Abstract
In this paper we report the identification and characterization of a DNA region containing putative nif genes and belonging to a Burkholderia endosymbiont of the arbuscular mycorrhizal fungus Gigaspora margarita. A genomic library of total DNA extracted from the fungal spores was also representative of the bacterial genome and was used to investigate the prokaryotic genome. Screening of the library with Azospirillum brasilense nifHDK genes as the prokaryotic probes led to the identification of a 6,413-bp region. Analysis revealed three open reading frames encoding putative proteins with a very high degree of sequence similarity with the two subunits (NifD and NifK) of the component I and with component II (NifH) of nitrogenase from different diazotrophs. The three genes were arranged in an operon similar to that shown by most archaeal and bacterial diazotrophs. PCR experiments with primers designed on the Burkholderia nifHDK genes and Southern blot analysis demonstrate that they actually belong to the genome of the G. margarita endosymbiont. They offer, therefore, the first sequence for the nif operon described for Burkholderia. Reverse transcriptase PCR experiments with primers designed on the Burkholderia nifH and nifD genes and performed on total RNA extracted from spores demonstrate that the gene expression was limited to the germination phase. A phylogenetic analysis performed on the available nifK sequences placed the endosymbiotic Burkholderia close to A. brasilense.
Arbuscular mycorrhizal (AM) fungi are one of the most widespread microbial communities of the rhizosphere, since they establish symbiotic associations with the roots of about 80% of plant species (38). They are of very ancient origin. Fossil and molecular data demonstrate that they first appeared 400 to 350 million years ago, suggesting that the origin of land plants may be closely correlated to their symbiosis with AM fungi (36, 41).
The success of mycorrhizas in evolution is mainly due to the central role their fungi play in the capture of nutrients from the soil in almost all ecosystems (43). As a consequence, AM fungi are crucial determinants of plant biodiversity, ecosystem variability, and the productivity of plant communities (46).
It has been demonstrated that the AM fungus Gigaspora margarita (BEG isolate 34) harbors a homogeneous population of endobacteria in its cytoplasm throughout its life cycle (9). The high number of bacterial cells (about 250,000 per spore) and their presence in successive generations (V. Bianciotto, unpublished data) suggest that they are an integral part of the fungal system. On the basis of 16S rDNA sequence analysis, they have been assigned to the genus Burkholderia (9), an extremely heterogeneous group which includes soil bacteria, plant growth promoting rhizobacteria, and human and plant pathogens (6, 43). Strains of Burkholderia cepacia can survive within vacuoles in different isolates of the protozoa Acanthamoeba (27). In addition, Burkholderia isolates from rhizosphere have been proposed but not definitively shown to fix nitrogen (18).
Our objectives were to determine whether the symbiotic Burkholderia spp. possess the nitrogen fixation genes and whether these genes are expressed. Nitrogenase, the enzyme responsible for fixation, consists of two components, component I (also called FeMo protein or dinitrogenase), an α2β2 tetramer encoded by the nifD and nifK genes, and component II (dinitrogenase reductase or Fe protein), a homodimer encoded by the nifH gene. Component I contains two unusual, rare metal clusters, one of which is the iron molybdenum cofactor (FeMo-co), regarded as the site of dinitrogen reduction (26). The two components are conserved in structure, function, and amino acid sequence through diazotrophs (15, 32). For this reason we wished to detect the nifH, nifD, and nifK genes. Since Burkholderia cannot be maintained on a cell-free medium, we took advantage of a genomic library constructed from G. margarita spores (BEG 34) that also contained the bacterial genome (45). As a probe we used two Azospirillum brasilense DNA fragments (16, 32; R. Fani, unpublished data) harboring the nifH and nifDK genes, respectively. The second fragment has been previously reported to hybridize strongly with the DNA of Burkholderia nitrogen-fixing strains (40). A DNA region from an endosymbiotic Burkholderia strain harboring the −24 to −12 promoter region and the nifHDK operon was identified, isolated, and molecularly characterized. Reverse transcriptase PCR (RT-PCR) experiments showed the expression of the nif genes in germinated spores. In addition, in order to compare these genes with those from free-living Burkholderia, a partial sequence of the nifK gene from Burkholderia vietnamiensis TVV75 was obtained.
MATERIALS AND METHODS
DNA extraction.
Two fungal isolates were used: G. margarita Becker et Hall (New Zealand isolate BEG 34) and Gigaspora rosea Nicolson et Schenck (BEG 9), which have been demonstrated to possess or not possess bacteria in their cytoplasm (10). About 50 spores of both of the AM fungal species were recovered from pot cultures of Trifolium repens L. by wet sieving (17). They were rinsed five times with sterile, filtered, distilled water; as a further step they were surface sterilized with 4% chloramine-T and 300 ppm of streptomycin for 30 min, treated with 14 U of DNase (Promega) at 37°C for 30 min, sonicated five times, and then rinsed seven times for 1 h total with sterile, filtered distilled water. The intact spores were incubated with the Live/Dead BacLight bacteria viability kit (Molecular Probes Europe BV) at room temperature in the dark for 15 min to visualize contaminant bacteria, according to the manufacturer's instructions. This kit contains a proprietary mixture of nucleic acid stains that distinguishes live bacteria from dead bacteria (9).
DNA was extracted with a slightly modified cetyltrimethylammonium bromide method, according to Henrion et al. (22). Extreme care was taken to avoid any contamination. All solutions were filter sterilized, and sterile procedures were used throughout the DNA extraction.
RNA extraction.
About 100 spores of G. margarita, treated as reported above, germinated in filter-sterilized distilled water at 28°C for 6 days. RNA was extracted using the Qiagen RNeasy mini kit (Promega) according to the manufacturer's instructions from either germinated or nongerminated spores of G. margarita.
Bacterial lysates.
B. vietnamiensis strain TVV75 (40) was grown on tryptic soy agar medium (Sigma) at 37°C. Bacterial lysates were prepared by placing the samples in filter-sterilized distilled water at 92°C for 5 min and on ice for 2 min. Centrifugation at 14,000 × g followed, and the supernatant was collected and used in PCRs.
Screening of the genomic library.
A genomic library was constructed into the BamHI site of Lambda DASH II starting from total DNA extracted from G. margarita spores (including the Burkholderia endosymbiont DNA) and was partially digested with the restriction endonuclease Sau3AI (45). It was first established whether this library was representative of the bacterial genome. Calculations with Clarke and Carbon's equation (13) indicated that 2,000 primary recombinants are needed to represent the Burkholderia genome with 99% probability. To calculate this value it was considered that in each spore of G. margarita there are 2,000 to 3,000 fungal nuclei (7) of about 108 bp each (23) and 250,000 bacteria with a genome of approximately 106 bp (9). Therefore, about 30% of recombinant clones in the library should be prokaryotic. Since the number of total clones in the library was 65,000 (45), the number of prokaryotic clones should be about 22,000, thus making this library largely representative of the Burkholderia genome.
PCR products were separated by electrophoresis on 1.2% agarose gel in 1× Tris-borate-EDTA (TBE) running buffer. Approximately 4 × 104 Lambda DASH II (Stratagene) bacteriophage particles were plated onto NZY agar medium (per liter: 5 g of NaCl, 2 g of MgSO4, 5 g of yeast extract, 10 g of casein hydrolysate [pH 7.5], 15 g of agar) using Escherichia coli XL1-Blue MRA (P2) as the host (Stratagene). After incubation for 6 to 8 h at 38°C, the plates were overlaid for 2 min at room temperature with Hybond N+ nylon membranes (Amersham) (4 min in the case of duplicate filters). DNA was denatured and fixed by autoclaving the filters for 5 min at 120°C. The filters were then hybridized with a PstI-SmaI fragment of 1.7 kbp from plasmid pAFG1 (16) containing the entire nifD and the 5′-proximal region of the nifK genes of A. brasilense strain Sp7 (15, 30) and with an EcoRI-PstI fragment of about 1.5 kbp from plasmid pAFT4 (R. Fani, unpublished data) containing the entire nifH gene of A. brasilense strain Sp7. The probes were labeled using a chemiluminescence system (ECL Direct DNA labelling and detection system, Amersham). The filters were prehybridized with hybridization solution (0.5 M NaCl, 5% blocking reagent in ECL hybridization buffer) at 42°C for 1 h. Hybridization was performed overnight at 42°C. Then filters were washed twice for 20 min each at 42°C with 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.4% sodium dodecyl sulfate, 1 M urea, and finally, twice for 5 min each with 2× SSC at room temperature. An X-ray film (Hyperfilm ECL; Amersham) was exposed to the membranes for 3 h and developed as recommended by the manufacturer. Single positive plaques were identified after secondary and tertiary screening of positive areas using both probes. Positive plaques obtained using the nifDK probe were cut out from the agar plates and amplified. The DNA from one was purified with a Qiagen Lambda mini kit (Promega). The DNA was treated with 30 U of each of the restriction endonucleases EcoRI, SalI, NotI, and XbaI (Promega) which cleaved close to the BamHI site at 37°C for 3 h. The reaction products were analyzed by 0.8% agarose gel electrophoresis in TBE (4.5 mM Tris-borate, 1 mM EDTA) buffer.
Southern blotting analysis.
DNA fragments were transferred onto Hybond N+ membranes (Amersham) using standard procedures (33), and hybridizations were performed with the chemiluminescence detection system.
Subcloning and DNA sequencing.
Two SalI fragments of 800 bp and 1,500 bp giving a positive signal when hybridized to the nifDK probe were purified from agarose gel using the Qiaex II gel extraction kit (Promega) following the manufacturer's instructions. They were subcloned into the XhoI site of pZErO-1 plasmid vector (Invitrogene) using the Zero Background Cloning kit (Invitrogen) according to the manufacturer's instructions.
DNA sequencing was carried out by dideoxy sequencing (34) with the vector primers M13f and M13r, using the Sequenase kit (Pharmacia, Bridgewater, Mass.). Subsequent sequencing was done automatically using fluorescent dye-linked internal primers on the phage DNA and an Applied Biosystems model 370A DNA sequencer (Genome Express Society, Grenoble, France) and an Applied Biosystems model 373 stretch with XL upgrade DNA sequencer (Service de Sequencage, Université de Laval, Quebec, Canada). The sequence analysis was performed using the McVector program (IBI, New Haven, Conn.). (See “Nucleotide sequence accession numbers” below.)
Analysis of sequence data.
Amino acid and nucleotide sequences were retrieved from the GenBank, EMBL, and PIR databases. The ClustalW program (1) was used for pairwise and multiple alignments using default gap penalties. The structure and organization of the nifH, nifD, and nifK genes were deduced from the data available from the microorganisms available in data banks. BLAST (1) probing of the DNA and protein databases was performed with the tBLASTn, BLASTn, and BLASTP programs.
PCR and RT-PCRs.
PCRs were carried out on DNA extracted from the spores of G. margarita and G. rosea and on the washing solution of the spores, in a final volume of 20 μl containing 2 μl of DNA preparation, 250 mM each deoxynucleoside triphosphate, 2 μl of 10× buffer (Dynazyme; Celbio), 0.7 mM each primer, and 2.5 U of Dynazyme Taq polymerase (Celbio). The following amplification program was run in a Hybaid thermal cycler: 1.5 min at 95°C (1 cycle); 1.5 min at 95°C, 1 min at 62°C, and 2 min at 72°C (30 cycles); and a final elongation at 72°C for 10 min. PCR products were analyzed by electrophoresis on 1.2% agarose gel run in 1× TBE buffer (33). The following primers each amplified a 760-, 630-, and 1,080-bp fragment: nifHf (5′-GGCAAGGGCGGTATCGGCAAGTC-3′), nifHr (5′-CCATCGTGATCGGGTCGGGATG-3′), nifDf (5′-TGCACTTCACCTCGGATTTCCAGG-3′), nifDr (5′-CCTTGATGCTGTCGTCGAACAGAGC-3′), nifKf (5′-AAGGCTGCGTCGCCTATTACCG-3′), and nifKr (5′-GTCGCGCTCCAGATATTTGCC-3′).
PCR also was carried out on the washing solution of the spores using bacterial universal primers 704f and 1495r (9). Amplification of the B. vietnamiensis strain TVV75 nifK partial sequence was carried out by using primers nifKf and nifKr and by changing the annealing temperature (55°C instead of 62°C). The nucleotide sequence was determined via direct sequencing (Genome Express Society). (See “Nucleotide sequence accession numbers” below.)
RT-PCR experiments were carried out on RNA extracted from germinated and nongerminated spores of G. margarita using the Qiagen OneStep RT-PCR kit (Promega), according to the manufacturer's instructions, in a final volume of 25 μl containing 2 μl of RNA preparation, 10 mM each deoxynucleoside triphosphate, 5 μl of 5× OneStep RT-PCR buffer, 0.7 mM each primer, and 1 μl of OneStep RT-PCR enzyme mix. The amplification program was the same as described above, and the primers used were nifHf, nifHr, nifDf, nifDr, and the two additional primers BLOf and BLOr, which were specifically designed to amplify the 16S rDNA region of the endosymbiotic Burkholderia (9). Samples without reverse transcriptase were used as negative controls. RT-PCR products were analyzed by agarose gel electrophoresis in a 1× TBE running buffer (33). The amplified products were purified from agarose gel using the QIAEXII gel extraction kit (Promega) following the manufacturer's instructions. The nucleotide sequence was determined via direct sequencing (Genome Express Society).
Phylogenetic analysis.
Preliminary multiple alignments of amino acid sequences were generated with the program ClustalW (42) using default gap penalties. The selection of characters eligible for the construction of phylogenetic trees was optimized by comparing all sections of all the available NifK alignments with comprehensive inventories of significant binary alignments obtained by BLAST probing of the DNA and protein databases with representative archaeal and bacterial sequences (19). Phylogenetic trees were constructed using the distance matrix (DM) and maximum-parsimony (MP) methods. MP analysis used the PROTPARS program of the Phylogeny Inference Package (PHYLIP), version 3.57c (Department of Genetics, University of Washington, Seattle, Wash.); the PHYLIP programs SEQBOOT, PROTPARS, and CONSENSE were used to derive an MP tree which has replicated in 100 bootstraps. Evolutionary distances between all pairs of taxa (DM analysis) were calculated with the program PROTDIST of the PHYLIP 3.57c package, which estimates the number of expected amino acid replacements per position using a substitution model based on the Dayhoff 120 matrix; the resultant distant matrix was then used to construct a neighbor-joining tree with the program NEIGHBOR. The PHYLIP programs SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE were used (in that order) to derive a consensus tree based on 100 bootstrap replications of the original alignment.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been assigned GenBank accession numbers AF194084 (see “Subcloning and DNA sequencing” above) and AF175210 (see “PCR and RT-PCRs” above).
RESULTS
Isolation of DNA fragments harboring the endosymbiont Burkholderia nifHDK genes.
Low-stringency screening of 40,000 plaques of the genomic library of G. margarita with the A. brasilense nifDK and nifH probes gave 30 and 40 positive areas, respectively. Two single positive plaques were isolated following secondary and tertiary screening. They were designated λ1NIF and λ4NIF, respectively. The phage DNA of λ1NIF was purified and digested with four restriction enzymes (EcoRI, SalI, NotI, XbaI) that cleaved close to the cloning site. A Southern blotting experiment using the A. brasilense nifDK genes as a probe revealed the presence of strong hybridization signals (data not shown), suggesting that the recombinant phage harbored Burkholderia sequences homologous to the A. brasilense nifDK genes. Two SalI DNA fragments of about 800 and 1,500 bp, which gave strong hybridization signals with the nifDK probe, were purified and subcloned into the pZErO plasmid vector, and their nucleotide sequences were determined (not shown). A primer-walking strategy was adopted to obtain the nucleotide sequence of the flanking regions of the two SalI fragments.
The same primers were also used to determine the nucleotide sequences of the DNA of λ4NIF that gave a strong hybridization signal with the A. brasilense nifH probe. Results showed that the DNA inserts of λ1NIF and λ4NIF partially overlapped, as shown in Fig. 1. In this way the nucleotide sequence of a 6,413-bp region was obtained (accession number AF194084).
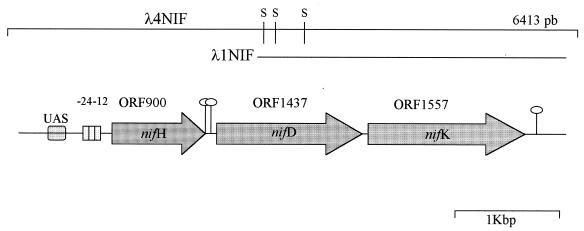
FIG. 1.
Schematic representation of the DNA region containing the putative Burkholderia nifHDK operon. Symbols: stem-loops, putative transcription terminators; UAS, upstream activator sequence; S, recognition site for SalI; −24−12 is the putative promoter recognized by ς54. The two lines represent the λ4NIF DNA insert and the overlapping λ1NIF sequence.
The analysis of the entire nucleotide sequence revealed the presence of three open reading frames (ORFs) of 900, 1,437, and 1,557 bp (hereinafter ORF900, ORF1437, and ORF1557). After tBLASTn and BLASTP searches, the putative amino acid sequences showed very low E values with the NifH and NifD-NifK Fe-Mo proteins from archaeal and bacterial diazotrophs (see below). These data confirmed that the DNA inserts harbored by λ1NIF and λ4NIF actually contained sequences encoding putative NifH, NifD, and NifK proteins, in agreement with hybridization data. The codon usage was in agreement with the GC contents of the three ORFs (63.0, 59.9, and 60.7%, respectively), similar to the GC content of the entire region (60.1%), with most codons ending with G or C.
The analysis of the region immediately upstream of the three ORFs revealed the presence of Shine and Dalgarno sequences (AGGAG, AGGAGG, and AAGGAA, respectively) located 9 bp from the ATG codon of each ORF. A sequence (CTGGCAC-N5-GTGCA) showing consensus with the −24 to −12 nif promoters (20) was found 80 nucleotides upstream of the nifH (ORF900) ATG codon. Moreover, two overlapping regions, identical to the upstream activated sequences of NifA-regulated genes (12) and characterized by the consensus sequence 5′-TGT-N10-ACA-3′, were found upstream of the putative Burkholderia nifH promoter region (Fig. 1). This suggested that the Burkholderia nifHDK genes might be subject to a regulation similar to that of other diazotrophs.
In the 192-bp nifH-nifD intergenic region, two repeated and inverted sequences that could act as putative Rho-dependent transcription terminators were found at positions 2092 and 2158, whereas no −24 to −12 sequences were found (Fig. 1). On the contrary, in the 34-bp nifD-nifK intergenic region, no sequence resembling transcription terminators was found. Finally, a putative strong Rho-independent transcription terminator (ΔG at 25°C of −90.5 kcal/mol) was found 67 bp downstream from the end of ORF1557 (nifK).
From these data we suggest that the three ORFs are arranged in an operon and that their transcription starts at the −24 to −12 promoter upstream from the nifH gene. A BLASTn analysis revealed that the nucleotide sequence of the three ORFs exhibited the highest degree of sequence identity to the corresponding A. brasilense genes (accession number M64344). On the contrary, the 1,000 bp downstream from ORF1557 (nifK) did not reveal the presence of ORFs (Fig. 1) and did not display a significant degree of identity with any sequence available in databases.
Analysis of the amino acid sequence of the Burkholderia NifH, NifD, and NifK proteins.
The three ORFs encoded putative proteins of 300, 479, and 518 amino acids, with molecular masses of 32,330, 53,938, and 58,012 Da, respectively. Amino acid sequences of the three proteins were then compared with those available in databases. A BlastP search revealed that the proteins encoded by ORF900, ORF1437, and ORF1557 exhibited sequence similarity (data not shown) to all the available archaeal and bacterial NifH, NifD, and NifK protein sequences, respectively.
A ClustalW pairwise comparison of the Burkholderia NifH, NifD, and NifK amino acid sequences with available orthologous archaeal and bacterial sequences determined their degrees of sequence identity and similarity. Results showed that the Burkholderia NifH, NifD, and NifK proteins exhibited the highest degree of sequence similarity to the A. brasilense NifH, NifD, and NifK (Fig. 2).
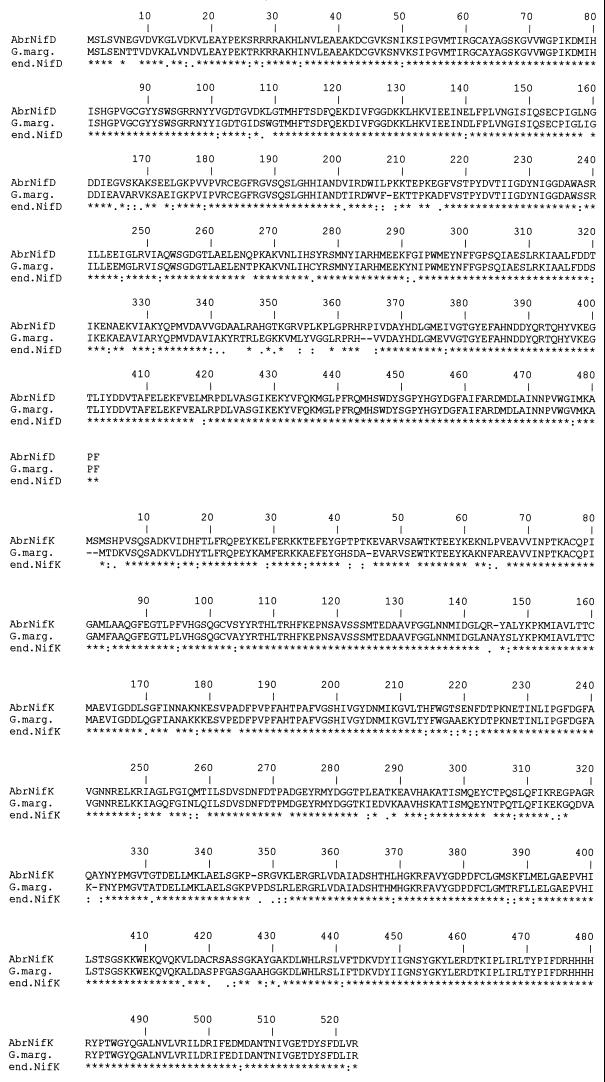
FIG. 2.
ClustalW alignment of the amino acid sequences deduced from the Burkholderia nifH, nifD, and nifK genes and from the A. brasilense orthologous genes. The amino acids are indicated by single-letter codes. Gaps were introduced for optimal alignment. Identical residues between two sequences are indicated by an asterisk (∗); similar residues are indicated by one dot (low similarity) or two dots (high similarity).
A ClustalW multialignment of all the available NifH, NifD, and NifK sequences was used to identify conserved regions and to detect the degrees of sequence similarity and identity between the Burkholderia NifH, NifD, and NifK proteins and the available orthologous proteins. The Burkholderia NifH protein contained four cysteine residues located at positions 39, 86, 98, and 132 and found at the corresponding positions in all known NifH proteins (39). Comparison of the α subunit sequences of component I of nitrogenase showed that the Burkholderia NifD protein contained five conserved cysteine residues at positions 62, 88, 154, 183, and 275 (12). As previously shown for other NifK proteins, the Burkholderia NifK protein contained three conserved cysteine residues at positions 75, 100, and 158 (5).
Amplification and sequencing of B. vietnamiensis TVV75 nifK partial sequence.
Primers nifKf and nifKr designed on the endosymbiont Burkholderia nifK sequence were used to amplify the corresponding region of B. vietnamiensis strain TVV75. Analysis of the 1,032-bp amplification fragment revealed that it encodes a putative polypeptide showing a very high degree of sequence similarity with the available NifK proteins (data not shown). ClustalW pairwise comparison of this partial sequence with the orthologous gene products revealed that the highest degrees of sequence identity and similarity were shown with the Bradyrhizobium japonicum NifK protein (68.9% identity; 86.6% considering amino acids with similar characteristics) rather than with the endosymbiont Burkholderia NifK (58.4% identity and 78.8% similarity).
Phylogenetic analysis.
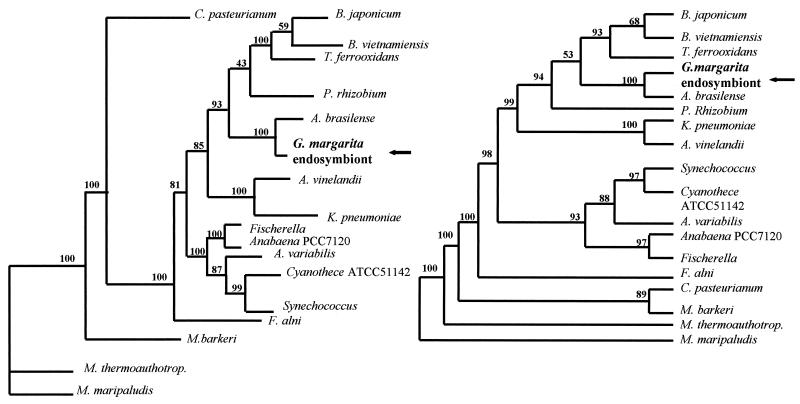
A ClustalW multialignment followed by visual inspection and manual editing of the NifK sequences reduced the complete sequences to a 322-amino-acid length where regions of ambiguous sequence alignment were excluded. Phylogenetic trees of the NifK amino acid sequences were inferred from the selected sequences by the DM and MP methods. Both the DM and MP analyses gave the same results; as shown in Fig. 3, both endosymbiont Burkholderia and B. vietnamiensis TVV75 belonged to a clearly separated group, which also includes A. brasilense, Parasponia rhizobium, B. japonicum, and Thiobacillus ferrooxidans. In agreement with the ClustalW results, endosymbiont Burkholderia was clearly placed closer to A. brasilense than to B. vietnamiensis TVV75, which is closer to B. japonicum.
FIG. 3.
Neighbor-joining (left) and maximum-parsimony (right) bootstrap consensus trees generated from the NifK alignment.
Amplification of Burkholderia nif genes from Gigaspora spores.
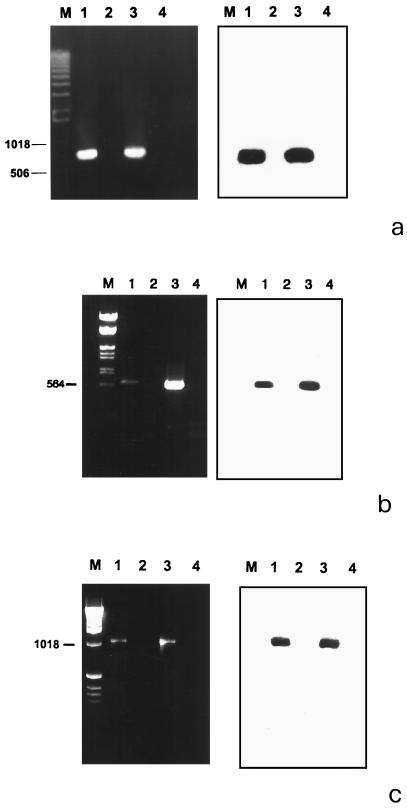
The sequences encoding the putative NifH, NifD, and NifK proteins were used to design three sets of primers for PCR experiments performed on DNA extracted from the spores of G. margarita and G. rosea. Unlike G. margarita isolates, G. rosea does not contain bacteria in its cytoplasm and therefore represents a negative control (10). The expected amplified fragments of about 760 bp (Fig. 4a), 630 bp (Fig. 4b), and 1,080 bp (Fig. 4c) were obtained from the G. margarita DNA. The results were further confirmed by Southern blotting experiments using as a probe the DNA of λ1NIF and λ4NIF amplified with the primers designed on the Burkholderia nifH, nifD, and nifK genes. By contrast no amplification product was obtained with DNA extracted from G. rosea (Fig. 4a to c).
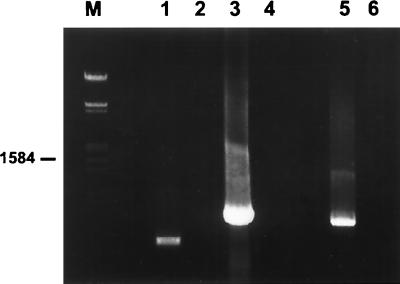
FIG. 4.
Agarose gel (left) and corresponding Southern blot (right) of PCR products amplified by using primers specific for the nifH (a), nifD (b), and nifK (c) genes. Lane 1, DNA from the spores of G. margarita; lane 2, DNA from the spores of G. rosea; lane 3, DNA from positive Lambda DASH phage; lane 4, no DNA. DNA markers (sizes are in base pairs) are shown on the left.
To exclude the presence of contaminating bacteria on the sporal surface at the end of the sterilization procedures, the intact spores of G. margarita and G. rosea were stained with the Live/Dead BacLight kit and observed in confocal microscopy as described by Bianciotto et al. (9). In both cases, the resulting spore surface was free from any bacterial contamination (data not shown). In addition, PCR experiments were carried out on the washing solution of G. margarita and G. rosea spores, using both the Burkholderia nif primers and the universal bacterial primers 704f and 1495r (9). As expected, universal bacterial primers amplified a DNA fragment of the expected size of about 790 bp, confirming the presence of contaminant bacteria on the surface of both of the fungal species prior to the spore sterilization protocol. No amplification product was obtained using the primers designed on the Burkholderia nif genes (data not shown).
RT-PCR experiments.
RT-PCR experiments were performed on the RNA extracted from nongerminated and germinated spores of G. margarita. Using the two sets of primers nifHf/nifHr and nifDf/nifDr, an amplified product of the expected size was obtained on RNA extracted from germinating spores. No amplified product was obtained from the RNA of nongerminated spores (data not shown) or from the RT negative controls. An amplified fragment of the expected size was also obtained using the primers BLOf and BLOr, specifically designed on the endosymbiont Burkholderia 16S rDNA sequence (9), on the RNA of germinated and nongerminated spores (Fig. 5).
FIG. 5.
Agarose gel of RT-PCR products amplified by using primers specific for the endosymbiont Burkholderia 16S ribosomal gene and the nifH and nifD genes. Lane 1, RNA from the germinated spores of G. margarita amplified with primers BLOf and BLOr; lane 2, no RT; lane 3, RNA from the germinated spores of G. margarita amplified with primers nifHf and nifHr; lane 4, no RT; lane 5, RNA from the germinated spores of G. margarita amplified with primers nifDf and nifDr; lane 6, no RT. A DNA marker (size in bp) is shown on the left.
The amplified products obtained with the nifHD genes and 16S rDNA primers were purified and sequenced. A nucleotide sequence of about 200 bp was obtained for all PCR products, showing a 100% sequence identity to the nifH, nifD, and 16S ribosomal genes of the endosymbiont Burkholderia.
DISCUSSION
The present paper demonstrates that the unculturable endosymbiotic Burkholderia harbored by G. margarita, an obligate mycorrhizal fungus, contains the nifHDK operon in its genome. To our knowledge, this is the first nif operon described for a bacterium of the genus Burkholderia. At least the nifH and nifD genes are expressed in the germinated spores of the fungal host. This is the first evidence in favor of the hypothesis that a fungus which improves P uptake (21) might also benefit from nitrogen fixation through a symbiosis with a diazotroph.
Identification and characterization of the nif genes in Burkholderia.
Unculturable bacteria may be successfully identified by using 16S rDNA isolated directly from natural samples (2, 28). Genes different from those encoding 16S or 23S rRNA, however, have rarely been investigated in unculturable micro-organisms. Nevertheless, highly conserved genes, such as those encoding the nitrogenase complex (nifH, nifD, and nifK), can be investigated from unculturable bacteria by using total DNA extracted from a given environment and by amplifying them via PCR. Following this approach and starting from a mixed DNA preparation, Ueda et al. (44) detected 23 nifH sequences belonging to different bacteria.
In our experiments, Burkholderia nif genes were identified and characterized by using a genomic library constructed from G. margarita spores (45) and representative of the endosymbiont Burkholderia genome. Bacterium-free genomic libraries can be constructed only by following special procedures, as demonstrated by Hosny et al. (24). We were able to sequence the nifHDK operon from the fungal-bacterial genomic library. Analysis of a 6,413-bp DNA region revealed that it harbors genes coding for putative proteins showing a very high degree of sequence similarity with three proteins of the nitrogenase complex, NifH, NifD, and NifK. The three proteins exhibited the highest degree of sequence similarity to FeMo NifD and NifK and to Fe NifH, suggesting that the Burkholderia nitrogenase should require a FeMo cofactor for its function. Moreover, the presence of a strong Rho-independent transcription terminator downstream from nifK and the absence of sequences resembling a −24 to −12 promoter in the short (34-bp) intergenic region suggested that they belong to the same transcriptional unit. The presence of two putative termination stem-loops located in the nifH-nifD intergenic region could be the signal for the production of nifH, nifDK, and nifHDK transcripts, suggesting a nitrogenase expression regulated at the level of initiation and termination of transcription (16).
The symbiont Burkholderia NifH, NifD, and NifK proteins showed the highest degree of sequence similarity to A. brasilense, a nitrogen-fixing rhizosphere bacterium often associated with maize (16, 35) and with diazotrophs involved in symbiotic relationships with legume plants, whereas the lowest degree of sequence similarity was shared with free-living bacterial and archaeal diazotrophs (31). The endosymbiont Burkholderia is more related to A. brasilense than to B. vietnamiensis. The apparent paradox of nif gene sequence similarity between unrelated species may be explained by horizontal gene transfer. Hurek et al. (25) found a high divergence of the nifH gene sequence in the genus Azoarcus. This situation is responsible for the deviation from the 16S ribosomal gene-based phylogeny. The divergence between the two Burkholderia NifK sequences might also be the consequence of a horizontal gene transfer which occurred between microorganisms belonging to distant taxa, as already demonstrated (4, 37).
nifHDK genes belong to the genome of the endosymbiont Burkholderia.
Southern blotting and PCR experiments on spores harboring (G. margarita) or not harboring (G. rosea) bacteria using specific primers designed on the Burkholderia nifHDK sequences provide evidence that the nifHDK genes characterized in this work actually belong to the genome of the G. margarita endosymbiont. The presence or absence of endosymbiotic bacteria in the two species was confirmed by morphological observations as well as by PCR experiments with universal and specific primers (9, 10).
It is known that rhizobacteria colonize the rhizosphere of AM plants (3) and may adhere to the surface of AM fungi during their extraradical phase (8). An obvious concern is that the DNA sequences may belong to such bacteria. However, these bacteria isolated from the rhizosphere, irrespective of their origin (endogenous or inoculated), have never been demonstrated to colonize the cytoplasm of living spores and hyphae. They are very different from the intracellular Burkholderia (29). In addition, they are easily detached from the sporal surface according to our cleaning procedures. By contrast, the presence of Burkholderia in successive fungal generations in vitro, as well as in situ hybridization experiments with the specific intracellular Burkholderia primers (10), confirm that they are an integral part of the fungal system.
Even if our results do not show directly that the sequences are derived from endosymbiotic bacteria, they surely show that bacteria are internal to the spores and that related fungi that lack bacteria lack the sequences. Taken as a whole, the results, coupled with the fact that nif genes are unknown for eukaryotes, strongly suggest that the nifHDK genes belong to the genome of the endosymbiotic Burkholderia.
Conclusion. This is the first demonstration that an AM fungus contains symbiotic intracellular bacteria possessing nif genes which are expressed at least during the germination of the spores. The significance of this finding (the potential capacity of a mycorrhizal fungus to fix nitrogen through a specific bacterial population) lies in the interest of such a combination for sustainable agriculture. The natural system consisting of mycorrhizal fungi and nitrogen-fixing bacteria may be an excellent “biofertilizer” with which to expand crop production and minimize the negative impact of chemical fertilizers.
ACKNOWLEDGMENTS
This research was supported by the EU GENOMYCA project, QLK5-CT-2000-01319.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programme. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amman R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrade G, Mihara K L, Linderman R G, Bethlenfalvay G J. Bacteria from rhizosphere soils of different arbuscular mycorrhizal fungi. Plant Soil. 1997;192:71–79. [Google Scholar]
- 4.Arber W. The generation of variation in bacterial genomes. J Mol Evol. 1995;40:7–12. [Google Scholar]
- 5.Arnold W, Rump A, Klipp W, Priefer U B, Puhler A. Nucleotide sequence of a 24,206-base-pair DNA fragment carrying the entire nitrogen fixation gene cluster of Klebsiella pneumoniae. J Mol Biol. 1988;203:715–738. doi: 10.1016/0022-2836(88)90205-7. [DOI] [PubMed] [Google Scholar]
- 6.Bevivino A, Tabacchioni S, Chiarini L, Carusi M V, Del Gallo M, Visca P. Phenotypic comparison between rhizosphere and clinical isolates of Burkholderia cepacia. Microbiology. 1994;140:1069–1077. doi: 10.1099/13500872-140-5-1069. [DOI] [PubMed] [Google Scholar]
- 7.Bianciotto V, Bonfante P. Quantification of the nuclear DNA content of two arbuscular mycorrhizal fungi. Mycol Res. 1993;96:1071–1076. [Google Scholar]
- 8.Bianciotto V, Minerdi D, Perotto S, Bonfante P. Cellular interactions between arbuscolar mycorrhizal fungi and rhizosphere bacteria. Protoplasma. 1996;193:123–131. [Google Scholar]
- 9.Bianciotto V, Bandi C, Minerdi D, Sironi M, Tichy H V, Bonfante P. An obligately endosymbiotic fungus itself harbors obligately intracellular bacteria. Appl Environ Microbiol. 1996;62:3005–3010. doi: 10.1128/aem.62.8.3005-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianciotto V, Lumini E, Lanfranco L, Minerdi D, Bonfante P, Perotto S. Detection and identification of bacterial endosymbionts in arbuscular mycorrhizal fungi belonging to the family Gigasporaceae. Appl Environ Microbiol. 2000;66:4503–4509. doi: 10.1128/aem.66.10.4503-4509.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brigle K E, Newton W E, Dean D R. Complete nucleotide sequence of the Azotobacter vinelandii nitrogenase structural gene cluster. Gene. 1985;37:37–44. doi: 10.1016/0378-1119(85)90255-0. [DOI] [PubMed] [Google Scholar]
- 12.Buck M, Miller S, Drummond M, Dixon R. Upstream activator sequences are present in the promoters of nitrogen fixation genes. Nature. 1986;320:375–378. [Google Scholar]
- 13.Clarke L, Carbon J. A colony bank containing synthetic ColE1 hybrids representative of the entire Escherichia coli genome. Cell. 1976;9:91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- 14.Dean D R, Jacobson M R. Biochemical genetics of nitrogenase. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 763–784. [Google Scholar]
- 15.de Zamaroczy M, Delorme F, Elmerich C. Regulation of transcription and promoter mapping of the structural genes for nitrogenase (nifHDK) of Azospirillum brasilense Sp7. Mol Gen Genet. 1989;220:88–94. doi: 10.1007/BF00260861. [DOI] [PubMed] [Google Scholar]
- 16.Fani R, Allotta G, Bazzicalupo M, Ricci F, Schipani C, Polsinelli M. Nucleotide sequence of the gene encoding the nitrogenase iron protein (nifH) of Azospirillum brasilense and identification of a region controlling nifH transcription. Mol Gen Genet. 1989;220:81–87. doi: 10.1007/BF00260860. [DOI] [PubMed] [Google Scholar]
- 17.Gerdemann J W. Spores of mycorrhizal Endogone species extracted from soil by wet sieving and decanting. Trans Br Mycol Soc. 1963;46:235–244. [Google Scholar]
- 18.Gillis M, Van Van T, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kerster K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 19.Gribaldo S, Cammarano P. The root of the universal tree of life inferred from anciently duplicated genes encoding components of the protein-targetting machinery. J Mol Evol. 1998;47:508–516. doi: 10.1007/pl00006407. [DOI] [PubMed] [Google Scholar]
- 20.Gussin G N, Ronson C W, Ausubel F M. Regulation of nitrogen fixation genes. Annu Rev Genet. 1986;20:567–591. doi: 10.1146/annurev.ge.20.120186.003031. [DOI] [PubMed] [Google Scholar]
- 21.Harrison M J, van Buuren M L. A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature. 1995;378:626–629. doi: 10.1038/378626a0. [DOI] [PubMed] [Google Scholar]
- 22.Henrion B, Le Tacon F, Martin F. Rapid identification of genetic variation of ectomycorrhizal fungi by amplification of ribosomal RNA genes. New Phytol. 1992;122:289–298. doi: 10.1111/j.1469-8137.1992.tb04233.x. [DOI] [PubMed] [Google Scholar]
- 23.Hosny M, Gianinazzi-Pearson V, Dulieu H. Nuclear DNA content of 11 fungal species in Glomales. Genome. 1998;41:422–428. [Google Scholar]
- 24.Hosny M, van Tuinen D, Jacquin F, Fuller P, Zhao B, Gianinazzi-Pearson V, Franken P. Arbuscular mycorrhizal fungi and bacteria: how to construct prokariotic DNA-free genomic libraries from the Glomales. FEMS Microbiol Lett. 1999;170:425–430. [Google Scholar]
- 25.Hurek T, Egener T, Reinhold-Hurek B. Divergence in nitrogenases of Azoarcus spp., Proteobacteria of the β subclass. J Bacteriol. 1997;179:4172–4178. doi: 10.1128/jb.179.13.4172-4178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Rees D C. Crystallographic structure and functional implications of the nitrogenase molybdenum-iron protein from Azotobacter vinelandii. Nature. 1992;360:553–560. doi: 10.1038/360553a0. [DOI] [PubMed] [Google Scholar]
- 27.Marolda C L, Hauroder B, John M A, Michel R, Valvano M A. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology. 1999;145:1509–1517. doi: 10.1099/13500872-145-7-1509. [DOI] [PubMed] [Google Scholar]
- 28.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 29.Perotto S, Bonfante P. Bacterial associations with mycorrhizal fungi: close and distant friends in the rhizosphere. Trends Microbiol. 1997;5:496–501. doi: 10.1016/S0966-842X(97)01154-2. [DOI] [PubMed] [Google Scholar]
- 30.Quiviger B, Franche C, Luftalla G, Rice D, Haselkorn R, Elmerich C. Cloning of a nitrogen fixation nif gene cluster of Azospirillum brasilense. Biochimie. 1982;64:495–502. doi: 10.1016/s0300-9084(82)80165-x. [DOI] [PubMed] [Google Scholar]
- 31.Rheinold-Hurek B, Hurek T. Life in grasses: diazotrophic endophytes. Trends Microbiol. 1998;6:139–144. doi: 10.1016/s0966-842x(98)01229-3. [DOI] [PubMed] [Google Scholar]
- 32.Ruvkun G B, Ausubel F M. Interspecies homology of nitrogenase genes. Proc Natl Acad Sci USA. 1980;77:191–195. doi: 10.1073/pnas.77.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5466. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrank I S, Zaha A, de Araujo E F, Santos D S. Construction of a gene library from Azospirillum brasilense and characterization of a recombinant containing the nif structural genes. Braz J Med Biol Res. 1987;20:321–330. [PubMed] [Google Scholar]
- 36.Simon L, Bousquet J, Levesque R C, Lalonde M. Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature. 1993;363:67–68. [Google Scholar]
- 37.Smith J M, Dawson C G, Spratt B G. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 38.Smith S E, Read D J. Mycorrhizal symbiosis. London, England: Academic Press; 1997. [Google Scholar]
- 39.Souillard N, Magot M, Possot O, Sibold L. Nucleotide sequence of regions homologous to nifH (nitrogenase Fe protein) from the nitrogen-fixing archaebacteria Methanococcus thermolithotrophicus and Methanobacterium ivanovii: J. Mol Evol. 1988;27:65–76. doi: 10.1007/BF02099731. [DOI] [PubMed] [Google Scholar]
- 40.Tabacchioni S, Visca P, Chiarini L, Bevivino A, Di Serio C, Fancelli S, Fani R. Molecular characterization of rhizosphere and clinical isolates of Burkholderia cepacia. Res Microbiol. 1995;46:531–542. doi: 10.1016/0923-2508(96)80559-6. [DOI] [PubMed] [Google Scholar]
- 41.Taylor T N, Remy W, Hass H, Kerp J. Fossil arbuscular mycorrhizae from the early devonian. Mycologia. 1995;87:560–573. [Google Scholar]
- 42.Thompson J D, Higgins D G, Gibson T J. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight-matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tipper J, Ingham E, Cove J H, Todd N, Kerr K G. Survival and multiplication of Burkholderia cepacia within respiratory epithelial cells. Clin Microbiol Infect. 1998;4:450–459. [Google Scholar]
- 44.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Buuren M, Lanfranco L, Longato S, Minerdi D, Harrison M J, Bonfante P. Construction and characterization of genomic libraries of two endomycorrhizal fungi: Glomus versiforme and Gigaspora margarita. Mycol Res. 1999;103:955–960. [Google Scholar]
- 46.van der Heijden M G, Klironomos J N, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders I R. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 1998;396:69–71. [Google Scholar]