucidate the mechanism by which LDLR expression was upregulated by PA, we investigated the effect of ten transcription factors involved in lipid metabolism on the activity of LDLR promoters. Dual-luciferase reporter assay revealed that SREBP2 had the most binding ability to LDLR promoter compared to other transcription factors (Figure 2A). Furthermore, the promoter activity of LDLR showed a dose-dependent manner with the increase of SREBP2 (Figure 2B). To further explore the SREBP2-binding sites in the LDLR promoter, we predicted the binding sites by JASPAR and found that −1890 ∼ −1881 bp (site a) and −1290 ∼ −1299 bp (site b) of LDLR promoter were predicted as two potential binding sites (Figure 2C). Mutation analysis demonstrated that the activity of mutational LDLR promoter at site a was inhibited compared to wild-type LDLR promoter (Figure 2D), indicating that SREBP2 mainly binds to site a (GCGGTGTGAC) of LDLR promoter to regulate LDLR transcription. Furthermore, the mRNA and protein levels of SREBP2 in the liver were significantly increased by PO diets (Figures 2E and 2F). In vitro, the mRNA and protein levels of SREBP2 in fish hepatocytes were significantly upregulated by PA (Figures S2A and S2B). To detect whether PA affected LDLR expression through SREBP2, fish hepatocytes were treated with PA for 12 h after interference with si-SREBP2. SREBP2 expression upregulated by PA was inhibited by si-SREBP2 (Figure 2G). As expected, SREBP2 knockdown successfully inhibited LDLR mRNA levels upregulated by PA in fish hepatocytes (Figure 2H). Thus, PA regulated LDLR transcription through SREBP2.
Figure 2.
SREBP2 activated by TNFα increases the transcription of LDLR
(A) LDLR promoter activity was analyzed by dual-luciferase reporter after co-transfection of LDLR promoter with PCS2+ vector or 10 transcription factors involved in lipid metabolism in HEK293T cells. Data are presented as mean ± SEM, n = 4, one-way ANOVA, ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) LDLR promoter activity was analyzed by dual-luciferase reporter after co-transfection of LDLR promoter with PCS2+ or PCS2+SREBP2 vectors in different doses (range: 0–600 ng/μL) in HEK293T cells. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗p < 0.05, ∗∗∗p < 0.001.
(C) The schematic diagram shows that two predicted SREBP2-binding sites (a and b) contained at LDLR promoter were mutated.
(D) LDLR promoter activity was analyzed by dual-luciferase reporter after co-transfection of wt/mut LDLR promoters with PCS2+ or PCS2+SREBP2 vectors in HEK293T cells. Data are presented as mean ± SEM, n = 4, one-way ANOVA, ∗∗∗p < 0.001.
(E and F) The expression levels of SREBP2 mRNA and protein were analyzed by RT-qPCR and Western blot in the liver of fish fed with FO, SO, OO, and PO diets. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01.
(G and H) SREBP2 and LDLR mRNA expressions were detected by RT-qPCR in fish hepatocytes interfered with control siRNA or si-SREBP2 for 24 h after treatment with PA for 12 h. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(I and J) Plasma and liver TNFα contents were detected in fish fed with FO, SO, OO, and PO diets. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01.
(K–O) Genes (TNFα, IL-1β, MMP9, COX-2, and VCAM-1) involved in inflammatory response were analyzed by RT-qPCR in the liver of fish fed with FO, SO, OO, and PO diets. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ns = not significant.
(P) Genes (TNFα, IL-6, NF-κB, MMP9, VCAM-1, COX-2, SREBP2, and LDLR) were analyzed after fish hepatocytes were treated with TNFα for 24 h. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗p < 0.05, ∗∗p < 0.01, ns = not significant.
(Q and R) Western blot analysis of SREBP2 in nucleus and LDLR protein expressions in fish hepatocytes treated with TNFα for 24 h. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗p < 0.05.
(S) TG content in fish hepatocytes was analyzed after TNFα treatment for 24 h. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗p < 0.05. FO, fish oil; SO, soybean oil; OO, olive oil, PO, palm oil; PA, palmitic acid. See also Figure S2.
PA-induced TNFα stimulates the activation of SREBP2
Next, we explored the mechanism of SREBP2 expression upregulated by PA. To our knowledge, PA plays a key role in the inflammatory response, while pro-inflammatory factor TNFα can activate SREBP2 protein. Thus, we assumed that TNFα might mediate the effects of PA on SREBP2. To verify the supposition, the TNFα content in plasma and liver was analyzed using an ELISA kit. Compared to the FO diets, the PO diets significantly increased the TNFα content in plasma and liver (Figures 2I and 2J). Results from RT-qPCR displayed that TNFα, IL-1β, and MMP9 were upregulated in the PO diets, while no differences were observed in COX-2 and VCAM-1 among all diets (Figures 2K–2O). In vitro, compared to DHA, PA increased the expression of genes involved in inflammatory response (TNFα, IL-1β, IL-6, COX-2, and MMP9), while no differences were observed in VCAM-1 expression (Figures S2C–S2H). These results confirmed PO diets/PA-induced inflammatory response in the liver of fish. Notably, the expressions of SREBP2 and genes related to the inflammatory response (TNFα, IL-1β, NF-κB, and VCAM-1) were significantly upregulated after fish hepatocytes were treated with 10 ng/mL TNFα for 24 h (Figure 2P). The activated form of SREBP2 protein in the nucleus was increased by TNFα (Figure 2Q). Meanwhile, LDLR expression (Figures 2P and 2R) and TG content (Figure 2S) in fish hepatocytes were increased by TNFα. Overall, these results suggested that PA-induced TNFα promoted SREBP2 into the nucleus to regulate LDLR transcription.
Identification and characterization of LDLR-AS as a long-chain non-coding RNA in fish
In this study, we also tested whether the LDLR expression is regulated by new regulators such as lncRNA. To discover a functional lncRNA associated with LDLR, we performed RNA-seq analysis on the liver from fish fed with FO, SO, OO, and PO diets. After obtaining the transcripts, we used three forecasting software CPC, txCdsPredict, and CNCI, and a Pfam database to predict the encoding capacity of the transcript to distinguish between mRNA and lncRNA. The transcript was considered to be mRNA or lncRNA when at least three of the four methods were consistent. Finally, 6924 novel lncRNAs and 9370 mRNAs were obtained in the liver (Figures 3A and 3B, Data S1). To validate the RNA-seq data, five candidate lncRNAs and five candidate mRNAs were selected for validation by RT-qPCR. Nine of ten genes detected by RT-qPCR were consistent with RNA-seq analysis (Figures S3A–S3J). The validation results confirmed the high accuracy of the transcriptomic analysis. Importantly, a novel lncRNA LXLOC_014005 was identified as a cis-regulator of LDLR through transcriptomic analysis.
Figure 3.
Screening and identification of lncRNA LDLR-AS associated with LDLR
(A and B) Venn diagram was used for identifying novel lncRNAs and mRNAs expressed in the liver of fish fed with FO, SO, OO, and PO diets.
(C) The schematic diagram of positional relationship between LDLR-AS and LDLR on the chromosome of large yellow croaker. “+” on the left side of strand represents the positive strand; “-” represents the negative strand. Red arrows indicate transcription direction, and two blocks with colors (light blue and yellow) represent the transcripts of LDLR and LDLR-AS, respectively.
(D) The localization of LDLR-AS in fish hepatocytes was evaluated by nuclear/cytosolic fractionation, followed by RT-qPCR analysis. U6 and GAPDH were used as internal references of nuclear and cytoplasmic RNA, respectively. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗∗p < 0.01.
(E) The subcellular distribution of Cy3-tagged LDLR-AS was visualized by RNA Fluorescent in situ hybridization (FISH) in fish hepatocytes (Scale bars, 10 μm). Nuclear were stained by DAPI.
(F) Correlation between LDLR-AS and LDLR FPKM expression from transcriptome sequencing in the liver from fish fed with FO, SO, OO, and PO diets. The result was analyzed using Pearson correlation analysis (n = 12, Pearson correlation analysis, p < 0.05).
(G and H) The tissue distribution of LDLR and LDLR-AS in large yellow croaker was analyzed by RT-qPCR. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗∗p < 0.01, ∗∗∗p < 0.001.
(I) RT-qPCR results of the expression of LDLR-AS in the liver of fish fed with FO, SO, OO, and PO diets. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01. FO, fish oil; SO, soybean oil; OO, olive oil, PO, palm oil. See also Figure S3.
To further identify lncRNA LXLOC_014005, we first used the SMART RACE assay to obtain the full-length sequences and size of LXLOC_014005 (Figure S3M) and found that the total length of LXLOC_014005 transcript was 1217 nt (more than 200 nt). Also, agarose gel electrophoresis in the liver and skin tissues confirmed the transcript size of LXLOC_014005 (Figure S3N). To test the expression abundance of LXLOC_014005, absolute expression was performed in the liver and skin tissues. The copy number of LXLOC_01400 was higher in the liver than skin (Figure S3O). Subsequently, LXLOC_014005 sequence was retrieved in the NCBI database. We found that this lncRNA was in the reverse strand of the LDLR gene and was 1496 bp away from LDLR (Figure 3C). Given its positional relationship with LDLR, we named it as LDLR-AS (antisense LDLR). Interestingly, conservation analysis using a BLAST search revealed that the LDLR-AS nucleotide sequence of this species had a high identity (85%) with LR699037.1 sequence close to LDLR in Nibea albiflora (Figure S3P) but did not show conservation in mammals, including humans and mouse, suggesting that it might be a fish-specific lncRNA. Since the function of lncRNA is closely related to the subcellular location, LDLR-AS distribution in fish hepatocytes was analyzed. Lnclocator website (Cao et al., 2018) predicted that LDLR-AS was a cytoplasm-enriched lncRNA (Figure S3K). Indeed, both nuclear/cytoplasmic RNA separation and RNA fluorescence in situ hybridization (FISH) assays showed that LDLR-AS was mainly located in the cytoplasm of fish hepatocytes (Figures 3D and 3E), suggesting that LDLR-AS might exert a post-transcriptional regulation function.
In general, lncRNA could regulate the expression of adjacent genes through the cis role. To explore the relationship between LDLR-AS and LDLR, Pearson correlation between LDLR-AS and LDLR was conducted based on FPKM obtained by RNA-seq. Correlation coefficient of LDLR-AS and LDLR was 0.6383 (more than 0.6), indicating that LDLR-AS might positively regulate LDLR (Figure 3F, Data S2). To test whether LDLR-AS and LDLR have similar tissue distribution, we determined the tissue distribution of LDLR and LDLR-AS. The highest expression of LDLR was observed in liver, followed by skin and gill, while the lowest expression of LDLR was found in the spleen, head kidney, heart, brain, and eye (Figure 3G). Similar to the tissue distribution of LDLR, the highest expression of LDLR-AS was observed in liver, followed by skin, while the lowest expression of LDLR-AS was found in the spleen, brain, heart, head kidney, muscle, intestine, and eye (Figure 3H). Results from RT-qPCR displayed that LDLR-AS expression was higher in three vegetable oil diets (SO, OO, and PO) than in the FO diets (Figure 3I). In vitro, OA and PA significantly upregulated the expression of LDLR-AS compared to DHA (Figure S3L). The results were consistent with the LDLR expression pattern in vivo and in vitro described above.
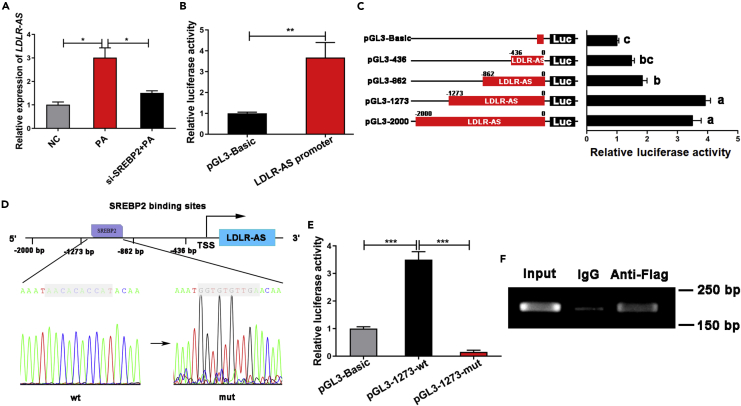
SREBP2 promotes the transcription of LDLR-AS
Given the fact that LDLR-AS was adjacent to LDLR, we guessed whether SREBP2 also modulated the expression of LDLR-AS. Firstly, we detected the effect of SREBP2 knockdown on LDLR-AS. Indeed, SREBP2 knockdown inhibited increased LDLR-AS expression induced by PA (Figure 4A). To determine the mechanism by which SREBP2 regulated LDLR-AS expression, we performed a series of double-luciferase reporter assays. The results showed that SREBP2 significantly increased the promoter activity of LDLR-AS (Figure 4B). Furthermore, activities of truncated LDLR-AS promoters (−862 and −436 bp) were significantly decreased compared to untruncated LDLR-AS promoters (−2000 bp), indicating that the SREBP2 responsive element was at −1273 ∼ −862 bp from LDLR-AS promoter (Figure 4C). To further test the SREBP2-binding site in LDLR-AS promoter, sequence analysis at −1273 ∼ −862 bp was performed and an SREBP2-binding site (AACACACCAT) was discovered. When the SREBP2-binding site (AACACACCAT) in the LDLR promoter was mutated, the LDLR promoter lost activity (Figures 4D and 4E). Meanwhile, the ChIP assay confirmed that SREBP2 could bind at −1273 ∼ −862 bp from the LDLR-AS promoter (Figure 4F). These results indicated that SREBP2 initiated the transcription of LDLR-AS.
Figure 4.
SREBP2 promotes the transcriptional level of LDLR-AS
(A) LDLR-AS expression was detected by RT-qPCR in fish hepatocytes interfered with control siRNA or si-SREBP2 for 24 h after treatment with PA for 12 h. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗p < 0.05.
(B) LDLR-AS promoter activity was analyzed by dual-luciferase reporter after co-transfection of LDLR-AS promoter with PCS2+ or PCS2-SREBP2 vectors in HEK293T cells. Data are presented as mean ± SEM, n = 4, Student’s t-test, ∗∗p < 0.01.
(C) LDLR-AS promoter activity was analyzed by dual-luciferase reporter after co-transfection of four truncated LDLR-AS promoters with PCS2+ or PCS2-SREBP2 vectors in HEK293T cells. Data are presented as mean ± SEM, n = 4, one-way ANOVA.
(D) The schematic diagram shows that predicted SREBP2-binding site contained at −1273 ∼ −862 bp of LDLR-AS promoter was mutated.
(E) The activities of wt/mut LDLR-AS promoter were analyzed by dual-luciferase reporter after co-transfection of wt/mut LDLR-AS promoters with PCS2+ or PCS2-SREBP2 vectors in HEK293T cells. Data are presented as mean ± SEM, n = 4, one-way ANOVA, ∗∗∗p < 0.001.
(F) ChIP-PCR assay of SREBP2-binding region at −1273 ∼ - 862 bp from LDLR-AS promoter using IgG or Flag antibody.
LDLR-AS affects hepatic triglyceride content by regulating LDLR expression
Next, we tested the effect of LDLR-AS on the TG levels and LDLR expression. The LDLR-AS and LDLR expression and triglyceride levels were analyzed after fish hepatocytes were transfected with pcDNA3.1 or pcDNA3.1-LDLR-AS plasmids by electroporation for 48 h. LDLR-AS expression was increased by about 50-fold in fish hepatocytes electroporated with pcDNA3.1-LDLR-AS (Figure 5A), suggesting that LDLR-AS was successfully overexpressed. Overexpression of LDLR-AS significantly upregulated LDLR expression (Figures 5B and 5C) and TG levels (Figure 5D). Conversely, three LDLR-AS antisense oligonucleotides (ASO) were transiently transfected into fish hepatocytes to knock down LDLR-AS expression. LDLR-AS expression was knocked down by ASO3 effectively (Figure S4A). Thus, we choose ASO3 for further experiments. To determine the effect of LDLR-AS knockdown on LDLR expression, fish hepatocytes were transiently transfected with ASO3 for 48 h. We found that LDLR-AS knockdown significantly inhibited LDLR expression (Figures S4B and S4C). To further verify the functional role of LDLR-AS on LDLR and triglyceride levels in vivo, fish fed with PO diets were injected intraperitoneally with ASO3 of LDLR-AS for 48 h. Unsurprisingly, about 70% knockdown of LDLR-AS strongly suppressed LDLR mRNA and protein levels (Figures 5E–5G). Plasma and liver TG in the PO groups were significantly decreased compared with those in the NC group (Figures 5H and 5I). Furthermore, the result of liver Oil Red O staining showed that LDLR-AS knockdown reduced lipid accumulation induced by PO diets, which was consistent with TG levels after LDLR-AS knockdown (Figure 5J). These results confirmed the possibility that LDLR-AS increased the triglyceride levels in the liver by upregulating LDLR expression.
Figure 5.
LDLR-AS affects hepatic triglyceride content by regulating LDLR expression
(A) Overexpression of LDLR-AS was detected by RT-qPCR after pcDNA3.1 or pcDNA3.1-LDLR-AS plasmids were electroporated into fish hepatocytes for 48 h pcDNA3.1 plasmid was used as a negative control. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗∗∗p < 0.001.
(B and C) RT-qPCR and Western blot assays were used to evaluate the expressions of LDLR in fish hepatocytes after LDLR-AS overexpression. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗p < 0.05.
(D) TG content in fish hepatocytes was analyzed after LDLR-AS overexpression. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗p < 0.05.
(E) LDLR-AS expression was knocked down in the PO diets group by injecting control siRNA (NC) or LDLR-AS ASO3 (ASO3). Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗p < 0.05.
(F and G) The expression levels of LDLR mRNA and protein were analyzed by RT-qPCR and Western blot after LDLR-AS knockdown. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗p < 0.05, ∗∗p < 0.01.
(H and I) Plasma and liver TG levels were detected after LDLR-AS knockdown. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗p < 0.05.
(J) Lipid accumulation in the liver was observed by Oil Red O staining after LDLR-AS knockdown (Scale bars, 100 μm). See also Figure S4.
LDLR-AS recruits hnRNPR to the 5′-UTR of LDLR to enhance LDLR mRNA stability
To determine the detailed mechanism by which LDLR-AS regulated LDLR expression, we captured interacting proteins with LDLR-AS by RNA pulldown assay. Biotinylated LDLR-AS sequence was incubated with total proteins extracted from fish hepatocytes. Interacting proteins with LDLR-AS were pulled down with streptavidin and analyzed by SDS-PAGE and silver staining. Subsequently, the distinct bands between antisense and sense LDLR-AS groups were analyzed by mass spectrometry. RNA pulldown assay revealed that heterogeneous nuclear ribonucleoprotein R (hnRNPR, around 82 kDa) expression in the treatment group was significantly different from the control. Thus, hnRNPR was identified as the potential binding protein with LDLR-AS (Figure 6A, Data S3). To test the binding region between hnRNPR and LDLR-AS, we predicted the potential protein-RNA interacting region using the catRAPID website. The result predicted that hnRNPR had a strong binding ability with 350–450 bp of LDLR-AS (Figure 6B). To further verify the interacting region, a ChIP-qPCR assay was conducted. Indeed, hnRNPR could bind to 350–450 bp of LDLR-AS (Figure 6C). These results confirmed that LDLR-AS and hnRNPR could form a protein-RNA complex. HnRNPR-targeted siRNAs were transfected into fish hepatocytes to knock down hnRNPR expression for 48 h. Knockdown of hnRNPR was confirmed at both RNA (Figure 6D) and protein (Figure 6E) levels. Notably, hnRNPR knockdown inhibited the LDLR expression (Figure 6E), while no difference was found in the LDLR-AS expression (Figure 6F). HnRNPR belongs to a subfamily of heterogeneous nuclear ribonucleoprotein (hnRNPs). Although some members of hnRNPs have a known role in modulating mRNA stability, the function of hnRNPR is rarely explored. Given that cytoplasmic LDLR-AS might play a role in post-transcription regulation, we investigated whether the LDLR-AS-hnRNPR complex regulated LDLR mRNA stability. We used actinomycin D to block de novo transcription of mRNA and detected the effect of hnRNPR on LDLR mRNA. HnRNPR knockdown reduced LDLR mRNA stability in fish hepatocytes (Figure 6G). These results indicated that the LDLR-AS-hnRNPR complex regulated the stability of LDLR mRNA at the post-transcription level. HnRNPs modulate mRNA stability by binding the untranslated regions of target RNA. Thus, the potential interacting region between hnRNPR and LDLR containing a 146 bp 5′-UTR region was predicted using the catRAPID website. The result predicted that hnRNPR had a strong binding ability with the 5′-UTR of LDLR (Figure 6H). To substantiate this, a dual-luciferase reporter assay was conducted after HEK293T cells were transfected with pGL3-5′-UTR or PCS2-hnRNPR plasmids. Dual-luciferase reporter assay showed that relative luciferase activity was higher in the treatment group than in the control group, which suggested that the 5′-UTR of LDLR was involved in the interaction with hnRNPR (Figure 6I). Meanwhile, the ChIP-qPCR assay revealed that hnRNPR could bind to the 5′-UTR of LDLR (Figure 6J). These results supported that LDLR-AS bound and recruited hnRNPR to LDLR 5’-UTR RNA regions to enhance mRNA stability.
Figure 6.
LDLR-AS interacts with hnRNPR to stabilize LDLR mRNA
(A) Silver staining of proteins pulled down with biotinylated LDLR-AS RNA and a negative control with antisense sequences. Blue triangle denotes the band identified as hnRNPR by mass spectrometry.
(B) Interaction profiles for LDLR-AS interaction with hnRNPR were predicted using the catRAPID website.
(C) ChIP-qPCR assay was performed to measure the binding ability of hnRNPR to LDLR-AS. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗∗∗p < 0.001.
(D) RT-qPCR results of hnRNPR knockdown in fish hepatocytes transfected with si-hnRNPR#1/2/3 for 24 h. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗p < 0.05.
(E) Western blot of LDLR and hnRNPR protein in fish hepatocytes transfected with si-hnRNPR#1 for 48 h. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01.
(F) RT-qPCR results of LDLR-AS expression in fish hepatocytes transfected with si-hnRNPR#1/2/3 for 24 h. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ns = not significant.
(G) RT-qPCR analysis was used to evaluate the stability of LDLR mRNA after fish hepatocytes were transfected with si-hnRNPR#1 for 24 h, then further exposed to 5 μg/mL actinomycin D for 24 h. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗∗p < 0.01.
(H) Interaction profiles for LDLR at the 5′-UTR interaction with hnRNPR were predicted using the catRAPID website.
(I) The luciferase activity of 5′-UTR of LDLR mRNA was detected in HEK293T cells transfected pGL3-Basic, pGL3-5′UTR, or PCS2+hnRNPR vectors. Data are presented as mean ± SEM, n = 3, one-way ANOVA, ∗∗p < 0.01, ∗∗∗p < 0.001. (J) ChIP-qPCR assay was performed to measure the binding ability of hnRNPR to the 5′-UTR of LDLR. Data are presented as mean ± SEM, n = 3, Student’s t-test, ∗∗p < 0.01.
Discussion
Our previous studies demonstrate that vegetable oils, including soybean oil, palm oil, and olive oil, can lead to TG accumulation in the liver and then induce hepatic steatosis in fish (Li et al., 2019a, 2019b; Liao et al., 2015; Zhu et al., 2018). In this study, the results from the transmission electron microscope and Oil Red O staining confirmed that fish fed with vegetable oils exhibited abnormal lipid accumulation in the liver. LDLR is the plasma LDL receptor and regulates hepatic lipid homeostasis (Yang et al., 2020a; 2020b). However, the mechanism by which LDLR is involved in TG metabolism remains unclear. Here, we demonstrated that PA could lead to hepatic TG accumulation by enhancing LDLR expression. Most specifically, PA markedly increased both mRNA and protein levels of LDLR. However, LDLR knockdown reversed hepatic TG deposition induced by PO diets rich in PA, which suggested that LDLR was involved in PA-mediated TG accumulation. In fish hepatocytes, fluorescently labeled LDL was colocalized with lipid droplets, indicating that LDL endocytosed by LDLR might be involved in TG esterification. Meanwhile, the co-incubation of LDL with PA augmented the number of lipid droplets in fish hepatocytes. Similar to our findings, a study has also shown that the uptake of LDL contributes to TG synthesis in primary hepatocytes (Minahk et al., 2008). These results suggested that PA-mediated LDLR promotes lipid accumulation by increasing cellular uptake of LDL.
Next, we explored precise molecular mechanisms by which PA elevated LDLR expression. SREBP2, a transcription factor, belongs to the family of sterol regulatory element-binding proteins, which controls the synthesis of triglycerides, cholesterol, and fatty acids (Engelking et al., 2018). Our luciferase assays found that SREBP2 increased the activity of LDLR promoter, and a conserved sequence motif (GCGGTGTGAC) in LDLR promoter was identified as the key binding site of SREBP2. However, SREBP2 knockdown inhibited LDLR expression induced by PA, suggesting that SREBP2 regulated the transcription of LDLR under PA. A previous study reveals that PA contributes to the development of inflammation (Hu et al., 2021), while the pro-inflammatory factor TNFα can activate the SREBP2 protein in human macrophages (Kusnadi et al., 2019). Thus, we assumed that TNFα might mediate the effects of PA on SREBP2. In the present study, TNFα levels in the liver were increased in the PO diets and increased TNFα enhanced SREBP2 protein expression in the nucleus. More importantly, LDLR expression in fish hepatocytes was increased by TNFα. These results suggested that PA-induced TNFα promoted SREBP2 into the nucleus to regulate LDLR transcription.
Increasing evidence shows that lncRNAs, having crucial regulatory roles in TG metabolism, are widely studied in mammals (Lan et al., 2019; Li et al., 2015; Wang et al., 2020). However, the role of lncRNAs on lipid metabolism in fish still needs to be explored. In this study, through a combination of the transcriptome, bioinformatics, and cell biological approaches, we successfully identified an unannotated antisense lncRNA, termed LDLR-AS. Conservation analysis of LDLR-AS revealed that the LDLR-AS nucleotide sequence of this species had a high identity (85%) with LR699037.1 sequence close to LDLR in N. albiflora, but was not conserved with mammals, including humans and mice, suggesting that it might be a fish-specific lncRNA.
Antisense lncRNAs, transcribed from the opposite strand of protein-coding genes (Rothzerg et al., 2021), have been ascribed roles in gene regulation involving histone methylation (Tian et al., 2021), mRNA stabilization (Han et al., 2020), and protein translation (Carrieri et al., 2015; Gu et al., 2019). LncRNA LDLR-AS, located in the opposite strand of the LDLR gene, was mainly distributed in the cytoplasm, suggesting that LDLR-AS might exert a post-transcriptional regulation. Interestingly, similarly to the expression pattern of LDLR, PA increased the expression of LDLR-AS through the transcription factor SREBP2. Functionally, LDLR-AS overexpression increased LDLR expression and TG content, whereas LDLR-AS knockdown in the PO diets remarkably decreased LDLR expression and relieved TG accumulation. In summary, these results revealed that LDLR-AS upregulated by SREBP2 had a positive regulation on LDLR expression and then increased TG accumulation.
Previous studies have shown that lncRNAs, although they do not encode proteins, can bind to intracellular proteins to perform physiological functions (Dong et al., 2021; Higashi et al., 2022; Munschauer and Vogel, 2018; Sun et al., 2018). Mechanistically, RNA pulldown and ChIP assays showed that LDLR-AS interacted with hnRNAR physically. HnRNPR belongs to the heterogeneous nuclear ribonucleic proteins (hnRNPs) family, which takes part in various biological processes, including transcription, pre-mRNA splicing, RNA modification, transport, localization, and translation (Kwon et al., 2021; Ryu et al., 2021; Thibault et al., 2021). A study reveals that the stability of LDLR mRNA can be controlled by a network of hnRNPs in different processes of mRNA decay (Li et al., 2009) because LDLR mRNA with a half-life of 45–60 min is labile. For example, hnRNPI directly binds to the LDLR 3′-UTR to induce LDLR mRNA decay, indicating that hnRNPI is an mRNA decay-promoting factor (Li et al., 2009). Conversely, our study found that hnRNPR promoted the LDLR mRNA stability through binding to the LDLR 5′-UTR. Consistent with our data, hnRNPR also enhances MHC class I mRNAs stability (Reches et al., 2016). These results suggested that the hnRNPR-LDLR-AS complex stabilized LDLR mRNA level.
In conclusion, our results unveil a crucial regulatory function of LDLR in hepatic TG accumulation in fish. LDLR expression upregulated by PA resulted in hepatic TG accumulation through the uptake of LDL. Mechanistically, we highlight the importance of SREBP2 and lncRNA LDLR-AS in LDLR function. On one hand, SREBP2 induced by PA enhances the transcription of LDLR directly. On the other hand, SREBP2-mediated LDLR-AS could recruit hnRNPR to LDLR 5′-UTR and then increase the LDLR mRNA stability at the post-transcription level. Therefore, these results provide a theoretical basis that SREBP2 or lncRNA LDLR-AS might be used as effective targets for treating hepatic TG accumulation in fish.
Limitations of the study
Our study demonstrated that diets rich in palmitic acid (PA) activated LDLR expression, thereby augmenting hepatic TG accumulation in fish. On one hand, SREBP2 promoted the transcription of the LDLR gene in the nucleus. On the other hand, LncRNA LDLR-AS transcription activated by SREBP2 could recruit hnRNPR to LDLR 5′-UTR and then increased the LDLR mRNA stability at the post-transcription level. However, LncRNA LDLD-AS sequences are not conserved in mammals, including humans and mice. Hence, future research should investigate whether fish-specific lncRNA LDLR-AS affects LDLR expression in humans and mice or whether there are specific lncRNAs that regulate LDLR expression in humans and mice.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal Anti-LDLR | Proteintech | Cat#10785-1-AP; RRID:AB_2281164 |
| Rabbit polyclonal Anti-SREBP2 | Proteintech | Cat#28212-1-AP; RRID:AB_2881091 |
| Rabbit polyclonal Anti-hnRNPR | Proteintech | Cat#15018-1-AP; RRID:AB_2120506 |
| Rabbit polyclonal Anti-Histone 3 | Abcam | Cat#ab1791 |
| Rabbit Anti-β-actin | Cell Signaling Technology | Cat#4970; RRID:AB_2223172 |
| Normal Rabbit IgG | Cell Signaling Technology | Cat#2729; RRID:AB_1031062 |
| Rabbit monoclonal Anti-DYKDDDDK Tag | Cell Signaling Technology | Cat#14793; RRID:AB_2572291 |
| Bacterial and virus strains | ||
| Escherichia coli-DH5α | Trans-Gen Biotech | CAT#CD201-01 |
| Chemicals, peptides, and recombinant proteins | ||
| Docosahexaenoic acid (DHA) | Matreya LLC | Cat#1136 |
| Linoleic acid (LA) | Matreya LLC | Cat#1024 |
| Oleic acid (OA) | Matreya LLC | Cat#1022 |
| Palmitic acid (PA) | Matreya LLC | Cat#1014 |
| Dil-LDL | AngYuBio | Cat#AY-1504 |
| Recombinant TNFα | MedChemExpress | Cat#HY-P7303 |
| Actinomycin D | GLPBIO | Cat#GC16866 |
| Critical commercial assays | ||
| Chromatin Immunoprecipitation Assay Kit | Beyotime | Cat#P2078 |
| Fluorescent in Situ Hybridization Kit | RiboBio | Cat#C10910 |
| RNA pull-down Kit | GENESEED | Cat#P0201 |
| T7 Biotin Labeled RNA Synthesis Kit | GENESEED | Cat#R0402 |
| Fast Silver Stain Kit | Beyotime | Cat#P0017S |
| TNFα ELISA | Nanjing Jiancheng | Cat#H052-1 |
| Deposited data | ||
| The RNA sequence data | This paper | GEO: GSE186012 |
| Experimental models: Cell lines | ||
| Human: HEK293T | ATCC | Cat#CRL-3216 |
| Large yellow croaker: primary hepatocytes | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Juvenile large yellow croaker | Aquatic Fingerlings Limited Co., Ltd. of Xiangshan Harbor (China) | N/A |
| Oligonucleotides | ||
| siRNA and primer sequences, see Table S2 | This study | N/A |
| Recombinant DNA | ||
| pcDNA3.1 | Thermo Fisher Scientific | Cat#V79020 |
| pGL3-Basic | YouBio | Cat#VT1554 |
| PCS2+ | KeLeiBio | kl-zl-0892 |
| Software and algorithms | ||
| GraphPad Prim 7.0 | GraphPad Software | https://www.graphpad.com/ |
| Image J | NIH | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Qinghui Ai (qhai@ouc.edu.cn).
Materials availability
Materials generated in this study will be made available on request and may require a material transfer agreement.
Experimental model and subject details
Diet formulation, fish, and feeding procedure
Four iso-nitrogenous (42% crude protein) and iso-lipidic (12% crude lipid) experimental diets were formulated, including FO (fish oil as a dietary fat source), SO (soybean oil as a dietary fat source), OO (olive oil as a dietary fat source) and PO (palm oil as a dietary fat source). The fatty acid profile of the four experimental diets were shown in detail in Table S1. Four-month-old large yellow croaker, including the male and female, was purchased from Aquatic Fingerlings Limited Company of Xiangshan Harbour (Ningbo, China). Prior to the feeding trial, fish were hand-fed twice daily (05:30 and 17:30) to acclimatized the experimental diets and conditions for 2 weeks. After the acclimation, fish of similar sizes (initial body weight: 15.87 ± 0.14 g) were randomly distributed into 24 floating sea cages (1.0 × 1.0 × 2.0 m, L × W × H) at a rate of 60 fish per cage. Fish were fed twice daily to apparent satiation for 10 weeks. Each diet was randomly assigned to four replicate cages. During the feeding trial, fish were reared under the following conditions: water temperature ranged from 22 to 30°C, dissolved oxygen content from 5.5 to 7.0 mg/L, salinity ranged from 25.0 to 32.0 g/L, pH from 7.0 to 7.3, respectively. At the end of the experiment, liver tissues from 9 fish each cage were sampled for the analysis of lipid content, histochemical observation, mRNA and protein expression assays, and RNA sequencing.
In the present study, all experimental procedures performed on fish were conducted in strict accordance with the Management Rule of Laboratory Animals (Chinese Order No. 676 of the State Council, revised 1 March 2017).
Cell culture and treatment
The cells, including primary hepatocytes from large yellow croaker and HEK293t, were used in this study. Liver from four-month-old large yellow croaker were removed and placed in sterile phosphate buffer (PBS, Biological Industries, Israel) containing penicillin and streptomycin (cat. no. P1400, Solarbio, China). After washing with Dulbecco’s modified Eagle medium/Ham’s F12 medium (1:1) (DMEM/F12, Biological Industries), liver tissue was chopped into 1-mm3 slices and digested with 0.25% trypsin (Thermo Fisher Scientific, USA) for 10 min. After neutralization with DMEM/F12 medium containing fetal bovine serum (FBS, Biological Industries), the cell precipitate was suspended in complete medium composed of DMEM/F12 medium supplemented with 10% FBS, 100 U penicillin and 100 g/mL streptomycin. The cell suspension was inoculated into a six-well culture plate and incubated at 28°C. HEK293t were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin. Fish hepatocytes were incubated with media containing palmitic acid (PA), oleic acid (OA), linoleic acid (LA), or docosahexaenoic acid (DHA) for 8 h, respectively. siRNAs were transfected into fish hepatocytes for SREBP2, LDLR, LDLR-AS, and hnRNPR knockdown. 5 μg pcDNA 3.1-LDLR-AS plasmid was electroporated into 1 × 106 fish hepatocytes for overexpression LDLR-AS. Fish hepatocytes were exposed to 50 μg/mL LDL to activate LDLR.
Method details
Injection of ASO and dsRNA for RNA interference
Double-stranded RNA (dsRNA) of LDLR and antisense oligonucleotide (ASO) of LDLR-AS was synthesized from GenePharma (China) and Integrated BiotechSolutions (China), respectively. At the end of the feeding trial (10 weeks), fish were starved for 24 h. 0.5 nmol/g dsRNA or ASO were injected intraperitoneally into the fish fed with FO and PO diets. After 48 h post-injection, the liver was sampled for RNA and protein extraction and histological analysis, while the plasma was collected to determine TG. Detailed sequences were listed in Table S2.
Histological analysis
Liver OCT sections were stained with Oil Red O to assess lipid content. Briefly, OCT sections were fixed in 4% formaldehyde. After three times wash in PBS, OCT sections were incubated in freshly prepared Oil Red O solution for 1 h at 37°C. Sections were rinsed in 60% isopropanol for 30 s, followed by washing with PBS before microscopic examination.
Plasmid construction
All plasmids were constructed with a recombinant method. The open reading frame (ORFs) of the LDLR gene was constructed into a pcDNA3.1-GFP vector for subcellular localization assay. The full length of LDLR-AS was cloning into a pcDNA3.1-Basic vector for overexpression and RNA pull-down assays. The sequence of Flag-tagged hnRNPR was constructed into a PCS2+ vector for ChIP-PCR. All plasmids were confirmed by DNA sequencing at Sangon Biotech.
RNA isolation and real time qPCR
Total RNA was isolated from tissues and cells using Trizol reagent (TaKaRa Biotechnology, Dalian, China) according to the manufacturer’s protocol. Total RNA was reverse transcribed with a PrimerScript RT-PCR kit (Takara Biotechnology, Dalian, China). Real time qPCR was conducted using a ChamQ Universal SYBR qPCR Master Mix kit (Vazyme Biotech Co., Ltd, Nanjing, China) protocol with a CFX real-time instrument (Bio-rad, Hercules, CA, USA). The absolute expression levels of genes were quantified as copy number per microgram of oligo-dT primed cDNA according to the curves of plasmid standards. The relative expression was analyzed using the 2−ΔΔCt method. GAPDH and U6 were used as house-keeping genes to normalize the nuclear/cytosolic fractionation experiment of LDLR-AS. β-actin was used as a normalizing control in all the other RT-qPCR experiments. All siRNA and specific primers were shown in Table S2.
RNA sequencing
RNA sequencing was performed on 12 liver samples of fish fed with FO, SO, OO, and PO diets, respectively. Total RNAs were extracted using TRIzol reagent (Invitrogen, United States USA) according to the manufacturer’s protocol. The quantity and quality of total RNA were determined by using the NanoDrop 2000 (Thermo Fisher Scientific, USA) and the Agilent 2100 Bioanaylzer (Agilent Technologies, USA, respectively. Ribosomal RNA of the 12 RNA samples was removed using the Ribo-Zero™ rRNA Removal Kit (Epicentre, USA). The remained RNAs were fragmented using Ambion Fragmentation Solution (Termo Fisher Scientifc, USA). Then double-stranded cDNA synthesis is performed with the incorporation of dUTP in the second strand. Finally, the libraries were sequenced using the BGISEQ-500 platform.
Identification of lncRNA
The expression levels of the transcripts were normalized to fragments per kilobase of exon model per million mapped reads (FPKM) using Cuffdiff. The prediction of lncRNA was performed according to previous research. The transcripts with length ≤200 and FPKM <0.5 in each sample were removed using shell scripts. Only non-coding classes (like i, j, u, x, and o) were retained using Cuffcompare. The coding potential of the remaining transcripts was evaluated using Coding Noncoding Index (CNCI) software (https://github.com/www-bioinfo-org/CNCI), coding potential calculator (CPC) software (http://cpc.cbi.pku.edu.cn/), txCdsPredict (http://hgdownload.soe.ucsc.edu/admin/jksrc.zip) and Pfam database (http://pfam.xfam.org/). All transcripts with CNCI score >0, CPC score >0, and txCdsPredict score >500 were discarded. Pfam database was used to ensure that predicted transcripts did not contain protein-coding domains. The transcript just was determined to be lncRNA when at least three of the above four methods were consistent.
Quantitative real-time PCR validation
Total RNA was extracted and reverse transcribed as described above. For quantitative analysis, the relative expression level of lncRNAs and mRNAs was normalized to internal invariant control, β-actin. The expression of each lncRNA or mRNA was represented as log2 (fold change) by using the 2−ΔΔCt method. The PCR primers for the five selected mRNAs (including HMGCR, HMGCS, LXRΑ, CYP7A1, and SRB1) and the five lncRNAs (including LXLOC_025972, LXLOC_008729, LXLOC_019532, LXLOC_031003, and LXLOC_023163) were designed using Primer 5.0. All primers used in the present study were shown in Table S2.
Western blotting
Total proteins were extracted from livers and hepatocytes of fish using RIPA lysis buffer with protease inhibitors and phosphatase inhibitors. Protein concentrations were determined with a BCA Protein Assay Kit (Beyotime Biotechnology, China) according to the manufacturer’s instructions. After standardization, the samples were separated by SDS-PAGE and transferred to 0.45 μm PVDF membranes (Millipore, USA). Membranes were blocked with 5% nonfat dry milk in TBST for 2 h and were then incubated with primary antibodies overnight. Primary antibodies specific to LDLR (#10785-1-AP, Proteintech), SREBP2 (#28212-1-AP, Proteintech), hnRNPR (#15018-1-AP, Proteintech), PLCB1 (#DF6726, Affinity), β-actin (#ab8227, abcam), GAPDH (#ab9485, abcam), Histone 3 (#4499, Cell signaling Technology) were used. The blots were then incubated with goat anti-rabbit secondary antibody (#A0208, Beyotime) and visualized using an ECL kit (#WP20005, Invitrogen). The intensity of target bands was quantified using the Image J Pro 1.52 software (National Institutes of Health, Bethesda, Maryland, USA), and then normalized to that of Histone 3 (H3), β-actin, or GAPDH.
Dual-luciferase reporter assay
The LDLR promoter (wt) was constructed into the pGL3-Basic vector, while CDS region of 10 transcription factors (SREBP2, FOXO3, ChREBP, PPARγ, FXR, LXRα, PPARα, NF-κB, C/EBPα, and HNF4α) was cloned into PCS2+ vector. The SREBP2 binding sites in LDLR promoter from large yellow croaker were predicted with JASPAR (http://jaspar.genereg.net). The LDLR promoters containing site mutated (a or b) was also constructed into the pGL3-Basic vector. 1 × 104 HEK293T cells were plated in each well of 24-well plates. After 12 h, HEK293T cells in each well were transfected with 200 ng reporter vector, 600 ng ectopic expression vector, and 10 μg pRL-TK vector using the Lipofectamine 2000 (Invitrogen). The pGL3-Basic was used as a control group and pRL-TK vector was used to standardize the expression level. After 12 h of transfection, we replaced the transfection medium with a complete culture medium. At 48 h post culture, the cells were washed twice with PBS and then lysed with 100 μL 1× cell lysis buffer at room temperature for 10 min. Finally, luciferase activity of all samples was measured using the Double-Luciferase Reporter Assay kit (TransGen, China) according to the manufacturer’s instructions. The firefly luciferase activity was normalized to Renilla activity. The results were expressed as fold induction compared to negative control.
Rapid amplification of cloned cDNA ends (RACE)
RNA was extracted from hepatocyte of large yellow croaker. The 5′ and-3′ RACE was performed using the HiScript-TS 5’/3′ RACE Kit (Vazyme, China) according to the manufacturer’s instructions. Briefly, 1 μg of purified RNA was used to synthesize the first-strand cDNA. The PCR amplification of 5′ and-3′ end was performed using 5′ and-3′ gene-specific primers (GSP). The cDNA samples (50 μL) were diluted 50-fold, and 5 μL was used for nested PCR reactions. The PCR products were detected by agarose gel electrophoresis and sequenced. Primer sequences used in RACE were given in Table S2.
Subcellular fraction extraction
1 × 107 fish hepatocytes were incubated with 50 ng/mL TNFα (BioVision) for 12 h and collected to extracted nuclear and cytoplasmic protein extraction using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific). For nuclear and cytoplasmic RNA separation, 1 × 106 fish hepatocytes were collected and extracted using Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Canada) according to the manufacturer’s instructions. Quantitative real-time PCR was used to determine the expression of LDLR-AS, with U6 and GAPDH used as internal references of nuclear and cytoplasmic RNA.
Fluorescence in situ hybridization (FISH)
FISH experiment was performed using the Fluorescent in Situ Hybridization Kit (RiboBio, China) according to the manufacturer’s instructions. Fish hepatocytes were fixed in 4% formaldehyde for 10 min at room temperature, and then were permeabilized in PBS plus 0.5% Triton X-100 for 5 min at 4°C. The Cells were prehybridized for 30 min at 37°C before hybridization. Hybridization was carried out using Cy3-labeled LDLR-AS probe mix at 37°C overnight. Finally, the cells were incubated with DAPI for 10 min. Images were visualized in the confocal microscopy (Zeiss, German).
RNA pull-down assay
To identify the interacting protein with LDLR-AS in fish hepatocytes, RNA pull-down was performed. Firstly, biotin-labeled RNAs were prepared in vitro with a GSTM T7 Biotin Labeled RNA Synthesis Kit (Geneseed, China), and purified using the E.Z.N.A.TM Micro-Elute RNA Clean-up Kit (OMEGA, USA) after treatment with RNase-free DNaseI. Then, sense or antisense of biotin-labeled LDLR-AS was incubated with streptavidin magnetic beads for 30 min at 4°C, and added cell lysates (1 × 107) lysed with cold capture buffer to isolate the RNA-protein complexes. The complexes were washed with wash buffer three times for 5 min each time at 4°C. After washes, the pull-down complexes were eluted by denaturation in 50 μL loading buffer for 10 min at 100°C and subjected to SDS–PAGE. The gel was stained with a Fast Silver Stain Kit (Beyotime, China) according to the manufacturer’s instructions. The silver-stained bands that were significantly different between sense and antisense were cut and subjected to mass spectrometric analysis. The LC-MS/MS analysis was performed with an TripleTOF 5600 + mass spectrometer (SCIEX, USA) equipped with a Tempo Ninterferano HPLC system. The mass spectra were retrieved using the ProteinPilot software (V4.5) in the Larimichthys crocea RefSeq protein database.
Chromatin immunoprecipitation (ChIP)
HEK293T cells were transfected with 5 μg pcDNA3.1-LDLR-AS or pGL3-LDLR promoter (containing a 146 bp 5′ UTR region) and 5 μg PCS2+hnRNPR-Flag using the Lipofectamine 2000 (Invitrogen). After 24 h transfection, cells fixed with 1% formaldehyde for 10 min, and quenched with 1 X glycine solution for 5 min. cells were lysed in SDS lysis buffer supplemented with 1 mM PMSF on ice for 10 min after washed with cold PBS three times. Then chromatin DNA extracted was sonicated for eight cycles (25% output, 5 s with 10 s between bursts) and sheared into 200 to 400 bp. The DNA lysate was incubated with 2 μg anti-Flag (#14793, CST) or anti-IgG antibody (#2729, CST) overnight on a rotating platform at 4°C, followed by the addition of 40 μL protein-A + G agarose beads for 1 h by rotation. The complexes were eluted by vortexing in elution buffer (1% SDS, 1 mM NaHCO3) twice per 5 min at room temperature. Final concentration of 0.54 M NaCl was added to the immunoprecipitates and heated 65°C in water bath for 8 h to reverse the cross-linking. Finally, the DNA was purified and then subjected to PCR and RT-qPCR. The values from the immunoprecipitated samples were normalized to that from the input DNA. Primers used are listed in Table S2.
Quantification and statistical analysis
Data are presented as the mean ± SEM. The statistical analyses between two groups were performed with Student t-test, and comparisons between multiple groups were performed with one-way ANOVA in SPSS 19.0 software. The level of significance was set at ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. The number of replicates for each experiment are indicated in the figure legends.
Acknowledgments
This research was financially supported by the Scientific and Technological Innovation of Blue Granary (grant number: 2018YFD0900402), the Key Program of National Natural Science Foundation of China (grant number: 31830103), the National Science Fund for Distinguished Young Scholars of China (grant number: 31525024), Leading Talent of Technological Innovation of Ten-Thousands Talents Program (grant number: CS31117200001), and the Agriculture Research System of China (grant number: CARS47-11).
Author contributions
Q.A., K.M., and X.C. conceptualized and managed the study. X.C. and X.W. generated the data. X.C., X.L., W.F., and X.W. analyzed the data. X.C. drafted the manuscript. Q.A. reviewed and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure diversity in experimental samples through the selection of the cell lines.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104670.
Supplemental information
Data and code availability
-
•
All relevant data are within the manuscript and Supplementary Material. The accession number for the RNA sequencing data reported in this paper is GEO: GSE186012.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ananthanarayanan M. A novel long noncoding RNA regulating cholesterol and bile acid homeostasis: a new kid on the block and a potential therapeutic target? Hepatology. 2016;64:16–18. doi: 10.1002/hep.28525. [DOI] [PubMed] [Google Scholar]
- Barale C., Melchionda E., Morotti A., Russo I. PCSK9 biology and its role in atherothrombosis. Int. J. Mol. Sci. 2021;22:5880. doi: 10.3390/ijms22115880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Pan X., Yang Y., Huang Y., Shen H.B. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34:2185–2194. doi: 10.1093/bioinformatics/bty085. [DOI] [PubMed] [Google Scholar]
- Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., Pesce E., Ferrer I., Chen Z. Progress and prospects of long noncoding RNAs in lipid homeostasis. Mol. Metab. 2015;5:164–170. doi: 10.1016/j.molmet.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Progress and prospects of long noncoding RNAs in lipid homeostasis. Mol. Metab. 2015;5:164–170. doi: 10.1016/j.molmet.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Du J., Cui K., Fang W., Zhao Z., Chen Q., Mai K., Ai Q. Acetyl-CoA derived from hepatic mitochondrial fatty acid β-oxidation aggravates inflammation by enhancing p65 acetylation. iScience. 2021;24:103244. doi: 10.1016/j.isci.2021.103244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Chen Q., Li Y., Xiang X., Xu W., Mai K., Ai Q. Activation of the farnesoid X receptor (FXR) suppresses linoleic acid-induced inflammation in the large yellow croaker (Larimichthys crocea) J. Nutr. 2020;150:2469–2477. doi: 10.1093/jn/nxaa185. [DOI] [PubMed] [Google Scholar]
- Dong K., Shen J., He X., Hu G., Wang L., Osman I., Bunting K.M., Dixon-Melvin R., Zheng Z., Xin H., et al. CARMN is an evolutionarily conserved smooth muscle cell-specific lncRNA that maintains contractile phenotype by binding myocardin. Circulation. 2021;144:1856–1875. doi: 10.1161/circulationaha.121.055949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelking L.J., Cantoria M.J., Xu Y., Liang G. Developmental and extrahepatic physiological functions of SREBP pathway genes in mice. Semin. Cell Dev. Biol. 2018;81:98–109. doi: 10.1016/j.semcdb.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P., Chen X., Xie R., Xie W., Huang L., Dong W., Han J., Liu X., Shen J., Huang J., Lin T. A novel AR translational regulator lncRNA LBCS inhibits castration resistance of prostate cancer. Mol. Cancer. 2019;18:109. doi: 10.1186/s12943-019-1037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Cao D., Sha J., Zhu X., Chen D. LncRNA ZFPM2-AS1 promotes lung adenocarcinoma progression by interacting with UPF1 to destabilize ZFPM2. Mol. Oncol. 2020;14:1074–1088. doi: 10.1002/1878-0261.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi M., Ikehara T., Nakagawa T., Yoneda M., Hattori N., Ikeda M., Ito T. Long noncoding RNAs transcribed downstream of the human β-globin locus regulate β-globin gene expression. J. Biochem. 2022;171:287–294. doi: 10.1093/jb/mvab130. [DOI] [PubMed] [Google Scholar]
- Hu M., Zhang D., Xu H., Zhang Y., Shi H., Huang X., Wang X., Wu Y., Qi Z. Salidroside activates the AMP-activated protein kinase pathway to suppress nonalcoholic steatohepatitis in mice. Hepatology. 2021;74:3056–3073. doi: 10.1002/hep.32066. [DOI] [PubMed] [Google Scholar]
- Kusnadi A., Park S.H., Yuan R., Pannellini T., Giannopoulou E., Oliver D., Lu T., Park-Min K.H., Ivashkiv L.B. The cytokine TNF promotes transcription factor SREBP activity and binding to inflammatory genes to activate macrophages and limit tissue repair. Immunity. 2019;51:241–257.e9. doi: 10.1016/j.immuni.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon P.K., Kim H.M., Kang B., Kim S.W., Hwang S.M., Im S.H., Roh T.Y., Kim K.T. hnRNP K supports the maintenance of RORγ circadian rhythm through ERK signaling. FASEB J. 2021;35:e21507. doi: 10.1096/fj.202002076r. [DOI] [PubMed] [Google Scholar]
- Lan X., Wu L., Wu N., Chen Q., Li Y., Du X., Wei C., Feng L., Li Y., Osoro E.K., et al. Long noncoding RNA lnc-HC regulates PPARγ-mediated hepatic lipid metabolism through miR-130b-3p. Nucleic Acids. 2019;18:954–965. doi: 10.1016/j.omtn.2019.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeau P.F., Byun J.H., Platko K., Saliba P., Sguazzin M., MacDonald M.E., Paré G., Steinberg G.R., Janssen L.J., Igdoura S.A., et al. Caffeine blocks SREBP2-induced hepatic PCSK9 expression to enhance LDLR-mediated cholesterol clearance. Nat. Commun. 2022;13:770. doi: 10.1038/s41467-022-28240-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chen W., Zhou Y., Abidi P., Sharpe O., Robinson W.H., Kraemer F.B., Liu J. Identification of mRNA binding proteins that regulate the stability of LDL receptor mRNA through AU-rich elements. J. Lipid Res. 2009;50:820–831. doi: 10.1194/jlr.m800375-jlr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Ruan X., Yang L., Kiesewetter K., Zhao Y., Luo H., Chen Y., Gucek M., Zhu J., Cao H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21:455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Cui K., Fang W., Chen Q., Xu D., Mai K., Zhang Y., Ai Q. High level of dietary olive oil decreased growth, increased liver lipid deposition and induced inflammation by activating the p38 MAPK and JNK pathways in large yellow croaker (Larimichthys crocea) Fish Shellfish Immunol. 2019;94:157–165. doi: 10.1016/j.fsi.2019.08.062. [DOI] [PubMed] [Google Scholar]
- Li X., Ji R., Cui K., Chen Q., Chen Q., Fang W., Mai K., Zhang Y., Xu W., Ai Q. High percentage of dietary palm oil suppressed growth and antioxidant capacity and induced the inflammation by activation of TLR-NF-κB signaling pathway in large yellow croaker (Larimichthys crocea) Fish Shellfish Immunol. 2019;87:600–608. doi: 10.1016/j.fsi.2019.01.055. [DOI] [PubMed] [Google Scholar]
- Li Y., Tocher D.R., Pang Y., Du J., Xiang X., Mai K., Ai Q. Environmental adaptation in fish induced changes in the regulatory region of fatty acid elongase gene, elovl5, involved in long-chain polyunsaturated fatty acid biosynthesis. Int. J. Biol. Macromol. 2022;204:144–153. doi: 10.1016/j.ijbiomac.2022.01.184. [DOI] [PubMed] [Google Scholar]
- Liao K., Yan J., Mai K., Ai Q. Dietary olive and perilla oils affect liver mitochondrial DNA methylation in large yellow croakers. J. Nutr. 2015;145:2479–2485. doi: 10.3945/jn.115.216481. [DOI] [PubMed] [Google Scholar]
- Lin Y.K., Yeh C.T., Kuo K.T., Yadav V.K., Fong I.H., Kounis N.G., Hu P., Hung M.Y. Pterostilbene increases LDL metabolism in HL-1 cardiomyocytes by modulating the PCSK9/HNF1α/SREBP2/LDLR signaling cascade, upregulating epigenetic hsa-miR-335 and hsa-miR-6825, and LDL receptor expression. Antioxidants. 2021;10:1280. doi: 10.3390/antiox10081280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minahk C., Kim K.W., Nelson R., Trigatti B., Lehner R., Vance D.E. Conversion of low-density lipoprotein-associated phosphatidylcholine to triacylglycerol by primary hepatocytes. J. Biol. Chem. 2008;283:6449–6458. doi: 10.1074/jbc.m706995200. [DOI] [PubMed] [Google Scholar]
- Munschauer M., Vogel J. Nuclear lncRNA stabilization in the host response to bacterial infection. EMBO J. 2018;37:e99875. doi: 10.15252/embj.201899875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muret K., Désert C., Lagoutte L., Boutin M., Gondret F., Zerjal T., Lagarrigue S. Long noncoding RNAs in lipid metabolism: literature review and conservation analysis across species. BMC Genomics. 2019;20:882. doi: 10.1186/s12864-019-6093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reches A., Nachmani D., Berhani O., Duev-Cohen A., Shreibman D., Ophir Y., Seliger B., Mandelboim O. HNRNPR regulates the expression of classical and nonclassical MHC class I proteins. J. Immunol. 2016;196:4967–4976. doi: 10.4049/jimmunol.1501550. [DOI] [PubMed] [Google Scholar]
- Rothzerg E., Ho X.D., Xu J., Wood D., Märtson A., Kõks S. Upregulation of 15 antisense long non-coding RNAs in osteosarcoma. Genes. 2021;12:1132. doi: 10.3390/genes12081132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H.G., Jung Y., Lee N., Seo J.Y., Kim S.W., Lee K.H., Kim D.Y., Kim K.T. HNRNP A1 promotes lung cancer cell proliferation by modulating VRK1 translation. Int. J. Mol. Sci. 2021;22:5506. doi: 10.3390/ijms22115506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Hao Q., Prasanth K.V. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34:142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault P.A., Ganesan A., Kalyaanamoorthy S., Clarke J.P.W.E., Salapa H.E., Levin M.C. hnRNP A/B proteins: an encyclopedic assessment of their roles in homeostasis and disease. Biology (Basel). 2021;10:712. doi: 10.3390/biology10080712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian N.N., Zheng Y.B., Li Z.P., Zhang F.W., Zhang J.F. Histone methylatic modification mediates the tumor-suppressive activity of curcumol in hepatocellular carcinoma via an Hotair/EZH2 regulatory axis. J. Ethnopharmacol. 2021;280:114413. doi: 10.1016/j.jep.2021.114413. [DOI] [PubMed] [Google Scholar]
- Wang J., Xiang D., Mei S., Jin Y., Sun D., Chen C., Hu D., Li S., Li H., Wang Y., et al. The novel long noncoding RNA Lnc19959.2 modulates triglyceride metabolism-associated genes through the interaction with Purb and hnRNPA2B1. Mol. Metab. 2020;37:100996. doi: 10.1016/j.molmet.2020.100996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X.D., Peng Z.S., Gu H.M., Wang M., Wang G.Q., Zhang D.W. Regulation of PCSK9 expression and function: mechanisms and therapeutic implications. Front. Cardiovasc. Med. 2021;8:764038. doi: 10.3389/fcvm.2021.764038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Turchini G.M., Francis D.S., Liang M., Mock T.S., Rombenso A., Ai Q. Are fish what they eat? A fatty acid's perspective. Prog. Lipid Res. 2020;80:101064. doi: 10.1016/j.plipres.2020.101064. [DOI] [PubMed] [Google Scholar]
- Yang B., Zhou Y., Wu M., Li X., Mai K., Ai Q. ω-6 Polyunsaturated fatty acids (linoleic acid) activate both autophagy and antioxidation in a synergistic feedback loop via TOR-dependent and TOR-independent signaling pathways. Cell Death Dis. 2020;11:607. doi: 10.1038/s41419-020-02750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.X., Zhang M., Long S.Y., Tuo Q.H., Tian Y., Chen J.X., Zhang C.P., Liao D.F. Cholesterol in LDL receptor recycling and degradation. Clin. Chim. Acta. 2020;500:81–86. doi: 10.1016/j.cca.2019.09.022. [DOI] [PubMed] [Google Scholar]
- Yu Q., Zheng H., Zhang Y. Inducible degrader of LDLR: a potential novel therapeutic target and emerging treatment for hyperlipidemia. Vascul. Pharmacol. 2021;140:106878. doi: 10.1016/j.vph.2021.106878. [DOI] [PubMed] [Google Scholar]
- Zelcer N., Hong C., Boyadjian R., Tontonoz P. LXR regulates cholesterol uptake through IDOL-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Tan P., Ji R., Xiang X., Cai Z., Dong X., Mai K., Ai Q. Influence of a dietary vegetable oil blend on serum lipid profiles in large yellow croaker (Larimichthys crocea) J. Agric. Food Chem. 2018;66:9097–9106. doi: 10.1021/acs.jafc.8b03382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All relevant data are within the manuscript and Supplementary Material. The accession number for the RNA sequencing data reported in this paper is GEO: GSE186012.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.