Graphical abstract
Keywords: Sepsis, Exosomes, Proteomics, Transcriptomics, Metabolomics
Highlights
-
•
The study for the first time describes the profile of molecular dynamics in septic serum exosomes.
-
•
We provide a new direction into proteasome-mediated protein degradation in septic serum exosomes.
-
•
IL-10 delivery by septic exosomes may play a vital role in alleviation of AKI of CLP mice.
-
•
Septic serum exosomes participate in the modulation of sepsis by regulating vitamin metabolism.
-
•
The molecular mechanisms proposed in the study may provide helpful insights for the therapy of sepsis.
Abstract
Introduction
Sepsis is an infection-induced severe inflammatory disorder leading to multiple organ dysfunction. It remains a highly lethal condition for which early diagnosis and therapy achieve unsatisfactory results. Circulating exosomes containing biomarkers and mediators of sepsis have recently received attention, but the progress has been far from optimal.
Objectives
The present study focuses on the profiles of molecular dynamics in serum exosomes and explores the potential molecular mechanisms on serum exosomes during the process of sepsis.
Methods
We used high-performance liquid chromatography-tandem mass spectrometry and RNA-seq to detect the dynamic profiles of exosome proteins and RNAs (including mRNAs, lncRNAs and miRNAs) in serum exosomes from 3 healthy individuals and 9 septic patients at the different stages. Then integrative multiomics analyses were performed and the results were validated by qRT-PCR, LiquiChip assay and metabolomics analysis on mice subjected to cecal ligation and puncture (CLP) modeling.
Results
A total of 354 proteins, 195 mRNAs, 82 lncRNAs and 55 miRNAs were identified as differentially expressed molecules in serum exosomes from septic patients. Integrative multiomics analysis showed that exosome components were associated with cytokine storm, complement and clotting cascades, the endothelial barrier, 20S proteasome-dependent protein degradation and vitamin metabolism. Importantly, pretreatment with serum exosomes derived from mice subjected to CLP significantly restrained proinflammatory cytokine expression and alleviated tissue injury in septic mice. Further metabolomics analysis demonstrated that pretreatment with septic serum exosomes significantly affected the metabolites associated with vitamin digestion and absorption in CLP mice.
Conclusion
Our study for the first time describes the landscape of the molecular dynamics of serum exosomes during the development of sepsis and proposes some hypothetical molecular mechanisms by integrative multiomics analysis, which may provide helpful diagnostic and therapeutic insights for the ongoing battle against sepsis.
Introduction
Sepsis is a condition of life-threatening organ dysfunction caused by an infection-triggered systemic inflammatory response. Over the past decades, sepsis has remained a highly lethal condition that lacks dependable diagnostic methods and adequate evidence-based treatments [1]. Currently, the incidence of sepsis is estimated to be 270 cases per 100,000 persons per year with a mortality rate of approximately 26%, and the mortality of septic shock may be as high as 40% or even 60% [1], [2].
The molecular mechanisms underlying sepsis are complex and have not been fully elucidated. Previous studies have demonstrated that the severity of sepsis is related to a series of cell activation steps that lead to an overwhelming cytokine storm [3]. Inflammatory cytokines such as tumor necrosis factor-α (TNF-α), interleukin-1-β (IL-1β) and interleukin-6 (IL-6), which have been identified as molecular markers for evaluation of sepsis severity and prognosis, have been targeted in several clinical trials [4]. However, cytokine storm targeting has failed with disappointing results [3], [5]. For example, TNF-α antagonists, which have shown promise in animal models, have not been successfully translated to clinical practice [3].
Successful treatment of life-threatening sepsis is attributed to timely and appropriate antibiotic therapy, hemodynamic support, and adequate source control [6]. However, the ability of these measures to significantly improve the survival of septic patients is usually limited[7]. The current Surviving Sepsis Campaign guidelines merely suggest the use of procalcitonin for shortening/discontinuation of ongoing antimicrobial treatment and encourage the discovery of more markers, especially those reflecting renal dysfunction and coagulopathy[8], [9]. Clearly, more effective early diagnosis and therapeutic strategies for sepsis are urgently needed.
Exosomes are double-membrane microvesicles with diameters of 30 to 200 nm [10]. Exosomes with diverse contents, such as proteins and RNAs, are secreted from various types of cells, such as macrophages, epithelial cells and dendritic cells[11]. Recently, exosomes have been considered to play roles in multiple processes, including delivery of cell substances, immune regulation, extracellular signal transduction, etc.[12], [13], holding promise for future diagnostics and therapy [14]. After their membranes fuse with those of recipient cells, the vesicles deliver their cargos into the cytoplasm to induce a cellular response to environment[7]. Hoshino et al. demonstrated that integrin mediates the cellular uptake of exosomes, which contributes to Src activation and upregulation of S100 genes in resident cells at tropic sites of metastasis. These promigratory and proinflammatory signals then accelerate the extracellular immune response by recruiting bone marrow-derived myeloid cells (BMDMs) that further stimulate inflammation [15]. Kitai et al. reported that DNA-containing exosomes derived from breast cancer cells treated with topotecan (an antitumor chemotherapeutic drug) activate the cyclic GMP-AMP synthase (cGAS)-STING signaling pathway and thus reinforce antitumor immunity [16]. Additionally, human bone marrow mesenchymal stem cells (hBMSCs) have been demonstrated to alleviate liver inflammation in acute-on-chronic liver failure (ACLF) through exosomal miR-20a-5p/intracellular CXCL8 axis of hepatocytes[17].
Furthermore, exosomes are very stable; they resist rupture in the blood circulation by evading proteolysis and phagocytosis. This feature can be leveraged to extend the half-lives of therapeutic agents and to reduce immunogenicity [18]. Thus, the characteristics of exosomes are helpful for the diagnosis and treatment of clinical diseases. A recent study reported that maternal administration of engineered exosomes containing an NF-κB inhibitor reduced the fetal inflammatory response, restrained immune cell migration, alleviated histologic chorioamnionitis and delayed preterm delivery in a mouse model of infection [19]. Our previous study demonstrated that exosomes enriched with cytokines/chemokines played crucial roles in T cell differentiation, proliferation and chemotaxis during the sepsis process [20]. Pretreatment with exosomes derived from lipopolysaccharide (LPS)-challenged mice reduced the inflammatory response and prolonged the survival of mice subjected to cecal ligation and puncture (CLP). Although these findings demonstrate the therapeutic effects of exosomes on sepsis, the identities of their cargos and their potential molecular mechanisms during sepsis are largely unknown.
Profiling exosome cargos, including proteins, mRNAs, lncRNAs and miRNAs, can be helpful for identification of molecular markers for diagnosis and prognosis and for closure of knowledge gaps regarding sepsis. In this study, we performed a multiomics analysis on the cargos in exosomes from patients with sepsis of different stages to systematically elucidate the biological processes related to the development of sepsis. The data presented in this article may help to generate new perspectives on the pathophysiology of sepsis and ultimately provide insight to support identification of new therapeutic targets and screening of effective molecular prognostic markers for sepsis.
Materials and methods
Ethics statement
All experiments involving human patients were conducted according to the ethical policies and procedures approved by the Ethics Committee of the Third Affiliated Hospital of Southern Medical University, Guangzhou, China (No. 2020028). Informed consent was obtained from the septic patients and healthy donors in the study. In addition, all animal experiments were conducted according to the ethical policies and procedures approved by the Ethics Committee of the Use and Care of Animals, Southern Medical University (No. L2018235).
Serum sample collection
In Sepsis-3, the clinical criteria for sepsis is defined as infection plus a Sequential Organ Failure Assessment (SOFA) score equal to or>2 [21]. To systematically elucidate the biological functions and signaling mechanisms of serum exosomes during the progression of sepsis, we collected blood samples from patients who met the Sepsis-2 diagnostic criteria. According to the criteria of the American College of Chest Physicians/Society of Critical Care Medicine, the presence of suspected or confirmed infection together with two or more criteria for a systemic inflammatory response was considered to indicate sepsis; sepsis with organ dysfunction was diagnosed as severe sepsis; and sepsis with hypotension despite adequate fluid resuscitation along with the presence of perfusion abnormalities was defined as septic shock [22].
Isolation of serum exosomes
In the present study, ultracentrifugation was used to isolate serum exosomes following the protocol described previously [20]. In brief, serum samples were centrifuged at 2,000 × g for 20 min at 4 °C to remove large cell fragments or debris. The supernatant was obtained and centrifuged at 10,000 × g for 45 min at 4 °C to eliminate small cellular debris. Then, the supernatant was transferred to a new 1.5 mL sterile centrifuge tube and subjected to ultracentrifugation at 120,000 × g for 120 min at 4 °C to pellet the small vesicles. After discarding the supernatant, the pellet was resuspended in 4 mL of phosphate buffer saline (PBS) and filtered with a 0.22 μm filter to eliminate potential contaminants. Finally, serum exosomes were obtained by centrifugation at 120,000 × g for 120 min at 4 °C.
Characterization of exosomes
The pelleted exosomes were resuspended in 100 μL of PBS. The morphology and size of the isolated exosomes were observed via HT-7700 transmission electron microscopy (TEM) (Hitachi, Chiyoda-ku, Tokyo, Japan) and ZetaVIEW S/N 17–310 nanoparticle tracking analysis (NTA) (Particle Metrix, Inning am Ammersee, Munich, Germany). Specific markers for exosomes, including Alix (Cat#2171 s, CST, Danvers, MA, USA) and CD63 (Cat#ab217345, Abcam, Cambridge Biomedical Campus, Cambridge, UK), were detected by Western blot analysis as a routine procedure.
Protein extraction and digestion
The isolated exosomes were suspended and lysed with 8 mol/L urea (Cat#U4883; Sigma, St. Louis, MO, USA). The protein concentration was quantified with a bicinchoninic acid (BCA) protein assay kit (Cat#71285–3; Merck, Kenilworth, NJ, USA). For liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis, 50 μg of sample protein was reduced, alkylated and digested into peptides according to the protocol for filter-aided sample preparation (FASP) [23]. Briefly, each protein sample was transferred to a 10 kD filter tube (Cat#VNO1HO2; Sartorius Stedim Biotech GmbH, Goettingen, Germany) and centrifuged at 14,000 × g at 25 °C for 15 min. Then, the sample was washed by adding 300 μL of 8 mol/L urea in 0.1 mol/L NH4HCO3 (pH 8.5) and centrifuged to remove irrelevant substances in the sample. This procedure was repeated twice and was followed by three additional washes with 300 μL of 0.1 mol/L NH4HCO3 (pH 8.5) to clear the urea. Next, 5 μL of 0.5 mol/L dithiothreitol (DTT) in 250 μL of 0.1 mol/L NH4HCO3 solution was added to the filter, and the sample was incubated at 56 °C for 30 min. Afterwards, the sample was alkylated by adding 10 μL of 0.5 mol/L iodoacetamide (IAA) to the filter and incubated for 30 min in the dark. After centrifugation at 14,000 × g at 25 °C for 15 min, the filter was washed with 300 μL of 0.1 mol/L NH4HCO3. Finally, the sample was digested with trypsin (Cat#v5280; Promega, Madison, WI, USA) (trypsin:protein = 1:50) in 300 μL of 0.1 mol/L NH4HCO3 and incubated for 16 h at 37 °C. The digested peptides were collected after centrifugation at 14,000 × g and 25 °C for 15 min, and desalted through a C18 column (Cat#S181001; Agela Technologies, Torrance, CA, USA) according to the manufacturer’s protocol.
LC-MS/MS analysis for proteins
The desalted peptides were analyzed on a C18 column (50 µm × 15 cm, 3 µm) at 50 °C using an EASY-nLC1200 connected to an Orbitrap Fusion mass spectrometer (Thermo Scientific, Waltham, MA, USA). The peptide mixtures were separated by a linear gradient of 5–30% acetonitrile (ACN) with 0.1% formic acid (FA) at 300 nL/min for 48 min. The concentration was then linearly increased to 100% ACN over 7 min and maintained at 100% for 5 min. The column was re-equilibrated with 6 μL of 0.1% FA. For data-dependent acquisition (DDA), the source was operated at 2.1 kV. The DDA scheme included a full mass spectrometry (MS) survey scan from m/z 350 to m/z 1500 at a resolution (full width at half maximum, FWHM) of 120,000 (at m/z 200) with the automatic gain control (AGC) set to 2.0E5 (maximum injection time of 50 ms). Top-speed data acquisition was then performed at an FWHM of 30,000 with the AGC set to 5.0E4 [24].
Proteomics data analysis
The acquired DDA data were searched with MaxQuant[25] (v.1.6.10.43) against the reviewed human UniProt protein database (https://www.uniprot.org/). Oxidation on methionine residues and N-terminal acetylation were set as variable modifications, while carbamidomethylation on cysteine residues was set as a fixed modification. Up to two missed trypsin cleavages were allowed. A false discovery rate (FDR) cutoff of 0.01 was accepted to filter the identified candidate peptides based on a search in reverse-sequence decoy mode. The identified proteins with P-values < 0.05 and fold change (FC) values > 1.5 between two groups were considered differential expressed proteins (DEPs). To reveal the functions of the DEPs in sepsis, we performed Gene Ontology (GO) annotation and pathway analysis as previously described [26], [27]. In brief, GO annotation was performed to discover the gene regulatory networks based on hierarchical categories according to the molecular function, biological process, and cellular component terms of the DEPs. Pathway analysis was performed with the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) to characterize the enriched pathways for the DEPs. We used Cytoscape software [28] (v3.7.2) combined with the ClueGO plugin (v2.5.6) to visualize the results of the functional analysis. In addition, we constructed a protein–protein interaction (PPI) network of DEPs based on the STRING online database (v11.0) (http://string-db.org) and visualized the results with Cytoscape software.
RNA sequencing
Total RNA was extracted from exosomes with TRIzol according to the manufacturer’s protocol. Then, ribosomal RNA (rRNA) was depleted from total RNA with a Ribo-Zero™ rRNA removal kit (Cat#RZH1046; Illumia, San Diego, CA, USA). For preparation of RNA libraries, an NEBNext® Multiplex Small RNA Library Prep Set (Cat#E7330S; NEB, Ipswich, MA, USA) for miRNAs and an NEBNext® UltraTM RNA Library Prep Kit (Cat#E7530L; NEB, Ipswich, MA, USA) for lncRNAs were used according to the manufacturer’s instructions. Subsequently, paired-end sequencing (PE150) was performed for mRNAs and lncRNAs on the Illumina HiSeq 3000 platform, and single-end sequencing (SE50) for miRNAs was performed on the Illumina HiSeq 2500 platform. The Bioconductor packages EdgeR and DESeq were used for differential expression analyses of the RNA-seq read counts. EdgeR was used to calculate differentially expressed mRNAs and lncRNAs according to the criteria of an |log2FC| > 1 and an adjusted P-value < 0.05. DESeq was used to identify the differentially expressed miRNAs according to the same criteria: an |log2FC| > 1 and a P-value < 0.05.
Bioinformatic analysis of the RNA-seq data
The miRanda [29] (v3.3a) and RNAhybrid [30] (v2.1.1) software programs were used to predict target genes of the differentially expressed miRNAs. It is widely accepted that lncRNAs regulate target genes mainly through two types of processes: cis-regulation and trans-regulation. The potential target genes affected by cis-regulation were obtained by integrating the data on the differentially expressed lncRNAs and their adjacent (within 10 kb) differentially expressed mRNAs. For assessment of trans-regulation, the sequences of the differentially expressed lncRNAs and mRNAs were extracted for target gene identification. The Basic Local Alignment Search Tool was used to detect the complementary pairing relationships of RNA bases (e < 1E-5), and RNAplex was used to identify the potential target genes of the lncRNAs.
Integration analysis of multiomics datasets
Multivariate analysis was performed to integrate the data for the proteins, mRNAs, lncRNAs and miRNAs simultaneously. The aim of the analysis was to gain new insights that could not be obtained through analysis of each individual omics dataset. The analysis was conducted with the R package “mixOmics” (version 6.10.9), which is based on the Data Integration Analysis for Biomarker discovery using Latent variable approaches for Omics studies (DIABLO) framework. DIABLO [31] is a novel multiomics framework for integration of multiple datasets through a multivariate dimension reduction method.
Untargeted metabolomics analysis
Each serum sample (100 μL) was resuspended in an Eppendorf (EP) tube with 500 μL of prechilled 80% methanol and 0.1% FA, and then incubated on ice for 5 min and centrifuged at 15,000 × g and 4 °C for 20 min. The supernatant was diluted with LC-MS-grade water to a final concentration containing 53% methanol. The sample was subsequently transferred to a new EP tube for centrifugation at 15,000 × g and 4 °C for 20 min. Finally, the supernatant was injected into an ultrahigh-performance liquid chromatography (UHPLC)-MS/MS system that consisted of a Vanquish UHPLC system coupled with an Orbitrap Q ExactiveTM HF-X mass spectrometer (Thermo Scientific, Waltham, MA, USA).
The samples were injected into a Hypesil Gold column using a 17-min linear gradient at a flow rate of 0.2 mL/min. Eluent A (0.1% FA in water) and eluent B (methanol) were used for the positive polarity mode. For the negative polarity mode, 5 mmol/L ammonium acetate (pH 9.0) and methanol were used as eluents A and B, respectively. The solvent gradient was set as follows: 2% B, 1.5 min; 2–100% B, 12 min; 100% B, 14 min; 100–2% B, 14.1 min; and 2% B, 17 min. The Q ExactiveTM HF-X mass spectrometer was operated in positive/negative polarity mode with a spray voltage of 3.2 kV (kV), a capillary temperature of 320 °C, a sheath gas flow rate of 40 arb and an auxiliary gas flow rate of 10 arb. The raw data files based on UHPLC-MS/MS were processed using Compound Discoverer 3.1 to perform peak alignment, peak picking, and quantitation for each metabolite.
The metabolites were annotated using the KEGG database, the Human Metabolomics Database (HMDB) (https://hmdb.ca/) and the LIPIDMaps (https://lipidmaps.org/) database. We applied univariate analysis (t-test) to calculate the statistical significance (P-value). The metabolites with variable importance in the projection (VIP) values > 1, P-values < 0.05 and |log2FC| > 0.263 were considered to be differentially expressed metabolites. The functions of the differentially expressed metabolites and metabolic pathways were studied using the KEGG database.
CLP model
Eight- to ten-week-old male C57BL/6 mice were used for cecal ligation and puncture (CLP) modeling of sepsis following a procedure described previously [32]. In brief, the cecum was exposed by forming a 2-cm midline incision on the anterior abdomen and was ligated at a 1 cm site from the end of the cecum. Afterwards, the cecum was punctured twice with an 18-gauge needle between the ligation site and the end of the cecum, and a small amount of cecal content was extruded. The cecum was then placed back into the abdomen, and the incision was closed with wound clips. The sham group was treated in the same way as the CLP group except for the ligation and puncture of the cecum. All animals were fluid-resuscitated subcutaneously with 1 mL of sterile saline.
In vivo studies
First, exosomes in the blood were isolated from septic mice as described in our previous study [20]. C57BL/6 mice were pretreated with septic mouse exosomes of 5 μg/g body weight via tail vein injection (TVI). One hour later, the mice were subjected to CLP modeling. To clarify the effects of exosomes on the levels of inflammatory cytokines, blood was collected from the mice at 12 h after CLP modeling for analysis of cytokines with a LiquiChip system (Qiagen, Hilden, Germany). The gene levels of inflammatory cytokines, including IL-1β, IL-6, and TNF-α, in the tissues were quantitated by quantitative real-time polymerase chain reaction (qRT-PCR). The primers for the amplification of these genes were synthesized in Beijing Genomics Institute (BGI) of China (Table S1). The lung, liver and kidney tissue injuries were evaluated in septic mice via hematoxylin eosin (HE) staining. Briefly, the tissues were immersed in 10% formaldehyde overnight, embedded in paraffin and sliced into tissue sections with a thickness of 4 μm. The HE-stained tissue slices were observed under a NanoZoomer S360 (Hamamatsu Photonics, Naka-ku, Hamamatsu, Japan) for pathological evaluation.
Availability of the raw data
The raw proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX database (https://iprox.org/) with an accession number of PXD025311. The raw transcriptomics data are accessible at https://www.ncbi.nlm.nih.gov/bioproject under accession number PRJNA722382. The raw metabolomics data are accessible at https://www.ebi.ac.uk/metabolights/index with accession number MTBLS2706.
Statistical analysis
SPSS 23.0 (IBM, Armonk, NY) was used for statistical analysis. All results are presented as the mean ± standard error of the mean (SEM). Significant differences between groups that pass both normality and equal variance test were evaluated using the one-way ANOVA. Bonferroni post hoc test was performed for multiple comparisons and the unpaired two-tailed Student’s t-test was performed for comparison between 2 groups. Differences were considered statistically significant when P was < 0.05.
Results
Characterization of serum exosomes derived from septic patients
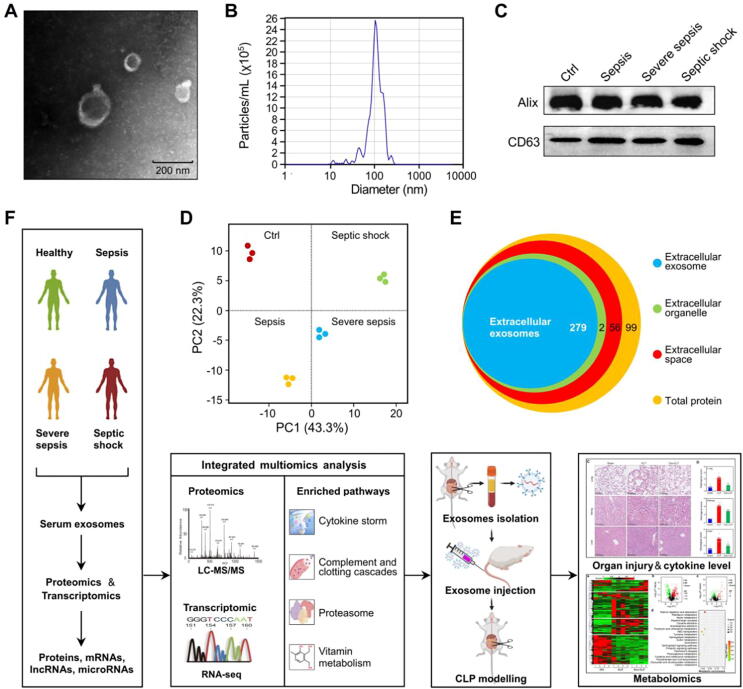
To characterize the exosomes isolated by ultracentrifugation from the serum of septic patients, we performed TEM, NTA and immunoblotting. We found that the isolated exosomes had a typical cup-shaped morphology (Fig. 1A) in TEM images, and the average exosome diameter was 105 ± 39 nm (Fig. 1B), consistent with previous reports [33], [34]. In addition, the immunoblotting results demonstrated that vesicles were positive for markers of exosomes, including Alix and CD63 (Fig. 1C).
Fig. 1.
Characterization of exosomes derived from the serum of healthy volunteers and septic patients. (A) The typical morphological characteristics of isolated exosomes were detected by TEM. (B) The size distribution of serum exosomes was determined by NTA. (C) Molecular markers for exosomes (Alix and CD63) were detected by Western blot analysis. (D) PCA of serum-derived exosomes from healthy volunteers and septic patients. (E) GO enrichment analysis was performed with cellular component terms to further confirm the quality and purity of the serum-derived exosome samples. (F) Overview of the experimental design for multiomics analysis of serum exosomes derived from septic patients.
To further confirm the quality and purity of the circulating exosomes, principal component analysis (PCA) and GO enrichment analysis for cellular components were conducted for the exosome proteome. PCA revealed distinct proteome clusters for serum exosomes derived from healthy volunteers and patients with sepsis, severe sepsis or septic shock (Fig. 1D). In terms of cellular components, we found that most exosome proteins were components of extracellular exosomes (Fig. 1E). In addition, we compared these proteins with the exosome database ExoCarta (http://exocarta.org/Archive/EXOCARTA_PROTEIN_MRNA_DETAILS_5.txt), and found that 84% serum exosome proteins identified in this study were shared by the ExoCarta database. Taken together, these results demonstrated that the quality of the exosomes isolated from septic patients was high and met the standard for the following multiomics analysis.
A schematic procedure for the study is shown in Fig. 1F. Serum samples were obtained from healthy volunteers or septic patients for isolation of serum exosomes. A DDA-based proteomics strategy and a second-generation high-throughput sequencing approach were used to quantitate exosome cargos, including proteins, mRNAs, lncRNAs and miRNAs. The data were analyzed with a series of bioinformatics tools for multiomics, and the results were verified experimentally.
Proteomic profiling and identification of DEPs in serum exosomes of patients with sepsis at different stages
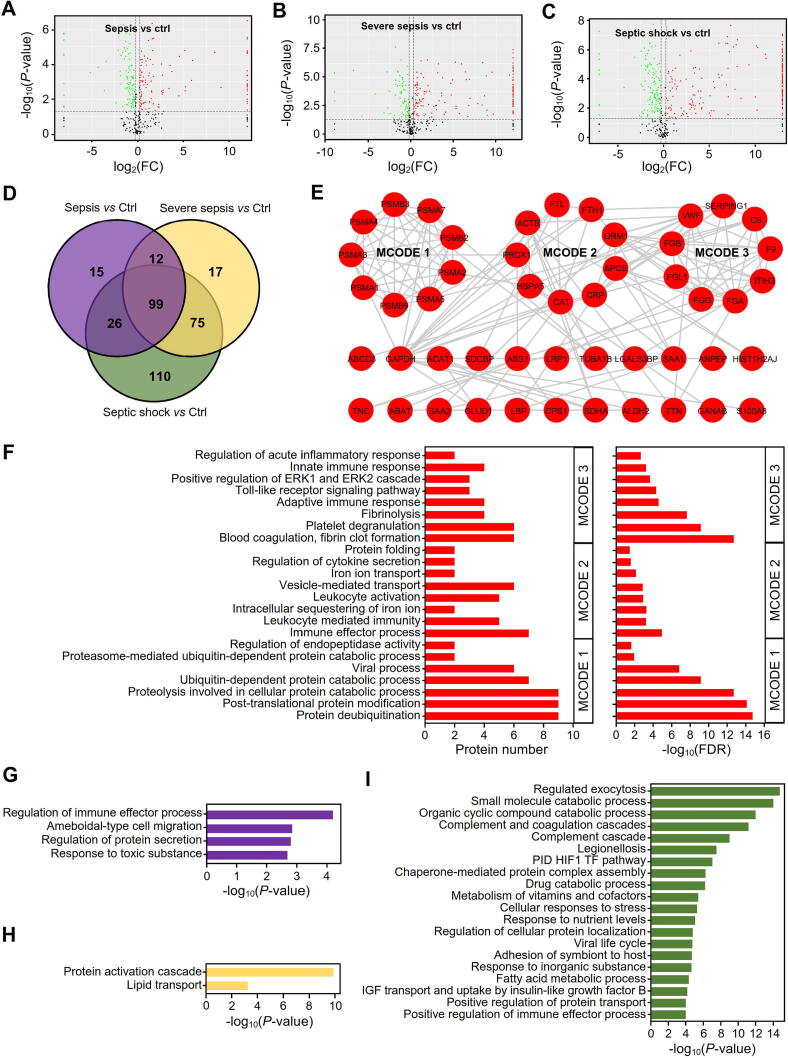
A DDA-based quantitative proteomics strategy was conducted to detect the dynamic profiles of exosome proteins during the progression of sepsis, and a total of 473 proteins were identified. We created volcano plots to further illustrate the DEPs in serum exosomes between healthy volunteers and patients with sepsis at different stages (Fig. 2A-C). Only proteins with P-values<0.05 and absolute FCs>1.5 in comparison with healthy volunteers were considered DEPs. We found that 152, 203 and 310 proteins were significantly dysregulated in exosomes derived from the serum of patients with sepsis (Fig. 2A), severe sepsis (Fig. 2B) and septic shock (Fig. 2C), respectively, that the number of DEPs tended to increase with the development of sepsis.
Fig. 2.
Proteomic characteristics and comparison of dysregulated proteins in exosomes derived from patients with sepsis at different stages. (A-C) Volcano plots of proteins in serum exosomes from patients with sepsis (A), severe sepsis (B) and septic shock (C). The black points represent proteins without significant changes, and the red and green points represent the upregulated and downregulated proteins with |log2FC| > 0.585 and P-values < 0.05 compared to those of the control group, respectively. (D) Venn diagram for the dysregulated proteins unique to the different stages of sepsis or shared among all stages of sepsis. (E) MCODE analysis of common upregulated proteins in sepsis. PPI networks were constructed based on STRING, and the hub subclasses of the network were filtered with MCODE in Cytoscape. (F) GO enrichment analysis on the three subclasses of protein interaction networks for common upregulated proteins in sepsis. (G-I) Metascape pathway and process enrichment for DEPs specifically dysregulated in serum exosomes from patients with sepsis (G), severe sepsis (H) and septic shock (I).
A Venn diagram was further constructed to identify the DEPs in serum exosomes during sepsis progression (Fig. 2D). We found that a total of 99 proteins were dysregulated throughout the entire period of sepsis progression, where some proteins such as peroxiredoxin-1 (PRDX1) were only present in septic patients but not the healthy which may be worth to do further verification as candidates for diagnostic markers. The protein–protein interaction (PPI) network for the 99 DEPs was analyzed with STRING, and we found that most of the upregulated proteins formed a tight interaction network (Fig. 2E). To further analyze the subnetworks for the upregulated proteins, we used the Molecular Complex Detection (MCODE) plugin (v1.5.1)[35] in Cytoscape and identified three protein complex modules (Fig. 2E), i.e., MCODE 1, MCODE 2 and MCODE 3, as key or hub modules.
Interestingly, we found that the proteins in MCODE 1 were significantly enriched in proteasome-related process terms, such as the proteasome-mediated ubiquitin-dependent protein catabolic process and proteolysis involved in cellular protein catabolic process terms (Fig. 2F). The MCODE 2 proteins were enriched mainly in the GO terms regulation of cytokine secretion, leukocyte activation and vesicle-mediated transport (Fig. 2F). For MCODE 3, the proteins were enriched primarily in inflammation and immune-related response terms, including the positive regulation of ERK1 and ERK2 cascade, Toll-like receptor signaling pathway, adaptive immune response and blood coagulation, fibrin clot formation terms, among others (Fig. 2F). In addition, KEGG pathway analysis was performed to investigate the functions of these modules (Table S2), and we found that the MCODE proteins participated in pathways related to the GO biological process terms. For example, the MCODE 1 proteins participated in proteasome, while the MCODE 3 proteins were enriched in the pathways of platelet activation and complement and coagulation cascades.
In addition to the 99 proteins dysregulated throughout the entire sepsis process, we also identified the DEPs with stage specificity. Fifteen proteins were associated with sepsis, 17 proteins were associated with severe sepsis and 110 proteins were associated with septic shock (Fig. 2D). Functional analysis of the DEPs was performed with Metascape (v3.5, http://metascape.org/) to explore the roles of exosomes secreted into the blood during different stages of sepsis (Fig. 2G-I). We found that the DEPs in the serum exosomes from septic patients were significantly enriched in the response to toxic substance, regulation of protein secretion and regulation of immune effector response, among others (Fig. 2G). For patients with severe sepsis, the exosome DEPs were enriched mainly in lipid transport and protein activation cascade (Fig. 2H), while the DEPs from septic shock patients significantly participated in the metabolism of vitamins and cofactors, complement and coagulation cascades, and regulated exocytosis, among other functions (Fig. 2I).
DEP profiles in serum exosomes from septic patients
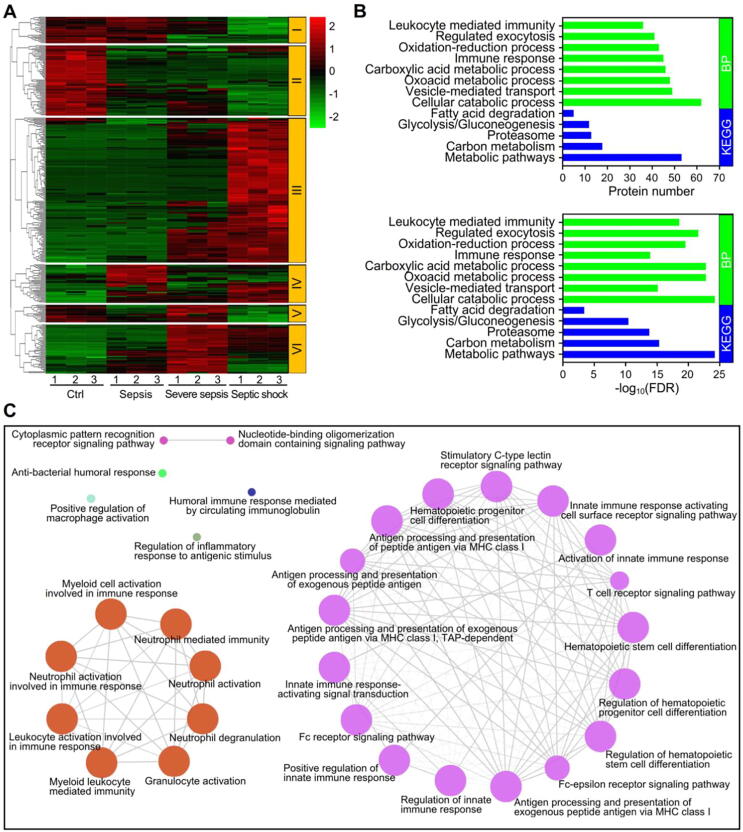
Clustering analysis was performed to reveal the dynamic profiles of DEPs during sepsis (Fig. 3A). Accordingly, six clusters were identified, of which cluster III was the largest. KEGG pathway analysis demonstrated that the proteins in cluster III were significantly enriched in the metabolic pathways, proteasome, and glycolysis/gluconeogenesis terms (Fig. 3B). GO enrichment analysis showed that the proteins were significantly associated with the immune response, vesicle-mediated transport and oxidation–reduction process biological process terms (Fig. 3B).
Fig. 3.
Profiling and functional identification of the dysregulated proteins in sepsis. (A) Clustering analysis of the significantly dysregulated proteins during the development of sepsis. In the color bar, red represents high expression, and green represents low expression. (B) Biological process terms from GO enrichment and KEGG pathway analyses of cluster III of the dysregulated proteins in sepsis. (C) Analysis of the immune system networks of the dysregulated proteins from cluster III with the ClueGO plugin in Cytoscape.
To further address the protein function of cluster III, we performed immune system analysis with the ClueGO plugin in Cytoscape and found that the proteins were involved mainly in immune-related responses (Fig. 3C). Interestingly, the proteins in cluster III formed two immune networks: the small network was associated with myeloid cell activation involved in the immune response and leukocyte activation, while the large network was associated with T cell receptor signaling pathway, Fc receptor signaling pathway and hematopoietic progenitor cell differentiation (Fig. 3C).
In addition, WikiPathway analysis demonstrated that the DEPs in cluster III were significantly associated with complement and coagulation cascades, vitamin B12 metabolism and folate metabolism (Fig. S1A), which further suggests that vitamin-related biological processes play a key role in sepsis, especially septic shock.
To investigate the relationships among the proteins in the six clusters, we constructed interaction networks with STRING and found that the DEPs in each cluster formed a highly connected interaction network (Fig. S1B). This finding indicated that the DEPs within each cluster shared some common features to contribute to a series of biological functions. To illuminate the differences in the biological terms enriched by each cluster, GO enrichment and KEGG pathway analysis were further performed on the DEPs in all clusters except cluster III (Fig. S1C, S1D). Intriguingly, we found that the proteins in cluster IV were significantly enriched in the biological process terms response to chemical, generation of precursor metabolites and energy, and regulation of cell migration, while those in cluster VI were enriched primarily in regulation of response to external stimulus, regulation of inflammatory response, immune response, etc. (Fig. S1C). In terms of signaling pathways, the proteins in cluster IV participated in valine, leucine and isoleucine degradation; synthesis and degradation of ketone bodies; pyruvate metabolism; etc. For cluster VI, the proteins were significantly enriched in the ECM-receptor interaction, protein digestion and absorption, and the PI3K-Akt signaling pathway, among others (Fig. S1D).
Notably, most of the clusters participated in complement and coagulation cascades (Fig. S1A and S1D). A heatmap was generated to determine the dynamic profiles of the DEPs with regard to the complement and coagulation cascades term. Interestingly, we found that different proteins showed different tendencies during the progression of sepsis (Fig. S1E).
Functional characterization of the consistently dysregulated proteins in serum exosomes from septic patients
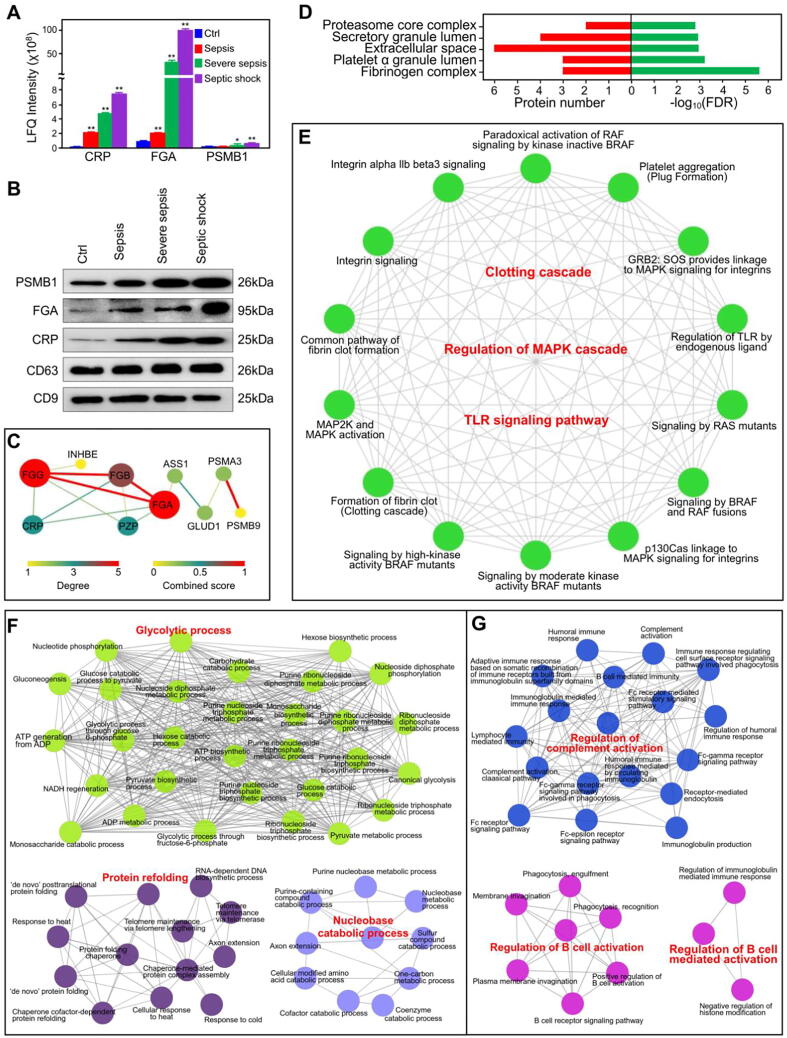
We first performed label-free quantification (LFQ) of C-reactive protein (CRP), fibrinogen alpha chain (FGA) and proteasome subunit β type-1 (PSMB1) with the proteomics data for serum exosomes from septic patients (Fig. 4A). To validate the proteomics results, we performed Western blotting with specific antibodies against these proteins and found the results to be consistent with the proteomics data (Fig. 4B).
Fig. 4.
Identification and functional characterization of the consistently up- or downregulated proteins. (A) The dysregulated proteins PSMB1, FGA and CRP in serum exosomes from septic patients were quantified by MS. The data represent three independent experiments (n = 3) and are expressed as the mean with SEM, *: P < 0.05, **: P < 0.01 compared with healthy group. (B) Western blot detection of the dysregulated proteins PSMB1, FGA and CRP in septic serum exosomes. (C) Interaction network of consistently up- or downregulated proteins. The size and color of each node represent the degree of interaction, and the size and color of each edge represent the combined score. (D) GO enrichment analysis of the cellular components of the consistently dysregulated proteins. (E) Network analysis of pathways enriched for the consistently dysregulated proteins with the ClueGO plugin in Cytoscape. (F, G) Networks of enriched biological processes in GO analysis for the proteins in serum exosomes that were specifically up- (F) and down- (G) regulated during septic shock.
To identify the DEPs associated with the severity of sepsis, we used the proteins that were consistently up- or downregulated in serum exosomes with the progression of sepsis to construct PPI networks. The results showed that most of the proteins were in a direct interaction network, in which there was only pregnancy zone protein (PZP) downregulated through sepsis progress (Fig. 4C). GO enrichment analysis showed that the consistently dysregulated proteins were associated mainly with the cellular component terms proteasome core complex, platelet alpha granule lumen and fibrinogen complex (Fig. 4D). We also found significant enrichment for the biological process terms plasminogen activation, platelet aggregation, and blood coagulation, fibrin clot formation (Fig. S2A). KEGG pathway analysis revealed that the consistently dysregulated proteins were associated with the biological function terms platelet activation, proteasome, and complement and coagulation cascades (Fig. S2B). Reactome analysis revealed that terms for clotting cascade-related pathways, including common pathway of fibrin clot formation term, were significantly enriched (Fig. S2C).
To illuminate the potential relationships among the biological processes and pathways enriched for these dysregulated proteins, we built an interaction network for the biological function terms with the ClueGO plugin in Cytoscape. The network showed close relationships among these functional terms, especially the clotting cascade, MAPK cascade and TLR signaling pathway terms (Fig. 4E). It is well established that the risk of death for septic patients increases dramatically when septic shock develops. Therefore, identification of serum exosome proteins that are specifically dysregulated in patients with septic shock but not in patients with other types of sepsis would be helpful. The proteins specifically upregulated in the serum exosomes of septic shock patients were significantly enriched in glycolytic process, protein refolding and nucleobase catabolic process biological process terms (Fig. 4F), while the proteins specifically downregulated at the stage of septic shock were associated with the regulation of complement activation, regulation of B cell activation and regulation of B cell-mediated activation biological process terms (Fig. 4G).
Genome-wide identification and functional characterization of the differential mRNAs in the serum exosomes of septic patients
In order to gain a comprehensive understanding of the potential roles of exosomes in sepsis, we performed RNA-seq experiments to clarify the transcriptomic variations among serum exosomes from patients with sepsis at different stages. Based on the criteria of a P-value < 0.05 and an |log2FC| > 1 in comparison with healthy volunteers, we identified 195 mRNAs, 82 lncRNAs and 55 miRNAs as differential RNAs in serum exosomes from patients with sepsis, severe sepsis and septic shock, including 2 mRNAs, 2 lncRNAs and 3 miRNAs that were dysregulated throughout the entire sepsis process and some differential expressed RNAs with stage specificity. For example, 55 mRNAs, 5 lncRNAs and 13 miRNAs were identified as dysregulated RNAs only in sepsis, whereas 72 mRNAs, 31 lncRNAs and 14 miRNAs were found to be specifically associated with septic shock.
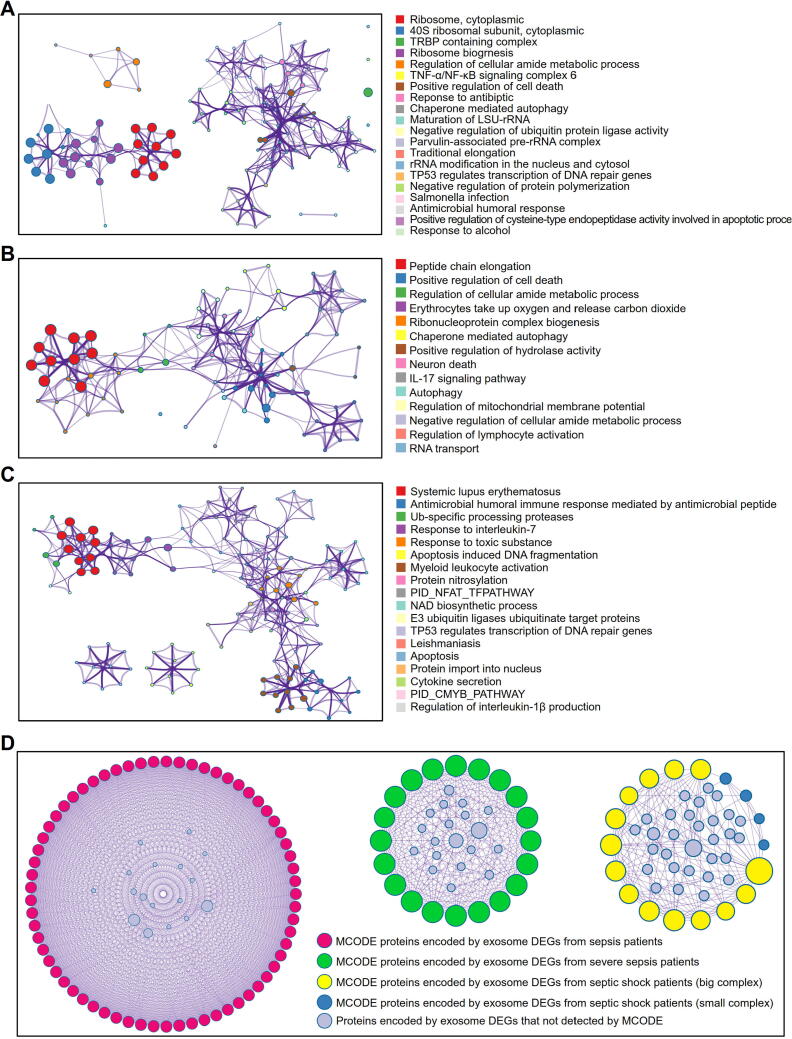
We identified 89 (2 up and 87 down), 61 (25 up and 36 down) and 91 (75 up and 16 down) differential mRNAs in serum exosomes from patients with sepsis, severe sepsis and septic shock, respectively, compared with exosomes from individuals in the healthy group. The Metascape tool was used to perform functional annotation of the differential mRNAs in exosomes and to explore the exosome-associated molecular mechanisms of sepsis. During the sepsis stage, terms such as negative regulation of ubiquitin protein ligase activity, antimicrobial humoral, and TNF-α/NF-κB signaling complex were significantly enriched (Fig. 5A). For severe sepsis, we found that the differential mRNAs in serum exosomes participated mainly in the IL-17 signaling pathway, neuron death and peptide chain elongation. (Fig. 5B). The differential mRNAs in serum exosomes from patients with septic shock were significantly enriched in function terms such as antimicrobial humoral immune response mediated by antimicrobial peptide, E3 ubiquitin ligases ubiquitinate target proteins, and cytokine secretion (Fig. 5C).
Fig. 5.
Functional characterization of differentially expressed mRNAs derived from serum exosomes of septic patients with Metascape. (A-C) Interaction networks of the enriched biological terms for the dysregulated mRNAs in serum exosomes from patients with sepsis (A), severe sepsis (B) and septic shock (C). Metascape enrichment networks for dysregulated mRNAs in exosomes were constructed by selecting each enriched term as a node and connecting the pairs of nodes with κ similarities > 0.3 to form a cluster with Cytoscape. Each node was colored by its cluster ID, and nodes that shared the same pathway were typically close to each other. (D) Interaction networks of proteins encoded by differentially expressed genes (DEGs) in serum exosomes from patients with sepsis at different stages. The networks were constructed with Metascape, and the modules of core genes from the PPI networks were filtered by MCODE in Cytoscape. The colored circles represent MCODE proteins encoded by DEGs of serum exosomes from patients with sepsis at different stages. The gray circles represent proteins encoded by DEGs of serum exosomes that were not detected by MCODE.
To further elucidate the potential relationships among the differential mRNAs, we constructed a molecular interaction network, and the core modules of this network were identified with the MCODE plugin in Cytoscape. As shown in Fig. 5D, we identified four hub clusters in the defined stages of sepsis. According to functional analysis of the clusters for sepsis and severe sepsis (Table S3), the hub mRNAs were significantly enriched in pathways associated with translation processes, such as peptide chain elongation, while for septic shock, the most important biological process-related terms were the HATs acetylate histones, HDACs deacetylate histones, and apoptosis-induced nucleosome positioning terms.
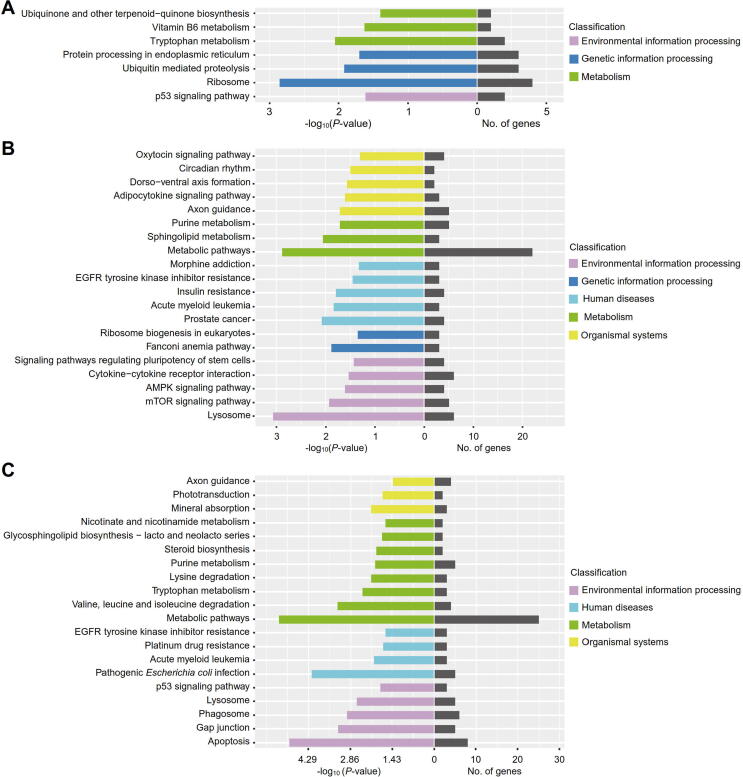
Dynamic profiles of lncRNAs in serum exosomes during the development of sepsis
Based on the cutoffs used for differential gene expression analysis, we identified 13 (1 up and 12 down), 46 (19 up and 27 down) and 49 (22 up and 27 down) lncRNAs that were dysregulated in serum exosomes from patients with sepsis, severe sepsis and septic shock, respectively. To elucidate the potential functions of these differential lncRNAs, we performed KEGG pathway analysis of their target genes, and the top 20 significant pathways are displayed. As shown in Fig. 6A, the ribosome, vitamin B6 metabolism, and ubiquitin-mediated proteolysis pathways were significantly enriched for the patients with sepsis. The differential lncRNAs in the serum exosomes of patients with severe sepsis were involved in immune-related pathways, such as the AMPK signaling pathway, the mTOR signaling pathway, ribosome biogenesis in eukaryotes and cytokine-cytokine receptor interaction (Fig. 6B). For septic shock patients, the significantly enriched terms were EGFR tyrosine kinase inhibitor resistance, purine metabolism, axon guidance and apoptosis (Fig. 6C). Interestingly, we found that the term insulin resistance was significantly enriched in both severe sepsis (Fig. 6B) and septic shock (P = 0.046), which was consistent with the clinical practice of administering intensive insulin therapy to septic patients [36], [37]. During the development of sepsis, ribosome-related pathways, such as ribosome biogenesis in eukaryotes, were significantly enriched, which suggests that serum exosomes play a role in the regulation of protein translation in patients with sepsis.
Fig. 6.
Pathway enrichment analysis for dysregulated lncRNAs in serum exosomes from septic patients. (A-C) KEGG pathway analysis for dysregulated lncRNAs in the serum exosomes of patients with sepsis (A), severe sepsis (B) and septic shock (C). The mRNAs targeted by the differentially expressed lncRNAs were used to perform KEGG pathway analysis according to the molecular interaction, reaction and relation networks for pathway mapping. The top 20 significant pathways are displayed.
To further investigate the functions of septic exosomes, we performed GO enrichment analysis on the differential lncRNAs in the serum exosomes of septic patients. The results showed that the genes targeted by the differential lncRNAs were highly enriched in metabolic process terms (Fig. S3). Consistent with this finding, previous studies have demonstrated that septic patients, especially those who develop septic shock, exhibit metabolic imbalances[38], [39].
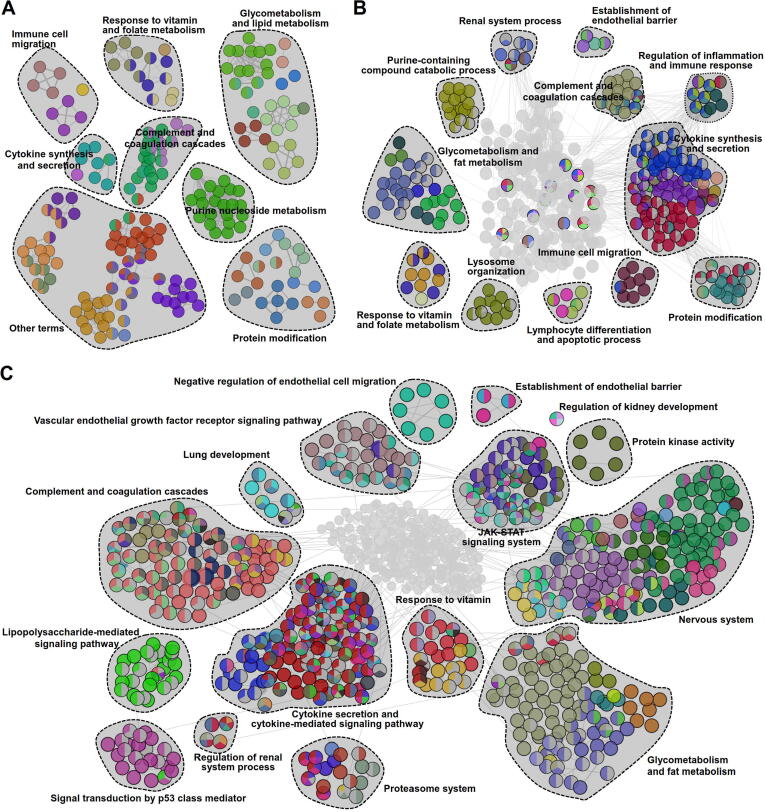
Atlas of circulating miRNAs in exosomes derived from septic patients
We identified 24 (3 up and 21 down), 23 (7 up and 16 down) and 26 (2 up and 24 down) differential miRNAs in serum exosomes from patients with sepsis, severe sepsis and septic shock, respectively, compared with exosomes from individuals in the healthy group (Fig. S4). To gain further insights into the biological functions of the miRNAs in serum exosomes, we performed functional enrichment analysis on the target genes of the differential miRNAs. The results showed that terms associated with cytokine synthesis and secretion, immune cell migration, complement and coagulation cascades, response to vitamin and folate metabolism, purine nucleoside metabolism, glycometabolism and lipid metabolism, and protein modification were enriched during the early stage of sepsis (Fig. 7A). For severe sepsis, the functional networks were composed of the same terms noted for sepsis in addition to terms not enriched at that stage, such as the regulation of inflammation and immune response, establishment of endothelial barrier, lymphocyte differentiation and apoptotic process, and renal system process terms (Fig. 7B).
Fig. 7.
Functional network map of genes targeted by dysregulated miRNAs in serum exosomes derived from septic patients. (A-C) Functionally grouped networks of enriched biological terms for dysregulated miRNAs in serum exosomes from patients with sepsis (A), severe sepsis (B) and septic shock (C). The functional networks were constructed with the ClueGO plugin in Cytoscape to reveal the connections and differences in the biological processes and signaling pathways associated with the genes that were targeted by the differentially expressed miRNAs in serum exosomes from septic patients. Each node represents an enriched functional term, and the related terms sharing similar associated genes were grouped to reduce redundancy.
For the septic shock stage, some new terms, including the proteasome system, JAK-STAT signaling system, vascular endothelial growth factor receptor signaling pathway, lung development, and negative regulation of endothelial cell migration terms, appeared in the networks (Fig. 7C).
Integrative analysis of proteomics and transcriptomics datasets derived from the serum exosomes of septic patients
Integration of data with multivariate methods helps with prediction of complex traits or phenotypes by decreasing the heterogeneity in multiomics data. To identify the most important omics variables contributing to sepsis, we used the mixOmics R package based on DIABLO, a multiomics integrative method that seeks common information across different data types through selection of a subset of molecular features while discriminating among multiple phenotypic groups.
A heatmap of the multiomics signature during sepsis exhibited significant correlations between some proteins and mRNAs in the first principal component (PC1) (Fig. 8A). To detect the potential roles of these molecules in PC1, we constructed interaction networks for them and found that most of them formed a complex network (Fig. S5).
Fig. 8.
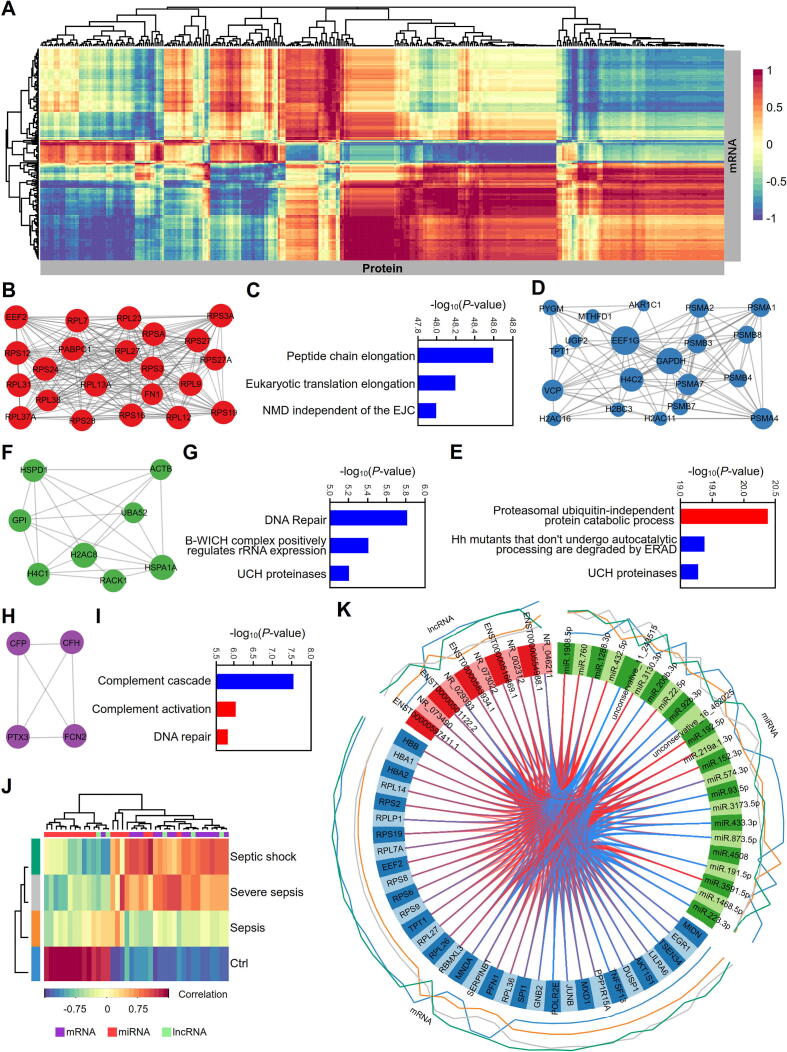
Integrative multiomics analysis of proteins, mRNAs, lncRNAs and miRNAs in the serum exosomes of septic patients. (A) Hierarchical clustering for canonical correlation analysis (CCA) of the mRNAs and proteins in serum exosomes from septic patients. The mixOmics R package was used to calculate the correlations between mRNA abundance (rows) and protein abundance (columns) in a pairwise fashion. (B-I) Identification and functional annotation of MCODE subclasses in the interaction networks for the hierarchical cluster described above. The interaction networks were created with the BioGrid, InWeb_IM and OmniPath databases. MCODE analysis was performed to identify densely connected components in the networks (B, D, F, H). Pathway and biological process enrichment analyses were applied to each MCODE component independently, and the three best-scoring terms by P-value were retained as the functional descriptions of the corresponding subclasses (C, E, G, I). The blue color represents the pathway enrichment analysis terms, and the red color represents the biological process terms from GO enrichment analysis. (J) CIM of the multiomics signatures for mRNAs, lncRNAs and miRNAs in the serum exosomes of septic patients. The map was constructed on Euclidian distance and complete linkage with the mixOmics R package for transcriptomic datasets including mRNAs, lncRNAs and miRNAs in serum exosomes from patients with sepsis at different stages. Grouped samples are represented in rows, and selected features on the first component are represented in columns. (K) Circos plot representing the correlations among mRNAs, lncRNAs and miRNAs in serum exosomes from patients with sepsis at different stages. The Circos plot was obtained by using the function circosPlot() from the package mixOmics and shows the positive (red line) and negative (blue line) correlations (|r| >0.7) between variables (edges inside the circle) and the average value of each variable (line profile outside the circle).
We further performed MCODE analysis to identify densely connected components, which revealed four hub clusters (Fig. 8B, 8D, 8F and 8H). The 2 largest clusters were composed mainly of ribosome-related and proteasome-related molecules (Fig. 8B and 8D). Functional analysis of the four clusters showed that exosomes in septic patients play essential roles in the progression of sepsis by regulating the translation system (Fig. 8C), the protein degradation system (Fig. 8E), DNA repair (Fig. 8G) and the complement cascade system (Fig. 8I). In addition, a clustered image map (CIM) was constructed with the mRNAs, lncRNAs and miRNAs in the serum exosomes of sepsis patients that were screened by DIABLO. The CIM depicted satisfactory classifications for different sepsis groups (Fig. 8J). The variables with the strongest correlations in the multiomics data were determined by analysis of the transcriptomics datasets with the mixOmics R package and displayed in Circos plots (Fig. 8K).
Regulation of cytokine secretion and tissue damage during sepsis by serum exosomes
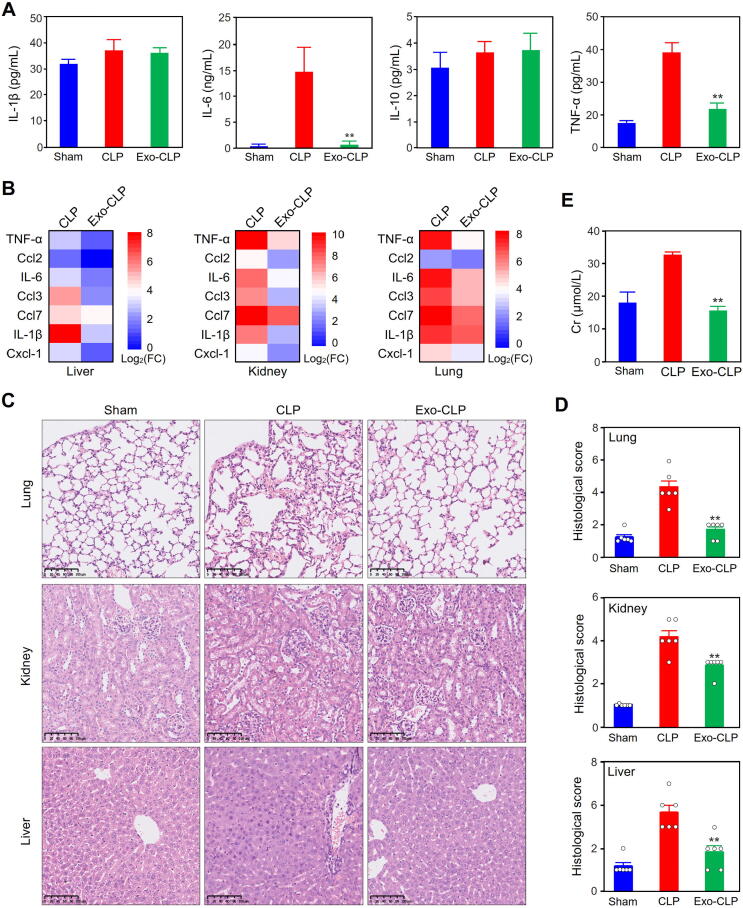
The multi-omics analysis results revealed that the differential cargos in serum exosomes from septic patients were significantly involved in the biological processes of cytokine synthesis and secretion (Fig. 2F, 5C, 6B and 7A-C). In addition, in the previous study, we found that the serum exosomes from the mice subjected to LPS-injection could significantly reduce the inflammatory response and prolong the survival of CLP mice[20]. Based on these results, we detected the effect of the serum exosomes from CLP mice on the expression of cytokines and chemokines in the blood and tissues, as well as organ injuries. Hence, we pretreated wild-type mice with exosomes derived from septic mice via TVI and then performed CLP modeling. Importantly, we found that the serum levels of cytokines such as IL-6 and TNF-α significantly decreased at 12 h after CLP modeling in mice pretreated with serum exosomes (Fig. 9A). We further characterized the gene expression profiles of inflammatory cytokines and chemokines in the liver, lung and kidney tissues through qRT-PCR (Fig. 9B). Consistently, pretreatment with exosomes derived from septic mice restrained the gene expression of cytokines and chemokines in these tissues in CLP mice.
Fig. 9.
Effects of exosomes from septic mice on cytokine expression and tissue injury in sepsis. (A) Effects of pretreatment with exosomes from septic mice on the levels of serum cytokines in CLP mice. The levels of IL-1β, IL-6, IL-10 and TNFα were detected with a LiquiChip system as described in the methods section. The data represent three independent experiments (n = 3) and are expressed as the mean with SEM. **: P < 0.01 compared with CLP group. (B) Effects of pretreatment with exosomes from septic mice on the gene expression of cytokines and chemokines in the tissues of CLP mice. The mRNA levels were detected by qRT-PCR (n = 3). The quantitative results for the CLP group and exo-CLP group were divided by those for the sham group, and the ratios were log2-transformed. (C, D) Effects of pretreatment with exosomes from septic mice on the histopathology of lung, kidney and liver tissues from CLP mice. HE staining (C) was performed with slides of lung, kidney and liver tissues to detect the morphological changes in CLP mice subjected to pretreatment with exosomes from septic mice. Pathological evaluation involved determining the histological score reflecting tissue injury in CLP mice (D). (E) Pretreatment with septic exosomes decreased the Cr levels in the serum of CLP mice.
Pathological examinations showed that the tissue injuries in the CLP group were attenuated by serum exosomes derived from septic mice (Fig. 9C). As expected, the histological scores for tissue damage were consistent with the morphological results (Fig. 9D). To test whether the morphological changes were associated with functional alterations, we detected serum creatinine (Cr) levels in CLP mice pretreated or not pretreated with serum exosomes from septic mice and found that the pretreatment significantly reduced Cr levels (Fig. 9E).
Serum metabolite profiles of septic mice preinjected with serum exosomes
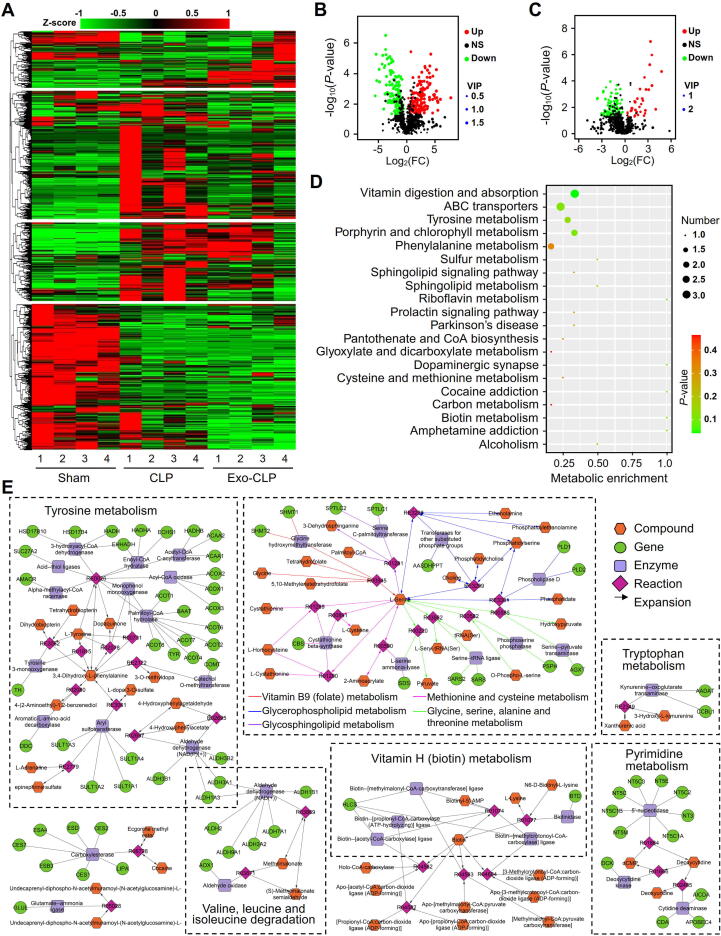
We utilized untargeted MS/MS to analyze the serum metabolites of CLP mice pretreated with exosomes derived from septic mice. The clustering heatmap showed that pretreatment with serum exosomes significantly influenced the metabolite profile, and at least four expression patterns were identified (Fig. 10A).
Fig. 10.
Metabolomic characterization of CLP mice pretreated with sepsis-associated exosomes. (A) Heat map and hierarchical clustering analysis of the serum metabolites of CLP mice. (B, C) Volcano plots for metabolites in CLP mice pretreated without (B) or with (C) septic exosomes. The red and blue dots represent significantly up- and downregulated metabolites, respectively, with VIP values > 1, |log2FC| > 0.263 and P-values < 0.05 in comparison with those in the sham group. (D) KEGG pathway analysis of the differential metabolites in the exo-CLP group and CLP alone group. (E) Model of the metabolic networks constructed with the differential metabolites in the exo-CLP group. The networks were generated with the Metscape plugin in Cytoscape. Genes and compounds in the network for each metabolic pathway were acquired with Metscape and the KEGG database.
Based on the cutoff set by a VIP value > 1, a P-value < 0.05 and an |log2FC| > 0.263, we identified 311 metabolites that were significantly dysregulated in the CLP group compared with the sham group (Fig. 10B); moreover, we identified 129 metabolites that were significantly up- or downregulated in the exosome-pretreated CLP group (exo-CLP group) in comparison with the CLP group (Fig. 10C). KEGG pathway analysis was performed for the 129 dysregulated compounds, and the results showed that the most significant term was vitamin digestion and absorption, demonstrating that serum exosomes play a crucial role in the progression of sepsis by regulating vitamin-related biological processes (Fig. 10D).
To illuminate the effects of serum exosomes from septic mice on metabolism in CLP mice, we constructed metabolic networks with the intersecting dysregulated metabolites from Fig. 10B and Fig. 10C with Metscape (v3.1.3)[40]. We found that the 39 differential metabolites shared in the volcano plots participated mainly in amino acid and vitamin metabolism-related processes, such as tyrosine metabolism; tryptophan metabolism; valine, leucine and isoleucine degradation; methionine and cysteine metabolism; glycine, serine, alanine and threonine metabolism; vitamin B9 (folate) metabolism; and vitamin H (biotin) metabolism (Fig. 10E).
Then, we surveyed the diseases that were associated with the metabolic network based on Medical Subject Heading (MeSH) terms. We found that 7 of the 39 differential metabolites had close connections to MeSH diseases, and most of them, including 3,4-dihydroxy-L-phenylalanine, L-serine, methylmalonate, biotin and xanthurenic acid, were associated with nutritional and metabolic diseases, vitamin B deficiency and folic acid deficiency. Importantly, the representative metabolite deoxycytidine was enriched in the terms bacterial infections, inflammation, sepsis, and systemic inflammatory response syndrome (Table S4).
Discussion
Recent studies have indicated that circulating exosomes are important mediators of systemic signaling events in sepsis [41], [42]. Via their different enveloped cargos, exosomes perform various biological functions during sepsis. Exosomes released from cells contain numerous components, including proteins, mRNAs, lncRNAs, and miRNAs, etc., that together mediate precise responses of cells to environmental changes or stress signals [1], [43]. Systematic studies on the components in serum exosomes from patients with sepsis at different stages will be helpful for elucidating the functions and related regulatory mechanisms of exosomes. In the present study, we performed integrative proteomics, transcriptomics and metabolomics analyses to landscape the molecular signatures of serum exosomes from individuals with sepsis and to promote a systematic understanding of the development of sepsis. Through multiomics analysis of the components in serum exosomes from patients with sepsis at different stages, we found that circulating exosomes are associated mainly with cytokine storm, complement and clotting cascades, the endothelial barrier, protein degradation and vitamin metabolism.
Many studies have confirmed that cytokine storm plays crucial roles in the development of sepsis, leading to tissue damage, organ failure and even death by disrupting the balance of the immune-inflammatory network [44], [45]. Our multiomics analysis revealed that serum exosomes from septic patients are closely related to cytokine synthesis and secretion. GO analysis revealed that the proteins that were co-upregulated in the serum exosomes of patients with sepsis at different stages were highly enriched in the regulation of cytokine secretion term (Fig. 2F), while Metascape analysis revealed that the differential mRNAs in serum exosomes from patients with septic shock participated in cytokine secretion-related processes (Fig. 5C). Interestingly, the genes targeted by the differential lncRNAs in serum exosomes from patients with severe sepsis were significantly enriched in the cytokine-cytokine receptor interaction pathway (Fig. 6B). Additionally, the differential miRNAs in serum exosomes from septic individuals were associated with the functional terms cytokine synthesis and secretion (Fig. 7A-C).
Our previous study has demonstrated that serum exosomes from septic mice contain abundant IL-10[20]. As a powerful immunomodulator, IL-10 exerts a protective effect against tissue damage through its strong anti-inflammatory capability [46]. For example, IL-10 has been reported to protect acute kidney injury (AKI) patients from ischemia-, cisplatin-, or ureteric obstruction-induced renal injury by reducing inflammatory cytokine production and immune cell infiltration[47], and treatment with engineered exosomes containing IL-10 significantly ameliorates renal tubular injury and inflammation in AKI patients through M2 macrophage polarization [48]. These data indicate that IL-10-enriched exosome treatment is a potential therapy for AKI in the context of sepsis.
Exosomes are lipid bilayer membrane vesicles decorated with multiple ligands and receptors, through which they interact with target cells, and have been implicated as carriers of biological macromolecules [49]. Importantly, we found that pretreatment with serum exosomes significantly downregulated the mRNA expression of TNF-α and IL-6 in lung, liver and kidney tissues from septic mice (Fig. 9B). The protective effects of serum exosomes against tissue injuries can be explained their transmission of biological signals to target cells through receptors on the cell membrane or through direct delivery of exogenous IL-10 protein into the target cells. Notably, the cytokine dynamics in the lung and liver tissues from CLP mice pretreated with exosomes from septic mice were different from those in the kidney tissues (Fig. 9B, S6A-C); this finding indicates that serum exosomes from septic mice exert different effects according to the complex characteristics of the target tissues in the context of sepsis. In addition to direct delivery of IL-10 protein in the serum exosomes, our study demonstrated that pretreatment with exosomes derived from septic mice alleviated kidney injury in CLP mice (Fig. 9C) possibly through upregulation of IL-10 mRNA levels in the kidneys (Fig. S6C).
To date, all clinical trials targeting specific cytokines, such as those utilizing neutralizing antibodies against TNF-α, have failed. Mortality due to sepsis is not merely attributable to a high level of one particular cytokine; rather, it is attributable to dynamic and complex inflammatory signatures. Our characterization of serum exosomes from septic patients may provide a foundation for further exploration of innovative approaches to suppress the overwhelming inflammatory cascade and thus to eventually attenuate tissue damage. Our multiomics data showed that the components in serum exosomes from septic patients were associated with the complement and clotting cascade (Fig. 2F and Fig. 7). Although the complement system is regarded as a protective system, overactivation of the system may be harmful or even lethal for the host in the case of septic shock.
Characterizing the complement and coagulating factors in serum exosomes may be helpful for understanding the mechanisms of complement and clotting activation in sepsis. In serum exosomes from septic patients, we identified not only proteins involved in activation of the classical complement pathway, such as C1R, C2, C3 and C7, but also proteins participating in alternative pathways, such as complement factors B (CFB), H (CFH) and I (CFI) (Fig. S1E). However, the different complement factors showed different dynamics during sepsis progression. For example, CFI levels peaked in the severe sepsis stage, while CFB and CFH levels were consistently decreased in the serum exosomes of septic patients, indicating that serum exosomes play different roles in complement activation during different sepsis stages.
Interestingly, we found that fibrinogen coagulation factors, including FGA, FGB and FGG, were upregulated, which highlights the potential role of exosomes from septic individuals in coagulation. In addition, some SERPIN family members showed different dynamics during sepsis development. Serpin peptidase inhibitor clade C (antithrombin) member 1 (SERPINC1), which restrains blood clotting [50], [51], was downregulated throughout sepsis. However, serpin peptidase inhibitor clade G (C1 inhibitor) member 1 (SERPING1) was upregulated in serum exosomes from septic patients. SERPING1 blocks plasma kallikrein and the activated form of factor XII and reduces the permeability of blood vessels, thus protecting tissues from damage by increasing blood perfusion [52], [53]. In addition, a primary inhibitor of tissue-type plasminogen activator (PLAT), serpin peptidase inhibitor clade E (nexin, plasminogen activator inhibitor type 1) member 1 (SERPINE1), was upregulated during septic shock, suggesting that serum exosomes from septic patients participate in downregulation of fibrinolysis and degradation of blood clots in the late stage of sepsis[54], [55].
Venn diagram analysis demonstrated that 99 proteins were dysregulated in serum exosomes throughout sepsis progression (Fig. 2D). Interestingly, actin levels increased, while gelsolin (GSN) levels decreased (Table S5), in serum exosomes during sepsis development. GSN is an actin-binding protein that has severing, capping and actin monomer-sequestering abilities[56]. When secreted into the circulatory system, GSN becomes part of the extracellular actin scavenger system to prevent actin polymerization and to remove toxic actin filaments released from necrotic cells into the blood [57]. It is believed that GSN is recruited to sites of tissue injury to scavenge the actin released by damaged cells, resulting in GSN depletion from plasma [58]. Previous studies had reported that a low level of GSN in plasma is a marker for poor prognosis of sepsis [59], and actin in extracellular vesicles could polymerize to microfilaments if there were absent in actin-binding proteins such as GSN[57]. Interestingly, increased actin levels and decreased GSN levels were observed in the serum exosomes of septic patients in the current study, suggesting that exosomal actin exists in polymers called microfilaments in sepsis.
The LC-MS/MS results demonstrated that 14 proteins, including 7 α and 7 β subunits of the 20S proteasome (Fig. S7A and S7B) and 3 subunits of the immunoproteasome (Fig. S7C and S7D), were significantly upregulated in exosomes from septic individuals. Consistent with this result, Western blot analysis showed that the levels of PSMB1, a β subunit of the 20S proteasome, were significantly increased in septic serum exosomes (Fig. 4B). Consistently, integrative multiomics analysis showed that proteasomal degradation-associated terms were significantly enriched during the progression of sepsis (Fig. 8E).
As the core of the ubiquitin proteasome system, the 20S proteasome has been revealed to exist in physiological body fluids in the contexts of various inflammatory and autoimmune diseases. Extracellular proteasomes have been identified to be closely correlated with the severity of disease and are of prognostic significance[60]. For example, platelet-derived exosomes have been reported to decrease CD36 levels in platelets and macrophages by enhancing ubiquitination and proteasome degradation, thus resulting in mitigation of the atherothrombotic process[61]. Lai RC et al. found that exosomes from mesenchymal stem cells (MSCs) contained functional 20S proteasome that were correlated with modest but significant reductions in the levels of oligomerized proteins in a mouse model of myocardial infarction[62]. A previous study has demonstrated that large amounts of exosome proteasomes can cause downregulation of IL-6 production in monocytes[63]. Importantly, we found that pretreatment with serum exosomes from septic mice significantly decreased the levels of cytokines such as IL-6 and TNF-α in the serum of CLP mice (Fig. 9A). Collectively, this evidence supports the hypothesis that serum exosomes from septic mice modulate proinflammatory cytokines through protein degradation in 20S proteasome, leading to alleviation of tissue damage. These findings provide new insight into the mechanism of proteasome-mediated protein degradation in serum exosomes during the development of sepsis.
It has been established that the complex, integrated immune system needs multiple specific vitamins, including folate and vitamins A, B6, B12, C, D, and E, which play vital roles in the immune response[64]. Some populations have inadequate dietary vitamin intake, and situations that increase vitamin requirements (e.g., infection and stress) further decrease the stores within the body[65]. The available evidence indicates that supplementation with multiple vitamins may improve immune function and reduce the risk of infection[66]. For example, vitamin D activates vitamin D receptors (VDRs) on macrophages in liver tissues, ameliorating liver inflammation, steatosis and insulin resistance[67].
Our proteomic data showed that the terms metabolism of vitamins and cofactors, vitamin digestion and absorption, and vitamin B12 metabolism were significantly enriched in the serum exosomes from septic patients (Fig. 2I, S1A and S1D). Recent studies have provided evidence supporting the immunoregulatory effects of vitamin B12, including modulation of TNF-α activity[68]. Consistently, the results of our transcriptomic analysis showed that the differential lncRNAs in the serum exosomes from septic patients participated in vitamin B6 metabolism (Fig. 6A). Doke et al. demonstrated that vitamin B6 is necessary for maintenance of cytokine levels and lymphoid function in the thymus and spleen in mice[69]. Furthermore, some terms associated with the response to vitamin and folate metabolism were enriched in the functional networks for the genes targeted by differential miRNAs in serum exosomes from septic patients (Fig. 7). Importantly, pretreatment with septic serum exosomes significantly affected the metabolites involved in vitamin digestion and absorption in CLP mice (Fig. 10D). Metscape analysis of the metabolic network demonstrated that the differential metabolites induced by pretreatment with serum exosomes from septic mice participated in vitamin B9 (folate) metabolism and vitamin H (biotin) metabolism (Fig. 10E). All the data above suggest that serum exosomes participate in the modulation of sepsis by regulating vitamin metabolism.
The results of our systematic analysis of multiomics data on serum exosomes will be helpful for illustrating the underlying mechanisms of the molecular interaction network involving dynamic cargos in serum exosomes collected from patients with sepsis at different stages. However, there are a few limitations. Firstly, only male mice were used in our study to reproduce CLP model. It’s interesting to answer the question that whether the findings from CLP model of male mice are consistent with the results from female mice, which might be an attractive topic in the future. Secondly, though methanol extraction has been widely used in metabolomics[70], [71], this method is mainly for polar metabolites and not good for lipid extraction and fatty acid assessment. Thirdly, the function analyses of the differentially expressed miRNAs in septic serum exosomes based on their predicted target genes rather than verification in the study. Lastly the sample size was relatively small, and most of the hypotheses were not fully validated by the experimental data. Future studies on large sample of serum exosomes in combination with experimental verification will further improve our understanding of the dose–response characteristics, pharmacokinetics, and potential toxicities of serum exosomes.
Conclusion
In summary, for the first time, we conducted systematic proteomic and transcriptomic analyses of serum exosomes from patients with sepsis of different severities and analyzed metabolic data from CLP mice pretreated with serum exosomes. Based on the multiomics data, we hypothesize that serum exosomes from septic mice exert their therapeutic effects through mechanisms involving cytokine storm suppression, complement and coagulation system activation, endothelial cell junction stabilization and vascular permeability reduction, 20S proteasome-mediated protein degradation, vitamin metabolism. Our study describes the dynamics of the molecular landscape of serum exosomes during the development of sepsis and proposes some hypothetical mechanisms for the therapeutic effects of serum exosomes based on multiomics analysis and experiments of CLP mice. Our findings may provide helpful diagnostic and therapeutic clues to support the ongoing battle against sepsis.
Compliance with Ethics requirement
This study was approved by the Ethics Committee of the Third Affiliated Hospital of Southern Medical University, Guangzhou, China (No. 2020028) and was performed in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national). Informed consent was obtained from the septic patients and healthy donors in the study.
CRediT authorship contribution statement
Lei Li: Conceptualization, Validation, Writing – original draft. Lin Huang: Methodology. Chenyang Huang: Methodology, Validation. Jia Xu: Validation, Visualization. Yukai Huang: Investigation, Resources. Haihua Luo: Resources. Xinya Lu: Validation. Shuyue He: Validation. Gang Yuan: Validation. Li Chen: Validation. Xue Han: Validation. Xusong Cao: Validation. Aolin Jiang: Validation. Cuiting Liu: . Junmin Shi: . Hong Yang: . Yong Jiang: Supervision, Conceptualization, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported by the National Natural Science Foundation of China ( nos. 82130063, 81971895 and 82002089), Special Support Plan for Outstanding Talents of Guangdong Province (no. 2019JC05Y340), and China Postdoctoral Science Foundation (no. 2020M672717).
Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.11.005.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Raeven P., Zipperle J., Drechsler S. Extracellular vesicles as markers and mediators in sepsis. Theranostics. 2018;8(12):3348–3365. doi: 10.7150/thno.23453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R., et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the global burden of disease study. Lancet. 2020;395(10219):200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chousterman B.G., Swirski F.K., Weber G.F. Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol. 2017;39(5):517–528. doi: 10.1007/s00281-017-0639-8. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhry H, Zhou J, Zhong Y, Ali MM, McGuire F, Nagarkatti PS, et al. Role of cytokines as a double-edged sword in sepsis. In Vivo 2013; 27(6):669-684. doi: NA. [PMC free article] [PubMed]
- 5.Fisher C.J., Slotman G.J., Opal S.M., Pribble J.P., Bone R.C., Emmanuel G., et al. Initial evaluation of human recombinant interleukin-1 receptor antagonist in the treatment of sepsis syndrome: a randomized, open-label, placebo-controlled multicenter trial. Crit Care Med. 1994;22(1):12–21. doi: 10.1097/00003246-199401000-00008. [DOI] [PubMed] [Google Scholar]
- 6.Chang R., Holcomb J.B. Choice of Fluid Therapy in the initial management of sepsis, severe sepsis, and septic shock. shock. 2016;46(1):17–26. doi: 10.1097/SHK.0000000000000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J., Wang Y., Li L. Functional significance of exosomes applied in sepsis: a novel approach to therapy. Biochim Biophys Acta Mol Basis Dis. 2017;1863(1):292–297. doi: 10.1016/j.bbadis.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 8.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhodes A., Evans L.E., Alhazzani W., Levy M.M., Antonelli M., Ferrer R., et al. Surviving Sepsis Campaign: international guidelines for management of sepsis and septic Shock: 2016. Intensive Care Med. 2017;43(3):304–377. doi: 10.1007/s00134-017-4683-6. [DOI] [PubMed] [Google Scholar]
- 10.Pegtel D.M., Gould S.J. Exosomes. Annu Rev Biochem. 2019;88(1):487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 11.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020; 367(6478):eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed]
- 12.Ferguson S.W., Nguyen J. Exosomes as therapeutics: the implications of molecular composition and exosomal heterogeneity. J Control Release. 2016;228:179–190. doi: 10.1016/j.jconrel.2016.02.037. [DOI] [PubMed] [Google Scholar]
- 13.Bielska E., May R.C. Extracellular vesicles of human pathogenic fungi. Curr Opin Microbiol. 2019;52:90–99. doi: 10.1016/j.mib.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Gezsi A., Kovacs A., Visnovitz T., Buzas E.I. Systems biology approaches to investigating the roles of extracellular vesicles in human diseases. Exp Mol Med. 2019;51(3):1–11. doi: 10.1038/s12276-019-0226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoshino A., Costa-Silva B., Shen T.-L., Rodrigues G., Hashimoto A., Tesic Mark M., et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitai Y., Kawasaki T., Sueyoshi T., Kobiyama K., Ishii K.J., Zou J., et al. DNA-containing exosomes derived from cancer cells treated with topotecan activate a sting-dependent pathway and reinforce antitumor Immunity. J Immunol. 2017;198(4):1649–1659. doi: 10.4049/jimmunol.1601694. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J., Gao J., Lin D., Xiong J., Wang J., Chen J., et al. Potential networks regulated by MSCs in acute-on-chronic liver failure: exosomal miRNAs and intracellular target genes. Front Genet. 2021;12 doi: 10.3389/fgene.2021.650536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X., Chen F., Zhang Q., Liu Y., You P., Sun S., et al. Salivary exosomal PSMA7: a promising biomarker of inflammatory bowel disease. Protein Cell. 2017;8(9):686–695. doi: 10.1007/s13238-017-0413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheller-Miller S., Radnaa E., Yoo J.K., Kim E., Choi K., Kim Y., et al. Exosomal delivery of NF-kappaB inhibitor delays LPS-induced preterm birth and modulates fetal immune cell profile in mouse models. Sci Adv. 2021;7(4):eabd3865 doi: 10.1126/sciadv.abd3865. [DOI] [PubMed] [Google Scholar]
- 20.Gao K., Jin J., Huang C., Li J., Luo H., Li L., et al. Exosomes derived from septic mouse serum modulate immune responses via exosome-associated cytokines. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cecconi M., Evans L., Levy M., Rhodes A. Sepsis and septic shock. Lancet. 2018;392(10141):75–87. doi: 10.1016/S0140-6736(18)30696-2. [DOI] [PubMed] [Google Scholar]
- 22.Xie J., Wang H., Kang Y., Zhou L., Liu Z., Qin B., et al. The epidemiology of sepsis in Chinese ICUs: a national cross-sectional survey. Crit Care Med. 2020;48(3):e209–e218. doi: 10.1097/CCM.0000000000004155. [DOI] [PubMed] [Google Scholar]
- 23.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 24.Wang F., Gong S., Wang T., Li L., Luo H., Wang J., et al. Soyasaponin II protects against acute liver failure through diminishing YB-1 phosphorylation and Nlrp3-inflammasome priming in mice. Theranostics. 2020;10(6):2714–2726. doi: 10.7150/thno.40128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11(12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 26.Chen R., Liu L., Xiao M., Wang F., Lin X. Microarray expression profile analysis of long noncoding RNAs in premature brain injury: A novel point of view. Neuroscience. 2016;319:123–133. doi: 10.1016/j.neuroscience.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 27.Li L., Zhang Y., Luo H., Huang C., Li S., Liu A., et al. Systematic identification and analysis of expression profiles of mRNAs and IncRNAs in macrophage inflammatory response. Shock. 2019;51(6):770–779. doi: 10.1097/SHK.0000000000001181. [DOI] [PubMed] [Google Scholar]
- 28.Otasek D., Morris J.H., Boucas J., Pico A.R., Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh A, Shannon CP, Gautier B, Rohart F, Vacher M, Tebbutt SJ, et al. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 2019; 35(17):3055-3062. doi: 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed]
- 32.Yan Z, Luo H, Xie B, Tian T, Li S, Chen Z, et al. Targeting adaptor protein SLP76 of RAGE as a therapeutic approach for lethal sepsis. Nat Commun 2021; 12(1):308. doi: 10.1038/s41467-020-20577-3. [DOI] [PMC free article] [PubMed]
- 33.Haraszti R.A., Didiot M.-C., Sapp E., Leszyk J., Shaffer S.A., Rockwell H.E., et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J Extracell Vesicles. 2016;5(1):32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y., Zhang C., Xiang P., Shen J., Sun W., Yu H. Exosomes derived from HeLa cells break down vascular integrity by triggering endoplasmic reticulum stress in endothelial cells. J Extracell Vesicles. 2020;9(1):1722385. doi: 10.1080/20013078.2020.1722385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bader G.D., Hogue C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunkhorst F.M., Engel C., Bloos F., Meier-Hellmann A., Ragaller M., Weiler N., et al. Intensive insulin therapy and pentastarch resuscitation in severe sepsis. N Engl J Med. 2008;358(2):125–139. doi: 10.1056/NEJMoa070716. [DOI] [PubMed] [Google Scholar]
- 37.Investigators C.S., Annane D., Cariou A., Maxime V., Azoulay E., D'Honneur G., et al. Corticosteroid treatment and intensive insulin therapy for septic shock in adults: a randomized controlled trial. JAMA. 2010;303(4):341–348. doi: 10.1001/jama.2010.2. [DOI] [PubMed] [Google Scholar]
- 38.Zhou B., Liu J., Zeng L., Zhu S., Wang H., Billiar T.R., et al. Extracellular SQSTM1 mediates bacterial septic death in mice through insulin receptor signalling. Nat Microbiol. 2020;5(12):1576–1587. doi: 10.1038/s41564-020-00795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ilaiwy A., ten Have G.A.M., Bain J.R., Muehlbauer M.J., O'Neal S.K., Berthiaume J.M., et al. Identification of metabolic changes in ileum, jejunum, skeletal muscle, liver, and lung in a continuous I.V. pseudomonas aeruginosa model of sepsis using nontargeted metabolomics analysis. Am J Pathol. 2019;189(9):1797–1813. doi: 10.1016/j.ajpath.2019.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao J, Tarcea VG, Karnovsky A, Mirel BR, Weymouth TE, Beecher CW, et al. Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics 2010; 26(7):971-973. doi: 10.1093/bioinformatics/btq048. [DOI] [PMC free article] [PubMed]
- 41.Murao A, Brenner M, Aziz M, Wang P. Exosomes in sepsis. Front Immunol 2020; 11:2140. doi: 10.3389/fimmu.2020.02140. [DOI] [PMC free article] [PubMed]
- 42.Hashemian S.M., Pourhanifeh M.H., Fadaei S., Velayati A.A., Mirzaei H., Hamblin M.R. Non-coding RNAs and exosomes: their role in the pathogenesis of sepsis. Mol Ther Nucleic Acids. 2020;21:51–74. doi: 10.1016/j.omtn.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singla D.K., Johnson T.A., Tavakoli D.Z. Exosome treatment enhances anti-Inflammatory M2 macrophages and reduces inflammation-induced pyroptosis in doxorubicin-induced cardiomyopathy. Cells. 2019;8(10):1224. doi: 10.3390/cells8101224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Copaescu A., Smibert O., Gibson A., Phillips E.J., Trubiano J.A. The role of IL-6 and other mediators in the cytokine storm associated with SARS-CoV-2 infection. J Allergy Clin Immunol. 2020;146(3):518–534.e1. doi: 10.1016/j.jaci.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., et al. Synergism of TNF-alpha and IFN-gamma triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184(1):149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouyang W., O'Garra A. IL-10 family cytokines IL-10 and IL-22: from basic science to clinical translation. Immunity. 2019;50(4):871–891. doi: 10.1016/j.immuni.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 47.Jin Y., Liu R., Xie J., Xiong H., He J.C., Chen N. Interleukin-10 deficiency aggravates kidney inflammation and fibrosis in the unilateral ureteral obstruction mouse model. Lab Invest. 2013;93(7):801–811. doi: 10.1038/labinvest.2013.64. [DOI] [PubMed] [Google Scholar]
- 48.Tang T.-T., Wang B., Wu M., Li Z.-L., Feng Y.e., Cao J.-Y., et al. Extracellular vesicle-encapsulated IL-10 as novel nanotherapeutics against ischemic AKI. Sci Adv. 2020;6(33) doi: 10.1126/sciadv.aaz0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tkach M., Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 50.Mulder R., Croles F.N., Mulder A.B., Huntington J.A., Meijer K., Lukens M.V. SERPINC1 gene mutations in antithrombin deficiency. Br J Haematol. 2017;178(2):279–285. doi: 10.1111/bjh.14658. [DOI] [PubMed] [Google Scholar]
- 51.Pasi K.J., Rangarajan S., Georgiev P., Mant T., Creagh M.D., Lissitchkov T., et al. Targeting of antithrombin in hemophilia A or B with RNAi therapy. N Engl J Med. 2017;377(9):819–828. doi: 10.1056/NEJMoa1616569. [DOI] [PubMed] [Google Scholar]
- 52.Cugno M., Zanichelli A., Foieni F., Caccia S., Cicardi M. C1-inhibitor deficiency and angioedema: molecular mechanisms and clinical progress. Trends Mol Med. 2009;15(2):69–78. doi: 10.1016/j.molmed.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 53.Zuraw B.L. Hereditary angioedema. N Engl J Med. 2008;359(10):1027–1036. doi: 10.1056/NEJMcp0803977. [DOI] [PubMed] [Google Scholar]
- 54.Lee M.H., Vosburgh E., Anderson K., McDonagh J. Deficiency of plasma plasminogen activator inhibitor 1 results in hyperfibrinolytic bleeding. Blood. 1993;81(9):2357–2362. doi: 10.1182/blood.V81.9.2357.2357. [DOI] [PubMed] [Google Scholar]
- 55.Fay W.P., Parker A.C., Condrey L.R., Shapiro A.D. Human plasminogen activator inhibitor-1 (PAI-1) deficiency: characterization of a large kindred with a null mutation in the PAI-1 gene. Blood. 1997;90(1):204–208. doi: 10.1182/blood.V90.1.204. [DOI] [PubMed] [Google Scholar]
- 56.Jankun J., Aleem A.M., Selman S.H., Skrzypczak-Jankun E., Lysiak-Szydlowska W., Grafos N., et al. Highly stable plasminogen activator inhibitor type one (VLHL PAI-1) protects fibrin clots from tissue plasminogen activator-mediated fibrinolysis. Int J Mol Med. 2007;20(5):683–687. doi: 10.3892/ijmm.20.5.683. [DOI] [PubMed] [Google Scholar]
- 57.Holliday L.S., Faria L.P., Rody W.J., Jr. Actin and actin-associated proteins in extracellular vesicles shed by osteoclasts. Int J Mol Sci. 2019;21(1):158. doi: 10.3390/ijms21010158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Messner C.B., Demichev V., Wendisch D., Michalick L., White M., Freiwald A., et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11(1):11–24.e4. doi: 10.1016/j.cels.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee P.S., Waxman A.B., Cotich K.L., Chung S.W., Perrella M.A., Stossel T.P. Plasma gelsolin is a marker and therapeutic agent in animal sepsis. Crit Care Med. 2007;35(3):849–855. doi: 10.1097/01.CCM.0000253815.26311.24. [DOI] [PubMed] [Google Scholar]
- 60.Diakonov E.E., Selenina A.V., Tomilin A.N., Tsimokha A.S. Evidences against vesicle-dependent trafficking and involvement of extracellular proteasomes into cell-to-cell communications. Biochem Biophys Res Commun. 2019;508(2):368–373. doi: 10.1016/j.bbrc.2018.11.152. [DOI] [PubMed] [Google Scholar]
- 61.Srikanthan S., Li W., Silverstein R.L., McIntyre T.M. Exosome poly-ubiquitin inhibits platelet activation, downregulates CD36 and inhibits pro-atherothombotic cellular functions. J Thromb Haemost. 2014;12(11):1906–1917. doi: 10.1111/jth.12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lai R.C., Tan S.S., Teh B.J., Sze S.K., Arslan F., de Kleijn D.P., et al. Proteolytic potential of the MSC exosome proteome: implications for an exosome-mediated delivery of therapeutic proteasome. Int J Proteomics. 2012;2012:1–14. doi: 10.1155/2012/971907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jia X., Chen J., Megger D.A., Zhang X., Kozlowski M., Zhang L., et al. Label-free proteomic analysis of exosomes derived from inducible hepatitis B virus-replicating HepAD38 cell line. Mol Cell Proteomics. 2017;16(4):S144–S160. doi: 10.1074/mcp.M116.063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maggini S., Pierre A., Calder P.C. Immune function and micronutrient requirements change over the life course. Nutrients. 2018;10(10):1531. doi: 10.3390/nu10101531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gombart A.F., Pierre A., Maggini S. A review of micronutrients and the immune system-working in harmony to reduce the risk of infection. Nutrients. 2020;12(1):236. doi: 10.3390/nu12010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kashiouris M.G., L'Heureux M., Cable C.A., Fisher B.J., Leichtle S.W., Fowler A.A. The emerging role of vitamin C as a treatment for sepsis. Nutrients. 2020;12(2):292. doi: 10.3390/nu12020292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dong B., Zhou Y., Wang W., Scott J., Kim KangHo, Sun Z., et al. Vitamin D receptor activation in liver macrophages ameliorates hepatic inflammation, steatosis, and insulin resistance in mice. Hepatology. 2020;71(5):1559–1574. doi: 10.1002/hep.v71.510.1002/hep.30937. [DOI] [PubMed] [Google Scholar]
- 68.Miller A., Korem M., Almog R., Galboiz Y. Vitamin B12, demyelination, remyelination and repair in multiple sclerosis. J Neurol Sci. 2005;233(1–2):93–97. doi: 10.1016/j.jns.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 69.Doke S., Inagaki N., Hayakawa T., Tsuge H. Effects of vitamin B6 deficiency on cytokine levels and lymphocytes in mice. Biosci Biotechnol Biochem. 1998;62(5):1008–1010. doi: 10.1271/bbb.62.1008. [DOI] [PubMed] [Google Scholar]
- 70.Zheng H., Perreau J., Powell J.E., Han B., Zhang Z., Kwong W.K., et al. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc Natl Acad Sci U S A. 2019;116(51):25909–25916. doi: 10.1073/pnas.1916224116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao Z., Xia W., Zhang X., Yuan H., Guan D., Gao L. Hepatotoxicity of nutmeg: a pilot study based on metabolomics. Biomed Pharmacother. 2020;131:110780. doi: 10.1016/j.biopha.2020.110780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.