Abstract
COVID-19 vaccines elicit humoral and cellular immune responses. Durable maintenance of vaccine-induced immunity is required for long-term protection of the host. Here, we examine activation and differentiation of vaccine-induced CD8+ T cells using MHC class I (MHC-I) multimers and correlations between early differentiation and the durability of CD8+ T cell responses among healthcare workers immunized with two doses of BNT162b2. The frequency of MHC-I multimer+ cells is robustly increased by BNT162b2 but decreases 6 months post-second vaccination to 2.4%–65.6% (23.0% on average) of the peak. MHC-I multimer+ cells dominantly exhibit phenotypes of activated effector cells 1–2 weeks post-second vaccination and gradually acquire phenotypes of long-term memory cells, including stem cell-like memory T (TSCM) cells. Importantly, the frequency of TSCM cells 1–2 weeks post-second vaccination significantly correlates with the 6-month durability of CD8+ T cells, indicating that early generation of TSCM cells determines the longevity of vaccine-induced memory CD8+ T cell responses.
Keywords: COVID-19, vaccine, SARS-CoV-2, CD8+ T cell, memory cell, MHC class I multimer
Graphical abstract
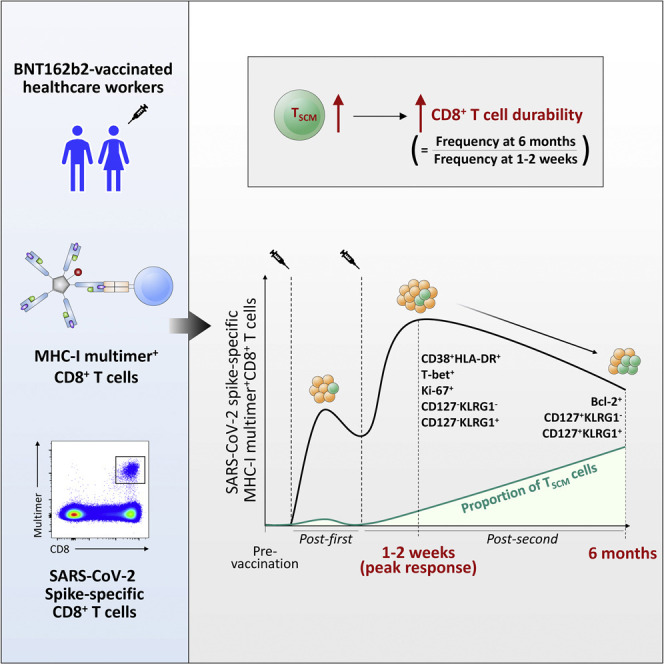
The longevity of SARS-CoV-2-specific CD8+ T cells elicited by BNT162b2 has yet to be fully understood. Jung et al. demonstrate that early generation of TSCM cells after vaccination determines the durability of spike-specific memory CD8+ T cells. This early generation of TSCM cells negatively correlates with age.
Introduction
Since the outbreak of the COVID-19 pandemic, prophylactic vaccines have been developed at an unprecedented pace and administered to large populations on a global scale. Although these vaccines were initially highlighted to induce robust immune responses against SARS-CoV-2 spike (S) protein (Anderson et al., 2020; Jackson et al., 2020; Sahin et al., 2020), recent reports have raised concerns regarding the decay of neutralizing antibody (nAb) titers (Levin et al., 2021; Shrotri et al., 2021). Furthermore, emerging variants of concern (VOC) reduce the neutralizing activity of vaccine-induced nAbs (Chen et al., 2021; Liu et al., 2021; Planas et al., 2021), resulting in an increase in breakthrough infections (Hacisuleyman et al., 2021; Kustin et al., 2021). Thus, vaccine-induced nAbs are neither durable nor sufficient for long-term protection against SARS-CoV-2, particularly major VOC, including the Delta and Omicron variants.
COVID-19 vaccines elicit not only humoral responses, but also T cell responses (Noh et al., 2021). T cells confer host protection by reducing viral titers and lung pathology even in the absence of nAbs following SARS-CoV-2 challenge in immunized mice (Zhuang et al., 2021). T cells also protect convalescent macaques against SARS-CoV-2 when nAb titers are suboptimal (McMahan et al., 2021). In patients with B cell depletion, SARS-CoV-2-specific CD8+ T cells contribute to host protection and survival (Bange et al., 2021). In addition, SARS-CoV-2-specific T cell responses elicited by natural infection or vaccination are rarely escaped by VOC because the majority of T cell epitopes are conserved across variants (Tarke et al., 2021). Thus, we need to understand vaccine-induced, SARS-CoV-2 S-specific T cell responses, including differentiation, memory generation, and durability.
A few studies have reported dynamic changes in S-specific T cells following mRNA vaccination (Goel et al., 2021; Mateus et al., 2021; Oberhardt et al., 2021; Painter et al., 2021). Vaccine-elicited T cells appear to develop a population of polyfunctional memory T cells in a proportion comparable with that of memory cells provoked by natural infection (Goel et al., 2021; Oberhardt et al., 2021; Painter et al., 2021). Vaccine-induced memory CD4+ T cells seem to mainly differentiate into CCR7+CD45RA− central memory T (TCM) and CCR7−CD45RA− effector/effector memory T (TEM) cells, whereas CD8+ T cells differentiate into TEM cells (Oberhardt et al., 2021; Painter et al., 2021). Importantly, vaccine-elicited S-specific CD4+ T cell responses are stably maintained (Goel et al., 2021; Mateus et al., 2021). However, the durability and decay rate of the CD8+ T cell responses remain controversial (Goel et al., 2021; Mateus et al., 2021).
Here, we conducted a longitudinal study to analyze the dynamic changes in SARS-CoV-2 S-specific CD8+ T cell responses following BNT162b2 first-second vaccination on a single-epitope level using MHC class I (MHC-I) multimer staining of serially collected blood samples from a cohort of healthcare workers. We examined the activation, differentiation, and durability of S-specific CD8+ T cells for up to 6 months post-second vaccination and investigated the impact of early stem cell-like memory T (TSCM) cell generation on the durability of vaccine-induced CD8+ T cell responses.
Results
S-specific CD8+ T cell responses are successfully elicited by BNT162b2 but wane over 6 months
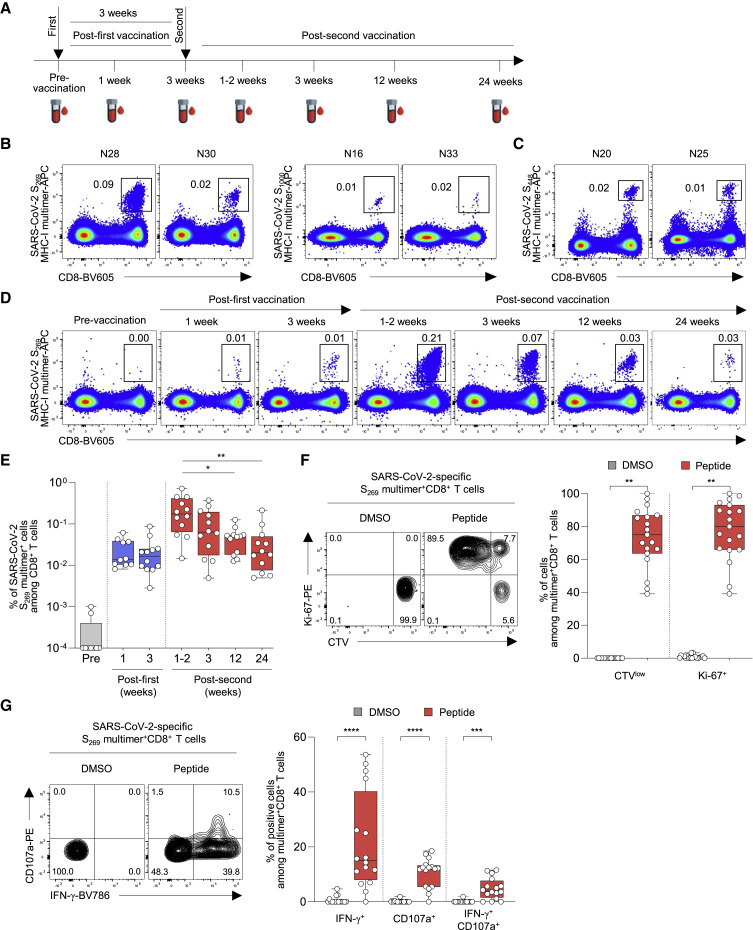
A total of 40 healthcare workers vaccinated with two doses of BNT162b2 without previous SARS-CoV-2 infection were enrolled in this study and peripheral blood obtained until 6 months post-second vaccination (Figure 1A). To directly detect S-specific CD8+ T cells, we stained peripheral blood mononuclear cells (PBMCs) from 23 HLA-A∗02(+) individuals using four different S-specific HLA-A∗0201 multimers (Figures S1A and S1B). S269-specific cells were detected in 12 individuals, and S1000-specific cells were detected in 3 of the 12 individuals who had S269-specific cells (Figure 1B). S996- and S1220-specific cells were not detected in any vaccinated individuals, although they were detected in individuals who had recovered from SARS-CoV-2 infection (Figure S1C). We also stained PBMCs obtained post-second vaccination from 13 HLA-A∗24(+) individuals using HLA-A∗2402 S448 multimers, and S448-specific cells were detected in two individuals (Figure 1C). The binding specificity of HLA-A∗0201 S269 multimer and HLA-A∗2402 S448 multimer was validated by HLA-matched and -mismatched staining of T cell lines (Figure S1D). In subsequent analyses, we focused on the 12 individuals who had S269-specific cells (Table S1).
Figure 1.
The frequency of SARS-CoV-2 S-specific MHC-I multimer+CD8+ T cells in BNT162b2-vaccinated individuals over time
PBMCs from BNT162b2-vaccinated individuals were analyzed by flow cytometry.
(A) Blood sampling schedule from pre-vaccination to 6 months post-second vaccination.
(B and C) Representative flow cytometry plots showing the frequency of (B) SARS-CoV-2 S269 and S1000 MHC-I multimer+CD8+ T cells from HLA-A∗02(+) individuals and (C) SARS-CoV-2 S448 MHC-I multimer+CD8+ T cells from HLA-A∗24(+) individuals in the gate of CD3+ T cells.
(D) Representative flow cytometry plots of the frequency of SARS-CoV-2-specific S269 multimer+CD8+ T cells from a single vaccinated individual over time.
(E) The frequency of SARS-CoV-2 S269 multimer+CD8+ T cells in vaccinated individuals (n = 12) from pre-vaccination to 24 weeks (6 months) post-second vaccination summarized in a box graph.
(F) CTV-labeled PBMCs were stimulated with SARS-CoV-2 S269 epitope (n = 19; samples obtained 6 months post-second vaccination) for 120 h. Left: representative flow cytometry plots showing PBMCs from a vaccinated individual stimulated with DMSO control and peptide. Right: summary box graph showing the frequency of CTVlow and Ki-67+ cells among SARS-CoV-2 S269 multimer+CD8+ T cells.
(G) PBMCs were stimulated with SARS-CoV-2 S269 epitope (n = 16; samples obtained 6 months post-second vaccination) for 6 h in the presence of anti-CD107a-PE antibody, and S269 multimer staining and IFN-γ ICS were performed. Left: representative flow cytometry plot showing PBMCs from a vaccinated individual stimulated with DMSO control and S269 peptide. Right: summary box graph showing the frequency of IFN-γ+, CD107a+, and IFN-γ+CD107a+ cells among SARS-CoV-2-specific S269 multimer+CD8+ T cells. Statistical analysis was performed using Kruskal-Wallis test with Dunns’ multiple comparison test (E) or paired Wilcoxon signed rank test (F and G) (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). CTV, CellTrace Violet; DMSO, dimethyl sulfoxide. See also Figure S1 and Table S1.
We stained PBMCs obtained from the 12 individuals during the course of vaccination and follow-up using S269 MHC-I multimer. The frequency of S269 multimer+ cells increased post-first vaccination with a peak at 1–2 weeks post-second vaccination, and then gradually decreased (Figures 1D and 1E). The frequency of S269 multimer+ cells significantly decreased 24 weeks (6 months) post-second vaccination compared with the peak (1–2 weeks post-second vaccination). We evaluated the functions of S269 multimer+ cells using PBMCs obtained 6 months post-second vaccination. In CellTrace Violet (CTV) dilution assays, S269 multimer+ cells exhibited robust proliferation based on CTV dilution and Ki-67 upregulation following 5-day ex vivo stimulation with S269 peptide (Figures 1F and S1E). When MHC-I multimer staining was combined with intracellular cytokine staining (ICS), interferon-γ (IFN-γ) production and degranulation were observed among S269 multimer+ cells upon S269 peptide stimulation, and the frequency of IFN-γ+CD107a+ cells ranged from 0% to 11.6% (mean 4.8%; Figures 1G and S1F). We validated MHC-I multimer-combined IFN-γ ICS by performing IFN-γ ICS with or without S269 multimer staining and found no difference in the frequency of IFN-γ+ cells between assays with and without S269 multimer staining (Figure S1G). In summary, BNT162b2 vaccination successfully elicits an S-specific CD8+ T cell response, but the response wanes over 6 months.
S-specific effector CD8+ T cells with activation phenotypes increase early after BNT162b2 vaccination
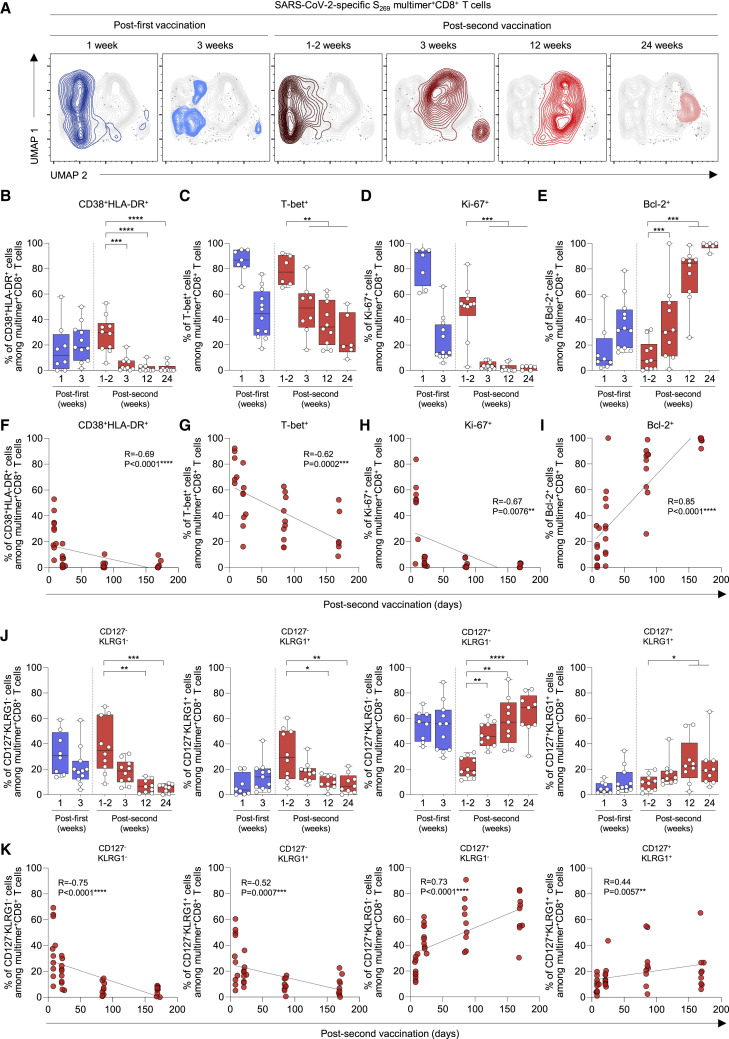
We investigated the dynamic changes in phenotypes of S269-specific CD8+ T cells during the course of vaccination and follow-up. First, we performed UMAP analysis integrating the expression of CD38, HLA-DR, CD127, KLRG1, CCR7, CD45RA, and CD95. The location of S269 multimer+ cells expressing each marker (Figure S2A) or exhibiting each memory subset (Figure S2B) is indicated in the UMAP plots. We found that the phenotypes of S-specific multimer+ cells strikingly changed from 3 weeks post-second vaccination (Figures 2A and S3). In the analysis of T cell activation markers CD38 and HLA-DR, the percentage of CD38+HLA-DR+ cells among S269 multimer+ cells peaked 1–2 weeks post-second vaccination, and then significantly decreased thereafter (Figures 2B and S3A). Similarly, the percentage of T-bet+ cells among S269 multimer+ cells peaked 1–2 weeks post-second vaccination, and then steadily decreased (Figures 2C and S3B). These data show that S-specific CD8+ T cells exhibit an activated effector phenotype early after the second vaccination and then progressively lose the phenotype of activated effector cells. We also examined the expression of Ki-67 and Bcl-2. The percentage of Ki-67+ cells among S269 multimer+ cells peaked 1–2 weeks post-first and post-second vaccination and significantly decreased from 3 weeks post-second vaccination (Figures 2D, S3C, and S3D). The percentage of Bcl-2+ cells among S269 multimer+ cells steadily increased after the second vaccination and reached a peak 6 months post-second vaccination (Figures 2E, S3C, and S3D). The percentage of CD38+HLA-DR+ or T-bet+ cells inversely correlated with the time after the second vaccination (Figures 2F and 2G), and the percentage of Ki-67+ and Bcl-2+ cells exhibited inverse and positive correlations with the time after the second vaccination, respectively (Figures 2H and 2I).
Figure 2.
Phenotypes of SARS-CoV-2 S269 MHC-I multimer+CD8+ T cells in BNT162b2-vaccinated individuals over time
(A) UMAP plot of SARS-CoV-2-specific S269 multimer+CD8+ T cells from multiple patients showing the clustering of the multimer+CD8+ T cells over time. Each time point consists of 4–5 patients with a total of 650–750 cells.
(B–E and J) The percentages of the indicated subpopulation among SARS-CoV-2-specific S269 multimer+CD8+ T cells in vaccinated individuals over time (B) (n = 11), (C) (n = 8), (D and E) (n = 11), (J) (n = 11) from 1 week post-first vaccination to 24 weeks (6 months) post-second vaccination summarized in box graphs.
(F–I and K) Correlation between the percentage of the indicated subpopulation of SARS-CoV-2-specific S269 multimer+CD8+ T cells and days post-second vaccination (F) (n = 11), (G) (n = 8), (H and I) (n = 11), (K) (n = 11) with black lines representing linear regression. Data from boxplot graphs are presented as median and interquartile range (IQR). Statistical analysis was performed using Mann-Whitney U test (B–E and J) or Spearman correlation test (F–I and K) with linear regression analysis (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). See also Figures S2 and S3.
Next, we examined the expression of CD127 and KLRG1 to more precisely phenotype the S269-specific CD8+ T cells (Figures S3E and S3F). The percentage of CD127−KLRG1− early effector and CD127−KLRG1+ short-lived effector cells among S269 multimer+ cells increased 1–2 weeks post-second vaccination and gradually decreased thereafter (Figures 2J and S3E), coinciding with the dynamics of CD38+HLA-DR+ and T-bet+ cells. In contrast, the percentage of long-lived CD127+KLRG1− cells among S269 multimer+ cells and normalized median fluorescence intensity of CD127 on S269 multimer+ cells remained relatively high after the first vaccination, decreased 1–2 weeks post-second vaccination, and progressively increased thereafter (Figure S3G). The percentage of CD127+KLRG1+ cells among S269 multimer+ cells remained relatively low despite a transient increase 12 weeks post-second vaccination. The percentage of CD127+KLRG1− and CD127+KLRG1+ cells positively correlated with the time after the second vaccination, whereas the percentage of CD127−KLRG1− and CD127−KLRG1+ cells was inversely correlated (Figure 2K). These results demonstrate sequential events from effector cell generation to memory cell differentiation after BNT162b2 vaccination.
TSCM cells are successfully developed by BNT162b2 vaccination
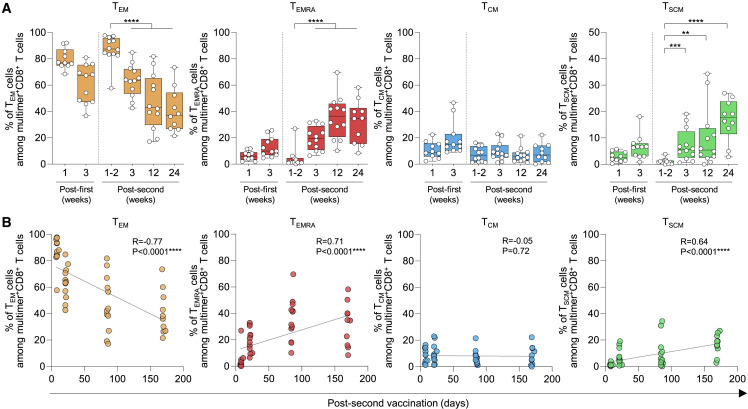
We further analyzed the differentiation of S269-specific CD8+ T cells by examining the expression of CCR7 and CD45RA (Figure S4A). The percentage of CCR7−CD45RA− TEM cells among S-specific multimer+ cells peaked 1–2 weeks post-second vaccination and significantly decreased thereafter, whereas the percentage of CCR7−CD45RA+ TEMRA cells was minimal 1–2 weeks post-second vaccination and significantly increased thereafter (Figures 3A and S4B). The percentage of TCM cells remained relatively low during the course of vaccination and follow-up. We also examined CCR7+CD45RA+CD95+ TSCM cells, which are known to have the capacity for self-renewal and multipotent differentiation (Gattinoni et al., 2011), and found that the percentage of TSCM cells among S269 multimer+ cells progressively increased after the second vaccination. We also compared the frequency of TSCM cells among S269 multimer+ cells in convalescent and vaccinated individuals at three different time points: 3–4 weeks, 11–13 weeks, and after 24 weeks post-symptom onset in the convalescent group or post-second vaccination in the vaccinated group. We found that the frequency of TSCM cells was significantly higher in vaccinated individuals at the early time point (3–4 weeks; Figure S4C). However, the difference disappeared at later time points. The percentage of TEM cells inversely correlated with the time after the second vaccination, and the percentage of TEMRA and TSCM cells positively correlated with the time after the second vaccination (Figure 3B).
Figure 3.
Differentiation kinetics of SARS-CoV-2 S269 MHC-I multimer+CD8+ T cells in BNT162b2-vaccinated individuals
(A) The percentages indicate subpopulations among SARS-CoV-2-specific S269 multimer+ memory CD8+ T cells in vaccinated individuals over time summarized in box graphs (n = 12).
(B) Correlation between the percentage of the indicated subpopulations of SARS-CoV-2-specific S269 multimer+ memory CD8+ T cells and days post-second vaccination (n = 12) with black lines representing linear regression. Data are presented as median and IQR. Statistical analysis was performed using Mann-Whitney U test (A) or Spearman correlation test with linear regression analysis (B) (∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). See also Figure S4.
TSCM cell generation early after the second vaccination predicts the 6-month durability of vaccine-induced memory CD8+ T cell responses
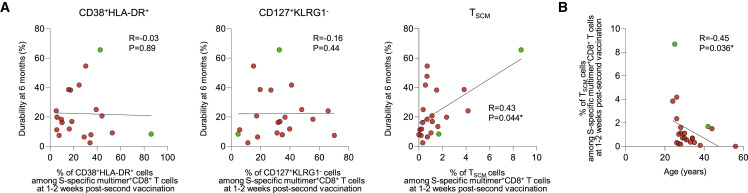
Finally, we analyzed whether T cell differentiation status early after vaccination determines the durability of vaccine-induced CD8+ T cell responses. We focused on 1–2 weeks post-second vaccination, when the frequency of S-specific multimer+ cells peaked with highly activated effector phenotypes. For this analysis, we increased the number of multimer+ individuals by including two donors who had HLA-A∗2402 S448 multimer+ cells (Figure 1C). We also enrolled an additional eight BNT162b2-vaccinated healthcare workers who had S269 multimer+ cells (Table S2). We performed correlation analyses between various T cell parameters 1–2 weeks post-second vaccination and the 6-month durability of S-specific CD8+ T cell responses. Among multiple T cell parameters, the percentage of TSCM cells among multimer+ cells significantly correlated with the 6-month durability of multimer+ cells, whereas the percentage of CD38+HLA-DR+ or CD127+KLRG1− cells did not (Figure 4A). In addition, the percentage of multimer+ TSCM cells among CD3+ T cells or total live cells 1–2 weeks post-second vaccination significantly correlated with the frequency of multimer+ cells among CD8+ T cells 6 months post-second vaccination (Figure S5A), although it did not correlate with the 6-month durability of multimer+ cells (Figure S5B). Moreover, the percentage of TSCM cells 1–2 weeks post-second vaccination inversely correlated with the age of vaccinated individuals (Figure 4B). We also performed correlation analysis between the frequency of multimer+ cells among CD8+ T cells 6 months post-second vaccination and the donor age (Figure S5C) or the percentage of TSCM cells among multimer+ cells 1–2 weeks post-second vaccination (Figure S5D) and found no significant correlation. In summary, the durability of BNT162b2-induced memory CD8+ T cell responses is determined by early generation of TSCM cells, which is inversely related to age.
Figure 4.
Correlation between the durability of SARS-CoV-2 S-specific MHC-I multimer+CD8+ T cells and TSCM cells
(A) Correlation between the durability of S269 (red) and S448 (green) multimer+CD8+ T cells and the percentage of CD38+HLA-DR+ (n = 20), CD127+KLRG1− (n = 20), and TSCM cells (n = 22) among multimer+CD8+ T cells 1–2 weeks post-second vaccination with black lines representing linear regression. Durability was calculated as follows: (the frequency of multimer+ cells among total CD8+ T cells 6 months post-second vaccination/the frequency of multimer+ cells among total CD8+ T cells 1–2 weeks post-second vaccination) × 100. Data are presented as IQRs.
(B) Correlation between the percentage of TSCM cells among multimer+CD8+ T cells 1–2 weeks post-second vaccination and the age of the individuals (n = 22) with black lines representing linear regression. Statistical analysis was performed using Spearman correlation test with linear regression analysis (∗p < 0.05). See also Figure S5 and Table S2.
Discussion
The BNT162b2 mRNA vaccine elicits S-specific humoral and cellular immune responses that contribute to host protection against SARS-CoV-2 (Collier et al., 2021; Goel et al., 2021; Sahin et al., 2021). However, the longevity and differentiation trajectory of BNT162b2-induced S-specific memory T cells remain poorly understood. In this study, we conducted a longitudinal analysis of BNT162b2-induced S-specific CD8+ T cell responses on a single-epitope level using MHC-I multimers until 6 months post-second vaccination, and found that early generation of TSCM cells significantly correlated with the 6-month durability of CD8+ T cell response.
TSCM cells have a phenotype of CD95+ naive-like memory cells in humans and possess the capacity for self-renewal and multipotency to reconstitute both effector and memory T cell subsets upon antigen re-challenge (Gattinoni et al., 2011). Their extreme lifespan stems from their long telomere and high telomerase activity (Ahmed et al., 2016). Previously, yellow fever virus (YFV)-specific TSCM-like CD8+ cells were shown to persist for more than a decade after YFV vaccination, with retained epigenetic signatures of effector molecules (Akondy et al., 2017; Fuertes Marraco et al., 2015). This indicates that the virus-specific clonal reservoir provided by the long-lasting TSCM cells allows for enhanced recall response upon virus re-exposure. Recently, SARS-CoV-2-specific TSCM cells were demonstrated after BNT162b2 vaccination (Guerrera et al., 2021). However, the relationship between CD8+ TSCM generation and the durability of vaccine-induced CD8+ T cell responses have not yet been elucidated. Our study shows that the early generation of TSCM cells inversely correlates with the age of vaccinated individuals, consistent with the number of CD8+ TSCM cells decreasing with age (Li et al., 2019).
In this study, the percentage of TEMRA cells, which are known as terminally differentiated cells (Mittelbrunn and Kroemer, 2021; Rufer et al., 2003), gradually increased among multimer+ cells after the second vaccination. A recent study showed enrichment of SARS-CoV-2 S-specific TEMRA cells upon repeated antigen exposure due to infection and two doses of BNT162b2 vaccination (Minervina et al., 2022). In addition, another study using clonal tracking of the SARS-CoV-2 multimer+CD8+ cells in convalescent individuals showed a progressive transition of SARS-CoV-2-specific TEM cells into TEMRA cells 1 year after COVID-19 (Adamo et al., 2022). These findings suggest that a proportion of TEMRA cells among SARS-CoV-2-specific CD8+ T cells increases after vaccination or natural infection. Interestingly, Adamo et al. (2022) recently showed that SARS-CoV-2-specific CD8+ TEMRA cells express TCF1 in convalescent individuals, although TEMRA cells are known to be terminally differentiated cells (Mittelbrunn and Kroemer, 2021; Rufer et al., 2003). Further examination of the functional and molecular characteristics of SARS-CoV-2-specific CD8+ TEMRA cells is needed.
In summary, we demonstrated that early generation of TSCM cells after BNT162b2 vaccination determines the durability of vaccine-induced memory CD8+ T cell responses, providing knowledge for the development of vaccines that induce durable cellular immunity.
Limitations of the study
Limitations of this study include the relatively small sample size of MHC-I multimer+ individuals. In addition, our results mainly relied on HLA-A∗0201 S269 multimer+ cells. Also, our study examined CD8+ T cells, but not CD4+ T cells. Further research is required to study BNT162b2-induced CD4+ T cell responses using MHC-II multimers. Moreover, although SARS-CoV-2 infection has been shown to activate tissue-resident T cells in the airways of COVID-19 patients (Szabo et al., 2021), whether mRNA vaccination is also capable of generating tissue-resident T cells remains to be elucidated. Finally, a more comprehensive comparison of CD8+ T cells elicited by vaccines based on diverse platforms, including mRNA, adenoviral vector, and recombinant protein, may be necessary to better understand COVID-19 vaccine-induced memory CD8+ T cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD28/49d (clone L293 and L25) | BD Biosciences | Cat# 347690; RRID: AB_647457 |
| BV421 mouse IgG1, κ isotype (clone ×40) | BD Biosciences | Cat# 562438; RRID: AB_11207319 |
| BV421 anti-human CD127 (clone HIL-7R-M21) | BD Biosciences | Cat# 562436; RRID: AB_11151911 |
| BV510 anti-human CD3 (clone UCHT1) | BD Biosciences | Cat# 563109; RRID: AB_2732053 |
| BV605 anti-human CD8 (clone SK1) | BD Biosciences | Cat# 564116; RRID: AB_2869551 |
| BV786 mouse IgG1, κ isotype (clone ×40) | BD Biosciences | Cat# 563330; RRID: AB_2869484 |
| BV786 anti-human CD38 (clone HIT2) | BD Biosciences | Cat# 563964; RRID: AB_2738515 |
| BV786 anti-human Ki-67 (clone B56) | BD Biosciences | Cat# 563756; RRID: AB_2732007 |
| BV786 anti-human CD3 (clone SK7) | BD Biosciences | Cat# 563800; RRID: AB_2744384 |
| BV786 anti-human IFN-γ (clone 4S.B3) | BD Biosciences | Cat# 563731; RRID: AB_2738391 |
| PE anti-human CD107a (clone H4A3) | BD Bioscience | Cat# 555801; RRID: AB_396135 |
| BB515 anti-human CD45RA (clone HI100) | BD Biosciences | Cat# 564552; RRID: AB_2738841 |
| FITC anti-human CD8 (clone RPA-T8) | BD Biosciences | Cat# 557085; RRID: AB_396580 |
| PerCP-Cy5.5 anti-human CCR7 (clone 150503) | BD Biosciences | Cat# 561144; RRID: AB_10562553 |
| PE anti-human CD95 (clone DX2) | BD Biosciences | Cat# 555674; RRID: AB_396027 |
| PE-CF594 anti-human CD14 (clone MφP9) | BD Biosciences | Cat# 562335; RRID: AB_11153663 |
| PE-CF594 anti-human CD19 (clone HIB19 | BD Biosciences | Cat# 562294; RRID: AB_11154408 |
| PE-Cy7 mouse IgG2a, κ isotype (clone MOPC-173) | BD Biosciences | Cat# 560906; RRID: AB_10561847 |
| PE-Cy7 anti-human CD8 (clone RPA-T8) | BD Biosciences | Cat# 557746; RRID: AB_396852 |
| BV421 anti-human Bcl-2 (clone 100) | BioLegend | Cat# 658709; RRID: AB_2563283 |
| PE anti-human Ki-67 (clone Ki-67) | BioLegend | Cat# 350504; RRID: AB_10660752 |
| PE-Cy7 anti-human T-bet (clone 4B10) | BioLegend | Cat# 644824; RRID: AB_2561761 |
| APC-Cy7 anti-human HLA-DR (clone L243) | BioLegend | Cat# 307618; RRID: AB_493586 |
| APC-Cy7 anti-human CD45RA (clone HI100) | BioLegend | Cat# 304128; RRID: AB_10708880 |
| PE-Cy7 anti-human KLRG1 (clone 13F12F2) | Invitrogen | Cat# 25-9488-42; RRID: AB_2573546 |
| Biological samples | ||
| Blood samples from individuals with BNT162b2 vaccination | Korea University Guro Hospital, Seoul National University Bundang Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| CD8 Microbeads, human | Miltenyi Biotec | Cat# 130-045-201 |
| FcR blocking reagent, human | Miltenyi Biotec | Cat# 130-059-901 |
| SARS-CoV-2 S269-277 YLQPRTFLL peptide | Mimotopes | Customized |
| SARS-CoV-2 S448-456 NYNYLYRLF peptide | Peptron | Customized |
| SARS-CoV-2 S996-1004 LITGRLQSL peptide | Peptron | Customized |
| SARS-CoV-2 S1220-1228 FIAGLIAIV peptide | Peptron | Customized |
| Recombinant human IL-2 | Peprotech | Cat# 200-22 |
| Fetal bovine serum | Corning | Cat# 35-015-CV |
| Lymphocyte separation medium | Corning | Cat# 25-072-CV |
| Dimethyl sulfoxide | Sigma-Aldrich | Cat# D8418 |
| GolgiPlug (Brefeldin A) | BD Biosciences | Cat# 51-2092KZ |
| GolgiStop (Monensin) | BD Biosciences | Cat# 51-2301KZ |
| Critical commercial assays | ||
| LIVE/DEAD red fluorescent reactive dye | Invitrogen | Cat# L34972 |
| CellTrace Violet cell proliferation kit | Invitrogen | Cat# C34557 |
| FoxP3/transcription factor staining buffer set | Invitrogen | Cat# 00-5523-00 |
| Software and algorithms | ||
| FlowJo software version 10.7 | FlowJo LLC | https://www.flowjo.com/ |
| Prism version 9 | Graphpad | https://www.graphpad.com/ |
| Other | ||
| APC YLQPRTFLL (SARS-CoV-2 S269) HLA-A∗0201 Pentamer | Proimmune | Cat# 4339 |
| APC LITGRLQSL (SARS-CoV-2 S996) HLA-A∗0201 Pentamer | Proimmune | Cat# 4323 |
| APC RLQSLQTYV (SARS-CoV-2 S1000) HLA-A∗0201 Pentamer | Proimmune | Cat# 4358 |
| APC FIAGLIAIV (SARS-CoV-2 S1220) HLA-A∗0201 Pentamer | Proimmune | Cat# 4321 |
| APC NYNYLYRLF (SARS-CoV-2 S448) HLA-A∗2402 Pentamer | Immudex | Customized |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Eui-Cheol Shin (ecshin@kaist.ac.kr).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects and sample collection
A total of 40 healthcare workers (27 females/13 males with a mean age of 34 years; range 24–56 years) who were immunized with the first and second doses of 30-μg of BNT162b2 vaccine (Pfizer-BioNTech, NY, USA) were enrolled in the study from Korea University Guro Hospital, Republic of Korea. All enrolled participants were healthy at the time of vaccination with no reported prior history of SARS-CoV-2 infection. In addition, they were screened for anti-SARS-CoV-2 spike IgG and confirmed to be negative. BNT162b2 vaccines were administered in two separate injections at an interval of 3 weeks (21 days) between the first and second vaccinations. Peripheral blood samples were serially collected from all enrolled participants following each vaccination: 1 week (days 7–9) and 3 weeks (days 18–21) after first vaccination and 1–2 weeks (days 7–11), 3 weeks (days 20–25), 12 weeks (days 84–87), and 24 weeks (days 166–173) after second vaccination. Information regarding which time points were tested for each vaccinated individual in each of the assays is summarized in Table S3. For the analysis in Figure 4, we recruited an additional eight HLA-A∗02(+) vaccinated individuals (8 females) who were confirmed to have S269 multimer+ cells from Seoul National University Bundang Hospital, Republic of Korea. This study was reviewed and approved by the institutional review boards of all participating institutions (Korea University Guro Hospital: 2021GR0099; Seoul National University Bundang Hospital: B-2102-669-302) and conducted in accordance with the principles of the Declaration of Helsinki. Informed consent was obtained from all participants.
PBMCs were isolated from all peripheral blood samples by density gradient centrifugation using Lymphocyte Separation Medium (Corning, NY, USA). Isolated PBMCs were cryopreserved at −196°C in fetal bovine serum (Corning) and 10% dimethyl sulfoxide (Sigma-Aldrich, St. Louis, USA) until use.
Method details
MHC class I multimer staining and multi-color flow cytometry
PBMCs were stained with APC-conjugated MHC class I multimers for 15 min at room temperature (RT) and washed. Cells were then stained with fluorochrome-conjugated antibodies specific for cell surface markers for 15 min at RT. Dead cells were excluded using LIVE/DEAD red fluorescent reactive dye (Invitrogen, Carlsbad, CA, USA). For intracellular staining, cells were fixed and permeabilized using the FoxP3 staining buffer kit (Invitrogen) prior to staining with fluorochrome-conjugated antibodies specific against intracellular markers for 30 min at 4°C. Multi-color flow cytometry was performed using the LSR II Flow Cytometer (BD Biosciences, San Jose, CA, USA) and data analyzed in FlowJo 10.7.1 (FlowJo LLC, Ashland, OR, USA).
Stimulation for intracellular cytokine staining
PBMCs were cultured in the presence of 5 μg/mL S269 peptide, along with 1 μg/mL anti-human CD28 and CD49d mAbs (BD Biosciences) and anti-CD107a-PE antibody for 6 h at 37°C in a 5% CO2 atmosphere. Brefeldin A (GolgiPlug, BD Biosciences) and monensin (GolgiStop, BD Biosciences) were added 1 h after the initial stimulation. Cells were stained with MHC class I multimers and antibodies for further analysis by flow cytometry according to the protocols described above.
Proliferation assay
PBMCs were labeled using the CellTrace™ Violet Cell Proliferation Kit (Invitrogen) according to the manufacturer’s instructions. Briefly, PBMCs were stained with CellTrace Violet at a concentration of 1.0 × 106 cells/mL for 20 min at 37°C. We then added 1% FBS in PBS (Corning) to the cells for 5 min at RT to quench unbound dye. Cells were washed and cultured in RPMI 1640 supplemented with 10% FBS and 1% penicillin-streptomycin (Corning) at a concentration of 5.0 × 105 cells per well in the presence of 5 μg/mL SARS-CoV-2 S overlapping peptide pools and 1 μg/mL anti-human CD28 and CD49d mAbs for 120 h. Stimulation with an equal concentration of DMSO in PBS was used as a negative control. After incubation, cells were harvested and stained with antibodies for analysis by flow cytometry according to the protocols described above.
Peptide-specific T-cell lines
CD8+ T cells were magnetically enriched using CD8 MicroBeads (Miltenyi Biotec, Auburn, CA, USA) according to the manufacturer’s instructions. Enriched CD8+ T cells were cultured in RPMI 1640 containing S269 (YLQPRTFLL), S448 (NYNYLYRLF), S996 (LITGRLQSL), or S1220 (FIAGLIAIV) peptides (1 μg/mL) and IL-2 (200 IU/mL) with irradiated autologous PBMCs for 2 weeks at 37°C. Cells were harvested on day 14 and stained with MHC class I multimers and antibodies for analysis by flow cytometry.
Quantification and statistical analysis
Statistical analyses were performed using Prism (version 9, GraphPad Software). Significance was set at p < 0.05. The Wilcoxon signed-rank test was used to compare data between two paired groups, and the Mann-Whitney U test was used to compare two unpaired groups. The Kruskal-Wallis test with Dunns’ multiple comparisons test was used to compare non-parametric data between multiple unpaired groups. The significance of correlation was assessed using the Spearman correlation test and the correlation was shown with black lines representing linear regression.
Acknowledgments
This work was supported by the Institute for Basic Science under project code IBS-R801-D2 (to E.-C.S.) and the Korea National Institute of Health and the Korea Disease Control and Prevention Agency (2021-ER2601-00 to K.-H.S. and 2021-ER2603-00 to J.Y.S.).
Author contributions
S.J., J.H.J., J.Y.N., S.-H.P., K.-H.S., J.Y.S., and E.-C.S. designed the research. J.Y.N., S.-Y.Y., J.J., E.S.K., H.B.K., H.J.C., W.J.K., K.-H.S., and J.Y.S. and collected clinical specimens and information. S.J., J.H.J., J.Y.N., and W.-J.K. performed the experiments. S.J., J.H.J., J.Y.N., and E.-C.S. analyzed the results. S.J., J.H.J., J.Y.N., K.-H.S., J.Y.S., and E.-C.S. wrote and edited the manuscript. All authors critically reviewed the manuscript and approved the final draft for submission.
Declaration of interests
The authors declare no competing interests.
Published: July 26, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111138.
Supplemental information
Data and code availability
All data supporting the findings of this study are available within the main manuscript and the supplementary files. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
References
- Adamo S., Michler J., Zurbuchen Y., Cervia C., Taeschler P., Raeber M.E., Baghai Sain S., Nilsson J., Moor A.E., Boyman O. Signature of long-lived memory CD8+ T cells in acute SARS-CoV-2 infection. Nature. 2022;602:148–155. doi: 10.1038/s41586-021-04280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed R., Roger L., Costa Del Amo P., Miners K.L., Jones R.E., Boelen L., Fali T., Elemans M., Zhang Y., Appay V., et al. Human stem cell-like memory T cells are maintained in a state of dynamic flux. Cell Rep. 2016;17:2811–2818. doi: 10.1016/j.celrep.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy R.S., Fitch M., Edupuganti S., Yang S., Kissick H.T., Li K.W., Youngblood B.A., Abdelsamed H.A., McGuire D.J., Cohen K.W., et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 2017;552:362–367. doi: 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange E.M., Han N.A., Wileyto P., Kim J.Y., Gouma S., Robinson J., Greenplate A.R., Hwee M.A., Porterfield F., Owoyemi O., et al. CD8(+) T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021;27:1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., Vanblargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier A.-R.Y., Yu J., McMahan K., Liu J., Chandrashekar A., Maron J.S., Atyeo C., Martinez D.R., Ansel J.L., Aguayo R., et al. Differential kinetics of immune responses elicited by covid-19 vaccines. N. Engl. J. Med. 2021;385:2010–2012. doi: 10.1056/nejmc2115596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes Marraco S.A., Soneson C., Cagnon L., Gannon P.O., Allard M., Abed Maillard S., Montandon N., Rufer N., Waldvogel S., Delorenzi M., Speiser D.E. Long-lasting stem cell-like memory CD8+ T cells with a naive-like profile upon yellow fever vaccination. Sci. Transl. Med. 2015;7:282ra48. doi: 10.1126/scitranslmed.aaa3700. [DOI] [PubMed] [Google Scholar]
- Gattinoni L., Lugli E., Ji Y., Pos Z., Paulos C.M., Quigley M.F., Almeida J.R., Gostick E., Yu Z., Carpenito C., et al. A human memory T cell subset with stem cell-like properties. Nat. Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrera G., Picozza M., D’Orso S., Placido R., Pirronello M., Verdiani A., Termine A., Fabrizio C., Giannessi F., Sambucci M., et al. BNT162b2 vaccination induces durable SARS-CoV-2 specific T cells with a stem cell memory phenotype. Sci. Immunol. 2021;6:eabl5344. doi: 10.1126/sciimmunol.abl5344. [DOI] [PubMed] [Google Scholar]
- Hacisuleyman E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., Schaefer-Babajew D.J., Dasilva J., Muecksch F., Gaebler C., et al. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl. J. Med. 2021;384:2212–2218. doi: 10.1056/nejmoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustin T., Harel N., Finkel U., Perchik S., Harari S., Tahor M., Caspi I., Levy R., Leshchinsky M., Ken Dror S., et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat. Med. 2021;27:1379–1384. doi: 10.1038/s41591-021-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., et al. Waning immune humoral response to BNT162b2 covid-19 vaccine over 6 months. N. Engl. J. Med. 2021;385:e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Yao D., Zeng X., Kasakovski D., Zhang Y., Chen S., Zha X., Li Y., Xu L. Age related human T cell subset evolution and senescence. Immun. Ageing. 2019;16:24. doi: 10.1186/s12979-019-0165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Dan J.M., Zhang Z., Rydyznski Moderbacher C., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minervina A.A., Pogorelyy M.V., Kirk A.M., Crawford J.C., Allen E.K., Chou C.-H., Mettelman R.C., Allison K.J., Lin C.-Y., Brice D.C., et al. SARS-CoV-2 antigen exposure history shapes phenotypes and specificity of memory CD8+ T cells. Nat. Immunol. 2022;23:781–790. doi: 10.1038/s41590-022-01184-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M., Kroemer G. Hallmarks of T cell aging. Nat. Immunol. 2021;22:687–698. doi: 10.1038/s41590-021-00927-z. [DOI] [PubMed] [Google Scholar]
- Noh J.Y., Jeong H.W., Kim J.H., Shin E.C. T cell-oriented strategies for controlling the COVID-19 pandemic. Nat. Rev. Immunol. 2021;21:687–688. doi: 10.1038/s41577-021-00625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt V., Luxenburger H., Kemming J., Schulien I., Ciminski K., Giese S., Csernalabics B., Lang-Meli J., Janowska I., Staniek J., et al. Rapid and stable mobilization of CD8(+) T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021;597:268–273. doi: 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., et al. Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54:2133–2142.e3. doi: 10.1016/j.immuni.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Rufer N., Zippelius A., Batard P., Pittet M.J., Kurth I., Corthesy P., Cerottini J.-C., Leyvraz S., Roosnek E., Nabholz M., Romero P. Ex vivo characterization of human CD8+ T subsets with distinct replicative history and partial effector functions. Blood. 2003;102:1779–1787. doi: 10.1182/blood-2003-02-0420. [DOI] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., et al. BNT162b2 vaccine induces neutralising antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- Shrotri M., Navaratnam A.M.D., Nguyen V., Byrne T., Geismar C., Fragaszy E., Beale S., Fong W.L.E., Patel P., Kovar J., et al. Spike-antibody waning after second dose of BNT162b2 or ChAdOx1. Lancet. 2021;398:385–387. doi: 10.1016/S0140-6736(21)01642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.A., Dogra P., Gray J.I., Wells S.B., Connors T.J., Weisberg S.P., Krupska I., Matsumoto R., Poon M.M.L., Idzikowski E., et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity. 2021;54:797–814.e6. doi: 10.1016/j.immuni.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z., Lai X., Sun J., Chen Z., Zhang Z., Dai J., Liu D., Li Y., Li F., Wang Y., et al. Mapping and role of T cell response in SARS-CoV-2–infected mice. J. Exp. Med. 2021;218:e20202187. doi: 10.1084/jem.20202187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the main manuscript and the supplementary files. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.