Abstract
EDTA is a chelating agent, widely used in many industries. Because of its ability to mobilize heavy metals and radionuclides, it can be an environmental pollutant. The EDTA monooxygenases that initiate EDTA degradation have been purified and characterized in bacterial strains BNC1 and DSM 9103. However, the genes encoding the enzymes have not been reported. The EDTA monooxygenase gene was cloned by probing a genomic library of strain BNC1 with a probe generated from the N-terminal amino acid sequence of the monooxygenase. Sequencing of the cloned DNA fragment revealed a gene cluster containing eight genes. Two of the genes, emoA and emoB, were expressed in Escherichia coli, and the gene products, EmoA and EmoB, were purified and characterized. Both experimental data and sequence analysis showed that EmoA is a reduced flavin mononucleotide-utilizing monooxygenase and that EmoB is an NADH:flavin mononucleotide oxidoreductase. The two-enzyme system oxidized EDTA to ethylenediaminediacetate (EDDA) and nitrilotriacetate (NTA) to iminodiacetate (IDA) with the production of glyoxylate. The emoA and emoB genes were cotranscribed when BNC1 cells were grown on EDTA. Other genes in the cluster encoded a hypothetical transport system, a putative regulatory protein, and IDA oxidase that oxidizes IDA and EDDA. We concluded that this gene cluster is responsible for the initial steps of EDTA and NTA degradation.
EDTA is a synthetic chelating agent that has a variety of uses in cleaners, water treatment plants, metal processing, and paper bleaching (41). It can cause mobilization of radionuclides and heavy metals (8, 28). Such mobilization increases the exposure of humans to toxic heavy metals and radionuclides. EDTA is not removed by conventional sewage treatment procedures and is recalcitrant in the environment (1, 44). The removal of EDTA can occur via photodegradation of EDTA-Fe(III) in surface waters (17, 18). Noncomplexed EDTA or EDTA complexed with other metals is not sensitive to photodegradation (24, 30). Although EDTA is recalcitrant, it can be degraded in the environment (7, 26). Belly et al. (4) observed slow degradation of EDTA in an aerated lagoon. Tiedje (45, 46) and Bolton et al. (7) reported slow biodegradation of EDTA in sediments and soils. Three pure cultures of microorganisms have been isolated that are able to degrade EDTA under aerobic conditions: the gram-negative bacterium BNC1 (33, 34), Agrobacterium sp. strain ATCC 55002 (22), and strain DSM 9103 (51).
The EDTA monooxygenase has been purified and characterized in strains BNC1 and DSM 9103. In BNC1, an EDTA monooxygenase oxidizes EDTA to ethylenediaminetriacetate (ED3A) and glyoxylate (19, 36). In DSM 9103, a similar enzyme oxidizes EDTA to ED3A and then to ethylenediaminediacetate (EDDA) (51). Both EDTA monooxygenases are reduced flavin mononucleotide (FMNH2)—utilizing monooxygenases that rely on NAD(P)H:flavin mononucleotide (FMN) oxidoreductases to supply FMNH2. However, the genes encoding EDTA-degrading enzymes have not been cloned and sequenced. In this study, we report the cloning, sequencing, and characterization of a gene cluster from bacterium BNC1 that is involved in the degradation of EDTA and nitrilotriacetic acid (NTA).
MATERIALS AND METHODS
Bacterial strains and plasmids.
The plasmids used or constructed in this study are listed in Table 1. The EDTA-degrading bacterium BNC1 was obtained from Bernd Nörtemann (Technical University of Braunschweig, Braunschweig, Germany). The cells were grown in a mineral medium containing 0.3 g of Na2EDTA · 2H2O per liter and 0.25 g of glycerol per liter (33). The medium was modified for growing uninduced cells by replacing EDTA with 0.15 g of NH4Cl per liter. Escherichia coli strain Invα was used as the host for plasmid pCR2.1 (Invitrogen, Carlsbad, Calif.), strain DH5α was used for pBluescript II KS+ (Stratagene, La Jolla, Calif.), strains Nova Blue and BL21(DE3) were used for pET30-LIC (Novagen, Madison, Wis.), strain JM109 was used for pTrc99A (Pharmacia, Alameda, Calif.), and strain MRA/P2 was used for phage λDASHII (Stratagene). E. coli cells were grown in Luria-Bertani medium (40). Kanamycin at 30 μg/ml and ampicillin at 50 μg/ml were added to the media when required.
TABLE 1.
Strains and vectors used in this study
| Strains and vectors | Genotype or description | Reference or source |
|---|---|---|
| E. coli Nova Blue | endA1 hsdR17(rK12−mK12+) supE44 thi-1 recA1 gyrA96 relA1lac [F′ proA+B+ lacIqZΔM15::Tn10 (Tcr)] | Novagen |
| E. coli JM109 | e14− (McrA−) recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Stratagene |
| E. coli BL21(DE3) | F−ompT [lon] hsdSB (rB− mB−) λDE3 lysogen, T7 DNA polymerase | Novagen |
| E. coli InvαF′ | F′ endA1 recA1 hsdR17 (rK−mK+) supE44 thi-1 gyr A96 relA1 φ80lacZΔM15 Δ(lacZYA-argF) U169 | Invitrogen |
| E. coli MRA/P2 | D(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-lgyrA96 relA1 lacc (P2 lysogen)c | Stratagene |
| BNC1 | Wild type | 33 |
| pBluescript II KS+ | fl/ColE1 ori, lacZα, Apr | Stratagene |
| pET30 | fl, T7 lac promoter, Kmr | Novagen |
| pCR2.1 | fl/ColE1 ori, lacZα, Kmr, Apr | Invitrogen |
| pTrc99A | trc promoter, pBR322 ori, Apr | Pharmacia |
| λDASHII/BamHI | red+ gam+, T3 and T7 promoters, double digested with BamHI and XhoI | Stratagene |
| pTA71-1 | pCR2.1 harboring a DNA fragment containing emoA | This study |
| λ1-1 | 14- to 15-kb fragment containing emoA in λ phage | This study |
| λ2-1 | 16- to 17-kb fragment containing emoA in λ phage | This study |
| pB1 | 3.1-kb BamHI fragment of λ1-1 insert in pBluescript | This study |
| pE1 | 6-kb EcoRI fragment of λ1-1 insert in pBluescript | This study |
| pE2 | 2.2-kb EcoRI fragment of λ1-1 insert in pBluescript | This study |
| pE16 | 1.1-kb EcoRI fragment of λ1-1 insert in pBluescript | This study |
| pE17 | 1.8-kb EcoRI fragment of λ1-1 insert in pBluescript | This study |
| pH2 | 7-kb HindIII fragment of λ1-1 insert in pBluescript | This study |
| pH7 | 4.1-kb HindIII fragment of λ1-1 insert in pBluescript | This study |
| pEmoA | 2.4-kb EcoRI fragment containing emoA in pTrc99A | This study |
| pEmoB | NdeI-NotI emoB PCR product in pET30 | This study |
Gene cloning.
The EDTA monooxygenase was purified from bacterium BNC1 by following the previously reported method (36). Purified protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (21) and then electroblotted onto a polyvinylidene difluoride membrane (27, 29) for N-terminal sequence determination. The N-terminal sequence was analyzed by automated microsequencing utilizing Edman degradation on an ABI 476A protein analyzer (PE Biosystems, Norwalk, Conn.) by the Nucleic Acid/Protein Service Unit of the University of British Columbia (Vancouver, Canada).
Two sets of degenerate 17-bp primers were designed to correspond to each end of the N-terminal sequence of EDTA monooxygenase. A PCR product of the expected size was obtained using BNC1 genomic DNA as template for 30 cycles with a thermal profile of 30 s at 94°C, 30 s at 40°C, and 10 s at 72°C. The PCR product was ligated into pCR2.1 (Invitrogen) and transformed into E. coli Invα. Colonies were screened for the insert, and the correct insert was identified by sequencing.
The genomic DNA of BNC1 was isolated by a standard method (40). To construct the genomic library, 25 μg of BNC1 genomic DNA was partially digested with BamHI. The DNA was ligated into the vector λDASHII/BamHI (Stratagene), and the constructs were packaged in vitro with Gigapack III XL packaging extract (Stratagene). E. coli MRA/P2 cells were infected with the packaging products to generate the BNC1 genomic library. Probe labeling by random priming, Southern blotting, plaque lifts, hybridizations, and isolation of phage DNA were performed by standard methods (3, 40). Restriction enzymes and T4 DNA ligase were purchased from Promega (Madison, Wis.). Shrimp alkaline phosphatase was purchased from Amersham (Piscataway, N.J.). Small-scale plasmid preparations were done using the Qia-Spin column kit (Qiagen, Chatsworth, Calif.). Transformation of E. coli was done by high-voltage electroporation (10).
DNA sequencing and sequence analysis.
DNA cycle-sequencing reactions using dye-labeled dideoxynucleotides (PE Biosystems) were done following the manufacturer's instructions. T3 and T7 primers were used for the initial reactions for cloned DNA fragments in pBluescript. Subsequent reactions used synthetic oligonucleotides identical or complementary to sequenced segments (37). The samples were analyzed by the Laboratory for Biotechnology and Bioanalysis at Washington State University, Pullman, Wash. The primary DNA sequence data were analyzed and assembled using GCG version 10 (Wisconsin Package, Genetics Computer Group, Madison, Wis.). The Basic Local Alignment Search Tool (BLAST) network service (2) was used to search for homologous DNA and protein sequences.
Overexpression of emoA and emoB.
For the overexpression of emoA, a 2.4-kb EcoRI fragment containing emoA was cloned into pTrc99A, creating a nonfusion construct pEmoA. The plasmid was electroporated into competent E. coli JM109 cells. The orientation of the insert was tested by HindIII digestion. The cells were grown to an optical density of 0.4 at 600 nm and induced by 1 mM isopropyl-β-d-thiogalactoside at 37°C for 3 h. For emoB overexpression, the DNA was amplified using primers EmoB-1 (5′-GAT-GAC-GAC-GAC-CAT-ATG-ACC-TAC-TCC-3′) with an introduced NdeI site (underlined) and EmoB-2 (5′-TCA-AGT-GAT-GTG-CGG-CCG-CGC-GCG-3′) with an introduced NotI site (underlined). The primer EmoB-2 overlapped the last 12 bases of emoB so that the gene continued to include a His-tag coding region on the vector. PCR amplification was performed with a profile of 30 s at 95°C, 45 s at 52°C, and 1.5 min at 72°C for 30 cycles. The PCR product was cut with NdeI and NotI and then ligated into the plasmid pET30-LIC previously digested with NdeI and NotI to produce plasmid pEmoB. The plasmid was electroporated into E. coli Nova Blue. Then the plasmid was isolated from Nova Blue cells and electroporated into E. coli BL21(DE3). The transformed BL21(DE3) cells were grown in Luria-Bertani medium at 37°C and induced with 1 mM isopropyl-β-d-thiogalactoside for protein production.
Purification of overexpressed proteins.
Overexpressed EmoA was purified from cell-free extracts of E. coli JM109 carrying pEmoA following the previously described procedure (36). The C-terminal fusion of EmoB was purified in one step, using an Ni2+-NTA-agarose matrix according to the manufacturer's instruction (Qiagen). The cell-free extract containing the EmoB fusion was incubated with 20 mM imidazole and the Ni2+-NTA-agarose matrix for 1 h. The mixture was loaded into a small column. The matrix was washed first with the loading buffer containing 20 mM imidazole and then with the buffer containing 80 mM imidazole. The target proteins were then eluted with the same buffer containing 250 mM imidazole. The purity of proteins was tested (SDS-PAGE) (21). The native molecular weight of EmoB was analyzed using a high-performance liquid chromatography (HPLC) system (Waters, Milford, Mass.) equipped with a size-exclusion column Biosep Sec S-3000, (Phenomenex, Torrance, Calif.). The sample was eluted with 20 mM potassium phosphate (KPi) buffer (pH 7) with 150 mM NaCl at 0.5 ml min−1. Size-exclusion molecular weight markers (Sigma Chemical Co., St. Louis, Mo.) were used to construct a standard curve.
Enzyme assays.
The activity of EmoB was analyzed in a reaction mixture containing 20 mM KPi, 10 μM FMN and an appropriate amount of the enzyme in a total volume of 1 ml. The reaction was started by addition of NADH to a final concentration of 200 μM. The reaction was immediately analyzed spectrophotometrically. The oxidation of NADH was monitored as the decrease of absorbance at 340 nm, using an Ultrospec 4000 spectrophotometer (Pharmacia). One unit of enzyme activity was defined as the amount of EmoB required to catalyze the consumption of 1 μmol of NADH per min. The optimal pH for EmoB activity was determined using 40 mM KPi buffer ranging from pH 6.2 to 7.8, and the optimal ionic strength for EmoB was determined using the pH 7 KPi buffer with 0 to 250 mM NaCl. The following substrate ranges were used for the determination of kinetic parameters: 0.5 to 20 μM for FMN and flavin adeninedinucleotide (FAD) and 25 to 300 μM for NADH.
The activity of EmoA was analyzed as previously described (36) by monitoring the production of glyoxylate. The standard reaction mixture contained 250 μM EDTA, 500 μM MgCl2, 10 μM FMN, 225 U of catalase, 0.013 U of EmoA, and 0.078 U of EmoB in 500 μl of 20 mM HEPES buffer (pH 7.8). The reaction was started by addition of NADH to a concentration of 2 mM. One unit of EmoA activity was defined as the amount of the enzyme required to produce 1 μmol of glyoxylate per min under the assay condition. For the completion of EDTA oxidation, the same amount of NADH was added three more times at approximately 1-h intervals.
Analysis of EmoA reaction end products.
Two independent methods were employed to analyze the end products of the EmoA reaction. A standard reaction mixture was used for both methods. In the first method, previously described procedures (5, 9) were modified to monitor the amount of EDTA and EDDA in the samples with an HPLC system (Waters). The sample was mixed, prior to injection, with (CH3COO)2Cu to a final concentration of 1 mM and injected onto a C8 column (250 by 4.6 mm; particle size, 5 μm; Phenomenex) heated to 40°C. The samples were eluted by an isocratic mobile phase of 200 mg of (CH3COO)2Cu per liter–1 g of dodecyltrimethylammonium bromide per liter–10 ml of CH3COOH per liter (pH 5.2) at a flow rate of 1 ml/min. In the second method, mass spectrometry was used for the end product identification. The samples were directly injected into a Finnigan LCQ mass spectrometer (Finnigan, Bremen, Germany) operated in the negative-ion mode. The typical electrospray source parameters were at a capillary voltage of 41 V, source voltage of 3,500 V, and capillary temperature of 200°C. EDTA, ED3A, and EDDA were all detectable by this method.
Reverse transcription-PCR analysis of emoA and emoB mRNAs.
RNA from BNC1 cells was isolated by a previously described phenol extraction method (40). Genomic DNA contamination was removed from the RNA preparation by using an RNeasy minikit (Qiagen) and following the manufacturer's instructions. Two sets of primers were designed for reverse transcription (RT)-PCR analysis. The first set, LY-17 and LY-18, was designed to span only the emoB gene. Primer sequences for emoB were 5′-GAC-TCG-CAA-GGC-AGG-CAT-ATT-CAC (LY-17 [bp 8205 through 8228]), and 5′-GT-TTA-TAC-GGA-CGG-AAC-GCT CAG (LY-18 [bp 8658 through 8636]). The second set, LY-21 and LY-23, was designed to span both emoA and emoB genes. Primer sequences for emoAB were 5′-ACC-AGG-TGC-TGC-AGG-AGA-AGG-ACT (LY-21 [bp 7638 through 7660]) and 5′-GGA-TCG-AGG-TCG-ATG-ACG-TGA-AT (LY-23 [bp 8245 through 8223]). PCRs were carried out using the OneStep RT-PCR kit (Qiagen) in a 25-μl volume containing 0.1 μg of template RNA and 20 pmol of each primer in the reaction mixture by following the manufacturer's instructions. After RT at 50°C for 30 min, the reaction mixtures were heated to 95°C for 15 min and given 35 cycles of 45 s at 94°C, 45 s at 52°C, and 2 min at 72°C, followed by the final extension at 72°C for 10 min. Negative-control reactions were done by performing normal PCR using the same thermal cycles.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been deposited in the GenBank database under accession number AF176664.
RESULTS
Cloning the EDTA-monooxygenase gene.
EDTA-monooxygenase was purified from cell-free extracts of BNC1, and its N-terminal amino acid sequence was determined to be MRKRRMYLVSWLNSSGVLPNSWNEGRH. Degenerate primers JPN1 [5′-ATG-CG(C/G)-AA(A/G)-CG(C/G)-CG(C/G)-AT-3′], corresponding to amino acids Met-1 through Met-6, and JPN3 [5′-TC-(A/G)TT-CCA-(C/G)(C/G)(A/T)-(A/G)TT-(C/G)GG-3′], corresponding to amino acids Glu-24 through Pro-19, were designed. PCR resulted in many bands on an agarose gel. A second round of PCR using the 71-bp band as a template resulted in a dominant 71-bp band. Reaction products were then cloned into plasmid pCR2.1. Clone pTA71-1 contained the sequence of the correct size, and its translated amino acid sequence matched the determined N-terminal sequence. The insert was cut out and used as probe 71-1. The probe hybridized to a single fragment of genomic DNA digested by selected restriction enzymes: a 3.1-kb BamHI fragment, a 2.4-kb EcoRI fragment, a 4-kb HindIII fragment, and a 1-kb PstI fragment. This result showed that the probe was specific. The probe hybridized to two plaques in the BNC1 genomic library, λ1-1 and λ2-1. Digestion of the DNA inserts by BamHI, EcoRI, and HindIII was performed. Agarose gel electrophoresis revealed similar band patterns for both clones, corresponding to the results from Southern analysis of the genomic DNA. It was estimated that λ1-1 had an insert of 14 to 15 kb and that λ2-1 had an insert of 16 to 17 kb. The insert fragments of λ2-1 were subcloned into pBluescript. The plasmid clones included a 3.1-kb BamHI fragment (pB1), a 6-kb EcoRI fragment (pE1), a 2.2-kb EcoRI fragment (pE2), a 1.1-kb EcoRI fragment (pE16), a 1.8-kb EcoRI fragment (pE17), a 7-kb HindIII fragment (pH2), and a 4.1-kb HindIII fragment (pH7). The DNA sequences of all the fragments were determined. Because of the overlapping of the fragments, the entire phage insert sequence was assembled.
Sequence analysis of emoA and adjacent ORFs.
The organization of the open reading frames (ORFs) identified in the phage insert is shown in Fig. 1. The coding region of emoA was from positions 6701 through 7990 and encoded a protein of 430 amino acids with a calculated molecular weight of 47,330, which was close to that estimated from the relative mobility of EmoA on SDS-PAGE (36). The translated N-terminal amino acid sequence of EmoA matched exactly with the determined N-terminal amino acid sequence of EDTA monooxygenase, confirming that emoA encodes EDTA monooxygenase. A good Shine-Delgarno sequence, GGAGG, was located seven bases upstream from the start codon of emoA. A promoter sequence could not be found. A computer search with BLAST revealed that the amino acid sequence of EmoA was similar to several FMNH2-utilizing monooxygenases. When the entire sequences of these proteins were compared by the GCG Gap program, the percentages of similarity/identity were as follows: 46.2/37.8 for NTA monooxygenase of Chelatobacter heintzii ATCC 29600 (20, 53), 45.6/34.6 for pristinamycin IIA synthase SnaA of Streptomyces pristinaespiralis (6), and 39.6/31.9 for DszA of Rhodococcus sp. strain IGTS8, which participates in conversion of dibenzothiophene to 2-hydroxybiphenyl (38).
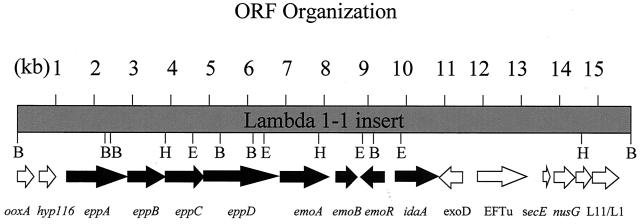
FIG. 1.
The restriction sites and gene organization of the 15,603-bp DNA fragment. BamHI (B), HindIII (H), and EcoRI (E) target sites are indicated. Orientation of the arrows indicates the direction of gene transcription. The genes represented by black arrows may directly participate in EDTA degradation.
The DNA sequences upstream and downstream of emoA were also analyzed. Upstream of emoA, six ORFs were identified. A set of ORFs directly upstream from emoA code for a hypothetical transport system. These genes are closely related to the genes of oligopeptide permease systems like oppABCDF of E. coli (31, 39). Thus, they were designated eppABCD for encoding a putative permease system. The gene eppA is similar to mppA encoding a periplasmic-binding protein (35). EppA is a 593-amino-acid protein and has a putative signal peptide (amino acids 1 to 26), indicating that it is a periplasmic protein. The gene eppB encodes a protein of 315 amino acid residues similar to OppB. The gene eppC codes for a 308-amino-acid protein similar to OppC. The next gene immediately upstream of emoA is eppD, which encodes a 610-amino-acid protein similar to OppD. The GCG Motifs program identified two ATP-binding “p-loops” within EppD. The sequence analysis showed that eppABCD genes code for a putative ATP-binding cassette-type transport system. The stop codon of eppD overlapped the start codon of emoA, suggesting that these genes may be cotranscribed with emoA. Upstream of eppA, an ORF designated hyp116 which spans positions 589 to 936 encoded a 116-amino-acid protein with unknown function. Upstream of hyp116, an incomplete ORF spanning positions 1 through 526 and similar to the C terminus of d-nopaline dehydrogenase was designated ooxA.
Several ORFs were identified downstream of emoA. The sequence from positions 8106 through 8696 encoded a 197-amino-acid protein with a calculated molecular weight of 20,930. It was most similar to an NADH-dependent FMN reductase from Pseudomonas putida (16). This ORF was designated emoB. The sequence from position 8733 through 9353 encoded a putative regulatory protein in the opposite orientation to emoA and emoB. This ORF encoded a 206-amino-acid protein with a calculated molecular weight of 23,729. It was similar to ntaR from the NTA monooxygenase operon (20, 53). This ORF was designated emoR. The sequence from position 9777 to 10889 encoded an ORF designated idaA (Y. Liu, T.-M. Louie, and L. Xun, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. Q395, 1999; Y. Liu, T.-M. Louie, J. Payne, J. Bohuslavek, H. Bolton, Jr., and L. Xun, submitted for publication). It encoded a protein of 371 amino acid residues, with a calculated molecular weight of 39,025. This sequence was similar to glycine oxidase yjbR (32). The sequence from positions 11515 through 10865 encoded a 217-amino-acid protein with a calculated molecular weight of 23,313. This ORF, designated exoD, was similar to ORF203 of Caulobacter crescentus, which may be a membrane protein involved in exopolysaccharide synthesis (14). The sequence from positions 11837 through 13009 encoded an EF-Tu, which was 91% identical to the EF-Tu from Agrobacterium tumefaciens (42). The sequence from positions 13515 through 13715 encoded a 67-amino-acid protein with a calculated mass of 7,392 that had strong sequence similarities with the SecE preprotein translocase subunit of Thermotoga maritima and was designated secE (11). The sequence from 13752 through 14273 encoded a 174-amino-acid protein with a calculated mass of 19,569. It was similar to the NusG transcription termination protein from E. coli (11). The sequences from position 14413 through 14796 and 14844 through 15539 encoded ribosomal proteins L11 and L1, respectively. The L11 and L1 proteins were most similar to L11 from Serratia marcescens and to L1 from a citrus greening disease-associated bacterium (13).
Overexpression and purification of emoA.
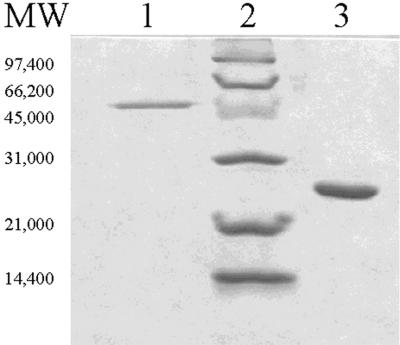
To confirm that emoA encodes EDTA monooxygenase, the gene was cloned into a pTrc99A expression vector to yield plasmid pEmoA. Expression of emoA in JM109 cells harboring pEmoA resulted in the production of about 1% soluble EmoA in the cell-free extracts of the cells grown at 37°C. The enzyme in the cell extract was active, and it was purified by previously described methods (36). Seventy-two micrograms of pure protein was purified from 75.6 mg of protein in cell-free extracts (Fig. 2) with 8% recovery of EDTA monooxygenase activity. The pure EmoA had a specific activity of 3.8 U (± a standard deviation of 0.42 U, the average of three measurements) for glyoxylate production in the presence of FMN, NADH, and EmoB. The enzyme obtained by this procedure was used for all analytical assays in this study.
FIG. 2.
SDS-PAGE of the purified EmoA and EmoB proteins. Lane 1 contains 0.6 μg of nonfusion EmoA, lane 2 contains molecular weight markers (low-range SDS-PAGE markers; Bio-Rad, Hercules, Calif.), and lane 3 contains 2 μg of EmoB (C-terminal fusion). The gel was stained with Gel Code Blue.
Overexpression, purification, and characterization of EmoB.
Overexpression of emoB in BL21(DE3) cells harboring pEmoB yielded highly soluble EmoB in the cells grown at room temperature. EmoB was purified from cell-free extracts using the Ni2+-NTA agarose column. The purification procedure yielded highly concentrated and purified EmoB (Fig. 2). The 1.24 mg of pure EmoB was obtained from 17.6 mg of protein in the cell-free extract, with 95% recovery of EmoB activity. The pure EmoB had a specific activity of 105 U (±13 U) for FMN reduction. The apparent molecular weight of the enzyme based on SDS-PAGE was around 25,000, which corresponded to the size calculated from the gene sequence with the C-terminal fusion. The apparent molecular weight based on HPLC analysis using a gel filtration column was between 45 and 55 kDa, indicating that EmoB is a dimer. The protein is an NADH-dependent FMN reductase. It can utilize NADH, but not NADPH, for FMN reduction. Besides FMN, EmoB can also reduce FAD, but not riboflavin. The rate of NADH consumption when reducing FAD was about 60% of the rate when reducing FMN. The optimum pH for the enzymatic activity was 7.0. The enzyme showed highest activity in 40 mM potassium phosphate buffer. The Km for NADH was 40 μM. The Km for FMN was 13.3 μM. The kcat for NADH was 47.7 s−1, based on the molecular mass of one subunit, which is 25,000 Da.
EDTA degradation by the EmoA-EmoB system.
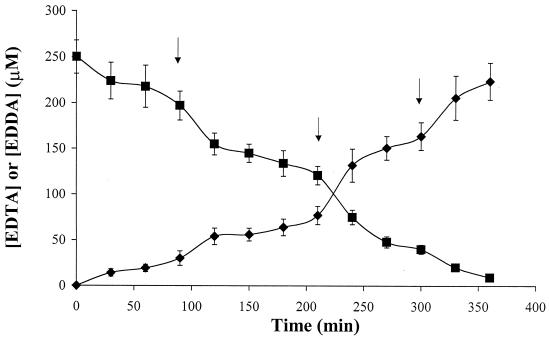
Enzymatic assays were performed with reactions containing both EmoA and EmoB in appropriate ratios. EmoA required metal ions in the reaction mixture. In the absence of metals, EmoA exhibited no activity against EDTA. Mg2+, Co2+, Cu2+, Zn2+, and Cd2+ promoted the highest EmoA activity. The enzyme showed no activity with Ni2+, Pb2+, and Ca2+. The amount of glyoxylate produced during the reaction was determined. It was shown that the molar ratio of glyoxylate production to EDTA consumption was close to 2. This result indicates that two acetyl groups were removed from EDTA during the reaction, suggesting that EDDA was the final end product. This was supported by HPLC and mass spectrometry analyses. HPLC analysis showed that the amount of EDTA eluted at 21 min decreased as the reaction progressed. The amount of EDDA eluted at 2.7 min steadily increased (Fig. 3). Since the HPLC method we used could not detect ED3A, mass spectrometry analysis of ED3A was used. In mass spectrometry, the only peak observed at time 0 was EDTA (m/z = 313). As the reaction progressed, a peak at m/z = 233 appeared after several minutes of incubation. After 30 min of incubation, a peak at m/z = 175 was detected. The peak at m/z = 233 corresponds to that of ED3A, which is the first reaction product from EDTA (36, 51). The peak at m/z = 175 represents EDDA. When the reaction was completed, the peaks at 313 and 233 disappeared, and only the peak at 175 was observed. Thus, the data indicated the oxidation of EDTA to ED3A and then to EDDA by EmoA.
FIG. 3.
The time course of EDTA degradation by EmoA/EmoB. Symbols: ⧫, EDDA accumulation; ■, EDTA degradation. The arrows indicate the subsequent addition of NADH. Error bars indicate the standard deviations for triplicate samples.
Analysis of the emoA and emoB expression in BNC1.
The specific activities of EmoA and NADH:FMN oxidoreductases in the cell-free extracts of induced and uninduced cells were compared. The specific activity of EmoA from induced cells was 0.174 U mg−1 of protein (±0.022 U), and the activity of EmoA from uninduced cells was 0.0055 U mg−1 of protein (±0.00026 U). These results showed a 31-fold increase in EmoA specific activity in induced cells. The specific activity of NADH:FMN oxidoreductases was 55 U mg−1 of protein (±2.34 U) from induced cells and was 17.6 U mg−1 of protein (±4.6 U) from uninduced cells. These data showed a mere 3.13-fold increase of NADH:FMN oxidoreductase activity in induced cells.
RT-PCR analysis of transcripts present in BNC1.
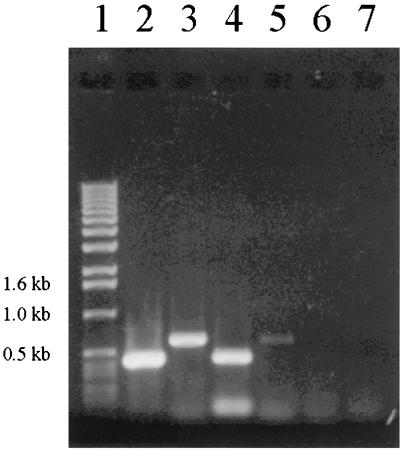
In order to show that emoA and emoB genes are cotranscribed and to further prove that emoB is induced by EDTA, we examined transcripts from BNC1 cells grown on EDTA and on NH4Cl as a control. Two sets of primers were designed, one spanning the emoB gene alone and the other spanning both genes. The expected RT-PCR product size for emoB alone was 453 bp, and for emoAB it was 607 bp. The PCR products were analyzed by agarose gel electrophoresis. When primers spanning emoB alone were used, approximately fourfold more product was obtained from EDTA-grown cells than from cells grown on NH4Cl (Fig. 4). When primers spanning both genes were used, approximately 35-fold more product was obtained from EDTA-grown cells than from NH4Cl-grown cells (Fig. 4). The ratios of product amounts were determined by analysis of diluted samples by agarose gel electrophoresis. No products were obtained from reaction mixtures when normal PCR was performed, showing that there was no genomic DNA contamination in the RNA samples.
FIG. 4.
RT-PCR of emo genes. Molecular markers are in lane 1 (1 kb DNA ladder; Lifetech, Rockville, Md.). Lane 2, emoB-induced cells; lane 3, emoAB-induced cells; lane 4, emoB-uninduced cells; lane 5, emoAB-uninduced cells; lane 6, emoB amplification by normal PCR (no reverse transcriptase)-induced cells; and lane 7, emoB amplification by normal PCR-uninduced cells.
DISCUSSION
Two proteins, EmoA and EmoB, were expressed in E. coli and purified. The purified EmoA was used to analyze the initial steps of EDTA degradation. This is the first report of purification and characterization of NADH:FMN oxidoreductase EmoB. The activity of EmoB was about 2.5-fold higher than that of Fre, which is a flavin reductase isolated from E. coli (54). The activity of EmoB was 105 U mg−1, whereas the activity of Fre was 40 U mg−1, when using NADH to reduce FMN. EmoB was found to be a dimer of identical subunits. EmoA depends on EmoB to generate FMNH2 as its cosubstrate. However, EmoB can be replaced by other flavin reductases (36), suggesting that the physical contact of the two enzymes is not required for the function of EmoA in vitro. Similar results were obtained for the EDTA monooxygenase and its flavin reductase of DSM 9103 (51) and for NmoA and NmoB of Chelatobacter heintzii ATCC 29600 (53).
EmoA and EmoB together oxidized EDTA to ED3A and then to EDDA. The function of EmoB is to generate FMNH2 for EmoA. For the complete oxidation of 250 μM EDTA to EDDA, the repeated addition of NADH to a final concentration of 8 mM was required. The data showed that only about 6% of FMNH2 produced by EmoB was used by EmoA for EDTA metabolism, representing significant uncoupling of the two enzyme activities under the assay conditions. The assay conditions were optimized for the determination of EmoA specific activity, and EmoA was the limiting factor. The excess amount of FMNH2 produced by EmoB is autooxidized by O2 to produce H2O2 (54). This explains the need for repeated addition of NADH for the completion of EDTA oxidation. Catalase was added to the reaction solution to remove H2O2.
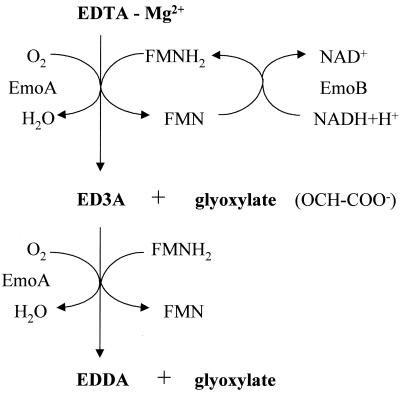
The first two steps of EDTA biodegradation by bacterium BNC1 were clarified in this study. As was reported previously, EmoA catalyzes the breaking of the N—C bond in EDTA in the presence of oxygen and FMNH2 to produce ED3A and glyoxylate (36). Our results demonstrated that EmoA also catalyzes the second degradation step, oxidizing ED3A to EDDA and glyoxylate by the same type of reaction (Fig. 5). These results correspond to the reactions catalyzed by the EDTA monooxygenase of strain DSM 9103 (51). In the beginning of that reaction, ED3A and glyoxylate are produced, which leads to the accumulation of ED3A. This short-term accumulation of ED3A was observed in our study by mass spectrometry, which showed only the peak of ED3A but no peak of EDDA after 5 min of incubation. Later in the reaction, ED3A was further degraded to EDDA by the enzyme system as detected by both HPLC and mass spectrometry. The transitory accumulation of ED3A indicates that ED3A is detached from EmoA before it is further metabolized. The presence of certain metal ions in the reaction mixture was essential for EDTA degradation although EDTA monooxygenase with the commercial NAD(P)H:FMN oxidoreductase of Photobacterium fischeri (Boehringer Mannheim Co., Indianapolis, Ind.) oxidized EDTA without any added divalent cations (36), probably due to the trace amount of certain cations present in the commercial product which allow this oxidation of EDTA by EDTA monooxygenase. On the basis of catalytic properties and identical N-terminal amino acid sequences, EmoA of BNC1 is very similar to the EDTA monooxygenase of DSM 9103 (51).
FIG. 5.
The EDTA degradation by EmoA in the bacterium BNC1.
There are similarities between the EDTA degradation by EDTA monooxygenases of BNC1 and DSM 9103 and NTA degradation by NTA monooxygenase of C. heintzii ATCC 29600 (48, 49, 51, 53). The NTA monooxygenase is also an FMNH2-utilizing enzyme, and it depends on an NADH:FMN oxidoreductase to generate FMNH2. The end products of NTA oxidation are iminodiacetate and glyoxylate, which indicates that the reaction mechanism is similar to that of EmoA. Moreover, there are sequence similarities between NTA monooxygenase and EmoA. Both enzymes require FMNH2 and oxygen, and both are polypeptides of about 47 kDa. There is also a substrate overlap, as EmoA degrades NTA as well as EDTA (36), but NTA monooxygenase degrades only NTA (48). Both enzymes belong to a family of FMNH2-utilizing monooxygenases, together with EDTA monooxygenase of the bacterium DSM 9103 (51), bacterial luciferase of Photobacterium fisheri (47), pristinamycin IIA synthase of S. pristinaespiralis (6, 43), and two other monooxygenases which participate in desulfurization of dibenzothiophene in Rhodococcus sp. strain IGTS8 (12, 23). A similar enzyme system was also reported in P. putida S-313. The ssuD and ssuE genes form a two-component FMNH2-dependent monooxygenase in the ssu operon, which plays a key role in organosulfur metabolism in this bacterium (16).
The sequence data indicated that in addition to emoA and emoB, six other genes in the gene cluster could participate in EDTA degradation. The group of four genes eppABCD could function as an ATP-binding cassette-type transport system, facilitating transport of EDTA into the cell. Since EDTA is a highly hydrophilic compound, it should require active transport across the cytoplasmic membrane. Direct evidence of an active transport for EDTA uptake has been recently reported (52), in which Witschel et al. showed that EDTA transport into the cells in strain DSM 9103 requires energy and is inducible. The regulatory gene emoR could also play a role in EDTA degradation. A regulatory gene is present in some other operons containing genes for similar FMNH2-utilizing monooxygenases. The NTA monooxygenase operon in C. heintzii ATCC 29600 contains the nmoR gene (20, 53), and an aryl desulfonation operon in P. putida S-313 contains regulatory gene asfR (50). Finally, the IDA oxidase gene idaA could participate in EDTA degradation by metabolizing the end product of EmoA (Liu et al., submitted).
Our sequence analysis, activity assays, and RT-PCR results indicate that the transport genes (eppABCD) and emoA and emoB are likely cotranscribed and regulated as an operon. The stop codon of the last gene of the putative permease system, eppD, overlaps the start codon of emoA. This suggests that the epp genes are transcribed together with emoA. The inducibility of the EDTA transport in the bacterium DSM 9103 (52) suggests that the genes encoding the EDTA transport proteins and EDTA monooxygenase may be coregulated as an operon. Such operons containing both transport and FMNH2-utilizing monooxygenase genes have been reported. The ssu operon, containing genes for an FMNH2-utilizing monooxygenase system and transport proteins involved in the metabolism of organosulfur compounds in P. putida, has been identified (16). The EDTA-degrading operon proposed here contains the putative EDTA permease genes eppABCD, EDTA monooxygenase gene emoA, the NADH:FMN oxidoreductase gene emoB, and the regulatory gene emoR. The RT-PCR showed that the genes of emoA and emoB are cotranscribed when growing on EDTA and emoB is also constitutively expressed by its own promoter. The genes emoB and emoA are separated by 116 bp. The RT-PCR data are in good agreement with the data from the activity assays. The EmoA activity was increased 31-fold, while the NADH-FMN oxidoreductase activity was increased by only 3.1-fold when the cells were grown on EDTA. The iminodiacetate (IDA) oxidase gene idaA is probably not a part of the operon, because it is positioned downstream from the regulatory gene emoR and it is separated from emoR by more than 400 bp. However, IDA oxidase can oxidize IDA to glycine and glyoxylate (Liu et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999) and EDDA to ethylenediamine (ED) and two glyoxylates (Liu et al., submitted). Therefore, the gene cluster contains all of the necessary genes to remove the chelating properties of EDTA and its metabolic intermediates and to degrade NTA to common metabolic intermediates. NTA is oxidized by EmoA to IDA, and then IDA is oxidized by IdaA to glycine and glyoxylate, while EmoA oxidizes EDTA to ED3A and then to EDDA, which is oxidized to ED by IdaA.
Little is known about the phylogeny of the bacterium BNC1. Based on the polyamine pattern, strain BNC1 seems to belong to the alpha subclass of proteobacteria (33). The sequences of some genes in the gene cluster reported in this study can reveal the relationships of BNC1 to other bacteria. In addition to rRNA genes, EF-Tu can be used as a phylogenetic marker (25). The bacterium BNC1 appears to be closely related to A. tumefaciens, on the basis of sequence similarities of the EF-Tu genes. If bacterium BNC1 belongs to the genus Agrobacterium, it is also related to the EDTA-degrading Agrobacterium sp. (22). Although the genes in this gene cluster are involved in the degradation of chelating agents, the GC content of the cluster does not suggest that the cluster is acquired from another organism. The EDTA-degrading cluster has GC content of 61.49%, which corresponds to CG contents of the most closely related bacteria. A. tumefaciens has GC content of 57 to 63%, and Agrobacterium rhizogenes has 59 to 63% GC content (15).
ACKNOWLEDGMENTS
Jan Bohuslavek and Jason W. Payne contributed equally to this work.
This work was supported by the Natural and Accelerated Bioremediation Research Program, Office of Biological and Environmental Research, U.S. Department of Energy. Pacific Northwest National Laboratory is operated for the Department of Energy by Battelle Memorial Institute under contract DE-AC06-76RLO 1830.
REFERENCES
- 1.Alder A C, Siegrist H, Gujer W, Giger W. Behavior of NTA and EDTA in biological wastewater treatment. Water Res. 1990;24:733–742. [Google Scholar]
- 2.Altschul S F, Lipman D J. Protein database searches for multiple alignments. Proc Natl Acad Sci USA. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel M F. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1993. [Google Scholar]
- 4.Belly R T, Lauff J J, Goodhue C T. Degradation of ethylenediaminetetraacetic acid by microbial populations from an aerated lagoon. Appl Microbiol. 1975;29:787–794. doi: 10.1128/am.29.6.787-794.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergers P J M, DeGroot A C. The analysis of EDTA in water by HPLC. Water Res. 1994;28:639–642. [Google Scholar]
- 6.Blanc V, Lagneaux D, Didier P, Gill P, Lacroix P, Crouzet J. Cloning and analysis of structural genes from Streptomyces pristinaespiralis encoding enzymes involved in the conversion of pristinamycin IIB to pristinamycin IIA (PIIA): PIIA synthase and NADH:riboflavin 5′-phosphate oxidoreductase. J Bacteriol. 1995;177:5206–5214. doi: 10.1128/jb.177.18.5206-5214.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolton H., Jr Biodegradation of synthetic chelates in subsurface sediments from the southeast coastal plain. J Environ Quality. 1993;22:125–132. [Google Scholar]
- 8.Cleveland J M, Rees T F. Characterization of plutonium in Maxey Flats radioactive trench leachates. Science. 1981;200:1506–1509. doi: 10.1126/science.212.4502.1506. [DOI] [PubMed] [Google Scholar]
- 9.DeJong J, Van Polanen A, Driessen J J M. Determination of ethylenediaminetetraacetic acid and its salts in canned mushrooms by reversed phase ion-pair liquid chromatography. J Chromatogr. 1991;553:243–248. [Google Scholar]
- 10.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downing W L, Sullivan S L, Gottesman M E, Dennis P P. Sequence and transcriptional pattern of the essential E. coli secE-nusG operon. J Bacteriol. 1990;172:1621–1627. doi: 10.1128/jb.172.3.1621-1627.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray K A, Pogrebinski O S, Mrachko G T, Xi L, Monticello D J, Squires C H. Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat Biotechnol. 1996;14:1705–1709. doi: 10.1038/nbt1296-1705. [DOI] [PubMed] [Google Scholar]
- 13.Jagoueix S, Bove J-M, Garnier M. The phloem-limited bacterium of greening disease of citrus is a member of the α subdivision of proteobacteria. Int J Syst Bacteriol. 1994;44:379–386. doi: 10.1099/00207713-44-3-379. [DOI] [PubMed] [Google Scholar]
- 14.Janakiraman R S, Brun Y V. Transcriptional and mutational analysis of the rpoN operon in Caulobacter crescentus. J Bacteriol. 1997;179:5138–5147. doi: 10.1128/jb.179.16.5138-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan D C. Family III: Rhizobiaceae, Conn 1938. In: Holt J G, Krieg N R, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. [Google Scholar]
- 16.Kahnert A, Vermeij P, Wietek C, James P, Leisinger T, Kertesz M A. The ssu locus plays a key role in organosulfur metabolism in Pseudomonas putida S-313. J Bacteriol. 2000;182:2869–2878. doi: 10.1128/jb.182.10.2869-2878.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kari F G, Giger W. Modeling the photochemical degradation of ethylenediaminetetraacetate (EDTA) in the river Glatt. Environ Sci Technol. 1995;29:2814–2827. doi: 10.1021/es00011a018. [DOI] [PubMed] [Google Scholar]
- 18.Kari F G, Hilger S U, Canonica S. Determination of the reaction quantum yield for the photochemical degradation of Fe(III)EDTA—implications for the environmental fate of EDTA in surface waters. Environ Sci Technol. 1995;29:1008–1017. doi: 10.1021/es00004a022. [DOI] [PubMed] [Google Scholar]
- 19.Kluner T, Hempel D C, Nörtemann B. Metabolism of EDTA and its metal chelates by whole cells and cell-free extracts of strain BNC1. Appl Microbiol Biotechnol. 1998;49:194–201. [Google Scholar]
- 20.Knobel H, Egli T, Van Der Meer J R. Cloning and characterization of the genes encoding nitrilotriacetate monooxygenase of Chelatobacter heintzii ATCC 29600. J Bacteriol. 1996;178:6123–6132. doi: 10.1128/jb.178.21.6123-6132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Lauff J J, Steele D B, Coogan L A, Breitfeller J M. Degradation of the ferric chelate of EDTA by a pure culture of an Agrobacterium sp. Appl Environ Microbiol. 1990;56:3346–3353. doi: 10.1128/aem.56.11.3346-3353.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei B, Tu S-C. Gene overexpression, purification, and identification of a desulfurization enzyme from Rhodococcus sp. strain IGTS8 as a sulfide/sulfoxide monooxygenase. J Bacteriol. 1996;178:5699–5705. doi: 10.1128/jb.178.19.5699-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockhart H B, Blakeley R V. Aerobic photodegradation of Fe(III)-(ethylenedinitrilo)tetraacetate (ferric EDTA): implications for natural waters. Environ Sci Technol. 1975;9:1035–1038. doi: 10.1080/00139307509437453. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig W, Schleifer K-H. Phylogeny of bacteria beyond the 16S rRNA Standard. ASM News. 1999;65:752–757. [Google Scholar]
- 26.Madsen E L, Alexander M. Effects of chemical speciation on the mineralization of organic compounds by microorganisms. Appl Environ Microbiol. 1985;50:342–349. doi: 10.1128/aem.50.2.342-349.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 28.Means J L, Crerar D A, Duguid O J. Migration of radioactive wastes: radionuclide mobilization by complexing agents. Science. 1978;200:1477–1481. doi: 10.1126/science.200.4349.1477. [DOI] [PubMed] [Google Scholar]
- 29.Moos M, Nguyen N Y, Liu T Y. Reproducible high yield sequencing of proteins electrophoretically separated and transferred to an inert support. J Biol Chem. 1988;263:6005–6008. [PubMed] [Google Scholar]
- 30.Natarajan P, Endicott J F. Photoredox behavior of transition metal-ethylenediaminetetraacetate complexes: a comparison of some group VIII metals. J Phys Chem. 1973;77:2049–2054. [Google Scholar]
- 31.Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 32.Nishiya Y, Imanaka T. Purification and characterization of a novel glycine oxidase from Bacillus subtilis. FEBS Lett. 1998;438:263–266. doi: 10.1016/s0014-5793(98)01313-1. [DOI] [PubMed] [Google Scholar]
- 33.Nörtemann B. Total degradation of EDTA by mixed cultures and a bacterial isolate. Appl Environ Microbiol. 1992;58:671–676. doi: 10.1128/aem.58.2.671-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nörtemann B. Biodegradation of EDTA. Appl Microbiol Biotechnol. 1999;51:751–759. doi: 10.1007/s002530051458. [DOI] [PubMed] [Google Scholar]
- 35.Park J T, Raychaudhuri D, Li H, Normark S, Mengin-Lecreulx D. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate. J Bacteriol. 1998;180:1215–1223. doi: 10.1128/jb.180.5.1215-1223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Payne J W, Bolton H, Jr, Campbell J C, Xun L. Purification and characterization of EDTA monooxygenase from the EDTA-degrading bacterium BNC1. J Bacteriol. 1998;180:3823–3827. doi: 10.1128/jb.180.15.3823-3827.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Payne J W. Cloning and characterization of a gene cluster involved in EDTA degradation from bacterium BNC1. M.S. thesis. Pullman, Wash: Washington State University; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piddington C S, Kovacevich B R, Rambosek J. Sequence and molecular characterization of a DNA region encoding the dibenzothiophene desulfurization operon of Rhodococcus sp. strain IGTS8. Appl Environ Microbiol. 1995;61:468–475. doi: 10.1128/aem.61.2.468-475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudner D Z, Ledeaux J R, Ireton K, Grossman A D. The spoOK locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J Bacteriol. 1991;173:1388–1398. doi: 10.1128/jb.173.4.1388-1398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Frisch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Silanpaa M. Environmental fate of EDTA and DTPA. Rev Environ Contam Toxicol. 1997;152:85–111. doi: 10.1007/978-1-4612-1964-4_3. [DOI] [PubMed] [Google Scholar]
- 42.Sylvanen A-C, Amiri H, Jamal A, Anderson S G E, Kurland C G. A chimeric disposition of the elongation factor genes in Rickettsia prowazeki. J Bacteriol. 1996;178:6182–6199. doi: 10.1128/jb.178.21.6192-6199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thibaut D, Ratet N, Bisch D, Faucher D, Debussche L, Blanche F. Purification of the two enzyme system catalyzing the oxidation of the d-proline residue of pristinamycin IIB during the last step of pristinamycin IIA biosynthesis. J Bacteriol. 1995;177:5199–5205. doi: 10.1128/jb.177.18.5199-5205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thom N S, Agg A R. The breakdown of synthetic organic compounds in biological processes. Proc R Soc London Ser B. 1975;189:347–357. doi: 10.1098/rspb.1975.0061. [DOI] [PubMed] [Google Scholar]
- 45.Tiedje J M. Microbial degradation of ethylenediaminetetraacetate in soils and sediments. Appl Environ Microbiol. 1975;30:327–329. doi: 10.1128/am.30.2.327-329.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiedje J M. Influence of environmental parameters on EDTA biodegradation in soils and sediments. J Environ Quality. 1977;6:21–26. [Google Scholar]
- 47.Tu S-C, Mager H I X. Biochemistry of bacterial bioluminescence. Photochem Photobiol. 1995;62:615–624. doi: 10.1111/j.1751-1097.1995.tb08708.x. [DOI] [PubMed] [Google Scholar]
- 48.Uetz T, Schneider R, Snozzi M, Egli T. Purification and characterization of a two-component monooxygenase that hydroxylates nitrilotriacetate from “Chelatobacter” strain ATCC 29600. J Bacteriol. 1992;174:1179–1188. doi: 10.1128/jb.174.4.1179-1188.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uetz T, Egli T. Characterization of an inducible, membrane-bound iminodiacetate dehydrogenase from Chelatobacter heintzii ATCC 29600. Biodegradation. 1993;3:423–434. [Google Scholar]
- 50.Vermeij P, Wietek C, Kahnert A, Wuest A, Kertesz M A. Genetic organization of sulphur-controlled aryl desulphonation in Pseudomonas putida S-313. Mol Microbiol. 1999;32:913–926. doi: 10.1046/j.1365-2958.1999.01398.x. [DOI] [PubMed] [Google Scholar]
- 51.Witschel M, Nagel S, Egli T. Identification and characterization of the two-enzyme system catalyzing oxidation of EDTA in the EDTA-degrading bacterial strain DSM 9103. J Bacteriol. 1997;179:6937–6943. doi: 10.1128/jb.179.22.6937-6943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Witschel M, Egli T, Zehnder A J B, Wehrli E, Spycher M. Transport of EDTA into cells of the EDTA-degrading bacterial strain DSM 9103. Microbiology. 1999;145:973–983. doi: 10.1099/13500872-145-4-973. [DOI] [PubMed] [Google Scholar]
- 53.Xu Y, Mortimer M W, Fisher T S, Kahn M L, Brockman F J, Xun L. Cloning, sequencing, and analysis of a gene cluster from Chelatobacter heintzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH:flavin mononucleotide oxidoreductase. J Bacteriol. 1997;179:1112–1116. doi: 10.1128/jb.179.4.1112-1116.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xun L, Sandvik E R. Characterization of 4-hydroxyphenylacetate 3-hydroxylase (HpaB) of Escherichia coli as a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl Environ Microbiol. 2000;66:481–486. doi: 10.1128/aem.66.2.481-486.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]