Abstract
The contagious SARS-CoV-2 has had a tremendous impact on the life and health of many communities. It was first rampant in early 2019 and so far, 539 million cases of COVID-19 have been reported worldwide. This is reminiscent of the 1918 influenza pandemic. However, we can detect the infected cases of COVID-19 by analysing either X-rays or CT, which are presumably considered the least expensive methods. In the existence of state-of-the-art convolutional neural networks (CNNs), which integrate image pre-processing techniques with fully connected layers, we can develop a sophisticated AI system contingent on various pre-trained models. Each pre-trained model we involved in our study assumed its role in extracting some specific features from different chest image datasets in many verified sources, such as (Mendeley, Kaggle, and GitHub). First, for CXR datasets associated with the CNN trained model from the beginning, whereby is comprised of four layers beginning with the Conv2D layer, which comprises 32 filters, followed by the MaxPooling and afterwards, we reiterated similarly. We used two techniques to avoid overgeneralization, the early stopping and the Dropout techniques. After all, the output was one neuron to classify both cases of 0 or 1, followed by a sigmoid function; in addition, we used the Adam optimizer owing to the more improved outcomes than what other optimizers conducted; ultimately, we referred to our findings by using a confusion matrix, classification report (Recall & Precision), sensitivity and specificity; in this approach, we achieved a classification accuracy of 96%. Our three integrated pre-trained models (VGG16, DenseNet201, and DenseNet121) yielded a remarkable test accuracy of 98.81%. Besides, our merged models (VGG16, DenseNet201) trained on CT images with the utmost effort; this model held an accurate test of 99.73% for binary classification with the (Normal/Covid-19) scenario. Comparing our results with related studies shows that our proposed models were superior to the previous CNN machine learning models in terms of various performance metrics. Our pre-trained model associated with the CT dataset achieved 100% of the F1score and the loss value was approximately 0.00268.
Keywords: CXR&CT chest COVID-19 images integration of three pre-trained CNN models Fine-tuning, Image processing, Performance evaluation, Artificial intelligence
Abbreviations: X-ray,CXR, energic high frequency electromagnetic radiation; CNNs, Convolutional Neural Networks; CT, Computed Tomography; Covid-19, Coronavirus disease of 2019; RT-PCR, Reverse Transcription Polymerase Chain Reaction; SARS_COV_2, Severe acute respiratory syndrome coronavirus; AI, Artificial Intelligence; DL, Deep Learning; ML, Machine Learning; ANNs, Artificilal Neural Networks; ReLU, Rectified Linear Unit; Conv2D, 2D Convolutional Layer
1. Introduction
COVID-19 first dispersed in China and later expanded to other communities [1].
We are confronting the most serious global public health emergency since the 1918 influenza pandemic. Shortly afterwards the primary causes of this transcendental respiratory viral ailment began in Wuhan and other states of China in late December 2019, COVID-19 hurriedly spread around the world, compelling the World Health Organization to announce this disease as a broad disease on March 11, 2020. From that moment on, SARS-CoV-2 has desolated multiple countries and overpowered countless healthcare frameworks.
Despite significant advances in the clinical examination that have recognized SARS-CoV-2, constraining the proceeding spread of this infection and its variations is still an issue of concern. As COVID-19 proceeds, the world is not as safe as it once was and the threat of global annihilation has become genuine.
Similar to other RNA infections, COVID-19 acclimatizes to its modern human hosts and is inclined to hereditary growth with improved transformation over time that occurs in mutant variations [2].
Universally, at 17:13 Central European Time and March 2022, the WHO has meticulously reported 445,096,612 confirmed cases of COVID-19 and 5,998,301 deaths. As of March 5, 2022, an add-up to 10,704,043,684 antibody measurements had been performed [3]. Coronavirus infection has influenced lung health since its inception.
For this exceptional reason, specialists are monitoring patients’ oxygen levels with an oximeter; therefore, they might immediately recognise any abnormalities [4].
However, PCR is not an adequate test in consequence of cases that have been rocketing. Correspondingly, a reliable test is necessary to perceive infected individuals immediately during isolation or segregation. The radiological assessment encompasses either CXR or computed tomography (CT) images. Radiological deviations concerning the standard have been determined [5].
CXRs are a great observation of COVID-19 lung symptoms, and their viable structural evaluations provide a precise strategy for anticipating the seriousness of lung illnesses [6].
CT plays a major part in identifying and combating COVID-19 infection, which is not a common symptom of COVID-19 infection in the lower lobes, but an inconsistent area of ground glass invasion. As the disease progresses, other signs become apparent, such as dormancy, air bronchogram, cell lung disease and air bubbles develop amid the course of the illness. Segment chest CT offers great sensitivity, but low specificity in determining COVID-19 [7].
We organised our paper according to the following steps: Section 2 outlines the datasets used in this study, Section 3 describes the methods used, Section 4 demonstrates the outcomes, and then the visualization is involved in Section 5.
In conclusion, section 6 summarizes the paper.
Considerable discussions have been carried out on the identification of chest illnesses using Deep Neural Network techniques. The most common approach is transfer learning. Many authors have associated CNN with various CXR or CT images to extract attributes from diverse layers of images [8]. However, the disease is still arduous to diagnose because of the lack of big data for training in neural networks. Data quality and quantity are important in developing the Covid-19 detection system. Most SARS-CoV-2 diagnostic studies are conducted based on CT or chest x-rays, as opposed to our study we discussed both. Another essential point is that most of them gained an average accuracy of 90%. We have striven to include the best scientific research with over 90% accuracy.
Related work done on CXR and CT chest images:
Following a review of several previous articles, we have included several previous studies in this article. We applied the Transfer Learning Model (CNN) to improve lung COVID-19 diagnosis based on the overall accuracy of the test and the minimal validation loss achieved while avoiding generalization. Below is a list of assorted previous studies in terms of the identification of SARS-CoV-2 using various machine-learning algorithms.
1.1. CXR research
Dongsheng et al. used in their experimental framework five transfer-learning models entitled “Fusion Model”: ResNet152, DenseNet201, VGG19, Xception, InceptionResNetV2 CNN to take out attribute information from SARS-CoV-2 X-ray. The authors replaced the FC layer of each network with a “Global average-pooling” layer that effectively dwindled the parameters of the training process. They assigned the Dropout to 0.5 [9].
Akter et al. suggested a modified MobileNetV2 employing the RMSProp optimizer, offering two strides instead of one for the residual layers. They set the image size to 224 × 224 pixels. However, they proposed other pre-trained CNN models and their modified model achieved the highest accuracy [10].
Hayat et al. employed CNN algorithms to construct the proposed neural network model and other transfer-learning models, getting maximum accuracy of 96% in VGGNET [11].
Roy et al. present two CNN models: VGG16 and MobileNet-V2, each with two types of optimizers: Adam and RMSProp. They conducted their results using two types of optimizers and two types of deep learning models. Even if the database is small, insufficient, and from a non-standard source [12].
SVM is a supervised machine learning algorithm for both regression and logistic regression problems. However, in many regards, it is challenging to harness this approach. The most paramount of which is the sufferance in using a good kernel; configuration and tuning of hyper-parameters. It is also difficult to explicate the final model.
It can be a very long-lasting training with big data, Panigrahi et al. resized the images according to the following dimensions (1024, 1024), which surely increases the training time and heavy load on the device [50]. Finally, the CNN algorithms are user-friendly and accurate way; therefore, we will get over 98%. Dissimilar to what the authors mentioned, in our paper, when amalgamating these networks (VGG16, DenseNet201, and DenseNet169) associated with the CT dataset [14]. Even though we attained 100% accuracy, we did not document it because of the inadequacy of acceptance of this experimental value; thereupon, we did not use a third network and had only two networks to leave a margin of error.
CNN has proven to surpass SVM in this subject area.
1.2. CT research
J.prabhu et al. used a dataset comprising 3873 scans and several models of deep learning they applied to this dataset for COVID-19 detection, including (MobileNet, NasNet, Xception, and EfficientNet).
Even though we implemented these models in our study, we excluded them from the findings because of misclassification [37].
While Shah et al. applied several deep learning models. The number of filters harnessed in the proposed model was large, which used 126 filters per layer (Conv2D), so their model named (CTnet-10) led up to negative results in accuracy [38].
Chouat et al. examined the effect of the training epoch number on the CT images. They had better take advantage of the early stopping technique. Trying several iterations is redundant, so this regularization technique provides an enormous advantage by halting the training as soon as the model starts Overfitting [40].
In the end, Islam et al. got the lowermost accuracy among the previous studies mentioned above using LenNet-5 in the final study where one of the neural network models was used, neglecting the effect of many parameters on the classification accuracy, so the classification result was sadly low, and the study was not sufficient at all [42].
RESEARCH MOTIVATION:
This study develops an integrated transfer-learning model to reduce misdiagnosis and delayed diagnosis with SARS-CoV-2 cases all over the extraction analysis process of attributes. Relying on the two available types of images available on the diverse certified websites.
Therefore, the emphasis was placed on two types of images, either X-radiation or CT.
We considered many available datasets in this paper to appeal to future researchers. We compared our study to the related studies quoted from well-known and certified websites to provide information about the main methods, techniques, most of the datasets, and CNN models used in identifying the disease. This facilitates the information retrieval process from this comprehensive paper for researchers.
We proved the best performance of each optimizer, neural network and hyper-parameters tuning in this paper for maximum classification accuracy.
2. Database
We fetched various sample images from different sources to verify the appropriateness of the proposed models. Classification scenarios are COVID-19/Normal 0/1 classes labelled for each, respectively. We did not clean any of the images from the dataset; we used all image formats within CXR datasets, including (PNG, JPEG, and JPG) formats. Considering our CT dataset format In contrast to all X-ray datasets, images were in the PNG format. Our datasets have already been augmented, so there was no need for augmentation techniques to avoid Overfitting. We have split in consonance with the two scenarios:
2.1. In the case of building the model from the beginning
We allocated a 0.5:0:5 ratio of the CXR dataset for training and test data, respectively. We also allocated 20% of the training data for the validation ratio. The ratio of the test set split was unreasonable because of a lack of hardware resources in the device used. The machine could not handle as many images in RAM. We used OpenCV to read and write images out of memory rather than files. The Tensor-flow GPU was not compatible with our machine. In this approach, we adjusted the dimensions of the images as follows (500, 500, 1).
2.2. In the case of hiring transfer-learning systems
We shared the dataset between the training, validation, and test sets, the training set is 70% of our datasets, the validation set is 20% of the 70% training set, and the test set is 30% of the dataset.
We resized the dimensions of the images in the transfer learning approach as follows (224, 224, 3).
Our dataset included 3050 images of X-rays divided into two folders; the first class has 1525 X-ray images of SARS-CoV-2 infected and the second one is composed of 1525 CXR images of healthy people. This dataset is available at Mendeley and is collected from the Kaggle repository and NIH dataset [13]. The second dataset comprises 865 images of COVID-19 cases and the average age of patients is approximately 50 years [46,49]. We combined these two datasets to create a larger combined dataset of 3915 images. The third dataset used holds two folders: COVID and healthy people of CT chest images with 1252 and 1230 images, respectively, which were gathered d in hospitals in São Paulo, Brazil [14]. The optimal ratio for data splitting for the training and test sets respectively in the pre-trained models’ approach is 70%:30%. We collected multiple datasets from various websites. (Mendeley, Kaggle, GitHub, and arXiv); therefore, we will include all of them in the reference section for the benefit of the researchers [[44], [45], [47], [48]].
3. Methodology
We illustrate the flow process of the research methodology used in this paper in Fig. 1 and Fig. 2 . We achieved our study in the following steps:
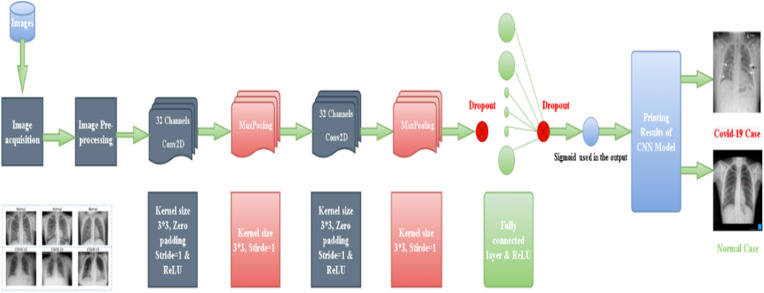
Fig. 1.
The architecture of the proposed-built-model-from-beginning.
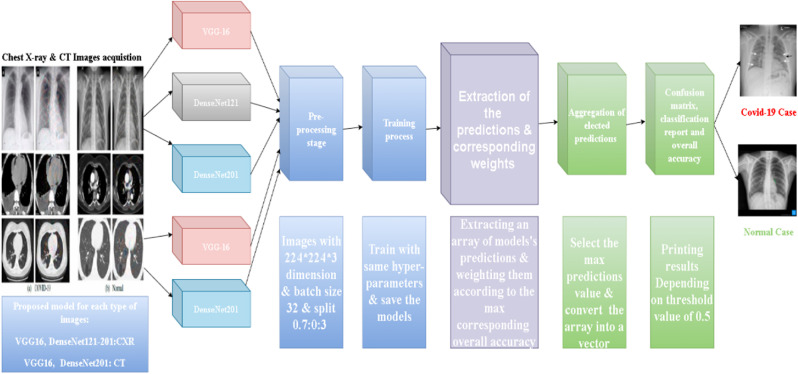
Fig. 2.
The architecture of the integrated transfer-learning models.
3.1. Image acquisition
We proposed two scenarios for CNN models:
3.1.1. Building the model from the beginning
In the image acquisition stage, we used the OpenCV library to read images with the help of the Pickle library. The input format for this architecture was 500 × 500 and there was one channel for grayscale images. We reshaped the data into (−1, 500, 500, 1).
3.1.2. Transfer learning model
These architectures accommodate CXR and CT chest images. The tensor is (224 × 224 × 3) three refers to RGB channels, and we invoked Keras preprocessing image for image processing. We set the class mode to binary mode. We also invoked the Pandas library to get the path and label using a concatenating function. In both cases, the set follows:
Labels 0 and 1 respectively symbolize COVID-19 and normal cases. The validation split ratio was 0.2. We normalized the training and test data to decimal values between (0 and 1). This scale is convenient for the values of the neural network whose output value fluctuates between zeros and ones, the array of pixel values passes through several multiplications between some activation functions and weights. We adjusted the batch size to 32.
We shuffled the data to avoid getting the same result in each training epoch. Not only that, but we performed mathematical operations easily, where vectorization and broadcasting are two executable essential concepts.
3.2. CNN system (built from the beginning)
CNNs are good at recognizing important properties without human surveillance. They are modelled after the structure of neurons in the brains of human beings. Each neuron in a layer in an FC neural system connects with all neurons in the corresponding layers, but CNN suggests that only the local patch of neurons is linked (Sparse Connectivity) [29].
One of its most crucial applications is a medical diagnosis [15].
These networks are effective methods that limit the processing of only those pixels within a region of interest, prioritizing them over other pixels. This forbids the excessive load on the neural network.
3.2.1. Conv2D layer
In our model, we employed 32 filters in each Conv2d layer. These filters apply masks to detect edges, which are the most vital characteristics of an image. The kernel size was a 2D matrix (3 × 3), but both the padding and stride were the same as the default. The result of our convolutional height and width is given as Eq. (1) [16].
3.2.2. Rectified linear unit (ReLU) layer
The hidden layer usually uses a ReLU activation function. All negative inputs are set to zero, which can be calculated from the equation Eq. (2) [29].
Some arithmetic operations have not affected neural networks when using a sigmoid activation function that uses exponential functions; as a result, it slows down the learning process.
3.2.3. Max-Pooling
Often, there are adaptations of lower-definition input functions that involve large or significant structural components, without small details of no value to the process [16]. In our model, the output of the Max-Pooling was 249, and the kernel size was (2 × 2).
Edges are distinguished by large integer values of pixel intensity in the image, so we use this technique to select the maximum values of pixels.
3.2.4. Dropout
Srivastava et al. proposed this strategy for handling a great issue. The main idea is to discard units arbitrarily (along with associations) from the neural network during training [17].
Thrown neurons are not part of the back or forward propagation. Conversely, neurons are used de novo to make anticipations in the test process [29].
This technology is in a way similar to how employees or workers usually find a suitable balance between work and daily living. Giving them a few days off might reduce their stress and anxiety. We set the Dropout value to 0.2 in our model.
3.2.5. Flatten
It maps the output of a matrix-shaped CNN to the input of a vector-shaped network that could be performed by flattening the data used.
3.2.6. Fully connected layer
Implementing a variety of image processing techniques, including edge detection, morphological processes, and other image processing techniques included in convolutional networks is so efficient; anyway, the role of traditional neural networks is to decide on the classification and segmentation of samples.
There are two types of identification in our study: zero if it meets the criteria for COVID-19 and one if it does not. We simply set the threshold value to 0.5. The final layer in our model comprised just one neuron that could classify the COVID-19/Normal scenario by using the sigmoid function in the output layer. We denote the sigmoid function as Eq. (3) .
We illustrated the architecture of our model built from the beginning in Fig. 2.
3.3. CNN system (transfer learning)
This system enhances the visualization of the target learner's basic domain through knowledge transfer [18].
Transfer learning is a machine learning method that uses a pre-trained system to learn new data. Transfer learning introduces a new database called “ImageNet” [19].
A variety of new techniques has been developed especially transfer-learning Networks [20].
ImageNet's defiance was launched in 2010. It can divide 100,000 experimental images into 1000 distinctive categories [21].
Transfer learning is a method of training a network on a particular dataset and then sending the learned properties (ImageNet's weights) to another network to train on other data [22].
In other respects, implementing transfer learning networks is very efficient, as well as fine-tuning the weights and the number of filters used. We also considered other hyper-parameters, such as compatibility with the number of inputs and outputs, the optimizer used, and where to stop training.
3.3.1. Neural networks with deep depth
Simyon and Zisserman developed the VDNN system at Oxford College. The number of parameters for a function is between 138 million and 144 million [23]. The architecture of the model includes a traditional network of thousands of neurons. In our work, we replaced thousands of neurons in the last layer with two neurons for binary classification. The problem we confront is not multiclass classification, so we do not require many neurons to classify the output; as a result, we replaced the Softmax function of the last layer with the sigmoid function.
3.3.1.1. VGG16
There are 13 Conv2D blocks and three completely associated layers; the total number of parameters is 138.4 M. This is a highly effective classification network in data science. This system could classify and separate the samples used in our study efficiently. The bottom accuracy achieved by this network was 96% at its first epoch. We attained an optimal accuracy of 100% at the last iteration.
3.3.1.2. VGG19
This network can classify samples satisfactorily, but merging this network with other networks has no significant progress in the classification process; the parameter count for this function is 143.7 million.
3.3.2. DenseNet
DenseNet-BC design with 4 dense layers and 224,224 image input size [24]. The ReLU used helps speed up learning by avoiding the zero-gradient issue. These networks are good at extracting specific features, and we specially segmented our samples. Each network extracted a few features from the images until accomplishing the highest possible accuracy of classification decisions.
3.3.2.1. DenseNet121
It follows the same structure as DenseNet at the transition layer and has [6,12,16,24] layers in 4 Dense blocks for each 1 × 1, 3 × 3 convolutional.
3.3.2.2. DenseNet201
This is also a model of Densely Connected CNN, so it follows the same structure as the DenseNet initially and in the transition layers. Its architecture has [6, 12, 64, 48] layers in the four dense blocks for each 1 × 1, 3 × 3 convolutional.
3.4. Hyper-parameters tuning
We tuned the hyper-parameters as specified in the next steps:
3.4.1. Fine-tuning for our proposed model (built from the beginning)
The model included many filters (Max or average pooling), and nonlinear activation layers. This section describes the general architectural design.
First, the model is composed of two Conv2D filters, followed by two activation functions of the Relu type and two Max-pooling layers. After that, we added a Dropout layer with a value of 0.2. In addition, we added a Flatten layer to get the input as it is, supported by a dense block of 32 neurons. In the next step, we included the Relu and once more; we involved a new additional Dropout layer after that and assigned it to the same previous multiplier of 0.2. To sum up, the output is a neuron to classify both cases of 0 or 1, ending up by using a sigmoid function.
3.4.2. Fine-tuning for (transfer learning models)
In our study, we inserted a 224x224 input image and the various model filters kept their original weights. We eliminated the last layer of each transfer-learning model and replaced it with one dense layer.
It is not appropriate for this study, which holds two categories, to classify them. We allocated the parameter weights provided by ImageNet Dataset. We have not trained the current layers of transfer-learning models. These layers are already trained and have pre-determined weights. At last, we need to flatten the resultant 2-Dimensional arrays for the ANN and reshape them into a one-dimensional. We used a dense block with one neuron and replaced the Softmax activation function with a sigmoid function. The veracious pre-trained models used in our experiments are (VGG16, DenseNet201, DenseNet169, and DensNet121).
3.4.3. Fine-tuning and integration of multiple models for accurate predictions
This step extracts a set of values from the model predictions. For each element in the array.
We multiplied a weight range from 0 to 1 by that element's value.
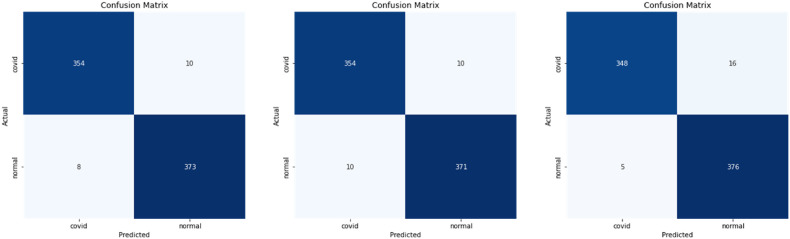
In the next step, we recollected the predictions of the models based on the largest value achieved from the predictions of our proposed models. Each network plays its role in extracting some specific features, for instance, Vgg16 misdiagnosed 18 cases: 10 false positives and 8 false negatives, so it was good at diagnosing both cases while DenseNet169 misdiagnosed 21 cases: 16 false positives and 5 false negatives. Even though it achieved a lower accuracy, it gets a better diagnosis for Normal cases and eventually combining the two approaches (DenseNet, and VGGNet) we got the perfect outcomes.
3.5. Model compile
It is a crucial function for the execution of the neural network processes [25]. After defining the concepts of the model, it is important to compile it by specifying some hyper-parameters [26]. Adam outperforms stochastic gradient descent (SGD) and RMSProp in terms of outcomes in quality improvement [27]. Most often, the approach between those antecedent enhancers was similar, but since the Adam optimization algorithm surpassed other optimizers, we conducted the findings considering this optimizer [28].
The cost function is scheduled in the last layer to calculate the expected loss during training in the CNN system.
This error discriminates between the true class and the expected one. The cross-entropy algorithm outputs the probability of an outcome: p∈ {0, 1} [29].
Since we are facing a binary classification problem, the enhancer that we provided “Adam” and the cost function “Binary cross-entropy”.
These workable selections of hyper-parameters have led to better classification findings than other ones.
3.6. Early stopping
It is a commonly used regularization technique to dispose of generalization during training (CNNs) [30].
We need to avoid the reckless choices in adjusting the number of iterations. To start by defining the first issue, if setting an insufficient number to iterate the training over our images in the dataset, we will encounter inadequate training, which is known as the “Under-fitting” concept. The second issue, choosing a huge number of iterations, leads to the generalization issue, so we would like to perform the right number of training iterations to yield the best results in the validation process.
Applying this technique to the transfer learning system is beneficial to check the performance of the validation loss at the end of each training iteration and diminish it as low as possible, whereas the validation accuracy values should be as high as possible.
We customized hyper-parameters for this function, so if the accuracy does not improve, the technique will accordingly halt the training process. These are determined based on specific criteria, such as the size of the data in each batch or the learning rate.
We specified the value of patience as five; subsequently, the learning rate was set to a lower value (ReduceLROnPlateau). In case, no enhancement is seen and the validation accuracy fluctuates; in parallel, we determined the patience parameter by the value of 3 and the number of epochs by 100, expecting in most cases that the training will not exceed almost 25 epochs.
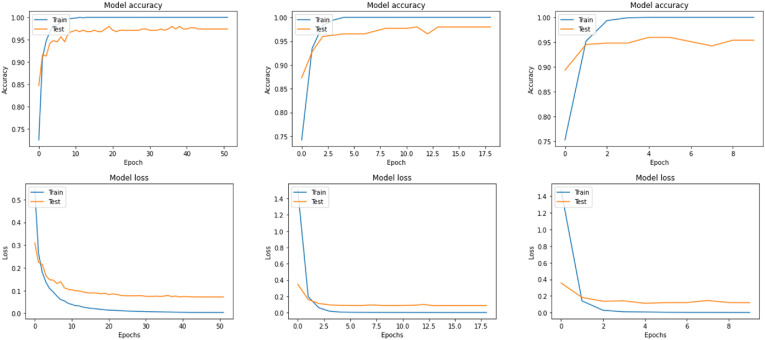
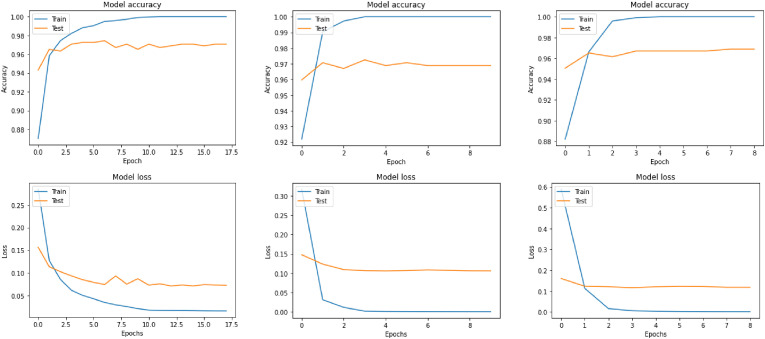
In this wise, after we had tuned all the previous hyper-parameters, we fitted our models. The following graphs, Fig. 3 and Fig. 4 , best show the training and test processes.
Fig. 3.
Accuracy, and Loss of VGG16, DenseNet201, and DenseNet169 respectively for CT [14].
Fig. 4.
Accuracy, and Loss of VGG16, DensNet201, and DensNet121 respectively for data in [13] + [46].
4. Results and discussion
In the analysis of results using measurement tools, precision, recall, and other metrics [31]. The assessment of high accuracy is recommended.
We used four concepts to define this criterion:
True Positives: This represents cases that have been infected by the Coronavirus and have been diagnosed as infected.
True Negatives: This represents cases that have not been infected by the Coronavirus and have been diagnosed as not infected.
False Positives: This represents cases that have been infected by the Coronavirus and have been diagnosed as not infected.
False Negatives: This represents cases that have been not infected by the Coronavirus and have been diagnosed as infected [32].
Finally, the results should be presented academically by researchers and those involved in a particular field.
4.1. Model evaluation
The score measurements included in the DL assignment play a crucial role in achieving improved identifiers. Making use of it to estimate the performance of the model.
4.1.1. Accuracy
This is a quantitative relationship between the number of correct diagnoses and the number of correct and incorrect diagnoses. It is the standard used to show the effectiveness and accuracy of the models and algorithms used. Eq. (4) .We proposed ensemble transfer deep learning (VGG16, DenseNet201, and DenseNet121) CNN models, and they scored noteworthy accuracy of 98.21, 97.78, and 97.95, respectively, with the maximum-elected value of their predictions, we obtained the overall result of 98.81% test accuracy. For CT chest images, the accuracy was 99.73% when combining the predictions of (VGG16, DensNet201). Another approach used, which is building-model-from-the-beginning, yielded an accuracy of 96%.
We included all results of our study in Table .1 and Table .2 .
Table 1.
Classification report of our models depending on training two datasets.
| Model | Image status | F1-score | Precision | Recall | Support | Overall accuracy | Test loss | Dataset |
|---|---|---|---|---|---|---|---|---|
| Integrated model1 | COVID | 1.00 | 1.00 | 0.99 | 364 | 99.73 | 0.00268 | [14] |
| Normal | 1.00 | 0.99 | 1.00 | 381 | ||||
| Integrated model2 | COVID | 0.99 | 1.00 | 0.98 | 698 | 98.81 | 0.01194 | [13] + [46] |
| Normal | 0.99 | 0.98 | 0.98 | 474 | ||||
| DensNet169 | COVID | 0.97 | 0.99 | 0.96 | 364 | 97.18 | 0.09095 | [14] |
| Normal | 0.97 | 0.96 | 0.96 | 381 | ||||
| VGG16 | COVID | 0.98 | 0.99 | 0.98 | 698 | 98.21 | 0.04855 | [13] + [46] |
| Normal | 0.98 | 0.97 | 0.98 | 474 | ||||
| COVID | 0.98 | 0.98 | 0.97 | 364 | 97.58 | 0.06493 | [14] | |
| Normal | 0.98 | 0.97 | 0.98 | 381 | ||||
| DenseNet201 | COVID | 0.98 | 0.98 | 0.98 | 698 | 97.78 | 0.05813 | [13] + [46] |
| Normal | 0.97 | 0.97 | 0.98 | 474 | ||||
| COVID | 0.97 | 0.97 | 0.97 | 364 | 97.32 | 0.07250 | [14] | |
| Normal | 0.97 | 0.97 | 0.97 | 381 | ||||
| VGG19 | COVID | 0.98 | 0.98 | 0.98 | 698 | 97.53 | 0.06570 | [13] + [46] |
| Normal | 0.97 | 0.97 | 0.97 | 474 | ||||
| COVID | 0.96 | 0.96 | 0.96 | 364 | 96.38 | 0.09129 | [14] | |
| Normal | 0.96 | 0.97 | 0.96 | 381 | ||||
| DenseNet121 | COVID | 0.98 | 0.99 | 0.98 | 698 | 97.95 | 0.06453 | [13] + [46] |
| Normal | 0.97 | 0.97 | 0.98 | 474 | ||||
| COVID | 0.96 | 0.97 | 0.95 | 364 | 95.97 | 0.10428 | [14] | |
| Normal | 0.96 | 0.95 | 0.97 | 381 |
Table 2.
Classification report of our models compared to other studies.
| Author | Architecture | Image status | F1-score | Precision | Recall | Overall accuracy | Dataset |
|---|---|---|---|---|---|---|---|
| This Study | Integrated model 1 | COVID | 1.00 | 1.00 | 0.99 | 99.73 | [14] |
| Normal | 1.00 | 0.99 | 1.00 | ||||
| [37] J.Prabhu | VGG16 | COVID | 0.98 | 1.00 | 0.96 | 97.68 | Kaggle 3873 |
| Normal | |||||||
| [38] Shah et al. | VGG19 | COVID | – | – | – | 94.52 | [39] |
| Normal | – | – | – | ||||
| [40] Chouat | VGGNet-19 | COVID | 86.5 | 88.5 | 86 | 87 | [41] |
| Normal | |||||||
| [42] Islam et al. | LeNet-5 | – | 87 | 85 | 89 | 86.06 | [43] |
| This Study | Integrated model 2 | COVID | 0.99 | 1.00 | 0.98 | 98.81 | [ [13]+ [46]] |
| Normal | 0.99 | 0.98 | 0.99 | ||||
| [10] Akter et al. | MobileNetV2 | COVID | 97 | 97 | 98 | 98 | [36] |
| Normal | |||||||
| [9] Dongsheng | Fusion model | COVID | 95.50 | 96.10 | 96.42 | 96 | Kaggle |
| Normal | |||||||
| [11] Hayat et al. | VGG16 | COVID | 0.94 | 0.93 | 0.95 | 0.95 | [34] |
| Normal | 0.98 | 0.98 | 0.98 | ||||
| [12] Roy et al. | MobileNetV2 | COVID | 0.94 | 0.99 | 0.90 | 0.93 | [35] |
| Normal | 0.91 | 0.85 | 0.98 |
4.2. Classification report
This report describes the important standards of measurement, such as sensitivity, precision or F1score.
4.2.1. Sensitivity
Used to count the proportion of infected cases that were predicted infected Eq. (5) .
4.2.2. Precision
It counts the proportion of infected cases divided by the total number of infected and uninfected cases that were accurately predicted by the model.Eq. (6)
4.2.3. F1-score
It calculates the measured ratio between the previous two concepts. Eq. (7) .
4.2.4. Specificity
Used to calculate the share of accurately identified negative designs. Eq. (8) [29].
We entitle our integrated model:
-
1.
Integrated model1:VGG16+DenseNet201
-
2.
Integrated model2:VGG16+DenseNet201+DensNet121.
Then, by implementing a radiograph or CT image, using various metrics, such as confusion matrix and then comparing our results with those from previous studies.
4.3. Confusion metric
This matrix usually summarizes the classification process using datasets. [33]; it lists the number of infected and healthy cases and shows us the failure of the model in each case of the diagnosis, considering an implicit comparison between the above four cases with a simple 2-by-2 matrix.
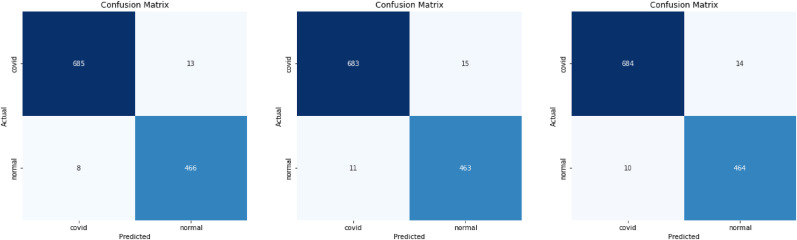
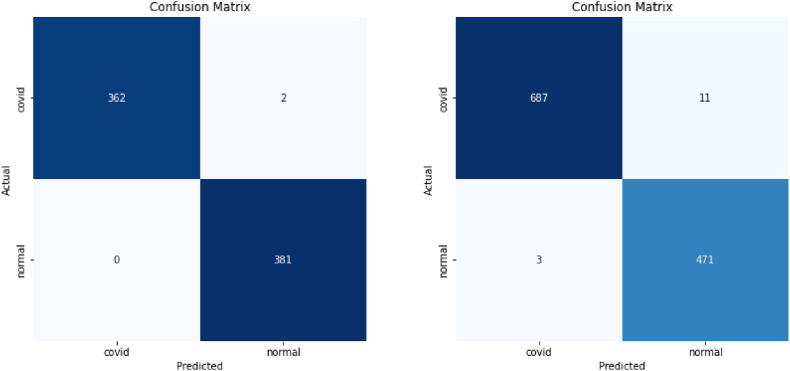
We show these matrices of our models in Fig. 6 , Fig. 7 , and Fig. 8 .
Fig. 6.
Confusion matrices VGG16, DenseNet201, DensNet169 respectively for CT data in [14].
Fig. 7.
Confusion matrices VGG16, DenseNet201, and DenseNet121 respectively for CT data in [13] + [46].
Fig. 8.
Confusion matrices integrated model 1, and integrated model 2for data in Ref. [14], [13] + [46] respectively.
Coronavirus disease usually develops within 8–10 days; the relevant feature of the disease is shown by the increase in the size of pathological changes on the x-ray images and their intensity compared to the normal images, besides the signs of emphysema.
The fifth section below visualizes the detection mechanism of the used neural networks for CXRs.
5. Visualization
This section covers the steps involved in applying image processing to X-ray images. In our study, we use 32 filters to process images that pass these filters during training, including morphological processes, denoising, brightness change, background deletion, and removal of objects that are considered noise and unimportant until the filters process the region of interest.
Identification of the boundaries and significant differences in the alveoli of the lungs between the infected and a healthy person.
We show the Filters used in Fig. 5 corresponding to the analysis process of images at each stage. As is apparent that the images become much darker.
Fig. 5.
Shows a depiction of the image processing steps carried out by the neural network.
6. Conclusion
Disclosure of SARS-CoV-2 in its early spread is an effective accomplishment that has helped limit the prevalence of SARS-CoV-2 infection in communities. In this paper, we present an integrated transfer-learning model based on three systems: VGG16, DenseNet201, and DenseNet121 for the discovery of CXR images. We integrated another model, which comprises two systems: VGG16, and DenseNet201 for the discovery of CT images. We set up our model built from zero entitled “model built from the beginning” to classify the chest images into two categories of COVID-19/Normal.
First, we had integrated Transfer-learning systems of the type (VGGNet, and DenseNet) and afterwards, we associated them with standard selected data from various verified open sources and secure online websites such as Mendeley, Kaggle, GitHub, and arXiv regardless of whether CT or CXR. Second, we considered the fine-tuning of the hyper-parameters.
Having set these parameters, Adam optimization algorithm and both (early stopping, and Dropout) techniques. In addition, setting the “ReLU” activation function as the default in the hidden layers and replacing the “Softmax” activation function in the output with the “sigmoid".
These parameters, techniques, and standard datasets which we selected alongside show the best performance in the science of data classification. We achieved the overall accuracy and F1score of 99.73% and 100% respectively by applying networks to CT images in PNG format. In addition, we also diagnosed most images correctly, and we conducted the study simultaneously on two platforms, Google Colab and Spyder.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Thesis Acknowledgement
Foremost, I would like to express my sincere gratitude to my advisor Dr Muhammad Mazen Almostafa for the continuous support of my M.D study and research, and his patience, motivation, enthusiasm, and immense knowledge. His guidance helped me in all the time of research and writing of this thesis. I could not have imagined having a better supervisor for my M.D study. I thank my entire fellow member for the stimulating discussions, for the sleepless nights we were working together before deadlines, and for all the fun we have had in the last four years. . Also, I would like to thank my family: my parents Abdulsalam Hamwi and Hasnaa Tarrab, for giving birth to me in the first place and supporting me spiritually throughout my life, as well as my sister I would like to thank her.
References
- 1.Wang W., Xu Y., Gao R., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cascella M., Rajnik M., Aleem A., Dulebohn S.C., Di Napoli R. StatPearls [internet]. Treasure island (FL) StatPearls Publishing; 2022 Jan. Features, evaluation, and treatment of coronavirus (COVID-19). 2022 feb 5. –. PMID: 32150360. [PubMed] [Google Scholar]
- 3.WHO Coronavirus (COVID-19) Dashboard. WHO 2022. https://covid19.who.int/
- 4.Sharma K. Here's what doctors want you to know—times of India, the times of India. 2021. Coronavirus: distressed breathing, lung involvement in COVID? Bombay, India. [Google Scholar]
- 5.Guan Wj, Ni Zy, Hu Y., Liang Wh, Ou Cq, He Jx, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasin R., Gouda W. Chest X-ray findings monitoring COVID-19 disease course and severity. Egypt J Radiol Nucl Med. 2020;51:193. doi: 10.1186/s43055-020-00296-x. [DOI] [Google Scholar]
- 7.Hefeda M.M. CT chest findings in patients infected with COVID-19: areview of literature. Egypt J Radiol Nucl Med. 2020;51:239. doi: 10.1186/s43055-020-00355-3. [DOI] [Google Scholar]
- 8.Rahib H. Deep convolutional neural networks for chest diseases detection. J. Healthc. Eng. 2018;4168538(1 Aug) doi: 10.1155/2018/4168538. Published online 2018 Aug 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji Dongsheng, Zhang Zhujun, Zhao Yanzhong, Zhao Qianchuan. Research on classification of COVID-19 chest X-ray image modal feature fusion based on deep learning. J. Healthc. Eng. 2021;2021 doi: 10.1155/2021/6799202. Article ID 6799202, 12 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akter S., Shamrat F.M.J.M., Chakraborty S., Karim A., Azam S. 13 Nov 2021. COVID-19 detection using deep learning algorithm on chest X-ray images published. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.H. O. Alasasfeh, T. Alomari and M. Ibbini, "Deep Learning Approach for COVID-19 Detection Based on X- Ray Images," 2021 18th International Multi-Conference on Systems, Signals & Devices (SSD), 2021, pp. 1-6, doi: 10.1109/SSD52085.2021.9429383.
- 12.D.Roy Competitive deep learning methods for COVID-19 detection using X-ray images journal of institution of engineers (india) Published online : 28 April 2021.
- 13.Asraf Amanullah, Islam Zabirul. Mendeley Data; 2021. COVID19, pneumonia and normal chest X-ray PA dataset”. V1. [DOI] [Google Scholar]
- 14.PlamenEduardo E.D. 2020. https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset SARS-COV-2 ct-scan dataset. [Online]
- 15.Mane D.T., Kulkarni U.V. A survey on supervised convolutional neural network and its major applications. Int. J. Rough. Sets. Data. Anal. 2017;4(3):71–82. [Google Scholar]
- 16.Brownlee J. Machine Learning Mastery; 2021. A gentle introduction to pooling layers for convolutional neural networks. [Google Scholar]
- 17.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from Overfitting. J Mach Learn Res. 2014;15(1):1929–1958. [Google Scholar]
- 18.Zhuang F., et al. A comprehensive survey on transfer learning. Proc IEEE. Jan. 2021;109(1):43–76. doi: 10.1109/JPROC.2020.3004555. [DOI] [Google Scholar]
- 19.Deng J., Dong W., Socher R., Li L.-J., Li Kai, Fei-Fei Li. IEEE conference on computer vision and pattern recognition. 2009. ImageNet: a large-scale hierarchical image database; pp. 248–255. 2009. [DOI] [Google Scholar]
- 20.Ebrahim M., Al-Ayyoub M., Alsmirat M.A. 10th international conference on information and communication systems. ICICS); 2019. Will transfer learning enhance ImageNet classification accuracy using ImageNet-pretrained models? pp. 211–216. 2019. [DOI] [Google Scholar]
- 21.Benbrahim H., Behloul A. International conference on artificial intelligence for cyber security systems and privacy. AI-CSP); 2021. Fine-tuned xception for image classification on tiny ImageNet; pp. 1–4. 2021. [DOI] [Google Scholar]
- 22.Ismail Fawaz H., Forestier G., Weber J., Idoumghar L., Muller P.-A. IEEE International Conference on Big Data (Big Data); 2018. Transfer learning for time series classification; pp. 1367–1376. 2018. [DOI] [Google Scholar]
- 23.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556. 2014..
- 24.Huang G., Liu Z., van, Weinberger Kilian Q. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2017): 2017:2261–2269. arxiv.org/abs/1608.06993. [Google Scholar]
- 25.Zeng X., Zhi T., Du Z., Guo Q., Sun N., Chen Y. IEEE 38th international conference on computer design. ICCD); 2020. ALT: optimizing tensor compilation in deep learning compilers with active learning; pp. 623–630. 2020. [DOI] [Google Scholar]
- 26.Ketkar N. Deep learning with Python. Apress; Berkeley, CA: 2017. Introduction to Keras. [DOI] [Google Scholar]
- 27.Zhang Z. IEEE/ACM 26th international symposium on quality of service (IWQoS) 2018. Improved Adam optimizer for deep neural networks; pp. 1–2. 2018. [DOI] [Google Scholar]
- 28.Yamashita R., Nishio M., Do R.K.G., et al. Convolutional neural networks: an overview and application in radiology. Insights Imag. 2018;9:611–629. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alzubaidi L., Zhang J., Humaidi A.J., et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data. 2021;8:53. doi: 10.1186/s40537-021-00444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corneanu C., Madadi M., Escalera S., Martinez A. 15th IEEE international conference on automatic face and gesture recognition (FG 2020) 2020. Explainable early stopping for action unit recognition. [Google Scholar]
- 31.Villmann T., Kaden M., Lange M., Stürmer P., Hermann W. IEEE symposium on computational intelligence and data mining (CIDM) 2014. Precision-recall-optimization in learning vector quantization classifiers for improved medical classification systems; pp. 71–77. 2014. [DOI] [Google Scholar]
- 32.Sammut C., Webb G.I., editors. Encyclopedia of machine learning. Springer; Boston, MA: 2011. True negative. [DOI] [Google Scholar]
- 33.Ting K.M. In: Encyclopedia of machine learning. Sammut C., Webb G.I., editors. Springer; Boston, MA: 2011. Confusion matrix. [DOI] [Google Scholar]
- 34.J. Paul Cohen, P. Morrison, and L. Dao, “COVID-19 image data collection,” arXiv. arXiv, Mar-2020.
- 35.https://www.kaggle.com. Accessed Sep 4 2020.
- 36.Kaggle. Available online:https://www.kaggle.com/tawsifurrahman/covid19-radiographydatabase?fbclid=IwAR0rw_prTvf9R0zInrJQkTFazeBaESxh3rB6otdrPdAWJDonEbIl2Nf6epk (accessed on 10 September 2021).
- 37.Kogilavani S.V., Prabhu J., Sandhiya R., Sandeep Kumar M., Subramaniam UmaShankar, Karthick Alagar, et al. Sharmila banu sheik imam, "COVID-19 detection based on lung ct scan using deep learning techniques. Comput Math Methods Med. 2022;2022 Feb 1;2022:7672196 doi: 10.1155/2022/7672196. Article ID 7672196, 13 pages, 2022, PMID: 35116074; PMCID: PMC8805449. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Shah V., Keniya R., Shridharani A., et al. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emerg Radiol. 2021;28:497–505. doi: 10.1007/s10140-020-01886-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J., Zhang Y., He X., Xie P. 2020. Covid-CT-dataset: a CT scan dataset about covid-19. arXiv:2003.13865. [Google Scholar]
- 40.Chouat I., Echtioui A., Khemakhem R., et al. COVID-19 detection in CT and CXR images using deep learning models. Biogerontology. 2022;23:65–84. doi: 10.1007/s10522-021-09946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.https://github.com/UCSD-AI4H/COVID-CT/tree/master/Images-processed.
- 42.Islam M.R., Matin A. 23rd international conference on computer and information technology (ICCIT) 2020. Detection of COVID 19 from CT image by the novel LeNet-5 CNN architecture; pp. 1–5. 2020. [DOI] [Google Scholar]
- 43.Yang X., He X., Zhao J., Zhang Y., Zhang S., Xie P. 2020. COVIDCT-dataset: a CT scan dataset about COVID-19. [Google Scholar]
- 44.El-Shafai Walid, Abd El-Samie, Fathi . Mendeley Data; 2020. Extensive COVID-19 X-ray and CT chest images dataset”. vol. 3. [DOI] [Google Scholar]
- 45.Patel P. Kaggle; 2020. Chest X-ray (COVID-19 & pneumonia)https://www.kaggle.com/prashant268/chest-xray-covid19-pneumonia [Google Scholar]
- 46.Cohen J.P. Github; 2020. COVID-19 image data collection.https://github.com/ieee8023/covid-chestxray-dataset online; last accessed July-13,2020. [Google Scholar]
- 47.Kermany D.S., Goldbaum M., Cai W., et al. Cell; 2018. Identifying medical diagnoses and treatable diseases by image-based deep learning.https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia [DOI] [PubMed] [Google Scholar]
- 48.Punn N. 2020. Covid-19 posteroanterior chest x-ray fused (cpcxr) dataset.https://github.com/nspunn1993/COVID-19-PA-CXR-fused-dataset [Google Scholar]
- 49.Cohen J.P. Github; 2020. COVID-19 image data collection.https://github.com/ieee8023/covid-chestxraydataset/blob/master/metadata.csv online; last accessed July-13,2020. [Google Scholar]
- 50.Effective clustering and accurate classification of the chest X-ray images of COVID-19 patients from healthy ones through the mean structural similarity index measure (DOI:10.13140/RG.2.2.33801.57441).