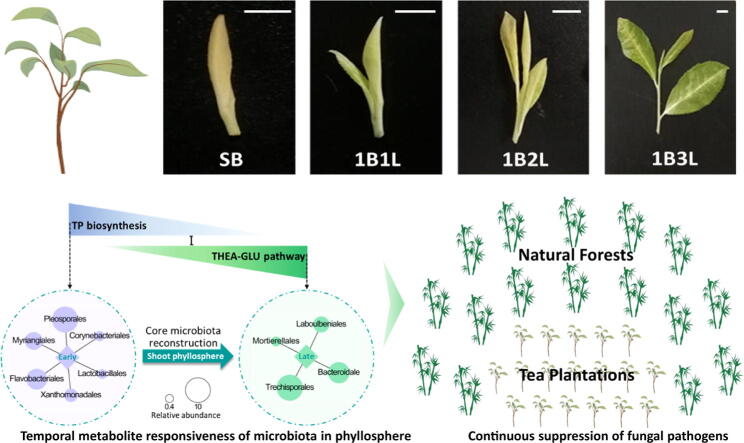
Graphical abstract
Keywords: Phyllosphere microbiota, Microbiome assembly, Co-adaption, Plant metabolites, Disease suppression
Highlights
-
•
Mechanistic insights into host-metabolite-driven microbiota assembly were obtained.
-
•
Tea plants can maintain a functional microbiota during shoot development.
-
•
The main drivers of microbial community assembly were identified.
-
•
Metabolite-responsive microbiota suppresses various tree pathogens in vitro and in vivo.
-
•
Establishment of tea plantations in the proximity of forests was linked to reduced disease incidence.
Abstract
Introduction
A broad spectrum of rhizosphere bacteria and fungi were shown to play a central role for health, fitness and productivity of their host plants. However, implications of host metabolism on microbiota assembly in the phyllosphere and potential consequences for holobiont functioning were sparsely addressed. Previous observations indicated that tea plants might reduce disease occurrence in various forests located in their proximity; the underlying mechanisms and potential implications of the phyllosphere microbiota remained elusive.
Objectives
This study aimed at deciphering microbiome assembly in the tea plant phyllosphere throughout shoot development as well as elucidating potential implications of host metabolites in this process. The main focus was to explore hidden interconnections between the homeostasis of the phyllosphere microbiome and resistance to fungal pathogens.
Methods
Profiling of host metabolites and microbiome analyses based on high-throughput sequencing were integrated to identify drivers of microbiome assembly throughout shoot development in the phyllosphere of tea plants. This was complemented by tracking of beneficial microorganisms in all compartments of the plant. Synthetic assemblages (SynAss), bioassays and field surveys were implemented to verify functioning of the phyllosphere microbiota.
Results
Theophylline and epigallocatechin gallate, two prevalent metabolites at the early and late shoot development stage respectively, were identified as the main drivers of microbial community assembly. Flavobacterium and Myriangium were distinct microbial responders at the early stage, while Parabacteroides and Mortierella were more enriched at the late stage. Reconstructed, stage-specific SynAss suppressed various tree phytopathogens by 13.0%-69.3% in vitro and reduced disease incidence by 8.24%-41.3% in vivo.
Conclusion
The findings indicate that a functional phyllosphere microbiota was assembled along with development-specific metabolites in tea plants, which continuously suppressed prevalent fungal pathogens. The insights gained into the temporally resolved metabolite response of the tea plant microbiota could provide novel solutions for disease management.
Introduction
Plants are commonly colonized by hundreds or even thousands of different microbes that are either localized in their endosphere or on their outer surfaces [1], [2], [3]. Similar to other organisms, it is assumed that plants have co-evolved with specific bacteria, fungi, archaea, and protists that have central implications for their host’s wellbeing [4], [5], [6]. Different host-microbiome models have demonstrated the importance of the presence of specific microorganisms for plant growth, development, productivity, and fitness [7], [8], [9], [10], [11]. Often, the beneficial functions exerted by microbes are due to their intimate impact on host metabolism [12], [13] or production of bioactive compounds that were traditionally believed to be of plant origin [14], [2].
Free-living microbial communities are known to respond to chemicals that are present in the local environment [15], [16]. Analogous observations were made with plant-associated microbial communities following exposure to agrochemicals [3]. Previously established models have shown that host plants can coordinate microbiome assembly through secretion of various chemical molecules, including endogenous phytohormones, signaling molecules, and secondary metabolites [17], [18], [19]. In terms of host-directed community assembly in root compartments (i.e. root endosphere and rhizosphere), it was discovered that host metabolites including flavonoids, terpenoids, and alkaloids modulate the microbial diversity and function in Arabidopsis [20], [21]. Less attention was paid to analogous processes in aerial parts of plants; however, the so-called phyllosphere, which harbors different niches for microbial colonization in above-ground tissues, is the largest biological surface on Earth and thus of high importance for global ecosystem integrity [3], [22]. The assembly of the phyllosphere microbiota is known to be substantially shaped by environmental factors, such as temperature, humidity, and UV radiation [22], [23]. In contrast to other plant compartments, the phyllosphere is often characterized by an increased occurrence of various secondary metabolites. The temporal dynamics in chemical diversity of different metabolites during plant development provides an unique environment for the inhabiting microbiota [4]. So far, implications of host metabolism on microbiota assembly in the phyllosphere and its potential consequences on functioning were sparsely addressed [13], [20], [24], [4], [25].
As one of the oldest trees on Earth, the tea plant (Camellia sinensis (L.) Kuntze) with a hundreds-to-thousands-years lifespan, is grown in over 60 countries in the world [26]. In China, the total area of tea plantations has increased from 18,440 km2 to 29,330 km2 during the last two decades. In addition to the traditional usage for tea leaf-derived beverages, tea plantations have also been established for eco-restoration of damaged forest areas. This is mainly due to empirical observations where tea plants were shown to contribute in the restoration of forest ecosystems by soil/water conservation, climate conditioning, species diversity enhancement and improvement of resilience towards different stressors [27]. During shoot development of tea plants, naturally occurring metabolites, e.g. polyphenols, theanine and caffeine, are known to underlie dynamic variations in terms of their concentration. These phyllosphere-specific metabolites are directly as well as indirectly involved in interactions with a wide array of pathogens and herbivores [28], [29], [30]. In addition to plant metabolites, phyllosphere microbial communities were recently found to harbor various responders to airborne foliar pathogens in vegetable plants [31]. Hence, we hypothesized that a co-adapted, functional microbiota is involved in the tea plants’ resistance to airborne foliar pathogens, and that it may have even broader ecological functions; especially in terms of the empirically observed reduction in disease occurrence of forest ecosystems following introduction of tea plants in their proximity [4], [32].
In the present study, we explored phyllosphere microbial community assembly during the shoot development stages of tea plants (C. sinensis cv. Xinan 4), and identified developmental-stage-specific metabolites that shape the phyllosphere microbiota. With further focus on the interaction between metabolite-responsive members of the microbiota and foliar pathogens that occur on the tea plants and the surrounding forests, we found that reconstructed synthetic assemblages (SynAss) exert strong pathogen suppressiveness. Our overall findings suggest that the host-driven assembly of the microbiota has potentially important implications on phyllosphere homeostasis of the host plant as well as surrounding ecosystems.
Results
Bacterial diversity increases during shoot development of tea plants
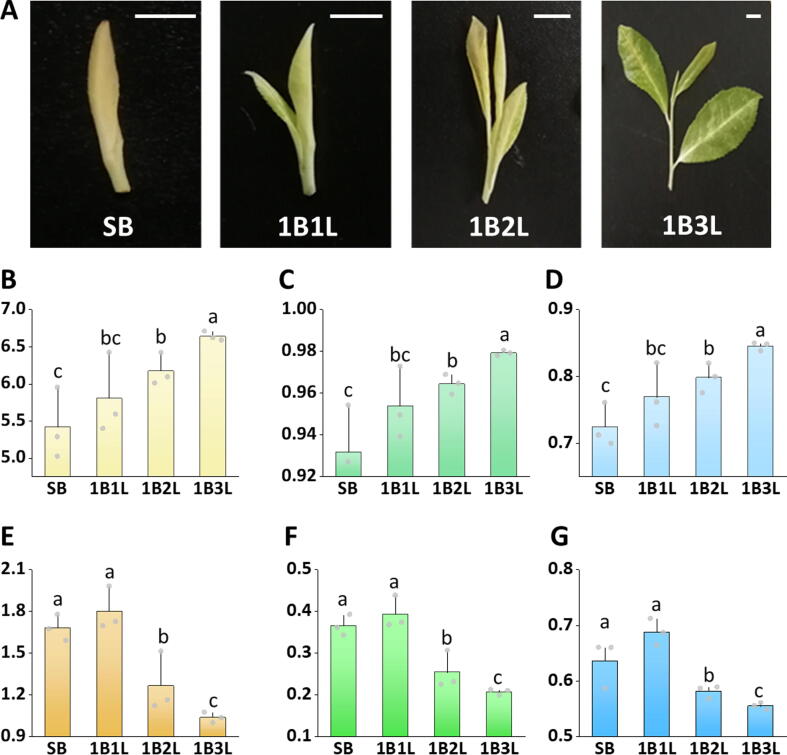
To investigate microbiome dynamics in the tea phyllosphere, we analyzed resident bacterial communities during the development of shoots at different representative stages, including the single bud stage (SB), one bud and one leaf stage (1B1L), one bud and two leaves stage (1B2L), and one bud and three leaves stage (1B3L) (Fig. 1A). Alpha diversity analyses indicated that the species richness of bacterial communities was not significantly altered during the SB to 1B3L period according to the Observed Species and Chao1 indices (Fig. S1; p>0.01). This was irrespectively of the remarkably increased Shannon and Simpson indices, which suggested a higher community evenness at the later stage of the shoot development (Fig. 1B&C; p<0.01). The increased community evenness was confirmed with complementary evaluation of Pielou’s evenness (Fig. 1D; p<0.01).
Fig. 1.
Temporal patterns of microbial community assembly during the shoot development of tea plants. The alpha diversity of the bacterial and fungal community was analyzed by using different indices. Four typical growth stages of tea plant shoots (A), including single bud (SB), one bud and one leaf (1B1L), one bud and two leaves (1B2L), and one bud and three leaves (1B3L) were implemented to assess bacterial (B-D) and fungal (E-G) community diversity by Shannon (B&E) and Simpson (C&F) indices. Community evenness was assessed by calculating Pielou’s evenness (D&G). Different letters with error bars indicate a significant difference according to one-way analysis of variance (ANOVA) with Tukey’s HSD test (p < 0.05). Values are means ± SD (shown as error bars; n = 3). Scale Bars: 0.5 cm.
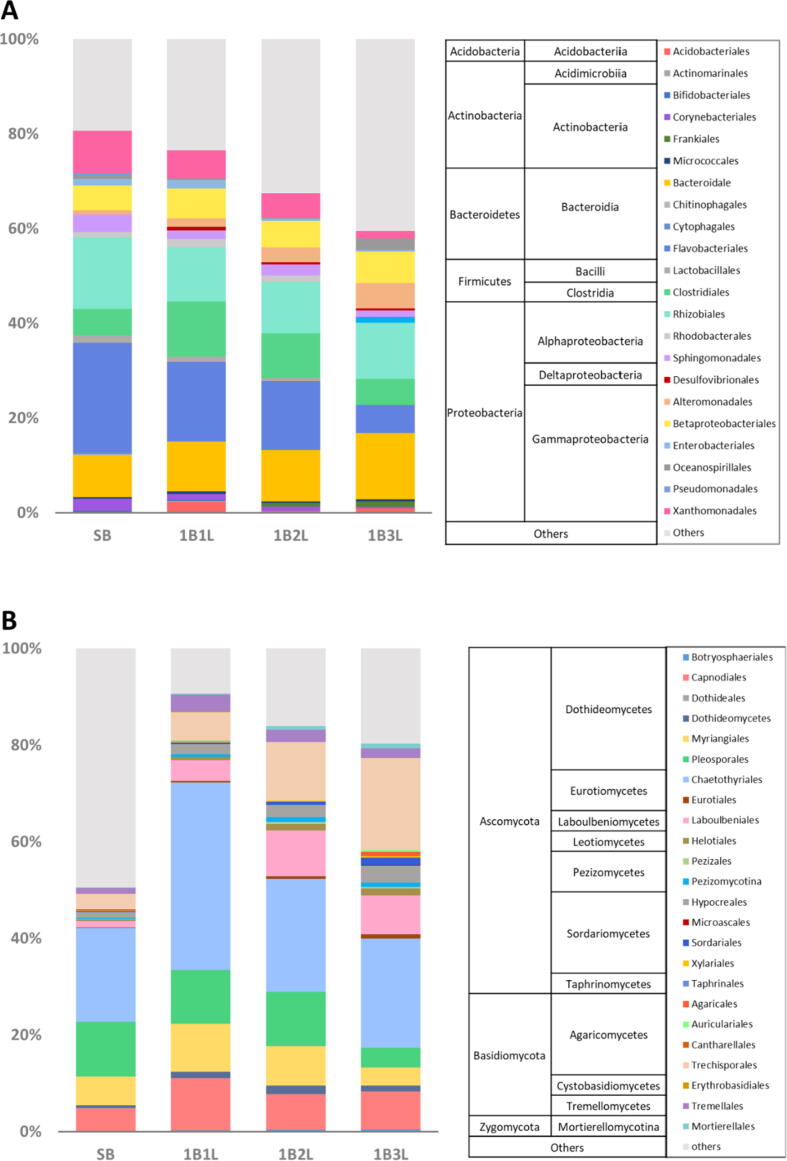
In terms of the bacterial structure at the phylum level, dominant phyla of the initial development stage followed a decreasing order of Proteobacteria (37.60%) > Bacteroidetes (35.98%) > Firmicutes (7.13%) > Actinobacteria (3.26%) > Acidobacteria (0.35%) (Fig. 2A, Table S1); when the buds developed to the 1B3L stage, the bacterial structure was found to be of a similar composition (Fig. S1, p>0.01) as in the earlier stages despite of the changes in the abundance of each phylum with Proteobacteria (31.93%) > Bacteroidetes (22.09 %)>Firmicutes (5.47%) >Actinobacteria (3.26%)>Acidobacteria (0.35%) (Fig. 2A, Table S1). It was also observed that the dominant bacterial phyla showed variable tendencies in terms of relative abundance during shoot development, in which only Bacteroidetes constantly declined until the 1B3L stage, while no significant changes were observed in other bacterial phyla (Fig. 2A). At order level, Flavobacteriales, Xanthomonadales, Lactobacillales and Corynebacteriales decreased with shoot development, while only the abundance of Bacteroidales increased (Fig. 2A). These observations suggest that all prevalent bacterial colonizers are subjected to relative abundance changes during shoot development which is accompanied by a global increase in diversity.
Fig. 2.
Temporal dynamics of microbial taxa during the shoot development of tea plants. The bacterial (A) and fungal (B) community was captured at four typical growth stages of tea plant shoots, including single bud (SB), one bud and one leaf (1B1L), one bud and two leaves (1B2L), and one bud and three leaves (1B3L) stages. Temporal dynamics of microbial community composition and structure were assessed at different taxonomic levels. The community compositions are summarized at the phylum, class, and order levels.
Fungal diversity in the tea plant phyllosphere reduces during shoot development
Previous studies have shown that bacterial community changes in plants are commonly accompanied by distinct alterations of the co-inhabiting fungal community [33]. In order to assess changes in the fungal community, the species richness was also analyzed during shoot development from SB to 1B3L. No significant changes were evident according to the Observed Species and Chao1 indices (Fig. S2; p>0.01). However, a decreasing tendency was observed for both the Shannon as well as the Simpson index (Fig. 1E&F; p<0.01). The lower diversity of the fungal community was mainly attributable to a decreased evenness at the later stages of shoot development (Fig. 1G). In a comparative assessment, an inverse temporal pattern was observed for the fungal community, in which phyllosphere fungal diversity decreased in parallel with increasing bacterial diversity (Fig. 1).
When the fungal community was assessed in more detail, it was shown that at the initial SB stage, shoots were predominately colonized by Ascomycota (88.70%), followed by the phyla Basidiomycota (4.65%) and Zygomycota (0.09%) (Fig. 2B, Table S2). After reaching the 1B3L stage, significant changes were observed for each phylum. For instance, Ascomycota decreased to 64.46%, while Basidiomycota and Zygomycota increased to 17.71% and 0.79%, respectively (Fig. 2B, Table S2). Interestingly, within the phylum Basidiomycota, certain taxa were found to substantially increase during shoot development (Fig. 2B). At the SB stage, the dominant orders were Chaetothyriales (19.45%) and Pleosporales (11.38%). When the 1B3L stage was reached, Chaetothyriales decreased to 17.94% but still remained as the most predominant order, while Pleosporales decreased to 3.31% and was replaced by Trechisporales (15.17%) (Fig. 2B). Moreover, while the majority of the fungal orders did not show constant tendencies along with shoot development, five orders were shown to be responsive. The abundance of Trechisporales, Laboulbeniales and Mortierellales constantly increased along with shoot development, while Myriangiales and Pleosporales decreased (Fig. 2B). Temporal patterns of microbial community assembly were characterized by different tendencies in bacterial and fungal communities, which indicated that development-stage-specific adaptions might be highly specific.
The core microbiota distinctively responds to development-stage-specific metabolites in the tea plant phyllosphere
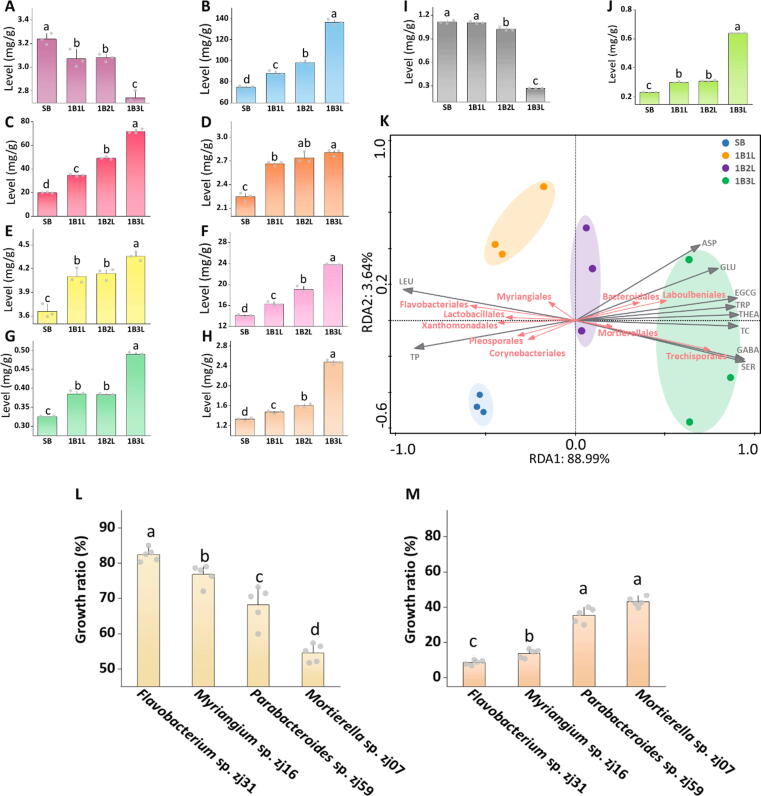
Analysis of metabolite profiles of tea shoots during the development stages SB to 1B3L, showed that typical primary metabolites of plants, soluble proteins (SP), and soluble sugars (SS) significantly decreased after the transition from SB to 1B1L stage (Fig. S3). During the subsequent development stages, SP remained at a steady level while SS first showed a decline and then a rise until 1B3L (Fig. S3). In the frame of detailed profiling of secondary metabolites, theophylline (TP) was found to be the only alkaloid that decreased significantly from SB to 1B3L (Fig. 3A, Fig. S4), whereas (-)-epigallocatechin gallate (EGCG) was identified to be one of the dominant catechins and increased by 3.8-fold during the assessed period, leading to a 1.8-fold increment of total catechins (TC) (Fig. 3B-C, Fig. S4). This was despite the fact that other alkaloids including caffeine (CAF) and theobromine (TB) and individual catechins did not show a clear tendency (Fig. S4). Moreover, concentrations of seven amino acids including ASP, GLU, THEA, TRP, SER, LEU and GABA showed specific trends during the development of the shoot (Fig. 3D-J), in which only LEU was constantly reduced (Fig. 3I), while the remaining other 12 amino acids showed alternations but without certain tendency (Fig. S5).
Fig. 3.
Association between key metabolites and the core microbiome of tea plants. Dynamics of key metabolites during the shoot development of tea plants were analyzed by using extracts of tea shoots collected at four representative growth stages (SB, 1B1L, 1B2L, and 1B3L). Significant concentration changes were shown for theophylline (TP, A), total catechins (TC, B), (-)-epigallocatechin gallate (EGCG, C), ASP (D), GLU (E), THEA (F), TRP(G), SER (H), LEU (I) and GABA (J). Different letters with error bars indicate a significant difference according to one-way analysis of variance (ANOVA) with Tukey’s HSD test (p < 0.05). Values are means ± SD (shown as error bars, n = 3). RDA showed the metabolites at diverse shoot development stages in association with the core microbiome (K). Gray arrows indicate different metabolites; red arrow lines indicate trends of distinct members of the core microbiome; development stage-specific clusters were highlighted by circles enclosed by an ellipse. Response of representative core microbiota members of tea plants to defense-associated secondary metabolites (L&M). Bacterial and fungal isolates representative for the core microbiota at the early and late stage of tea shoot development were exposed to early stage-enriched TP (L) and late stage-enriched EGCG (M) to confirm differing compatibility of the isolates with the plant’s secondary metabolites. Different letters with error bars indicate a significant difference according to one-way analysis of variance (ANOVA) with Tukey’s HSD test (p < 0.05). Values are means ± SD (shown as error bars; n = 5).
Redundancy analysis (RDA) was conducted to search for potential associations between host metabolism dynamics and the temporally-resolved core microbiome in tea plants (Fig. 3K, Dataset 2). The first constrained axis of the RDA explained 88.99% of the total variance, while the second explained only 3.64% (Fig. 3K, P = 0.022). The bacterial orders Flavobacteriales, Xanthomonadales, Lactobacillales, Corynebacteriales together with the fungal orders Myriangiales and Pleosporales showed a negative association with EGCG, TC, ASP, GLU, THEA, SER, TRP and GABA (P = 0.059 ∼ P<0.005, Dataset 2), but interestingly a positive association with LEU and TP (P<0.005, Dataset 2). This was in contrast to Bacteroidales, Trechisporales, Laboulbeniales, and Mortierellales that showed a reverse trend during the shoot development (Fig. 3K). Based on the observed microbial and metabolic parameters in the negative half of the first axis in the RDA, the TP- and LEU-enriched developmental stages of SB and 1B1L were predominated by Flavobacteriales, Xanthomonadales, Lactobacillales, Corynebacteriales, Myriangiales and Pleosporales. On the positive half of the first axis in the RDA, Bacteroidales, Trechisporales, Laboulbeniales, and Mortierellales were found as dominant taxa of the 1B2L and 1B3L stages (Fig. 3K).
For the further confirmation of the relationship between developmental-stage-specific microbiota and metabolites, Flavobacterium spp. and Myriangium spp. were implemented in a synthetic assemblage (SynAss) representing the early development stage, while Parabacteroides spp. and Mortierella spp. were included in a SynAss representing the late development stage. The SynAss taxa were representative for the core microbiota as inferred from microbiome analysis (Fig. 3K); other taxa that were identified in development-stage-dependent core microbiome, were not recoverable with the implemented isolation approaches. In order to include representative strains in the SynAss constructs, the most frequently occurring bacterial strains Flavobacterium sp. zj31 and Parabacteroides sp. zj59 together with the fungal strains Myriangium sp. zj16 and Mortierella sp. zj07 were selected from isolate libraries (Table S1 & S2). When exposed to tea plant metabolites, Flavobacterium and Myriangium were shown to be responsive to TP that is enriched in the phyllosphere during early development stages (Fig. 3L), while Parabacteroides and Mortierella were highly responsive to the late-stage-enriched EGCG (Fig. 3M). The differing responses to key metabolites of the host plant opened the question of whether the phyllosphere core microbiota can maintain its function in the dynamic micro-environment. We focused on disease suppression due to forgoing empirical observations of disease reduction when new tea plantations are established [27].
Development-stage-specific core microbiota protects tea plants and other tree species from prevalent fungal pathogens
The tea plantations that were implemented in the present study were established for eco-restoration in the proximity of bamboo (Bambusaceae), fir tree (Metasequoia glyptostroboides) and pine tree (Pinus massoniana Lamb.) forests, in which five prevalent foliar pathogens including bamboo pathogens (Stereostratum corticioides, Ceratosphaeria phyllostachydis), fir tree pathogens (Glomerella cingulata), pine tree (Pestalotiopsis disseminate, Pestalotiopsis funerea), together with tea plant pathogens (Gloeosporium theae-sinesis, Colletotrichum camelliae) were isolated.
To examine whether the development-stage-specific core microbiota of tea plants possesses sufficient antagonism against prevalent phytopathogens, we simulated interactions in vitro and in vivo between the identified tea phyllosphere microbes and foliar forest pathogens. Although we initially aimed to perform the experiments with sterile plants, we were not able to establish callus-based sterile tea seedlings in congruence with previous attempts [34], [35]. Nevertheless, by implementing in vitro experiments we confirmed that different degrees of antagonism were exerted by development-stage-specific taxa when they were individually tested against all aforementioned forests pathogens in a dual-interaction system (antagonistic rates from 34% to 89%, Fig. 4A). This indicated that the core microbiota representatives of all developmental stages of tea shoots exert certain antagonism towards the tested tree pathogens.
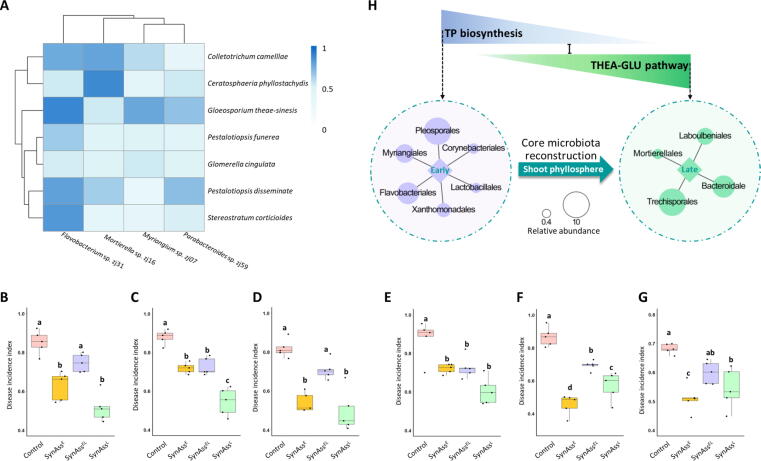
Fig. 4.
The metabolite-responsive core members of the tea plant’s microbiota suppress prevalent fungal pathogens. (A) Antagonistic activity of core metabolite-responsive isolates from the tea phyllosphere (Flavobacterium sp. zj31, Myriangium sp. zj16, Parabacteroides sp. zj59 and Mortierella sp. zj07) towards seven fungal forest pathogens, which included bamboo pathogens (Stereostratum corticioides and Ceratosphaeria phyllostachydis), fir tree pathogens (Glomerella cingulata), pine tree (Pestalotiopsis disseminate and Pestalotiopsis funerea), and tea plant pathogens (Gloeosporium theae-sinesis and Colletotrichum camelliae). For each member of the core microbiota, the antagonism towards seven phytopathogens was analyzed and the intensity depicted with a color gradient. Suppression of disease incidence by synthetic assemblages (SynAss) reconstructed with the core metabolite-responsive isolates in bamboo (B&C), fir (D), pine (E&G) and tea (G) in vivo. SynAssE was constructed using two isolated strains (Flavobacterium sp. zj31 and fungal strain Myriangium sp. zj16) representative for the early development stage; SynAssL was constructed using two isolated strains (Parabacteroides sp. zj59 and the fungal strain Mortierella sp. zj07) representative for the late development stage; SynAssEL was constructed by combining the four strains from SynAssE and SynAssL. Different letters with error bars indicate a significant difference according to one-way analysis of variance (ANOVA) with Tukey’s HSD test (p < 0.05). Values are means ± SD (shown as error bars; n = 5). (H) Schematic illustration of the enhanced resistance to fungal pathogens conferred by the tea plant’s development-synchronized microbiota. The TP- and THEA-GLU pathway-responsive core microbiota confer resilience during early and late shoot development, respectively.
Further experiments were performed to investigate whether synthetic assemblages (SynAss) consisting of different combinations of the same isolates show divergent activity. The bacterial strain Flavobacterium sp. zj31 and the fungal strain Myriangium sp. zj16 were included in the assemblage representing the early shot development stage (SynAssE). Two other isolates, the bacterium Parabacteroides sp. zj59 and the fungus Mortierella sp. zj07 were included in SynAssL, which represented the typical assembly at the late developmental stage. SynAssEL was generated by combining the four strains from SynAssE and SynAssL. Increased inhibition of seven common tree and forest pathogens was observed with SynAssE and SynAssL when compared to the individual isolates (Fig. S6). Interestingly, the inhibition efficiency was significantly decreased when the pathogens were exposed to SynAssEL (Fig. S6). We continued to explore the inhibition by including in vivo experiments. Here, a significant reduction of disease incidence in various tree species was confirmed after the introduction of SynAssE, SynAssL, and SynAssEL (Fig. 4B-G). Although not always significant, a clear tendency was observed that SynAssE and SynAssL were more effective than SynAssEL reflecting the finding from in vitro assays. The overall observations indicate that the development-specific core microbiota can sustain phyllosphere homeostasis by suppression of fungal pathogens, which is mostly shaped by the host’s THEA-GLU pathway and TP biosynthesis (Fig. 4H).
Potential interconnection of strong pathogen suppressiveness in the tea plant phyllosphere and disease reduction in local forests
Due to the observed in vitro and in vivo disease reduction by the core microbiota responsive to tea plant metabolites (Fig. 4&S6), we subsequently explored whether disease incidence is affected by establishment of tea plantations in the proximity of different forest ecosystems. Disease incidence surveys were performed in three different forest types. We analyzed forest disease incidence by evaluating a two-year field survey based on local forestry monitoring datasets. In the context of the bamboo pathogens (Stereostratum corticioides and Ceratosphaeria phyllostachydis) the fir tree pathogen (Glomerella cingulata) and the pine tree pathogens (Pestalotiopsis disseminate and Pestalotiopsis funereal) the disease incidence in the respective forests from 2018 to 2019 was assessed before and after tea plantation (C. sinensis cv. Xinan 4) as well as in reference forests without eco-restoration.
It was observed that the general fungal disease incidences was significantly reduced in all three representative forest ecosystems upon plantation of tea plants in their close proximity (Fig. S7). The tea plantations had a size of 2.3–3.7 ha, were 20 – 80 m away from the intact forest ecosystems (Fig. S7) and consisted of 1-year-old plants that were transplanted from local nurseries. In the bamboo forest (Bambusaceae), the pathogen incidence ranged from 1.12% to 4.98% in Hanghzou, Shaoxing, and Huzhou that are all located within Zhejiang Province (Fig. S8). In these sites that are characterized by identical soil properties and climatic conditions, the pathogen incidence was reduced to 0.20%-1.52% upon plantation of tea plants, with a maximum reduction rate of 82% that was observed in Huzhou (Fig. S8). In the other forest ecosystems dominated by pine trees (Pinus massoniana Lamb.) (Fig. S9) and fir trees (Metasequoia glyptostroboides) (Fig. S10), a similar reduction tendency was observed at sites where the tea plantations were implemented. These results suggest that the antagonistic core microbiota in the tea plant phyllosphere is potentially correlated with disease suppression in surrounding forests.
Tracking of beneficial microorganisms in important niches of the tea plant
To clarify whether the metabolite-responsive core microbiome is also present in other plant compartments besides developing shoots, mature shoots, mature leaves, roots, and rhizosphere soil were subjected to microbiome analyses. Plants generally show varying degrees of concordance in the microbiomes of different tissues and often a highly specific rhizosphere community that is characterized by a higher abundance of microorganisms [36]. In the present study, we observed that shoots and leaves shared 27 bacterial taxa, while only seven taxa were shared by the roots and rhizosphere soil (Fig. S11). This is in line with previous observations that have shown that spatially close niches with similar physicochemical properties share a higher proportion of common microorganisms [37]. We focused on the 50 most prevalent taxa in each eco-niche of C. sinensis and found that only four bacterial genera (Bradyrhizobium, Delfia, Rhodococcus, and Stenotrophomonas) were among the prevalent colonizers that could be found in all four compartments. We have conducted the same analysis to assess the mycobiome and found that here the core included more common taxa (9 taxa with a predominance of the order Trechisporales), while the number of shared taxa between shoots and leaves was substantially lower (9 taxa) than for bacteria (Fig. S12). In general, fungi showed a more even distribution of the prevalent taxa among the analyzed niches.
Subsequently we focused on localizing the previously identified development-specific antagonists of fungal pathogens. Three of the SynAss representatives (Flavobacterium, Myrangium and Parabacteroides) were found to be exclusive features of the phyllosphere (Fig. S11-12), while Mortierella was also highly abundant in the tea plant’s rhizosphere (Fig. S12). The findings highlight that tea plant establish and maintain phyllosphere-specific microbial communities. In case of Mortierella, it remains yet to be elucidated if it also plays an important role in shielding off phytopathogens from soil.
Discussion
Previous studies have indicated that plants have co-evolved with their microbiota in order to improve their adaptability to different environments [19]. While the consequences of the proposed co-evolution are evident, the underlying mechanisms by which plants shape and maintain a beneficial microbiota are still mostly elusive for phyllosphere compartments [38]. In the present study we observed temporal changes in the microbiome assembly of tea plant shoots. We identified distinct host metabolites as the main factors shaping bacterial and fungal communities in the plant’s phyllosphere. Neither the climate nor soil properties differ significantly among the three geographical sites located in the same province (Zhejiang) which is representative for tea plantation within China. The tea plant’s metabolism has been reported to show temporal variations during shoot development [39]. This guided us to focus on the temporal pattern of the tea plant’s metabolic traits and its potential impact on microbial assembly. Moreover, we wanted to deepen our understanding on how these complex interactions might affect tea plants and potentially the forest ecosystems closeby.
Among the characteristic secondary metabolites of tea leaves, polyphenols (mainly catechins), free amino acids represented by THEA, and purine alkaloids determine taste features as well as health-promoting effects of tea products. They also play crucial roles in protecting tea plants against biotic and abiotic stresses [30], [40]. However, their influence on the indigenous microbiota was not described so far. By implementing redundancy analyses in combination with metabolite-enrichment screening we found that TP and EGCG have distinct effects on the developmental-stage-specific core microbiota. TP belongs to a prevalent class of purine alkaloids, which are enriched at early development stages of tea plants and previously reported to act as a chemical defense to protect young tissues from pathogens and herbivores [41]. The TP-associated bacterial orders Flavobacteriales, Xanthomonadales, Lactobacillales, Corynebacteriales and the fungal orders Myriangiales, Pleosporales thus might support TP-based defense mechanisms against invasive fungal pathogens. During shoot development, the simultaneous increase of GLU and catechins is attributable to an simultaneous increase of THEA [42], which is the most abundant non-proteinogenic amino acid in tea plants and not only important for the umami taste of tea products, but also a favorable nitrogen source during growth of tea plants [43]. On the other hand, EGCG is the dominant and most representative antioxidant catechin during the shoot development and known to be indispensable for a prompt response to pathogen invasion in the above-ground tissues of tea plants [44], [45]. Microbes enriched by EGCG are potentially important for synergistic implications to maintain homeostasis of the phyllosphere microbiome threatened by plant pathogens.
Interestingly, we found contrary dynamics between TP biosynthesis and the THEA-GLU pathway during the shoot development, along with the involvement of the respective micro-environmental response of the microbiome. We thus deemed that TP biosynthesis negatively interacted with either the THEA-GLU pathway or the down-stream biosynthetic pathway of EGCG during the shoot development. The core microbiome composed of Bacteroidales, Trechisporales, Laboulbeniales, and Mortierellales likely responds to the progressively developing, catechin-enriched phyllosphere, whereas Flavobacteriales, Xanthomonadales, Lactobacillales, Corynebacteriales, Myriangiales and Pleosporales act as the core microbiota that is predominant in the earlier, TP-dominated phyllosphere, both of which involve in sustaining a lasting phyllosphere homeostasis by developmental-stage-specific synergistic impact on temporal suppression towards fungal pathogens.
Prevalent foliar pathogens of the forests in the proximity of newly established tea plantations were also detectable for a major part in the microbiome of the asymptomatic tea plants’ phyllosphere; however, they occurred in relatively low relative abundances (Dataset 1). Only Pestalotiopsis occurred with a relative abundance of 0.05%, while the other tree pathogens were even less abundant in the assessed phyllosphere fungal community. This implies that these pathogens are potentially transferable among the forest ecosystem and tea plants in the proximity (Fig. S7); However, their colonization of the phyllosphere is likely suppressed by the identified highly antagonistic core microbiota of tea plants, which was evidenced by in vivo and in vitro experiments. During the conducted 2-year survey we observed that disease incidence significantly decreased in different forest hosts including bamboo, fir, and pine trees after the introduction of tea plants. Subsequently we could show that a transfer of the antagonistic core microbiota in form of synthetic assemblages exerted the same protective effects. More profoundly, the assemblages of metabolite responsive SynAssE and SynAssL were more effective than SynAssEL as observed with tea plants. To some extent, these findings provide experimental evidences that reduced disease occurrence in forest ecosystems in proximity of tea plants is linked to the antagonistic core microbiota in the phyllosphere.
However, it remains elusive as of how the antagonistic core microbiota in the tea phyllosphere transfers to the forests. It is known that non-pathogenic microbes and pathogens are both vertically transmittable across various eco-niches within individual trees in other higher plants [14], [46], [47]. To clarify whether the core microbiota of the tea plant’s phyllosphere is eco-niche-specific or transmittable to other plant compartments, its occurrence was mapped in the whole plant. Within the identified core phyllosphere microbiota, the genus Mortierella was found to be highly abundant in the rhizosphere (Fig. S12). Although most plant-associated microorganisms not only show species and cultivar specificity but also a high tissue specificity [48], some are shared between compartments as shown in the current work.
Furthermore, we assume that a transmission of the core microbiota of the tea plant phyllosphere likely also occurs horizontally between tea plants and forests; prevalent forest pathogens were also found in the tea plants (Dataset 1). Previous works showed that disease epidemics in forest ecosystems can be largely attributed to vector- and environmental factor-mediated pathogen transmission routes among suitable hosts of the respective pathogens [49]. The plant phyllosphere in general provides an immense habitat for not only various pathogens and transient colonizers, but also beneficial microbes and their vectors as well [50], [51]. Therefore, the antagonistic core microbiota might simultaneously transmit from the tea plants to forest trees likewise prevalent forest pathogens transmit from forest trees to tea plants. The incorporation of tea plants is thus possibly linked to an introduction of a functional microbiota that can interfere with the occurrence of fungal pathogens in the forest ecosystems. However, further insights into the underlying molecular mechanisms of the phyllosphere microbiota-mediated crosstalk among tree species remain to be uncovered. This may provide an exploitable resource for the improvement of disease resilience of forest ecosystems in the future.
Conclusions and perspectives
In the frame of the detailed assessment of the role of host metabolites in shaping the core microbiota in the tea plant phyllosphere, we could show that a stable antagonistic potential against prevalent phytopathogens is maintained during shoot development stages. Distinct microbial responders to development-stage-specific metabolites were identified in both bacterial as well as fungal tea phyllosphere communities, of which the core taxa exerted antagonism against foliar tree pathogens in vivo and in vitro. Finally, a potential link between local alleviation of fungal disease incidence in adjacent forests and newly established tea plantations with a high antagonistic capacity in the phyllosphere was proposed. Most of the metabolite-responsive and pathogen-inhibiting core taxa identified were shown to be a specific feature of the tea plant’s phyllosphere; only certain taxa may also play a role in the plant’s rhizosphere. In conclusion, our findings suggest that the assembly of phyllosphere microbiota is subjected to temporal shaping by specific leaf metabolites during the plant development. Moreover, the developmental stage-specific core microbiota sustains phyllosphere homeostasis in turn by exerting suppression against prevalent fungal pathogens of tea plants and adjacent forest ecosystems.
Materials and methods
Sampling of tea plants
The tea phyllosphere samples were collected from plants grown at the single bud stage (SB), one bud and one leaf stage (1B1L), one bud and two leaves stage (1B2L), and one bud and three leaves stage (1B3L). At the 1B3L stage, the leaves, roots and rhizosphere soil samples were simultaneously collected at three sites of Zhejiang province, China, including Hangzhou, Huzhou and Shaoxing. A complete randomized block design was used for each sampling and all samples were immediately packed into sterilized bags and placed in a portable ice box immediately. We aimed to obtain a region-unspecific, core microbiome of tea plants that is only influenced by the development stage. Therefore, the samples collected at each site subjected to tea plantation in the same locations were evenly mixed and three biological replicates (one from each of the three regions) were included for each sample. All tea tissue samples were washed using sterile water to remove surface contaminants and stored in a freezer at − 80 °C for further experiments [14], [52], [53].
Analytical standards, solvents and instruments
The standards, solvents and analytical instruments used in this study are listed as follows: amino acids standards were obtained from TCI (Tokyo, Japan); all standards of the different catechins, gallic acid (GA), amino acids, and purine alkaloids were purchased from Sigma Chemical Co. (Missouri, USA). Acetonitrile, acetic acids and other organic solvents from Merck (chromatographic grade; Shanghai, China). The HPLC 1200 was acquired from Agilent Technology (San Diego, CA, USA), Milli-Q Water Purification System from Merck Millipore (Billerica, MA, USA).
Quantitative analysis of catechins, GA and purine alkaloids
Individual catechins (C, (+)-catechin; CG, (+)-catechin gallate; EC, (-)-epicatechin; ECG, (-)-epicatechin gallate; EGC, (-)-epigallocatechin; EGCG, (-)-epigallocatechin gallate; GC, (+)-gallocatechin; GCG, (+)-gallocatechin gallate), GA (gallic acid) and purine alkaloids (CAF, caffeine; TB, theobromine; TP, theophylline), and free amino acids (ASP, aspartic acid; THR, threonine; SER, serine; ASN, asparagine; GLU, glutamic acid; GLN, glutamine; ALA, alanine; VAL, valine; ILE, Isoleucine; LEU, leucine; TYR, tyrosine; PHE, phenylalanine; GABA, γ-aminobutyric acid; HIS, histidine; TYP, tryptophan; LYS, lysine; ARG, arginine; PRO, proline) were quantified according to the method described previously [54]. Briefly, catechins, GA and purine alkaloids were extracted with 70% methanol at 70 °C for 10 min, centrifuged, and subsequently separated in a Agilent HPLC equipped with Agilent-TC-C18 column (250 mm × 4.6 mm, 5.0 μm), whereas free amino acids were extracted with distilled water at 100 °C for 45 min, and then subjected to phenyl isothiocyanate derivatization. After that, their quantity was determined with an Agilent HPLC equipped with an ASB C18 analytical column (250 mm × 4.6 mm, 5.0 μm; Agela, China) and a diode array detector.
Quantitative analysis of soluble protein and sugar
The same extracts as described above were used to analysis of the contents of soluble protein (SP) and soluble sugar (SS). SP was determined according to the method reported previously using bovine serum albumin as the standard [55]; and SS was analyzed by using the anthrone-sulfuric acid method with glucose as standard [56].
Extraction of total community DNA
Tea plant tissues and rhizosphere samples were frozen with liquid nitrogen and thoroughly ground with a precooled sterile mortar before extraction of total DNA using the Fast DNA SPIN extraction kit (MP Biomedicals, USA) referring to the manufacturer’s instructions. Corresponding bulk soil samples were directly subjected to total DNA extraction. Total community DNA extracts were stored at −20 °C prior to further processing.
Preparation and sequencing of amplicon libraries
The quantity and quality of the extracted total community DNA was assessed using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA) and agarose gel electrophoresis, respectively. PCR amplification of bacterial 16S rRNA gene fragments (V3–V4 region) and the fungal ITS1 region was performed according to the Earth Microbiome guidelines [57]. Two consecutive PCR reactions were performed as previously described [58] and all PCR reactions were conducted in triplicates. Equimolar DNA concentrations of each barcoded amplicon sample were sent to Magigene Co., Ltd (Guangzhou, China) for next-generation sequencing. Following initial quality control and adapter ligation by the sequencing company, 16S rRNA gene fragment and ITS region amplicons were sequenced on an Illumina Hiseq 2500 platform.
Bioinformatics analyses and co-occurrence visualizations
The acquired data were analyzed with the QIIME1.9.1 pipeline [58], [59]. After removal of chimeric sequences, mitochondria and chloroplast reads (for 16S RNA gene amplicon data) or bacterial and archaeal reads (for ITS data), the remaining high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence identity with UCLUST [60]. Representative sequences were selected from each OTU using default parameters. Taxonomic classification of OTUs was conducted by BLAST searches with the representative sequence set against the SILVA v 128 and UNITE v7.2 databases using the best hit method [58]. An OTU table was generated to assess the abundance and taxonomy of each OTU in the respective samples. OTUs containing<0.001% of the total reads across all samples were discarded. To minimize the difference in the sequencing depth across samples, a rarefied OTU table was generated by averaging 100 evenly resampled OTU subsets under 90% of the minimum sequencing depth for further analysis. A microbial network was constructed based on the average abundance of predominant OTUs in the different samples within the QIIME pipeline. The resulting data was rendered with Cytoscape 3.6.1 for network visualization [61].
Isolation and identification of tea plant phyllosphere microbes
Tea phyllosphere samples collected above were used for the isolation of culturable phyllosphere bacteria and fungi. After transfer into a sterilized mortar, samples were ground with 10 mL of sterilized distilled water premixed with quartz sand. The resulting suspension was used for a serial dilution and as an inoculant. Agar plates of MW, PD, CM and Luria-Bertani (LB) were employed as a culture medium for bacteria and fungi. Plates were incubated for 5–10 days at 25 °C to obtain visible colonies. Subsequently, all culturable and distinguishable bacterial/fungal colonies were purified and identified using universal PCR primers for 16S rRNA gene and ITS region [14], [62].
Isolation and identification of forest and tea plant fungal pathogens
Diseased leaves from three forest types that are common in Zhejiang Province (bamboo, fir tree and pine tree) were used to isolate prevalent fungal pathogens. Isolation of fungal pathogens was conducted with Modified Winogradsky (MW), Potato Dextrose (PD) and Complete Medium (CM) with supplementation of agar (1.5%) [14]. Consequently, bamboo pathogens (Stereostratum corticioides, Ceratosphaeria phyllostachydis), fir tree pathogens (Glomerella cingulata), pine tree (Pestalotiopsis disseminate, Pestalotiopsis funerea) were isolated and identified. Additionally, two major pathogens of the tea plant including Gloeosporium theae-sinesis and Colletotrichum camelllae were isolated from (Camellia sinensis cv. Longjin 43). All pathogens were incubated in the dark at 25 °C during their growth phase.
Response of phyllosphere microbes to development-dependent plant metabolism
The isolated microbes were subjected to a specific assay to evaluate their adaption to EGCG and theophylline (TP) enriched phyllospheric microenvironments. Briefly, EGCG and TP was supplemented at levels present in the phyllosphere physiologically, 70 mg/g and 3.2 mg/g, respectively, while the control was supplemented with same volume of solvent only. The bacteria were cultured in liquid media on a shaker for 36 h and the cell densities quantified by optical measurements at 660 nm. The fungal isolates were grown on solidified media plates with 1.5% agar for 7 d and before the mycelial diameter was measured [63]. The adaption capacity was calculated by the growth ratio of the treated group in contrast to the control group.
Relationship between the core microbiota of tea plants and forest foliar pathogens in the tea phyllosphere
We intended to test the antagonistic relationship based on sterile tea seedlings, but were not able to break the previously described obstacle in obtaining callus-based sterile tea seedlings [34], [35]. Therefore, we relied on established in vitro experiments that served as simulations for the expected interactions between tea phyllosphere isolates and the forest pathogens, as well as a surface-sterilized tree seedling system.
In a dual-culture simulation system, single isolates and synthetic assemblages of metabolite-responsive microbes (SynAss) [14], [64] constructed from microbial isolates obtained in the frame of this study, were co-cultured with pathogens to measure antagonistic effects. Briefly, 5-mm plugs of mycelium from each pathogen were placed in the center of a fresh CM agar plate (9 cm). The single isolates or SynAss was transferred on a sterilized 5-mm paper disc and then symmetrically placed onto the pre-inoculated plate, at an equidistant distance of 2 cm from the central plug. Non-inoculated paper discs on the plate were implemented as controls. Incubation was carried out at 25 °C in the dark for 3–14 days depending on the growth characteristics of the utilized pathogens. The inhibition capacity of the single isolates and the SynAss was subsequently evaluated. The mycelium diameter of each fungal pathogen in the controls as well as in the treatments was measured and included in statistical analyses.
In addition, the introduction of respective SynAss was also done in vivo. Briefly, the inoculants were sprayed evenly by combining pathogens and SynAss and cultivated for 14–21 days (tree species-specific) in a greenhouse to observe the typical symptoms and calculate the disease incidences. The bacterial and fungal tea plant isolates were suspended in sterile water at 104 CFU/mL and 104 spore/mL, respectively, with the ratio at 1:1 (v/v), and then subjected to inoculation together with the prevalent fungal pathogens (104 spore/mL) in the relevant forest trees including bamboo (1-year-old juvenile seedlings), fir (2-year-old juvenile seedlings) and pine (2-year-old juvenile seedlings), and also tea plants (2-year-old juvenile seedlings) by spraying. The control was sprayed with the same volume of sterile water only. Each treatment included five seedlings, and the spray-inoculation was performed on 10 leaves for each tree. The average relative lesion area (RLA) was calculated using the Otsu method for evaluation of the disease incidence as described in a previous report [14].
Field survey on forest disease incidence
Forest disease field survey were carried out at three sites of Zhejiang province, China, including Hangzhou, Huzhou and Shaoxing. Briefly, three representative forest types from each location, including bamboo (Bambusaceae), fir tree (Metasequoia glyptostroboides) and pine tree (Pinus massoniana Lamb.) were selected and included in a two-year observation. During the first year, one site in close proximity of each forest was subjected to eco-restoration with tea plants (C. sinensis cv. Xinan 4; Fig. S9). In the second year, the pathogen incidence in the sites with and without incorporation of tea plants was again assessed. At each site, the assessment of the relative lesion area (RLA) of individual trees (50 trees per site) was performed and calculated using the Otsu method for evaluation of the disease incidence as previously reported [14].
Statistical analyses
Statistical analyses for significant differences between treatments were calculated and evaluated using the statistical program package SPSS (SPSS Inc, Chicago, IL; version 18.0). Redundancy analysis (RDA) using Canoco 5.0 and visualized by the implemented CANOCO CDW [65].
Data availability
The datasets generated during the presented study are publicly available in the Sequence Read Archive of NCBI under the BioProject accession PRJNA657458 and PRJNA622669.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (2021FZZX001-31), Programme for High-Level Talents Cultivation of Zhejiang University, and Strategic Research on “Plant Microbiome and Agroecosystem Health” (2020ZL008, Cao Guangbiao High Science and Technology Foundation, Zhejiang University).
Credit author statement
PX, MW and TC planned and designed research and experiments; XF, YM, HC, AX, WL, TL, YN and QM performed laboratory experiments and analyzed data; PX, XF, MW and TC wrote and edited the paper. PX, YW and MW acquired the funds. All authors read and approved the final manuscript.
CRediT author statement
Ping Xu: Conceptualization, Funding acquisition, Writing – original draft. Xiaoyan Fan: Data curation, Formal analysis, Writing – original draft. Yuxiao Mao: Data curation, Investigation, Formal analysis. Haiyan Cheng: Investigation, Formal analysis. Anan Xu: Data curation, Investigation, Formal analysis, Visualization. Wanyi Lai: Investigation, Formal analysis. Tianxing Lv: Investigation, Formal analysis. Yang Hu: Data curation, Investigation, Resources, Formal analysis. Yanxia Nie: Formal analysis, Visualization. Xuxia Zheng: Investigation. Qing Meng: Investigation. Yuefei Wang: Funding acquisition. Tomislav Cernava: Conceptualization, Methodology, Writing – original draft. Mengcen Wang: Conceptualization, Methodology, Funding acquisition, Writing – original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (2021FZZX001-31), Programme for High-Level Talents Cultivation of Zhejiang University, Strategic Research on “Plant Microbiome and Agroecosystem Health” (2020ZL008, Cao Guangbiao High Science and Technology Foundation, Zhejiang University). We appreciate Dr. Zhimin Sha (School of Agriculture and Biology, Shanghai Jiao Tong University) for her advice on statistics analysis. We also thank the staff of the local forest protection stations for their support during sample collection and surveys.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.10.003.
Contributor Information
Tomislav Cernava, Email: tomislav.cernava@tugraz.at.
Mengcen Wang, Email: wmctz@zju.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Hassani M.A., Duran P., Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6:58. doi: 10.1186/s40168-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsumoto H., Fan X., Wang Y., Kusstatscher P., Duan J., Wu S., et al. Bacterial seed endophyte shapes disease resistance in rice. Nat Plants. 2021;7(1):60–72. doi: 10.1038/s41477-020-00826-5. [DOI] [PubMed] [Google Scholar]
- 3.Wang M, Cernava T: Overhauling the assessment of agrochemical-driven interferences with microbial communities for improved global ecosystem integrity. Environmental Science and Ecotechnology 2020:100061. [DOI] [PMC free article] [PubMed]
- 4.Liu H., Brettell L.E., Singh B. Linking the Phyllosphere Microbiome to Plant Health. Trends Plant Sci. 2020;25(9):841–844. doi: 10.1016/j.tplants.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Ford SA, King KC: In Vivo Microbial Coevolution Favors Host Protection and Plastic Downregulation of Immunity. Mol Biol Evol, 38(4) (2021), 1330–1338. [DOI] [PMC free article] [PubMed]
- 6.Trivedi P., Leach J.E., Tringe S.G., Sa T., Singh B.K. Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18(11):607–621. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- 7.Turner T.R., James E.K., Poole P.S. The plant microbiome. Genome Biol. 2013;14(6):209. doi: 10.1186/gb-2013-14-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wassermann B., Cernava T., Muller H., Berg C., Berg G. Seeds of native alpine plants host unique microbial communities embedded in cross-kingdom networks. Microbiome. 2019;7(1):108. doi: 10.1186/s40168-019-0723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg G., Grube M., Schloter M., Smalla K. The plant microbiome and its importance for plant and human health. Front Microbiol. 2014;5:491. doi: 10.3389/fmicb.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cernava T., Aschenbrenner I.A., Soh J., Sensen C.W., Grube M., Berg G. Plasticity of a holobiont: desiccation induces fasting-like metabolism within the lichen microbiota. Isme J. 2019;13(2):547–556. doi: 10.1038/s41396-018-0286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qasim M., Islam S.U., Islam W., Noman A., Khan K.A., Hafeez M., et al. Characterization of mycotoxins from entomopathogenic fungi (Cordyceps fumosorosea) and their toxic effects to the development of asian citrus psyllid reared on healthy and diseased citrus plants. Toxicon. 2020;188:39–47. doi: 10.1016/j.toxicon.2020.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Helfrich E.J.N., Vogel C.M., Ueoka R., Schäfer M., Ryffel F., Müller D.B., et al. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nat Microbiol. 2018;3(8):909–919. doi: 10.1038/s41564-018-0200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashidoko Y. Studies on the metabolic regulation of denitrifying bacteria and phytopathogenic microorganisms using chemical agents found in chemical ecology-based phenomena. J Pesticide Sci. 2018;43(1):47–54. doi: 10.1584/jpestics.J17-04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan X., Matsumoto H., Wang Y., Hu Y., Liu Y., Fang H., et al. Microenvironmental Interplay Predominated by Beneficial Aspergillus Abates Fungal Pathogen Incidence in Paddy Environment. Environ Sci Technol. 2019;53(22):13042–13052. doi: 10.1021/acs.est.9b04616. [DOI] [PubMed] [Google Scholar]
- 15.Fan X., Fu Y., Nie Y., Matsumoto H., Wang Y., Hu T., et al. Keystone taxa-mediated bacteriome response shapes the resilience of the paddy ecosystem to fungicide triadimefon contamination. J Hazard Mater. 2021;417:126061. doi: 10.1016/j.jhazmat.2021.126061. [DOI] [PubMed] [Google Scholar]
- 16.Liao H, Li X, Yang Q, Bai Y, Cui P, Wen C, Liu C, Chen Z, Tang J, Che J et al: Herbicide Selection Promotes Antibiotic Resistance in Soil Microbiomes. Molecular Biology and Evolution 2021, 38(6): 2337–2350 [DOI] [PMC free article] [PubMed]
- 17.Sánchez-Cañizares C., Jorrín B., Poole P.S., Tkacz A. Understanding the holobiont: the interdependence of plants and their microbiome. Curr Opin Microbiol. 2017;38:188–196. doi: 10.1016/j.mib.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y., Bonkowski M., Shen Y.i., Griffiths B.S., Jiang Y., Wang X., et al. Root ethylene mediates rhizosphere microbial community reconstruction when chemically detecting cyanide produced by neighbouring plants. Microbiome. 2020;8(1):4. doi: 10.1186/s40168-019-0775-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen T., Nomura K., Wang X., Sohrabi R., Xu J., Yao L., et al. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature. 2020;580(7805):653–657. doi: 10.1038/s41586-020-2185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaparro J.M., Badri D.V., Vivanco J.M. Rhizosphere microbiome assemblage is affected by plant development. Isme J. 2014;8(4):790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang A.C., Jiang T., Liu Y.-X., Bai Y.-C., Reed J., Qu B., et al. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science. 2019;364(6440) doi: 10.1126/science:aau6389. [DOI] [PubMed] [Google Scholar]
- 22.Vorholt J.A. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10(12):828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 23.Compant S., Samad A., Faist H., Sessitsch A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J Adv Res. 2019;19:29–37. doi: 10.1016/j.jare.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collado M.C., Cernada M., Baüerl C., Vento M., Pérez-Martínez G. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes. 2012;3(4):352–365. doi: 10.4161/gmic.21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlechter R.O., Miebach M., Remus-Emsermann M.N.P. Driving factors of epiphytic bacterial communities: A review. J Adv Res. 2019;19:57–65. doi: 10.1016/j.jare.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cernava T., Chen X., Krug L., Li H., Yang M., Berg G. The tea leaf microbiome shows specific responses to chemical pesticides and biocontrol applications. Sci Total Environ. 2019;667:33–40. doi: 10.1016/j.scitotenv.2019.02.319. [DOI] [PubMed] [Google Scholar]
- 27.Qi D.-H., Guo H.-J., Sheng C.-Y. Assessment of plant species diversity of ancient tea garden communities in Yunnan Southwest of China. Agroforestry Syst. 2013;87(2):465–474. [Google Scholar]
- 28.Ayres M.P., Lombardero M.J. Lombardero MaJ: Assessing the consequences of global change for forest disturbance from herbivores and pathogens. Sci Total Environ. 2000;262(3):263–286. doi: 10.1016/s0048-9697(00)00528-3. [DOI] [PubMed] [Google Scholar]
- 29.Schuldt A., Hönig L., Li Y., Fichtner A., Härdtle W., Oheimb G., et al. Herbivore and pathogen effects on tree growth are additive, but mediated by tree diversity and plant traits. Ecol Evol. 2017;7(18):7462–7474. doi: 10.1002/ece3.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dou Q.P. Tea in Health and Disease. Nutrients. 2019;11(4):929. doi: 10.3390/nu11040929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo L., Zhang Z., Wang P., Han Y., Jin D., Su P., et al. Variations in phyllosphere microbial community along with the development of angular leaf-spot of cucumber. AMB Express. 2019;9(1) doi: 10.1186/s13568-019-0800-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grady K.L., Sorensen J.W., Stopnisek N., Guittar J., Shade A. Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-11974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venturi V., Keel C. Signaling in the Rhizosphere. Trends Plant Sci. 2016;21(3):187–198. doi: 10.1016/j.tplants.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Xia EH, Tong W, Wu Q, Wei S, Zhao J, Zhang ZZ, Wei CL, Wan XC: Tea plant genomics: achievements, challenges and perspectives. Hortic Res-England 2020, 7(1). [DOI] [PMC free article] [PubMed]
- 35.Mukhopadhyay M., Mondal T.K., Chand P.K. Biotechnological advances in tea (Camellia sinensis [L.] O. Kuntze): a review. Plant Cell Rep. 2016;35(2):255–287. doi: 10.1007/s00299-015-1884-8. [DOI] [PubMed] [Google Scholar]
- 36.Berg G., Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol. 2009;68(1):1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 37.Remus-Emsermann M.N.P., Lücker S., Müller D.B., Potthoff E., Daims H., Vorholt J.A. Spatial distribution analyses of natural phyllosphere-colonizing bacteria on Arabidopsis thaliana revealed by fluorescence in situ hybridization. Environ Microbiol. 2014;16(7):2329–2340. doi: 10.1111/1462-2920.12482. [DOI] [PubMed] [Google Scholar]
- 38.Hegazi N., Hartmann A., Ruppel S. The plant microbiome: Exploration of plant-microbe interactions for improving agricultural productivity. J Adv Res. 2019;19:1–2. doi: 10.1016/j.jare.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie H., Feng X., Wang M., Wang Y., Kumar Awasthi M., Xu P. Implications of endophytic microbiota in Camellia sinensis: a review on current understanding and future insights. Bioengineered. 2020;11(1):1001–1015. doi: 10.1080/21655979.2020.1816788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashihara H. Occurrence, biosynthesis and metabolism of theanine (γ-glutamyl-L-ethylamide) in plants: a comprehensive review. Nat Prod Commun. 2015;10(5):803–810. [PubMed] [Google Scholar]
- 41.Kemen A.C., Agler M.T., Kemen E. Host–microbe and microbe–microbe interactions in the evolution of obligate plant parasitism. New Phytol. 2015;206(4):1207–1228. doi: 10.1111/nph.13284. [DOI] [PubMed] [Google Scholar]
- 42.Xiong L., Li J., Li Y., Yuan L., Liu S., Huang J., et al. Liu Z: Dynamic changes in catechin levels and catechin biosynthesis-related gene expression in albino tea plants (Camellia sinensis L.) Plant Physiol Biochem. 2013;71:132–143. doi: 10.1016/j.plaphy.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 43.Dong C., Li F., Yang T., Feng L., Zhang S., Li F., et al. Wan X et al.: Theanine transporters identified in tea plants (Camellia sinensis L.) Plant J. 2020;101(1):57–70. doi: 10.1111/tpj.14517. [DOI] [PubMed] [Google Scholar]
- 44.Ullah C., Tsai C.-J., Unsicker S.B., Xue L., Reichelt M., Gershenzon J., et al. Salicylic acid activates poplar defense against the biotrophic rust fungus Melampsora larici-populina via increased biosynthesis of catechin and proanthocyanidins. New Phytol. 2019;221(2):960–975. doi: 10.1111/nph.15396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laitinen T., Morreel K., Delhomme N., Gauthier A., Schiffthaler B., Nickolov K., et al. A Key Role for Apoplastic H2O2 in Norway Spruce Phenolic Metabolism. Plant Physiol. 2017;174(3):1449–1475. doi: 10.1104/pp.17.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemanceau P, Barret M, Mazurier S, Mondy S, Pivato B, Fort T, Vacher C: Chapter Five - Plant Communication With Associated Microbiota in the Spermosphere, Rhizosphere and Phyllosphere. In: Advances in Botanical Research. Edited by Becard G, vol. 82: Academic Press; 2017: 101–133.
- 47.Abdelfattah A., Wisniewski M., Schena L., Tack A.J.M. Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environ Microbiol. 2021;23(4):2199–2214. doi: 10.1111/1462-2920.15392. [DOI] [PubMed] [Google Scholar]
- 48.Hardoim P.R., van Overbeek L.S., Berg G., Pirttilä A.M., Compant S., Campisano A., et al. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol Mol Biol Rev. 2015;79(3):293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Truitt L.L., McArt S.H., Vaughn A.H., Ellner S.P. Trait-Based Modeling of Multihost Pathogen Transmission: Plant-Pollinator Networks. Am Nat. 2019;193(6):E149–E167. doi: 10.1086/702959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vacher C., Hampe A., Porté A.J., Sauer U., Compant S., Morris C.E. The Phyllosphere: Microbial Jungle at the Plant-Climate Interface. Annu Rev Ecol Evol Syst. 2016;47(1):1–24. [Google Scholar]
- 51.Leach J.E., Triplett L.R., Argueso C.T., Trivedi P. Communication in the Phytobiome. Cell. 2017;169(4):587–596. doi: 10.1016/j.cell.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 52.Lundberg D.S., Lebeis S.L., Paredes S.H., Yourstone S., Gehring J., Malfatti S., et al. Rio TGd et al.: Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X., Fan X., Matsumoto H., Nie Y., Sha Z., Yi K., et al. Biotoxin Tropolone Contamination Associated with Nationwide Occurrence of Pathogen Burkholderia plantarii in Agricultural Environments in China. Environ Sci Technol. 2018;52(9):5105–5114. doi: 10.1021/acs.est.7b05915. [DOI] [PubMed] [Google Scholar]
- 54.Xu P., Su H., Jin R., Mao Y., Xu A., Cheng H., et al. Shading Effects on Leaf Color Conversion and Biosynthesis of the Major Secondary Metabolites in the Albino Tea Cultivar “Yujinxiang”. J Agric Food Chem. 2020;68(8):2528–2538. doi: 10.1021/acs.jafc.9b08212. [DOI] [PubMed] [Google Scholar]
- 55.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1-2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 56.MORRIS D.L. Quantitative Determination of Carbohydrates With Dreywood's Anthrone Reagent. Science (New York, NY) 1948;107(2775):254–255. doi: 10.1126/science.107.2775.254. [DOI] [PubMed] [Google Scholar]
- 57.Thompson L.R., Sanders J.G., McDonald D., Amir A., Ladau J., Locey K.J., et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551(7681):457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kusstatscher P., Zachow C., Harms K., Maier J., Eigner H., Berg G., et al. Microbiome-driven identification of microbial indicators for postharvest diseases of sugar beets. Microbiome. 2019;7(1) doi: 10.1186/s40168-019-0728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 61.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang M., Tachibana S., Murai Y., Li L., Lau S.Y.L., Cao M., et al. Indole-3-Acetic Acid Produced by Burkholderia heleia Acts as a Phenylacetic Acid Antagonist to Disrupt Tropolone Biosynthesis in Burkholderia plantarii. Sci Rep. 2016;6:22596. doi: 10.1038/srep22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang M., Hashimoto M., Hashidoko Y., Meijler M.M. Repression of tropolone production and induction of a Burkholderia plantarii pseudo-biofilm by carot-4-en-9,10-diol, a cell-to-cell signaling disrupter produced by Trichoderma virens. PLoS ONE. 2013;8(11):e78024. doi: 10.1371/journal.pone.0078024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y.-X., Qin Y., Bai Y. Reductionist synthetic community approaches in root microbiome research. Curr Opin Microbiol. 2019;49:97–102. doi: 10.1016/j.mib.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 65.Šmilauer P., Lepš J. edn. Cambridge University Press; Cambridge, United Kingdom; New York: 2014. Multivariate analysis of ecological data using Canoco 5, Second edition. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the presented study are publicly available in the Sequence Read Archive of NCBI under the BioProject accession PRJNA657458 and PRJNA622669.