Abstract
Oysters (Crassostrea virginica) were collected monthly from May 1998 to April 1999 from Mobile Bay, Ala., and analyzed to determine Vibrio parahaemolyticus densities at zero time and after 5, 10, and 24 h of postharvest storage at 26°C. After 24 h of storage at 26°C, oysters were transferred to a refrigerator at 3°C and then analyzed 14 to 17 days later. The V. parahaemolyticus numbers were determined by the most-probable-number procedure using alkaline phosphatase-labeled DNA probe VPAP, which targets the species-specific thermolabile hemolysin gene (tlh), to identify suspect isolates (MPN-VPAP procedure). Two direct plating methods, one using a VPAP probe (Direct-VPAP) and one using a digoxigenin-labeled probe (Direct-VPDig) to identify suspect colonies, were compared to the MPN-VPAP procedure. The results of the Direct-VPAP and Direct-VPDig techniques were highly correlated (r = 0.91), as were the results of the Direct-VPAP and MPN-VPAP procedures (r = 0.91). The correlation between the Direct-VPDig and MPN-VPAP results was 0.85. The two direct plating methods in which nonradioactive DNA probes were used were equivalent to the MPN-VPAP procedure for identification of total V. parahaemolyticus, and they were more rapid and less labor-intensive.
Vibrio parahaemolyticus is an enteric pathogen found in estuaries and various types of seafood throughout the world (1, 2, 13–15). V. parahaemolyticus infections can cause gastroenteritis in humans and are most frequently associated with consumption of raw or undercooked seafood and seafood recontaminated with the bacterium after cooking (19). Consumption of raw shellfish, primarily oysters, has been linked to four multistate V. parahaemolyticus illness outbreaks involving 650 reported cases in the United States since 1997 (Washington in 1997 and 1998; New York and Texas in 1998) (4–6). All patient isolates obtained from the 296 reported V. parahaemolyticus infections in Texas were serotype O3:K6, which commonly causes outbreaks in Asia but had not been identified previously in the United States (6). These outbreaks increased concern about V. parahaemolyticus densities in oysters and focused attention on the development of more efficient methods for environmental monitoring of this pathogen.
The Food and Drug Administration (FDA) Bacteriological Analytical Manual (BAM) most-probable-number (MPN) method (11) is most frequently used to enumerate V. parahaemolyticus in foods. The BAM-MPN method uses biochemical techniques to identify isolates and is time-consuming and labor-intensive. As an alternative, researchers recently described the use of nonradioactive DNA probes for identification of V. parahaemolyticus (17).
In the present study we compared two direct plating methods using nonradioactive DNA probes (Direct-VPAP and Direct-VPDig) with a modification of the BAM-MPN method in which confirmation of the identities of V. parahaemolyticus isolates was accomplished with a DNA probe (MPN-VPAP) targeting the species-specific thermolabile hemolysin gene (tlh) (21). In the Direct-VPAP method we used a tlh-alkaline phosphatase (tlh-AP)-labeled DNA probe, and in the Direct-VPDig method we used a tlh-digoxigenin-labeled DNA probe for identification of V. parahaemolyticus. Pathogenic V. parahaemolyticus strains contain additional hemolysin genes, designated the thermostable direct hemolysin (tdh) gene and the thermostable direct related hemolysin (trh) gene (18, 20). Since V. parahaemolyticus densities in oysters can vary with the season, salinity, temperature, and storage parameters (8), the methods were tested under a variety of conditions over a 1-year period.
MATERIALS AND METHODS
Oyster collection and handling.
Adult oysters (diameter, >7.82 cm; Crassostrea virginica) were harvested monthly from May 1998 through April 1999 by tonging in Mobile Bay, Ala. The salinity and temperature of the surface water in the harvest area were measured with a model 85 dissolved oxygen-conductivity meter (Yellow Springs Instrument Co., Yellow Springs, Ohio). At each sampling time, 12 oysters were chilled on ice and 60 to 80 oysters were held without icing at the ambient air temperature on the boat. The oysters were transported to the FDA Gulf Coast Seafood Laboratory on Dauphin Island, Ala., within 1 h of collection. The chilled oysters were analyzed within 2 h to obtain harvest (zero-time) levels of V. parahaemolyticus, and the remaining nonchilled oysters were placed in an incubator adjusted to 26°C.
Twelve of the oysters stored at 26°C were sampled and analyzed to determine V. parahaemolyticus densities at 5, 10, and 24 h after harvest. The current protocol for handling oysters from the Gulf of Mexico calls for refrigeration within 10 to 36 h, depending on the ambient water temperature at the time of harvest (10). A 24-h holding period at 26°C was chosen for this study in order to analyze oysters before, up to, and after the usual Gulf of Mexico oyster industry harvest times and refrigeration times. After 24 h, the remaining oysters were transferred to a refrigerator (3°C) and then analyzed 14 to 17 days later to simulate possible retail handling practices. The oysters were scrubbed, shucked, and mixed with an equal weight (1:1) of sterile phosphate-buffered saline (PBS) (7.65 g of NaCl per liter, 0.724 g of anhydrous Na2HPO4 [Sigma] per liter, 0.21 g of KH2PO4 [Sigma] per liter; pH 7.4) (11), and the mixture was blended for 90 s with a sterile Waring blender in preparation for analysis (9).
Enumeration by the MPN method.
The MPN method described in the FDA BAM (11) was used to estimate V. parahaemolyticus densities, except that a species-specific DNA probe targeting the tlh gene was used for identification (MPN-VPAP) (17) instead of biochemical utilization assays. This oligonucleotide probe conjugated with alkaline phosphatase (tlh-AP) was purchased from DNA Technology A/S (Aarhus, Denmark). Briefly, oyster homogenate was serially diluted, inoculated into a series of MPN tubes containing alkaline peptone water (11) (three tubes/dilution), and incubated for 16 to 18 h at 35°C, and then a loopful from each MPN tube showing growth was streaked onto a thiosulfate-citrate-bile salts (TCBS) agar plate. After 18 to 24 h of incubation at 35°C, suspect colonies from the TCBS agar (Difco) streak plates were transferred with sterile toothpicks into alkaline peptone water in individual wells of 96-well plates and incubated for 16 to 18 h at 35°C. Cells from the 96-well plates were transferred to Vibrio vulnificus agar (30 g of NaCl [Sigma] per liter, 10 g of cellobiose [Sigma] per liter, 20 g of proteose peptone [Difco] per liter, 0.06 g of bromthymol blue [Sigma] per liter, 25 g of agar [Difco] per liter) plates by using a 48-prong replicator (9). Colony lifts were prepared and tested for hybridization with the tlh-AP probe as described by McCarthy et al. (17) for confirmation of species identity. V. parahaemolyticus TX 2103 (a human stool sample isolate) and V. vulnificus MO6-24 (a human primary septicemia blood isolate) were used as positive and negative controls, respectively.
Direct-VPAP enumeration.
Aliquots of oyster homogenate (0.2 g of a 1:1 [wt/wt] preparation in PBS [equivalent to 0.1 g], taken directly from a blender, or 0.1-ml portions from subsequent 10-fold dilutions in PBS) were spread plated onto T1N3 (1% tryptone [Difco], 3% NaCl, 2% agar; pH 7.2) plates. After overnight incubation at 35°C, colony lift, hybridization, and colorimetric detection analyses were done as described previously for the tlh-AP probe (17). The nucleotide base sequence of the alkaline phosphatase-labeled DNA probe was the sequence from bases 904 to 927 of the species-specific V. parahaemolyticus tlh gene (accession number M36437) (3, 17, 21). After color development, colonies that hybridized with the tlh-AP probe were determined visually.
Direct-VPDig enumeration.
The V. parahaemolyticus species-specific tlh DNA fragments were synthesized with primers by PCR as described by Brasher et al. (3). Digoxigenin-labeled nucleotides were used to label the probe by the procedures of Boehringer Mannheim (The Genius System User's Guide for Filter Hybridization, version 2.9-92; Boehringer Mannheim Corp., Indianapolis, Ind.) and Weagant et al. (22). Nylon membranes (MagnaGraph; Osmonics-MSI, Minnetonka, Minn.) were placed onto tryptic soy agar (Difco) containing MgSO4 (TSAMS agar) (40 g of trytic soy agar per liter, 20 g of NaCl per liter, 1.5 g of MgSO4 [Sigma] per liter) plates and spread plated with the dilutions of oyster homogenate described above. The plates were incubated for 3 h at 35°C to repair sublethally injured cells, and the membranes were then transferred (with the inoculated side of each membrane up) to TCBS plates and incubated overnight at 40°C. Probe and membrane preparation, hybridization, and colorimetric detection were performed as described in The Genius System User's Guide for Filter Hybridization (Boehringer Mannheim Corp.), as outlined by McCarthy et al. (17) and Weagant et al. (22).
Statistical analyses.
Bacterial densities were converted to base 10 logarithms before being analyzed by Microsoft Excel and SAS. Twelve-month geometric means were determined for each analytical method. Samples with nondetectable colonies were assigned the minimum detectable density on the basis of the volume examined. The statistical methods used included linear regression analysis to compare correlations between the analytical methods and analysis of variance to compare differences between treatments (Direct-VPAP, Direct-VPDig, and MPN-VPAP). An alpha level (P < 0.05) was considered a minimum level of significance for each statistical method. Within-treatment comparisons will be described elsewhere.
RESULTS AND DISCUSSION
Regression analyses.
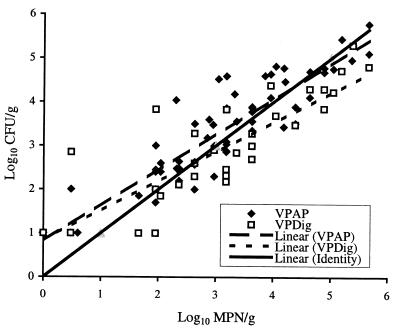
Figure 1 shows that there was close agreement between methods for enumerating V. parahaemolyticus in oysters under a variety of seasonal and storage conditions. It shows the regression lines and a line of identity for the two direct plating methods versus the MPN-VPAP method. The line of identity shows how the two direct plating methods compare with the MPN-VPAP procedure. The slopes of the two direct plating regression lines are almost identical (the slope of the Direct-VPAP regression line is 0.91, and the slope of the Direct-VPDig regression line is 0.94). The V. parahaemolyticus estimates obtained with the methods were highly correlated for either the Direct-VPAP and Direct-VPDig procedures or the Direct-VPAP and MPN-VPAP procedures (r = 0.91); the correlation between the Direct-VPDig and MPN-VPAP procedures was 0.85. Data from studies or monitoring programs obtained with any of these methods could be compared (i.e., for risk assessment). The differences between the direct plating and MPN methods appeared to be greatest at lower V. parahaemolyticus densities and may be attributed to different detection sensitivities. Because the MPN-VPAP method is more sensitive (3 MPN/g for a 0.1-g sample and 0.3 MPN/g for a 1-g sample) than the direct plating methods (10 CFU/g for a 0.1-g sample), use of this method is recommended when low V. parahaemolyticus densities are suspected (e.g., during the winter when water temperatures are lower). Under warm conditions, either direct plating method offers an alternative that is more rapid, economical, and less labor-intensive than the BAM-MPN procedure. Similar direct plating methods used for V. vulnificus have shown that direct plating methods are more precise than MPN analyses (9).
FIG. 1.
V. parahaemolyticus densities (log10 CFU/g or log10 MPN/g) in oysters analyzed by the Direct-VPAP, Direct-VPDig, and MPN-VPAP methods. The line of identity shows the points for which the results of all three methods would be identical.
In a previous study in which methods were compared, DePaola et al. (7) used resource-intensive biochemical tests to confirm the identities of suspect V. parahaemolyticus colonies and based direct plating estimates on only five suspect colonies per sample. The colony lift format used in this study eliminated the need to identify individual colonies and provided an efficient way to test all colonies on a plate.
Mean densities by method.
The 12-month geometric mean V. parahaemolyticus densities for the three methods for samples obtained at zero time and 5, 10, and 24 h and 14 to 17 days after harvest are shown in Table 1. These methods were tested by using oysters that were growing at wide ranges of temperature and salinity and there were no significant differences between method means at any time (P > 0.12) after either storage under warm conditions or long-term refrigeration. The counts ranged from <10 to 800 CFU/g or 0.9 to 900 MPN/g at harvest, depending upon the season. The levels of V. parahaemolyticus recovery in this study (i.e., Direct-VPAP 12-month geometric mean of 86 CFU/g) agree closely with those previously reported for Gulf Coast oysters (8). The mean V. parahaemolyticus density was 110 CFU/g in a seasonal survey when the hydrophobic grid membrane filter method was used, and the highest densities occurred in spring and summer (8).
TABLE 1.
Twelve-month mean V. parahaemolyticus values for the three methods for samples obtained at zero time and after 5, 10, and 24 h and 14 to 17 days
| Method | Log10 CFU/g or MPN/g
|
||||
|---|---|---|---|---|---|
| Zero time | 5 h | 10 h | 24 h | 14–17 days | |
| Direct-VPAP | 1.94 ± 0.18 (12)a | 2.58 ± 0.24 (12) | 3.35 ± 0.29 (12) | 4.72 ± 0.22 (12) | 3.94 ± 0.28 (11) |
| Direct-VPDig | 1.85 ± 0.25 (10) | 2.45 ± 0.32 (10) | NDb | 4.23 ± 0.50 (5) | 3.54 ± 0.25 (10) |
| MPN-VPAP | 1.57 ± 0.34 (10) | 2.27 ± 0.33 (12) | 3.25 ± 0.35 (12) | 4.39 ± 0.31 (12) | 3.55 ± 0.39 (12) |
Mean ± standard error. The numbers in parentheses are the numbers of observations. The method-hour interaction was not significant (P > 0.12).
ND, not determined.
The V. parahaemolyticus densities increased during storage at 26°C by 1.4 and 3 logs (12-month means) after 10 and 24 h, respectively. After 14 to 17 days of refrigeration at 3°C, the mean count decreased by only 0.9 log from the 24-h level, suggesting that long-term refrigeration may not substantially reduce the numbers of bacteria present in raw oysters. Johnson and Liston (12) observed similar decreases in V. parahaemolyticus densities in naturally contaminated oysters stored at 11°C (0.8-log reduction after 8 days) and 5°C (1.6-log reduction after 14 days).
The Direct-VPAP mean was slightly higher than the other two means at each time point. The Direct-VPDig method included a repair step on magnesium-supplemented TSAMS agar (16), but the levels of recovery were comparable to those obtained with the Direct-VPAP method. The repair step was performed to account for any cellular damage due to temperature or salinity either before harvest or during storage and to compare the bacterial counts obtained by this method with those obtained by the Direct-VPAP procedure. This repair step may be insufficient to overcome the subsequent inhibition on the selective TCBS medium. The selective components of TCBS medium include oxgall, sodium citrate, and an alkaline pH (pH 8.6). The T1N3 agar used with the Direct-VPAP method was not selective, but its high salt concentration may inhibit some competing microflora. While optimization studies were not conducted with T1N3 agar, this medium was simple and economical to prepare, required no repair step, limited colony spreading, and gave good levels of V. parahaemolyticus recovery under all experimental conditions. The Direct-VPAP method can be completed in 1 day, compared with 2 days for the Direct-VPDig method and 3 to 4 days for the BAM-MPN method.
In conclusion, recent V. parahaemolyticus illness outbreaks emphasized the need for rapid, quantitative methods for environmental monitoring of V. parahaemolyticus levels in the environment. Two direct plating methods (the Direct-VPAP and Direct-VPDig methods) using nonradioactive DNA probes were equivalent to the MPN-VPAP procedure and provided a faster alternative for V. parahaemolyticus enumeration and confirmation in oyster samples.
ACKNOWLEDGMENTS
We thank Jessica Jones and Tony Previto of the Gulf Coast Seafood Laboratory for their assistance with the project. SAS statistical analyses performed by Alfred B. Moore at Mississippi State University are gratefully acknowledged.
REFERENCES
- 1.Baross J, Liston J. Occurrence of Vibrio parahaemolyticus and related hemolytic vibrios in marine environments of Washington state. Appl Microbiol. 1970;20:179–186. doi: 10.1128/am.20.2.179-186.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartley C H, Slanetz L W. Occurrence of Vibrio parahaemolyticus in estuarine waters and oysters of New Hampshire. Appl Microbiol. 1971;21:965–966. doi: 10.1128/am.21.5.965-966.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brasher C W, DePaola A, Jones D D, Bej A J. Detection of microbial pathogens in shellfish with multiplex PCR. Curr Microbiol. 1998;37:1–8. doi: 10.1007/s002849900346. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Outbreak of Vibrio parahaemolyticus infections associated with eating raw oysters—Pacific Northwest, 1997. Morb Mortal Wkly Rep. 1998;47:457–462. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Outbreak of Vibrio parahaemolyticus infection associated with eating raw oysters and clams harvested from Long Island Sound—Connecticut, New Jersey, and New York, 1998. Morb Mortal Wkly Rep. 1999;48:48–51. [PubMed] [Google Scholar]
- 6.Daniels N A, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond R M, Thompson S, Wilson S, Bean N H, Griffin P M, Slutsker L. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis. 2000;181:1661–1666. doi: 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 7.DePaola A, Hopkins L H, McPhearson R M. Evaluation of four methods for enumeration of Vibrio parahaemolyticus. Appl Environ Microbiol. 1988;54:617–618. doi: 10.1128/aem.54.2.617-618.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePaola A, Hopkins L H, Peeler J T, Wentz B, McPhearson R M. Incidence of Vibrio parahaemolyticus in United States coastal waters and oysters. Appl Environ Microbiol. 1990;56:2299–2302. doi: 10.1128/aem.56.8.2299-2302.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DePaola A, Motes M L, Cook D W, Veazey J, Garthright W E, Blodgett R. Evaluation of an alkaline phosphatase-labeled DNA probe for enumeration of Vibrio vulnificus in Gulf Coast oysters. J Microbiol Methods. 1997;29:115–120. [Google Scholar]
- 10.Department of Health and Human Services Food and Drug Administration. Interstate Shellfish Sanitation Conference. Washington, D.C.: U.S. Department of Health and Human Services Food and Drug Administration; 1997. National shellfish sanitation program guide for the control of molluscan shellfish; pp. 53–55. [Google Scholar]
- 11.Elliot E L, Kaysner C A, Jackson L, Tamplin M L. Bacteriological analytical manual. 8th ed. Arlington, Va: Association of Official Analytical Chemists; 1995. Vibrio cholerae, V. parahaemolyticus, V. vulnificus, and other Vibrio spp; pp. 9.01–9.27. [Google Scholar]
- 12.Johnson H C, Liston J. Sensitivity of Vibrio parahaemolyticus to cold in oysters, fish fillets and crabmeat. J Food Sci. 1973;38:437–441. [Google Scholar]
- 13.Kaneko T, Colwell R R. Ecology of Vibrio parahaemolyticus in the Chesapeake Bay. J Bacteriol. 1973;113:24–32. doi: 10.1128/jb.113.1.24-32.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneko T, Colwell R R. Incidence of Vibrio parahaemolyticus in Chesapeake Bay. Appl Environ Microbiol. 1975;30:251–257. doi: 10.1128/am.30.2.251-257.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko T, Colwell R R. The annual cycle of Vibrio parahaemolyticus in Chesapeake Bay. Microb Ecol. 1978;4:135–139. doi: 10.1007/BF02014284. [DOI] [PubMed] [Google Scholar]
- 16.Ma-Lin C F A, Beuchat L R. Recovery of chill-stressed Vibrio parahaemolyticus from oysters with enrichment broths supplemented with magnesium and iron salts. Appl Environ Microbiol. 1980;39:179–185. doi: 10.1128/aem.39.1.179-185.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy S A, DePaola A, Cook D W, Kaysner C A, Hill W E. Evaluation of alkaline phosphatase- and digoxigenin-labeled probes for detection of the thermolabile hemolysin (tlh) gene of Vibrio parahaemolyticus. Lett Appl Microbiol. 1999;28:66–70. doi: 10.1046/j.1365-2672.1999.00467.x. [DOI] [PubMed] [Google Scholar]
- 18.Raimondi F, Kao J P, Fiorentini C, Fabbri A, Donelli G, Gasparini N, Rubino A, Fasano A. Enterotoxicity and cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin in in vitro systems. Infect Immun. 2000;68:3180–3185. doi: 10.1128/iai.68.6.3180-3185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rippey S R. Infectious diseases associated with molluscan shellfish consumption. Clin Microbiol Rev. 1994;7:419–425. doi: 10.1128/cmr.7.4.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirai H, Ito H, Hirayama T, Nakabayashi Y, Kumagai K, Takeda Y, Nishibuchi M. Molecular epidemiologic evidence for association of the thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun. 1990;58:3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi H, Hirano H, Kubomura S, Higashi K, Mizuguchi Y. Comparison of the nucleotide sequences of the genes for the thermostable direct hemolysin and the thermolabile hemolysin from Vibrio parahaemolyticus. Microb Pathog. 1986;1:425–432. doi: 10.1016/0882-4010(86)90004-5. [DOI] [PubMed] [Google Scholar]
- 22.Weagant S D, Jagow J A, Jinneman K C, Omiecinski C J, Kaysner C A, Hill W E. Development of digoxigenin-labeled PCR amplicon probes for use in the detection and identification of enteropathogenic Yersinia and shiga toxin-producing Escherichia coli from foods. J Food Prot. 1999;62:438–443. doi: 10.4315/0362-028x-62.5.438. [DOI] [PubMed] [Google Scholar]