Abstract
Mammalian bone is constantly metabolized from the embryonic stage, and the maintenance of bone health depends on the dynamic balance between bone resorption and bone formation, mediated by osteoclasts and osteoblasts. It is widely recognized that circadian clock genes can regulate bone metabolism. In recent years, the regulation of bone metabolism by non-coding RNAs has become a hotspot of research. MicroRNAs can participate in bone catabolism and anabolism by targeting key factors related to bone metabolism, including circadian clock genes. However, research in this field has been conducted only in recent years and the mechanisms involved are not yet well established. Recent studies have focused on how to target circadian clock genes to treat some diseases, such as autoimmune diseases, but few have focused on the co-regulation of circadian clock genes and microRNAs in bone metabolic diseases. Therefore, in this paper we review the progress of research on the co-regulation of bone metabolism by circadian clock genes and microRNAs, aiming to provide new ideas for the prevention and treatment of bone metabolic diseases such as osteoporosis.
Keywords: Circadian rhythm, Circadian clock gene, MicroRNAs, Bone metabolism
1. Introduction
Bone is an important component of the locomotor system. Bone health depends on a dynamic balance between bone resorption and bone formation determined by the regular metabolic activities of various bone cells. Bone metabolism is influenced and regulated by both mechanical and biological factors (Tong et al., 2019). Some of these factors, such as the production and secretion of various hormones and cytokines, show regular chronobiological properties, which can vary with circadian rhythms. The bone cells themselves also undergo rhythmic activity with the circadian alternation (Gonçalves and Meng, 2019). These effects can be explained by the existence of a time-measuring device, the circadian clock. It exists in all organisms and regulates internal physiology to adapt to changes in the external environment. Circadian clock genes have now been used as therapeutic targets for many diseases and this has led to the creation and development of chronotherapy (Ruan et al., 2021). The body needs to conform to its internal circadian clock to stay healthy, and abnormal circadian rhythms will negatively affect the functioning of its systems. Shift workers and mice exposed to reversed circadian conditions show osteoporotic features such as bone microstructure degeneration, reduced bone mass, decreased bone strength, and increased bone brittleness (Schilperoort et al., 2020). An abnormal circadian rhythm affects bone health, which again suggests that bone metabolism can be regulated by the circadian clock, and that altered expression of circadian clock genes is the underlying cause. However, the mechanism whereby circadian clock genes regulate bone metabolism is still unknown.
MicroRNAs (miRNAs) are endogenous small RNAs involved in post-transcriptional repression of messenger RNA (mRNA) in eukaryotic cells. Their regulation of gene expression is considered a third epigenetic mechanism in addition to histone modification and DNA methylation (Bartel, 2004; Michou, 2018). MiRNAs are widely involved in the regulation of various physiological and pathological processes in organisms, including bone metabolism (Taipaleenmäki, 2018). Numerous studies have shown that miRNAs can effectively regulate bone resorption and formation by targeting key factors in bone metabolic signaling pathways (Ma et al., 2019; Zheng et al., 2019; Zhang et al., 2021). As a result, targeted therapies involving miRNAs have gradually been applied to the field of bone metabolic diseases. Both circadian clock genes and miRNAs can participate in the regulation of bone metabolism, but the mechanisms whereby they regulate bone metabolism remain to be clarified. Therefore, in this review we analyze the progress made in research on circadian clock genes and miRNAs and their co-regulation in bone metabolism, aiming to provide a comprehensive new approach for the prevention and treatment of bone metabolic diseases.
2. Circadian clock genes and their biological functions
The circadian clock is an intrinsic rhythm that regulates various physiological and behavioral activities of mammals in accordance with the 24-h daily diurnal alternation. The circadian clock that regulates circadian rhythms can be divided into a central clock and a peripheral clock. It is located in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus and in some other brain regions and peripheral organs (Schibler et al., 2003). The production of two types of circadian clock depends on the transcription‒translation feedback loops of specific genes, which we refer to as circadian clock genes. The main circadian clock genes identified in mammals and Drosophila so far include: (1) core circadian clock genes such as brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like 1 (BMAL1), circadian locomotor output cycles kaput (CLOCK), neuronal PAS domain protein 2 (NPAS2), period (PER), and cryptochrome (CRY); (2) peripheral circadian clock genes such as nuclear receptor subfamily 1 group D member 1 (NR1D1)/REV-ERBα and NR1D2/REV-ERBβ, retinoid-related orphan receptor (ROR, including RORα, RORβ, and RORγ), timeless (TIM), differentiated embryo chondrocyte (DEC), Duffy-binding protein (DBP), and Nocturnin (NOCT). They encode proteins that constitute complex positive/negative regulatory loops. Positive elements activate transcription of downstream clock-controlled genes to promote rhythmic gene expression, playing important regulatory roles in organismal physiology, behavior, and even psychology (Schibler, 2005; Zvonic et al., 2007; Guo et al., 2009; An et al., 2020).
2.1. Core circadian genes BMAL1 and CLOCK and their biological functions
BMAL1 and CLOCK are positive regulators in the circadian rhythm system. BMAL1, a gene essential for the generation of 24-h periodic behavior, is predominantly located in the nucleus, where it encodes proteins with distinct circadian oscillations (Ray et al., 2020; Yang et al., 2020). A two-branch regulation model of circadian clock genes has been proposed, in which the BMAL1-centered circadian clock regulatory network in the body is divided into an autonomous response branch and a memory branch. When there is no clock dependent on BMAL1, the circadian clock is guided by light, but the body will rely on a BMAL1-dependent clock to "remember" time in the absence of light (Welz et al., 2019). BMAL1 often forms dimers with proteins encoded by other circadian clock genes (e.g., BMAL1-CLOCK, BMAL1-NPAS2), which bind to E-box or D-box elements located in the promoter of clock-controlled genes to activate their transcription (DeBruyne et al., 2007). CLOCK usually functions as a dimerization partner of BMAL1 by binding to BMAL1 (Menet et al., 2014). There are two known circadian rhythm complexes located in the nucleus: the BMAL1-CLOCK complex and the PER complex. The BMAL1-CLOCK positively drives the circadian rhythm system (Aryal et al., 2017).Studies suggested that BMAL1 and CLOCK in mammals such as mice and humans are involved in the regulation of various biological processes such as circadian rhythms, immunometabolism, hormone production, and cellular activities, as positive regulators of circadian clocks (Beker et al., 2019; Nagao et al., 2019; Alexander et al., 2020).
2.2. Core circadian genes CRY and PER and their biological functions
CRY (CRY1, CRY2) and PER (PER1, PER2, and PER3) are negative regulators of the circadian rhythm system. CRY and PER are phosphorylated by casein kinase 1δ (CK1δ) when their proteins accumulate to a critical level. They subsequently enter the nucleus to bind directly to the BMAL1-CLOCK protein dimer to form a complex that inhibits the transcription of CRY and PER themselves. This complex, called the PER complex, is another complex in the nucleus. It begins to accumulate at the onset of negative feedback and takes about 7 h to reach saturation and then negatively regulate circadian rhythms (Fogle et al., 2011; Aryal et al., 2017; Cao et al., 2017). CRY is considered as a blue photoreceptor in Drosophila due to its sensitivity to light, while its photoreceptor function in mammals has not been determined (Fogle et al., 2011). PER is circadian clock gene and is also thought to be tumor suppressor gene (Yang et al., 2009). Numerous studies have confirmed that CRY and PER regulate circadian rhythm and autoimmunity genes like BMAL1 and CLOCK. They also play important roles in hormone production, hypoxic response, and thermoregulation as negative regulators of the circadian clock (Chappuis et al., 2013; Cao et al., 2017; Kobayashi et al., 2017; Patke et al., 2017).
2.3. Peripheral circadian clock genes and their biological functions
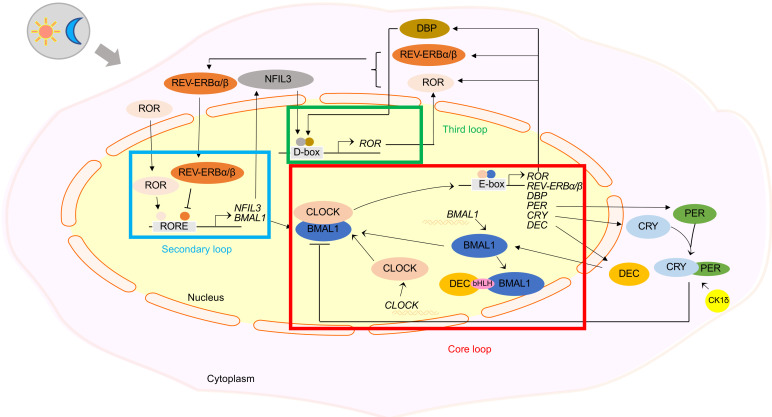
In addition to several core circadian clock genes, some other circadian clock genes play an integral role in the regulatory loop. We collectively refer to these other genes as peripheral circadian clock genes. Peripheral circadian clock genes include mainly REV-ERBα (NR1D1), ROR, TIM, DEC, DBP, and NOCT. Apart from the regulation of circadian rhythm, some of these genes have other functional commonalities due to their tissue specificity. For example, both REV-ERBα and RORα are involved in the development of airway inflammatory diseases and cancers (Gaertner et al., 2019; Durrington et al., 2020). However, their functions are not identical, as REV-ERBα plays a broad regulatory role in pathological situations of heart failure and cancer (Wang et al., 2020; Yue J et al., 2020), while RORα is currently considered a regulator of emotional disorders such as anxiety and depression (Guissart et al., 2018). Furthermore, these peripheral circadian clock genes can interact with core circadian clock genes and participate in the regulation of immune response, hypoxic response, energy metabolism, cell proliferation, and DNA replication (Lin et al., 2002; Liu et al., 2016, 2018; Sato et al., 2018; Kurien et al., 2019; Rageul et al., 2020). The expression of some peripheral circadian clock genes has been shown to serve as a reliable biomarker for the diagnosis and prognosis of certain diseases (Bianco et al., 2019; Rageul et al., 2020). Furthermore, some targeted drugs related to peripheral circadian clock genes have been developed to effectively treat some specific diseases in animals (Sulli et al., 2018). The peripheral clock circadian genes are not redundant in the circadian clock regulation system. On the contrary, they cooperate with the core circadian clock genes to greatly enrich the network of the circadian clock to regulate the metabolism of life activities. The cycle of the circadian clock system can be simply divided into three loops (Ruan et al., 2021), as shown in Fig. 1.
Fig. 1. A model for the positive and negative feedback loops formed by the interaction of circadian clock genes (modified from the study of Ruan et al. (2021)) . bHLH: basic helix-loop-helix; BMAL1: brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like 1; CK1δ: casein kinase 1δ; CLOCK: circadian locomotor output cycles kaput; CRY: cryptochrome; DBP: Duffy-binding protein; DEC: differentiated embryo chondrocyte; NFIL3: nuclear factor interleukin 3; PER: period; REV-ERBα/β: nuclear receptor subfamily 1 group D 1/2 (NR1D1/2); ROR: retinoid-related orphan receptor; RORE: ROR/REV-ERB-response element.
The relationship between circadian rhythm disorders and human diseases has received much attention. Many studies have focused on the mechanism of action of circadian clock genes in relation to cardiovascular diseases and tumor-related phenotypes, in which circadian clock genes were generally described as tumor suppressor genes (Jiang et al., 2016; Shi et al., 2022). Earlier studies pointed out that mice with defective circadian clock genes show reduced activity and their bones undergo pathological changes as they age. This showed that circadian clock genes not only regulate behavioral activities, but also play an important role in body metabolism, and obviously bone metabolism is one area of interest (Bunger et al., 2005; Jiang et al., 2016).
3. Regulation of bone metabolism by circadian clock genes
3.1. Bone metabolism
Bone metabolism is a dynamic and balanced process in which osteoclasts and osteoblasts coordinate with each other to break down and resorb aging bone and synthesize new bone to maintain normal bone mass and strength. In addition, bone mesenchymal stem cells (BMSCs) play an important role in bone metabolism because of their ability to differentiate into both osteoblasts and chondrocytes, while chondrocytes can be transformed into part of bone tissue. Therefore, as observed at the cellular level, cells involved in bone metabolism include osteoblasts, osteoclasts, BMSCs, and osteocytes (Khosla, 2013). At the molecular level, bone formation or bone resorption is generally determined by the expression of specific genes or proteins. The identified osteogenic markers include alkaline phosphatase (ALP), Runt-related transcription factor 2 (Runx2), osteocalcin (OCN), osteopontin (OPN), osteoprotegerin (OPG), collagen I (Coll I), and bone sialoprotein (BSP). The osteoclastic markers include tartrate-resistant acid phosphatase (TRACP) and nuclear factor of activated T cell cytoplasmic 1 (NFATc1). They regulate bone metabolism by mediating various signaling pathways such as Wnt/β-catenin, bone morphogenetic protein (BMP), OPG/receptor activator of nuclear factor-κΒ (NF-κB) (RANK)/RANK ligand (RANKL), and Notch. Bone metabolism is also affected by many other factors, including mechanical factors (such as mechanical stimulation caused by exercise) and biological factors (such as hormones and cytokines), and indirectly affected by drugs used to treat other systemic diseases on bone metabolism (Luan et al., 2019; Tong et al., 2019; Wang et al., 2021). A growing number of studies in recent years have shown that the balance of bone metabolism is closely related to circadian clock genes (Chan et al., 2021), so circadian clock genes are also one of the factors that affect bone metabolism.
3.2. Circadian clock genes in bone metabolism
Various types of bone cells have been shown to express circadian clock genes. The temporal programming of the expression of circadian clock genes is driven by timekeeping mechanisms that lead to the activity of rhythmic clock protein transcription factors (Feeney et al., 2016). At the cellular level, changes in the expression of various circadian clock genes caused by circadian rhythm disorders mainly cause the function or activity of bone cells to change the turnover of bone, rather than the quantity. The expression of genes in osteoclasts was found to be more rhythmic than that of osteoblasts in the regular alternation of light and dark throughout the day. However, osteoblasts and other bone cells share the same circadian oscillations in gene expression. This suggests that circadian rhythms exist in the activity of all types of bone cells and collectively promote rhythmic bone remodeling (Fujihara et al., 2014; Schilperoort et al., 2020). At the molecular level, the expression of some biomarkers related to bone formation and bone resorption maintains a characteristic oscillatory pattern with respect to the expression of circadian clock genes. For example, the expression of Bmal1 and Clock in mice tends to increase during the day and decrease at night, while Per and Cry show a decrease in expression during the day and an increase at night. Correspondingly, the expression of OPG rises during the day and falls during the night, while RANKL and cathepsin K (CTSK) show a daily decline and a nightly rise (Schilperoort et al., 2020). There may be a correlation between the expression of bone metabolic factors such as OPG and the expression of circadian clock genes in mice. In addition, a clinical study showed that serum levels of type 1 procollagen N-terminal prepropeptide (P1NP), an indicator of bone formation, were reduced after circadian rhythm disturbances (Swanson et al., 2017). Therefore, the evidence demonstrates that there is an inextricable link between changes in the expression of circadian clock genes caused by altered circadian rhythms and bone metabolism (Fig. S1).
3.2.1. Core circadian genes BMAL1 and CLOCK in bone metabolism
BMAL1 is expressed in bone and affects bone metabolic processes. Its abnormal expression is associated with skeletal diseases such as skeletal dysplasia, osteoarthritis, and osteoporosis (Chen et al., 2020). Recently, it has been shown that osteoclast-specific knockdown of Bmal1 in 10-week-old mice results in a phenotype with higher bone mass due to reduced osteoclast differentiation after two weeks of diurnal reversal (Samsa et al., 2016; Xu et al., 2016). In contrast, Zhou et al. (2018) suggested that Bmal1 deficiency accelerates osteoclast differentiation. This was demonstrated by a decrease in Bmal1 in the mandible of patients with skeletal mandibular hypoplasia (SMH) and a simultaneous decrease in OPG expression, which inhibits osteoclast differentiation. In addition, reduced mandibular bone mass and reduced OPG expression were observed in both a circadian disorder mouse model established by advancing the daily light time by 6 h for 4–8 weeks and the Bmal1 -/- mouse. The mechanism suggests that BMAL1 inhibits osteoclast differentiation by directly binding to the promoter of OPG and upregulating OPG expression, suggesting that Bmal1 deficiency accelerates osteoclast differentiation (Zhou et al., 2018). However, Tsang et al. (2019) suggest that bone mass in mice may be controlled to a greater extent by the intrinsic circadian clock in BMSCs rather than in osteoclasts. This is because knockout of Bmal1 in osteoclasts did not alter bone mass or trabecular architecture, and osteoclast differentiation was not affected in vitro compared to the knockout of Bmal1 in BMSCs. Several studies on Bmal1 and osteoclast differentiation have reached different conclusions. Differences in the age and sex of the mice selected, the manner in which the dysbiosis was modeled, the duration of the intervention, or the type of cells conditionally knocked out may be the reason. In addition, the indirect effect of BMSCs on osteoblast activity and the crosstalk between osteoblasts and osteoclasts may have been overlooked. For example, the expression of Bmal1 is suppressed in the BMSCs of type 2 diabetes mellitus (T2DM) rats. The ratio of RANKL/OPG and the expression of inhibitor κB (IκB), phosphorylated p65 (p-p65), and caspase-3 phosphorylated IκB (p-IκB) are increased. Thus, overall osteogenic capacity was reduced in T2DM rats compared to healthy rats. When Bmal1 is overexpressed, it restores the osteogenic capacity of BMSCs and inhibits osteoclast production and function by inhibiting the NF-κB signaling pathway (Li et al., 2018). This has been verified in the study of Mao et al. (2020) and provides a new direction for further exploring the mechanism of Bmal1 in diabetic osteoporosis. Overexpression of Bmal1 upregulates the expression of osteogenic genes including BMP2, Runx2, ALP, and OCN in BMSCs. When the effect of Bmal1 on osteogenic differentiation of BMSCs was analyzed alone, Bmal1 could promote osteogenic differentiation of BMSCs through the BMP2 signaling pathway (Huang ZF et al., 2020). The conclusion that Bmal1 can promote osteogenic differentiation by regulating the expression of BMP2 is also supported in osteoblasts and MC3T3-E1 (Min et al., 2016). The lack of Bmal1 in osteoblasts accelerates osteoclast-mediated bone resorption because of the interaction between osteoblasts and osteoclasts (Takarada et al., 2017). Apparently, there is also evidence indicating that Bmal1 in chondrocytes can regulate their activity and function, but the mechanism is unclear (Fu et al., 2019; Li et al., 2020).
Knockdown of CLOCK, a dimerization partner of BMAL1, in human BMSCs induced downregulation of BMAL1 and a reduction in the amplitude of oscillations without altering the cyclic oscillations. It also caused differentiation of BMSC to adipocytes and mildly suppressed expression of OCN, but this is not enough to conclude that knockdown of CLOCK significantly inhibits the osteogenic differentiation of BMSCs (Boucher et al., 2016). Knocking out the CLOCK alone may not lead to a dramatic effect on bone metabolism. An animal study with conditional deletion of Clock showed that protein disulfide isomerase family A member 3 (PDIA3), a circadian clock gene that encodes a protein for the 1,25-dihydroxy-vitamin D3 (1α,25(OH)2D3) receptor, can promote the differentiation of osteoblasts in mice by acting as a transcriptional activator of PDIA3 to further activate 1α,25(OH)2D3 and the downstream PKC signaling pathway (Yuan GS et al., 2017). However, the effect of CLOCK on human bone metabolism has not been directly investigated. Knockdown of CLOCK in human ovarian granulosa-like cell line, KNG cells, resulted in reduced levels of estradiol. The BMP system is also involved and synergistically regulates ovarian steroid hormone production (Nagao et al., 2019). Since estrogen levels are closely related to bone homeostasis, this pathway in which CLOCK regulates estrogen expression and then regulates bone metabolism may be a novel mechanism to investigate.
3.2.2. Core circadian clock genes CRY and PER in bone metabolism
CRY is light-sensitive circadian regulator in invertebrates, but whether it has light-sensitive effects in vertebrates is still being explored. Laser irradiation induces translocation of CRY1 from the cytoplasm to the nucleus and subsequently regulates extracellular calcification in mouse BMSCs. This promotes osteogenic differentiation and reduces lipogenic differentiation of BMSCs (Kushibiki and Awazu, 2009). Activating transcription factor 4 (ATF4) is a major regulator of osteoblast differentiation in growing mice. The expression of Cry1 is mediated by ATF4, which affects bone metabolism and bone strength in mice with chronic kidney disease (Pawlak et al., 2017). Further study of the mechanism revealed that Cry1 regulates cell proliferation and osteogenic differentiation in a way that is dependent on protein kinase B (PKB/AKT) and extracellular signal-regulated kinase (ERK) signaling pathways. This finding was based on the results of a cell counting kit-8 (CCK8) assay and mRNA expression of some osteogenic factors such as ALP after inhibiting the expression of Cry1 in C3H10 (Zhou et al., 2019). In pathological conditions such as arthritis, both mRNA and protein levels of CRY1 are significantly downregulated. After melatonin treatment of anti-type II collagen antibody-induced arthritis in mice, this downregulation is accompanied by degeneration of articular cartilage and bone structures, suggesting that Cry1 may be involved in the progression of melatonin-treated arthritis (Bang et al., 2012). The latest study found that the expression of CRY2, rather than CRY1,was significantly reduced in the cartilage of humans and mice with arthritis. Also, the knockout of Cry2 in mice showed a significant increase in the severity of pathological changes in cartilage, subchondral bone, and synovial tissue compared with the knockout of Cry1 (Bekki et al., 2020). Moreover, this study suggested that Cry2 may play an active role in maintaining dynamic homeostasis of the extrachondral matrix (ECM). Fifty-three differentially expressed genes, including Nr1d1, Nr1d2, DBP, and thyrotroph embryonic factor(Tef), which are all target genes of Cry2, were identified from RNA sequencing of knee cartilage in wild-type and Cry2 -/- mice. Gene Ontology (GO) and search tool for the retrival of interacting genes/proteins (STRING) databases were used to analyze the pathways and mechanisms by which these differentially expressed genes regulate osteoarthritis. A key protein, platelet-derived growth factor receptor α (PDGFRA), was found to belong to the angiogenic pathway, the circadian rhythm pathway and the ECM pathway (Bekki et al., 2020).
Previous studies have shown that mice exhibit high bone mass if they lack Per in their osteoblasts. Normal expression of Per in osteoblasts mediates leptin-dependent sympathetic inhibition of bone formation through inhibition of osteoblast proliferation and the expression of cyclin D1 (Fu et al., 2005). This suggests that Per negatively regulates bone formation. The speculation that Per does not directly regulate bone metabolism by altering gene expression in osteoblasts was corroborated in later studies in which Per2 mutant mice showed significantly higher bone formation rates compared to wild-type mice, but the parameters of osteogenic or osteolytic-specific genes did not differ. Therefore, Per2 mutation may lead to alterations in some other factors that can regulate osteoblast activity, and different levels of systemic and cell-autonomous regulatory mechanisms appear to regulate osteoblast activity (Maronde et al., 2010). Recently, functional studies of Per2 in BMSCs and osteoblasts derived from mice have shown that Per2 negatively regulates both osteogenic differentiation of BMSCs and the proliferative capacity of osteoblasts (Zhuo et al., 2018; Abe et al., 2019). In contrast, transfection experiments in human BMSCs revealed that knockdown of PER2 significantly promoted the lipogenic differentiation ability of BMSCs and mildly inhibited their osteogenic differentiation ability but had little effect, as did knockdown of CLOCK. The difference is that knockout of PER2 can also affect the activity of stem cells and reduce the cell migration ability of BMSCs (Boucher et al., 2016). Thus, although CRY and PER are both negative regulators of circadian rhythms, they play their respective roles in bone anabolism and catabolism, with CRY having mainly osteogenic effects and PER negatively regulating bone formation.
3.2.3. Peripheral circadian clock genes in bone metabolism
A functional study has shown that Rev-erbα affects the ability of BMSCs to proliferate and differentiate into osteoblasts, and that its agonist has the ability to inhibit osteoclastogenesis and prevent bone loss caused by ovariectomy (Song et al., 2018). Mechanistic studies revealed that the expression of some markers of osteoclastic differentiation and osteogenic differentiation was strongly influenced by the level of Rev-erbα (He et al., 2015; Kim et al., 2020). Inhibiting the expression of Rev-erbα in osteoclast and osteoblast precursor cells caused simultaneous elevated expression of osteoclastic factors such as TRACP and NFATc1, and osteogenic factors such as ALP and BSP. Overexpression of Rev-erbα in osteoblast and osteoblast precursor cells resulted in a simultaneous enhancement of osteoclastic and osteogenic differentiation, accompanied by the activation of the p38 mitogen-activated protein kinase (MAPK) signaling pathway. Therefore, it was inferred that Rev-erbα may negatively affect osteoclast and osteoblast differentiation through the p38 MAPK signaling pathway (He et al., 2015; Kim et al., 2020). Another gene, RORα,which has been poorly studied in relation to bone metabolism, is involved in the metabolic activity of osteoblasts. It stimulates the expression of osteogenic factors such as ALP and OCN in MG-63 cells or suppresses the inflammatory response (Benderdour et al., 2011). There is still a lack of evidence that RORα directly regulates bone metabolism. The expression of RORα showed some association with the Wnt/β-catenin signaling pathway in a mouse model of acute respiratory infection (Li et al., 2019). The Wnt/β-catenin signaling pathway is one of the most classical signaling pathways in bone metabolism. Therefore, whether RORα regulates bone metabolism through the Wnt/β-catenin signaling pathway might be investigated in the future. The relationship of another peripheral circadian clock gene, TIM, with bone metabolism has not been studied. While DEC is affected by BMP2, parathyroid hormone (PTH), and PTH-related protein (PTHrp) because of its tissue specificity, it is upregulated in both chondrocytes and BMSCs and regulates cell differentiation (Kato et al., 2014). There are many peripheral circadian clock genes that are less well studied and therefore less understood, such as nuclear factor interleukin 3 (Nfil3) and Noct. Nfil3 is expressed in osteoblasts in response to adrenergic receptor signaling and regulates the expression of BMP4, which in turn affects bone metabolism. Noct is regulated by the central circadian clock gene, although it is not part of the central core circadian clock complex. Its deletion promotes bone formation and rescues rosiglitazone-induced bone loss (Guntur et al., 2011; Hirai, 2018). In conclusion, functional and mechanistic studies have well demonstrated the regulatory role of peripheral circadian clock genes in bone metabolism, and different peripheral circadian clock genes may exert effects at different stages of bone metabolism.
The process of circadian clock genetic regulation of bone metabolism is very complex. On the one hand, circadian rhythm disturbances (e.g., shift work, sleep deprivation, fasting, or gene knockouts) are likely to cause bone and muscle dysfunction, but bone metabolism is affected to a greater extent by the presence of muscle‒bone crosstalk. On the other hand, circadian rhythm disorders lead to fluctuations in the levels of nutritional factors, hormones, and cytokines in the body, which can also have an impact on bone metabolism (Feskanich et al., 2009; Quevedo and Zuniga, 2010; Booth et al., 2013). In addition, the passage of the circadian temporal phase is often accompanied by the interaction of various endocrine hormones, cytokines, and non-coding RNAs such as miRNAs with bone metabolism. Therefore, the regulation of bone metabolism by circadian clock genes is not a direct and unidirectional process. Integrating new research on the contribution of miRNAs in studies of the influence of circadian rhythms on bone metabolism will bring new understanding of the mechanisms regulating bone metabolism.
4. Regulation of bone metabolism by miRNAs
4.1. Biological functions of miRNAs
MiRNAs are endogenous non-coding small RNAs about 22 nucleotides in length. They form RNA-induced silencing complexes (RISCs) when combined with ribonucleoprotein (RNP) complexes, and are capable of regulating the expression of downstream genes (Mohr and Mott, 2015). The main way by which miRNAs regulate gene expression is to degrade the mRNA of the target gene by guiding the Argonaute protein to the complementary site in mRNA. Another way is to inhibit the process of protein translation (Schirle et al., 2014). The expression of miRNAs themselves is influenced by many factors at the transcriptional, processing, and functional levels. The expression of target mRNAs is similarly regulated by miRNAs by means of epigenetic effects, promoter regulation, RNA processing and stability, and translation (Mohr and Mott, 2015). Ferrari et al. (2016) analyzed previous studies and found that miRNAs regulate physiological processes such as heart rate, vascular tone, and immune response by modulating purinergic receptors. Under pathological conditions, such as inflammatory responses in animal models, miR-147 was found to have a regulatory role (Kim et al., 2021). miR-122 was found to be involved in the regulation of ischemic responses in both human tissues and mouse models of liver ischemia (Ju et al., 2021). Numerous studies have also identified miRNAs that show great potential in the treatment of cancers and organ damages (Neudecker et al., 2016; Lee et al., 2020; Zhang et al., 2021). For example, delivery of miR-34a to various cancer models can exert significant inhibitory effects on tumor growth and survival (Liu et al., 2011; Kasinski et al., 2015). An increasing number of miRNAs have also gradually entered clinical studies in relation to different systemic diseases, but these studies are still in their early stages (Neudecker et al., 2016, 2017). Overall, miRNAs are widely involved in biological processes such as organism development, angiogenesis, cell proliferation, differentiation, and apoptosis as endogenous molecular regulators (Bartel, 2018).
4.2. MiRNAs in bone metabolism
Studies have confirmed that miRNAs play important roles in the differentiation and function of osteoblasts and osteoclasts (Table S1). Abnormal expression of miRNA leads to the development and progression of skeletal metabolic diseases (Sun YQ et al., 2019; Yuan et al., 2019). MiRNAs can regulate osteogenesis positively or negatively. miR-181a/b-1, miR-19b, and miR-96 promote osteogenesis and bone formation through the phosphatase and tensin homolog (PTEN)/phosphatidylinositide 3-kinase (PI3K)/AKT, PTEN/phospho-AKT (pAKT)/Runx2, and Wnt pathways, respectively. In addition, miR-378, miR-148b-3p, and many other miRNAs play active roles as positive regulators of osteogenesis in metabolic bone diseases such as osteoporosis, femoral head necrosis, and ectopic ossification (Zhang et al., 2018; Ma et al., 2019; Mollazadeh et al., 2019; Zheng et al., 2019; Sun et al., 2020). Other miRNAs play a negative regulatory role in osteogenesis or play a positive regulatory role in bone resorption. These include miR-23a and miR-1297, which inhibit osteogenic differentiation of periodontal ligament stem cells (PDLSCs) and BMSCs by targeting BMP, Wnt, and some other signaling pathways (Yuan Y et al., 2017, 2019; Wang et al., 2019; Zhang et al., 2019). Also, miR-155 can promote activation of osteoclast and bone resorption through both the AMPK and Wnt signaling pathways (Mao et al., 2019). However, a single miRNA can regulate multiple target genes, and can play different roles in the regulation of bone metabolism through different pathways. For example, knockdown of miR-223 can both reduce osteoclastogenesis and promote differentiation of osteoblasts, while overexpression of miR-223 has the dual role of both promoting and inhibiting the differentiation of osteoclasts (Xie et al., 2015).
Bone tissue is known to be sufficiently responsive to mechanical stimulation to enhance osteogenesis. It has been demonstrated that mechanical stress can mediate the involvement of miRNAs in the regulation of bone metabolism. For example, miR-214 can attenuate the osteogenic effects produced by mechanical stimulation. miR-103a also acts as a mechanosensitive miRNA directly targeting Runx2 to regulate osteogenic differentiation (Yuan Y et al., 2017, 2019). In recent years, in the field of bone metabolism research, there has been increasing interest in transorgan regulation, in which kidney‒bone crosstalk has been particularly popular (Hruska et al., 2017). MiRNAs, including miR-223, have been found to be the mediators of renal‒bone crosstalk (Colbert et al., 2017; Ulbing et al., 2017). Moreover, there is crosstalk between osteoblasts and osteoclasts, and bone formation and angiogenesis are coupled. Some miRNAs, referred to as "coupled miRNAs," are involved (Li et al., 2016; Zhu et al., 2018).
5. Co-regulation of bone metabolism by circadian clock genes and miRNAs
5.1. Interaction between circadian clock genes and miRNAs
Circadian clock genes can regulate a variety of behavioral activities and physiological and biochemical metabolic processes, but other regulatory factors may also be involved. For example, miRNAs have been shown to participate in the regulation of circadian rhythms (Xue and Zhang, 2018). With the continuous enrichment and improvement of various bioinformatics platforms, we can easily obtain the targeting relationship between miRNAs and circadian clock genes. For example, predictions from the latest version of TargetScanHuman (https://www.targetscan.org/vert_80) revealed that although no miRNAs with highly conserved binding sites that might target CRY1, DEC1, or NOCT were temporarily identified in human, mouse, or rat, and no miRNAs with highly conserved binding sites that might target Tim were identified in rats, there are still an enormous number of miRNAs that may target other circadian clock genes.
Many miRNAs that can target circadian clock genes have been experimentally confirmed. It was verified that BMAL1 can be directly targeted and regulated by miR-223, miR-219, miR-155, and miR-135b, and CLOCK can be targeted by miR-17-5p (Cheng et al., 2007; Curtis et al., 2015; Gao et al., 2016; Lou et al., 2017; Jiang et al., 2018). The relationships between PER, CRY, and miRNAs, on the other hand, often need to be explained by their interactions with BMAL1. For example, miR-219 is thought to be a target of BMAL1-CLOCK and it collaborates with miR-132 to activate transcription of PER through the BMAL1-CLOCK complex (Cheng et al., 2007). Surprisingly, studies of the interaction between peripheral circadian clock genes and miRNAs seem to have been more comprehensive and in-depth than those of core circadian clock genes. Rev-erbα was identified as the main circadian regulator of transcription of miR-122, and miR-122 was later found to regulate the rhythmic expression of Noct and RORα (Gatfield et al., 2009; Kojima et al., 2010). This indicates that there is a bidirectional pattern of regulation between circadian clock genes and miRNAs. Some other miRNAs have also been shown to target RORα. These include miR-1246, miR-7, miR-503-5p, and miR-137, which act synergistically to exert a potent inhibitory effect on RORα and thereby control the pathological development of diseases such as nonalcoholic steatohepatitis, hepatocellular carcinoma, and inflammation of brain tissue (Chai et al., 2020; Huang JL et al., 2020; Yue DX et al., 2020).
MiRNAs and circadian clock genes are increasingly being recognized as diagnostic and prognostic markers for some diseases. For example, miR-137 is considered a diagnostic candidate marker for autism spectrum disorders because it targets RORα in a specific way and affects the expression of autism-specific genes downstream of RORα (Devanna and Vernes, 2014). β-Catenin is an important factor in the Wnt signaling pathway. RORα can downregulate the expression of β-catenin to inhibit tumors, while miR-652 can activate the Wnt/β-catenin signaling pathway. Therefore, there is an antagonistic relationship between miR-652 and RORα to a large extent. Expression of miR-652 is closely associated with the tumor, node, metastasis (TNM) stage, so miR-652 is considered a possible prognostic marker for patients with gastric cancer (Sun et al., 2018; Li and Zou, 2019). RORα can also be considered a therapeutic target for diseases because it is targeted by specific miRNAs of some diseases if the function of RORα itself was analyzed from the perspective of the interaction between RORα and miRNAs. For example, a mechanism study found that miR-195 and lncRNA FGD5 antisense RNA 1 (FGD5-AS1) act as competitive endogenous RNA (ceRNA) to regulate the expression of RORα. In addition, miR-18a can target RORα and activate the NF-κB signaling pathway. miR-195 and miR-18a are miRNAs specific to patients with glioma and acute myocardial infarction. Therefore, RORα is considered a therapeutic target for these diseases (Cai et al., 2020; Jiang et al., 2020).
MiRNAs and circadian clock genes can also be simultaneously involved as mediators in the regulation of some biological processes. For example, exosomes derived from hypoxia-induced glioma are rich in miR-10a. Mechanistic studies have also revealed that hypoxia activates myeloid-derived suppressor cells (MDSCs) through the miR-10a/RORα/IκBα/NF-κB signaling pathway to exert a powerful immunosuppressive effect (Guo et al., 2018). Like RORα, TIM is related to the occurrence and development of cancer, and its interaction with miRNAs also shows advantages for anti-cancer treatments. The study by Zou et al. (2020) of miRNAs and TIM in breast cancer led to the identification of a very complex regulatory pathway: miR-5188 directly targets forkhead box protein O1 (FOXO1) and interacts with β-catenin in the cytoplasm to stimulate the Wnt signaling pathway and activate key regulators of the cancer system. TIM can induce the expression of miR-5188 by promoting c-Jun-mediated transcription, and then interacts with Sp1/c-Jun. Therefore, TIM regulates the progression of breast cancer through miR-5188-FOXO1/β-catenin-c-Jun, which provides a theoretical basis for the use of miR-5188 antagonists in the clinical treatment of breast cancer. Although it can be determined that TIM is regulated by miRNAs, it is important to note that TIM has several isoforms. The mRNA expressionof TIM-cold is controlled by miRNAs when it encodes TIM protein, while TIM-short and cold (TIM-sc)does not seem to be affected by miRNAs. In addition, miRNA sequencing suggests that miR-969 may be an enhanced regulator of TIM (Chen and Rosbash, 2016; Anduaga et al., 2019). However, further experiments are needed to confirm this. To sum up, circadian clock genes and miRNAs interact with each other and regulate many biological processes. Both can be candidate biomarkers for diagnosis and prognosis of many diseases.
5.2. Therapeutic potential of circadian clock genes and miRNAs in bone diseases
Both circadian genes and miRNAs can be used as diagnostic or prognostic markers for various diseases. The use of a small number of circadian clock genes and miRNAs as specific therapeutic targets for certain diseases has been discussed in a previous subsection. However, the main focus of most studies on the effects of circadian clock genes and miRNAs on bone metabolism has been to discern whether their regulatory effects are positive or negative. Their therapeutic effects on bone metabolic diseases are not yet clear enough, but their therapeutic potential should not be underestimated (Wu et al., 2019). An example is the concept of chronotherapeutics, induced by circadian rhythms, in which the concept of chronotherapy is used to tailor the dosing regimen to the patient's circadian rhythm in search of maximum efficacy and minimal side effects (Selfridge et al., 2016). Initial studies in a population of women with osteoporosis found that different timing of dosing affected the circadian rhythm of bone resorption markers (Luchavova et al., 2011). In the treatment of rheumatoid arthritis, the therapeutic effect of bedtime dosing was significantly enhanced compared to morning dosing (Arvidson et al., 1997; Buttgereit et al., 2013). MiRNA-elicited targeted therapies have shown initial success in the clinical treatment of cancer and cardiovascular disease. Studies related to such targeted therapies for bone disease are still being tested in animals, but are constrained by the lack of a safe and effective delivery system. However, the positive effects of miRNAs in animal experiments are also indicative of the therapeutic potential of miRNAs for bone disease (Sun X et al., 2019; Hu et al., 2020). Attention has been focused on the possible combined effects of chronotherapy and targeted therapies, and it has been proposed that targeting circadian genes using small molecule drugs to achieve therapeutic effects on bone diseases is worthy of study (Wu et al., 2019).
5.3. Regulation of bone metabolism by miRNAs through circadian clock genes
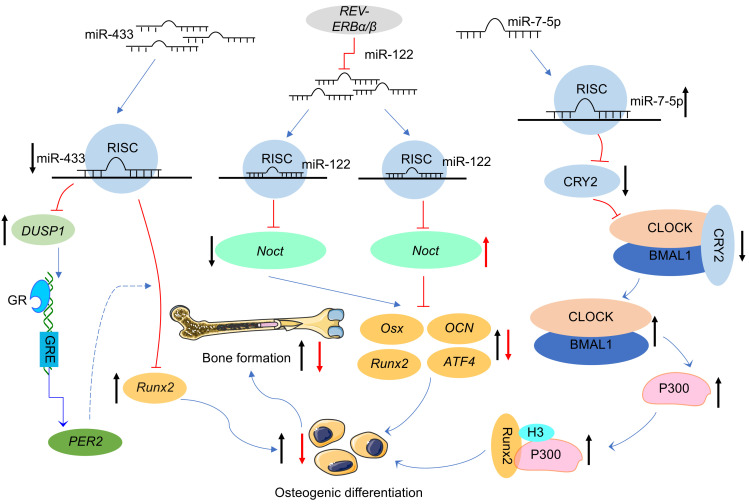
Smith et al. (2016) have demonstrated that miR-433 targets Runx2 to regulate bone metabolism, while predictions from miRanda suggest that it may also target binding to the core circadian clock gene PER2. The temporal phase of the peak expression of the core circadian clock gene PER2 is consistent with the peak mRNA and protein expression levels of Runx2, suggesting that Runx2 may be under the control of the circadian clock (Reale et al., 2013). However, luciferase reporter analysis showed that miR-433 was not directly targeted to PER2. Nevertheless, it cannot be inferred that there is no relationship between miR-433 and PER2 based on the above results alone. PER2 may be indirectly involved in the regulation of osteogenic gene expression by miR-433 throughdual specificity phosphatase 1(DUSP1) and glucocorticoid signaling. miR-433 can regulate the expression of the osteogenic gene Runx2 by regulating glucocorticoid signaling, and the glucocorticoid receptor can activate the expression of PER2 through the glucocorticoid response element in its intron (Smith et al., 2016). In addition, the peripheral circadian clock gene Rev-erbα, as mentioned above, has been identified as a major circadian regulator of miR-122 transcription, and miR-122 is able to regulate the rhythmic expression of Noct (Gatfield et al., 2009; Kojima et al., 2010). Combined with the relationship between Noct and bone metabolism, it is suggested that the Rev-erbα/miR-122/Noct pathway may also be one of the mechanisms by which circadian clock genes and miRNAs regulate bone metabolism.
All the above evidence suggests that circadian clock genes may be involved in the regulation of bone metabolism by miRNAs, and indicates that the regulatory mechanism of the miRNA/circadian clock gene/bone metabolism axis in bone metabolism has started to receive attention in the field of bone metabolism. However, the above studies have not been able to prove that circadian clock genes are the target genes of miRNAs to directly regulate bone metabolism. With the improvement of bioinformatics platforms and experimental techniques, recent studies have focused on the targeting relationship between miRNAs and circadian clock genes to deeply explore the regulatory mechanisms involved. It is surprising to find that knockdown of CRY2 is able to upregulate the expression of ALP, Runx2, collagen 1 (Colla1), OCN, and OPN to promote osteoblast differentiation in C3H10T1/2 cell lines. The target gene prediction website shows that miR-7-5p may target CRY2, which has been verified by luciferase reporter analysis. Overexpression of miR-7-5p is shown to significantly repress the expression of CRY2, and inhibition of CRY2 reduces its binding to CLOCK-BMAL1. This results in the release of a large amount of CLOCK-BMAL1 which then binds to the E-box in the P300 promoter region and stimulates transcription of P300. Finally, P300 promotes acetylation of histone 3 and forms a transcriptional complex with Runx2 to enhance osteogenesis. Thus, miR-7-5p targets CRY2 and promotes osteogenic differentiation through CLOCK/BMAL1/P300 signaling (Tang et al., 2020). Unfortunately, there have been few studies on the mechanisms involving circadian clock genes, miRNAs, and bone metabolism. The known mechanisms are shown in Fig. 2.
Fig. 2. Pathways of circadian clock genes and miRNAs regulating bone metabolism, modified from the study of Smith et al. (2016) and Tang et al. (2020) . ATF4: activating transcription factor 4; BMAL1: brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like 1; CLOCK: circadian locomotor output cycles kaput; CRY2: cryptochrome 2; DUSP1: dual specificity phosphatase 1; GR: glucocorticoid receptor; GRE: glucocorticoid receptor element; Noct: Nocturnin; Osx: osterix; OCN: osteocalcin; PER2: period 2; REV-ERBα/β: nuclear receptor subfamily 1 group D 1/2 (NR1D1/2); RISC: RNA-induced silencing complex; Runx2: Runt-related transcription factor 2.↑ ↑ : up-regulated;↓ ↓ : down-regulated.
6. Summary and outlook
In conclusion, bone metabolism is a very complex physiological metabolic activity that is influenced by multiple factors. Various functional and mechanistic studies have shown that core circadian clock genes such as BMAL1 and CLOCK, and peripheral circadian clock genes such as REV-ERB, play different roles in bone catabolism and anabolism. Specifically, BMAL1, CLOCK, and CRY play active roles in bone anabolism, while PER and NOCT are more dominant in bone catabolism. Some other circadian clock genes need further research to determine their roles in bone metabolism. The abnormal expression of circadian clock genes affects bone metabolism, and conversely, abnormal bone metabolism disrupts the expression of circadian clock genes. Moreover, the passage of the circadian phase is often accompanied by the interaction of various hormones, cytokines, and non-coding RNAs with bone metabolism, so the regulation of bone metabolism by circadian clock genes is not a direct and unidirectional process.
The latest research has focused on the circadian clock gene as a therapeutic target to treat autoimmune diseases. However, there has been no relevant research on whether the circadian clock gene can be targeted to treat bone metabolic diseases. Nowadays, there are more and more people working in shifts, and the susceptibility to bone metabolic diseases is enhanced by working around the clock. Therefore, we should firstly clarify the metabolic characteristics of various types of bone diseases, and then take the time of administration into account when using targeted therapy based on the mechanism of miRNA-mediated circadian clock genes to regulate the pathophysiological metabolism of bone. Combining chronotherapy and targeted therapy to achieve timely prevention or maximize the efficacy of treatments of bone metabolic diseases is a direction worthy of further study.
Supplementary information
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 81901430 and 81871835), the Guangdong Provincial Natural Science Foundation of China (No. 2022A1515010379), the Innovation Project from Department of Education of Guangdong Province (No. 2021KTSCX055), the Shanghai Frontiers Science Research Base of Exercise and Metabolic Health, and the Shanghai Key Laboratory for Human Athletic Ability Development and Support (Shanghai University of Sport) (No. 11DZ2261100), China.
Author contributions
Tingting LI: conceptualization, data curation, formal analysis, resources, writing-original draft, writing-review and editing. Shihua ZHANG: data curation, formal analysis, writing-review and editing. Yuxuan YANG: data curation, writing-review and editing. Lingli ZHANG: writing-review and editing. Yu YUAN: funding acquisition, supervision, writing-original draft, writing-review and editing. Jun ZOU: funding acquisition, supervision, writing-review and editing, project administration. All authors have read and approved the final manuscript.
Compliance with ethics guidelines
Tingting LI, Shihua ZHANG, Yuxuan YANG, Lingli ZHANG, Yu YUAN, and Jun ZOU declare that they have no conflicts of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Abe T, Sato T, Yoda T, et al. , 2019. The period circadian clock 2 gene responds to glucocorticoids and regulates osteogenic capacity. Regen Ther, 11: 199-206. 10.1016/j.reth.2019.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RK, Liou YH, Knudsen NH, et al. , 2020. Bmal1 integrates mitochondrial metabolism and macrophage activation. eLife, 9: e54090. 10.7554/eLife.54090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An K, Zhao H, Miao Y, et al. , 2020. A circadian rhythm-gated subcortical pathway for nighttime-light-induced depressive-like behaviors in mice. Nat Neurosci, 23(7): 869-880. 10.1038/s41593-020-0640-8 [DOI] [PubMed] [Google Scholar]
- Anduaga AM, Evantal N, Patop IL, et al. , 2019. Thermosensitive alternative splicing senses and mediates temperature adaptation in Drosophila . eLife, 8: e44642. 10.7554/eLife.44642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidson NG, Gudbjörnsson B, Larsson A, et al. , 1997. The timing of glucocorticoid administration in rheumatoid arthritis. Ann Rheum Dis, 56(1): 27-31. 10.1136/ard.56.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal RP, Kwak PB, Tamayo AG, et al. , 2017. Macromolecular assemblies of the mammalian circadian clock. Mol Cell, 67(5): 770-782.e6. 10.1016/j.molcel.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang J, Chang HW, Jung HR, et al. , 2012. Melatonin attenuates clock gene Cryptochrome1, which may aggravates mouse anti-type II collagen antibody-induced arthritis. Rheumatol Int, 32(2): 379-385. 10.1007/s00296-010-1641-9 [DOI] [PubMed] [Google Scholar]
- Bartel DP, 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116(2): 281-297. 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel DP, 2018. Metazoan microRNAs. Cell, 173(1): 20-51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beker MC, Caglayan B, Caglayan AB, et al. , 2019. Interaction of melatonin and Bmal1 in the regulation of PI3K/AKT pathway components and cellular survival. Sci Rep, 9: 19082. 10.1038/s41598-019-55663-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekki H, Duffy T, Okubo N, et al. , 2020. Suppression of circadian clock protein cryptochrome 2 promotes osteoarthritis. Osteoarthritis Cartilage, 28(7): 966-976. 10.1016/j.joca.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benderdour M, Fahmi H, Beaudet F, et al. , 2011. Nuclear receptor retinoid-related orphan receptor α1 modulates the metabolic activity of human osteoblasts. J Cell Biochem, 112(8): 2160-2169. 10.1002/jcb.23141 [DOI] [PubMed] [Google Scholar]
- Bianco JN, Bergoglio V, Lin YL, et al. , 2019. Overexpression of Claspin and Timeless protects cancer cells from replication stress in a checkpoint-independent manner. Nat Commun, 10: 910. 10.1038/s41467-019-08886-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth SL, Centi A, Smith SR, et al. , 2013. The role of osteocalcin in human glucose metabolism: marker or mediator? Nat Rev Endocrinol, 9(1): 43-55. 10.1038/nrendo.2012.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H, Vanneaux V, Domet T, et al. , 2016. Circadian clock genes modulate human bone marrow mesenchymal stem cell differentiation, migration and cell cycle. PLoS ONE, 11(1): e0146674. 10.1371/journal.pone.0146674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Walisser JA, Sullivan R, et al. , 2005. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis, 41(3): 122-132. 10.1002/gene.20102 [DOI] [PubMed] [Google Scholar]
- Buttgereit F, Mehta D, Kirwan J, et al. , 2013. Low-dose prednisone chronotherapy for rheumatoid arthritis: a randomised clinical trial (CAPRA-2). Ann Rheum Dis, 72(2): 204-210. 10.1136/annrheumdis-2011-201067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai XY, Zhang P, Wang S, et al. , 2020. LncRNA FGD5 antisense RNA 1 upregulates RORA to suppress hypoxic injury of human cardiomyocyte cells by inhibiting oxidative stress and apoptosis via miR-195. Mol Med Rep, 22(6): 4579-4588. 10.3892/mmr.2020.11558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Zhao X, Bai JW, et al. , 2017. Circadian clock cryptochrome proteins regulate autoimmunity. Proc Natl Acad Sci USA, 114(47): 12548-12553. 10.1073/pnas.1619119114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai C, Cox B, Yaish D, et al. , 2020. Agonist of RORA attenuates nonalcoholic fatty liver progression in mice via up-regulation of microRNA 122. Gastroenterology, 159(3): 999-1014.e9. 10.1053/j.gastro.2020.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WCW, Tan ZJ, To MKT, et al. , 2021. Regulation and role of transcription factors in osteogenesis. Int J Mol Sci, 22(11): 5445. 10.3390/ijms22115445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappuis S, Ripperger JA, Schnell A, et al. , 2013. Role of the circadian clock gene Per2 in adaptation to cold temperature. Mol Metab, 2(3): 184-193. 10.1016/j.molmet.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GJ, Tang QM, Yu SL, et al. , 2020. The biological function of BMAL1 in skeleton development and disorders. Life Sci, 253: 117636. 10.1016/j.lfs.2020.117636 [DOI] [PubMed] [Google Scholar]
- Chen X, Rosbash M, 2016. mir-276a strengthens Drosophila circadian rhythms by regulating timeless expression. Proc Natl Acad Sci USA, 113(21): E2965-E2972. 10.1073/pnas.1605837113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HYM, Papp JW, Varlamova O, et al. , 2007. MicroRNA modulation of circadian-clock period and entrainment. Neuron, 54(5): 813-829. 10.1016/j.neuron.2007.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert JF, Ford JA, Haeger SM, et al. , 2017. A model-specific role of microRNA-223 as a mediator of kidney injury during experimental sepsis. Am J Physiol Renal Physiol, 313(2): F553-F559. 10.1152/ajprenal.00493.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis AM, Fagundes CT, Yang GR, et al. , 2015. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1 . Proc Natl Acad Sci USA, 112(23): 7231-7236. 10.1073/pnas.1501327112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, Reppert SM, 2007. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci, 10(5): 543-545. 10.1038/nn1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanna P, Vernes SC, 2014. A direct molecular link between the autism candidate gene RORa and the schizophrenia candidate MIR137. Sci Rep, 4: 3994. 10.1038/srep03994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrington HJ, Krakowiak K, Meijer P, et al. , 2020. Circadian asthma airway responses are gated by REV-ERBα. Eur Respir J, 56(6): 1902407. 10.1183/13993003.02407-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney KA, Hansen LL, Putker M, et al. , 2016. Daily magnesium fluxes regulate cellular timekeeping and energy balance. Nature, 532(7599): 375-379. 10.1038/nature17407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari D, Bianchi N, Eltzschig HK, et al. , 2016. MicroRNAs modulate the purinergic signaling network. Trends Mol Med, 22(10): 905-918. 10.1016/j.molmed.2016.08.006 [DOI] [PubMed] [Google Scholar]
- Feskanich D, Hankinson SE, Schernhammer ES, 2009. Nightshift work and fracture risk: the nurses’ health study. Osteoporos Int, 20(4): 537-542. 10.1007/s00198-008-0729-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle KJ, Parson KG, Dahm NA, et al. , 2011. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science, 331(6023): 1409-1413. 10.1126/science.1199702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Patel MS, Bradley A, et al. , 2005. The molecular clock mediates leptin-regulated bone formation. Cell, 122(5): 803-815. 10.1016/j.cell.2005.06.028 [DOI] [PubMed] [Google Scholar]
- Fu SQ, Kuwahara M, Uchida Y, et al. , 2019. Circadian production of melatonin in cartilage modifies rhythmic gene expression. J Endocrinol, 241(2): 161-173. 10.1530/JOE-19-0022 [DOI] [PubMed] [Google Scholar]
- Fujihara Y, Kondo H, Noguchi T, et al. , 2014. Glucocorticoids mediate circadian timing in peripheral osteoclasts resulting in the circadian expression rhythm of osteoclast-related genes. Bone, 61: 1-9. 10.1016/j.bone.2013.12.026 [DOI] [PubMed] [Google Scholar]
- Gaertner VD, Michel S, Curtin JA, et al. , 2019. Nocturnal asthma is affected by genetic interactions between RORA and NPSR1 . Pediatr Pulmonol, 54(6): 847-857. 10.1002/ppul.24292 [DOI] [PubMed] [Google Scholar]
- Gao Q, Zhou L, Yang SY, et al. , 2016. A novel role of microRNA 17-5p in the modulation of circadian rhythm. Sci Rep, 6: 30070. 10.1038/srep30070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, le Martelot G, Vejnar CE, et al. , 2009. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev, 23(11): 1313-1326. 10.1101/gad.1781009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves CF, Meng QJ, 2019. Timing metabolism in cartilage and bone: links between circadian clocks and tissue homeostasis. J Endocrinol, 243(3): R29-R46. 10.1530/JOE-19-0256 [DOI] [PubMed] [Google Scholar]
- Guissart C, Latypova X, Rollier P, et al. , 2018. Dual molecular effects of dominant RORA mutations cause two variants of syndromic intellectual disability with either autism or cerebellar ataxia. Am J Hum Genet, 102(5): 744-759. 10.1016/j.ajhg.2018.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntur AR, Kawai M, Le P, et al. , 2011. An essential role for the circadian-regulated gene nocturnin in osteogenesis: the importance of local timekeeping in skeletal homeostasis. Ann N Y Acad Sci, 1237(1): 58-63. 10.1111/j.1749-6632.2011.06213.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JH, Cheng P, Yuan HY, et al. , 2009. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell, 138(6): 1236-1246. 10.1016/j.cell.2009.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XF, Qiu W, Liu QL, et al. , 2018. Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten pathways. Oncogene, 37(31): 4239-4259. 10.1038/s41388-018-0261-9 [DOI] [PubMed] [Google Scholar]
- He Y, Lin FW, Chen YQ, et al. , 2015. Overexpression of the circadian clock gene Rev-erbα affects murine bone mesenchymal stem cell proliferation and osteogenesis. Stem Cells Dev, 24(10): 1194-1204. 10.1089/scd.2014.0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai T, 2018. Regulation of clock genes by adrenergic receptor signaling in osteoblasts. Neurochem Res, 43(1): 129-135. 10.1007/s11064-017-2365-y [DOI] [PubMed] [Google Scholar]
- Hruska KA, Lanske B, Moe OW, 2017. Crosstalk between kidney and bone—bench to bedside. Bone, 100: 1-3. 10.1016/j.bone.2017.03.046 [DOI] [PubMed] [Google Scholar]
- Hu H, He XD, Zhang YZ, et al. , 2020. MicroRNA alterations for diagnosis, prognosis, and treatment of osteoporosis: a comprehensive review and computational functional survey. Front Genet, 11: 181. 10.3389/fgene.2020.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JL, Fu YP, Gan W, et al. , 2020. Hepatic stellate cells promote the progression of hepatocellular carcinoma through microRNA-1246-RORα-Wnt/β-Catenin axis. Cancer Lett, 476: 140-151. 10.1016/j.canlet.2020.02.012 [DOI] [PubMed] [Google Scholar]
- Huang ZF, Wei H, Wang X, et al. , 2020. Icariin promotes osteogenic differentiation of BMSCs by upregulating BMAL1 expression via BMP signaling. Mol Med Rep, 21(3): 1590-1596. 10.3892/mmr.2020.10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WL, Zhao SL, Jiang XH, et al. , 2016. The circadian clock gene Bmal1 acts as a potential anti-oncogene in pancreatic cancer by activating the p53 tumor suppressor pathway. Cancer Lett, 371(2): 314-325. 10.1016/j.canlet.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Jiang WL, Zhao SL, Shen J, et al. , 2018. The miR-135b-BMAL1-YY1 loop disturbs pancreatic clockwork to promote tumourigenesis and chemoresistance. Cell Death Dis, 9: 149. 10.1038/s41419-017-0233-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Zhou JP, Zhao JS, et al. , 2020. MiR-18a-downregulated RORA inhibits the proliferation and tumorigenesis of glioma using the TNF-α-mediated NF-κB signaling pathway. eBioMedicine, 52 : 102651. 10.1016/j.ebiom.2020.102651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C, Wang M, Tak E, et al. , 2021. Hypoxia-inducible factor-1α-dependent induction of miR122 enhances hepatic ischemia tolerance. J Clin Invest, 131(7): e140300. 10.1172/JCI140300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Kelnar K, Stahlhut C, et al. , 2015. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene, 34(27): 3547-3555. 10.1038/onc.2014.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kawamoto T, Fujimoto K, et al. , 2014. DEC1/STRA13/SHARP2 and DEC2/SHARP1 coordinate physiological processes, including circadian rhythms in response to environmental stimuli. Curr Top Dev Biol, 110: 339-372. 10.1016/B978-0-12-405943-6.00010-5 [DOI] [PubMed] [Google Scholar]
- Khosla S, 2013. Pathogenesis of age-related bone loss in humans. J Gerontol A Biol Sci Med Sci, 68(10): 1226-1235. 10.1093/gerona/gls163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Guaregua V, Chen XB, et al. , 2021. Characterization of a murine model system to study microRNA-147 during inflammatory organ injury. Inflammation, 44(4): 1426-1440. 10.1007/s10753-021-01427-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Kim JH, Kim I, et al. , 2020. Rev-erbα negatively regulates osteoclast and osteoblast differentiation through p38 MAPK signaling pathway. Mol Cells, 43(1): 34-47. 10.14348/molcells.2019.0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Morinibu A, Koyasu S, et al. , 2017. A circadian clock gene, PER2, activates HIF-1 as an effector molecule for recruitment of HIF-1α to promoter regions of its downstream genes. FEBS J, 284(22): 3804-3816. 10.1111/febs.14280 [DOI] [PubMed] [Google Scholar]
- Kojima S, Gatfield D, Esau CC, et al. , 2010. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS ONE, 5(6): e11264. 10.1371/journal.pone.0011264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurien P, Hsu PK, Leon J, et al. , 2019. Timeless mutation alters phase responsiveness and causes advanced sleep phase. Proc Natl Acad Sci USA, 116(24): 12045-12053. 10.1073/pnas.1819110116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushibiki T, Awazu K, 2009. Blue laser irradiation enhances extracellular calcification of primary mesenchymal stem cells. Photomed Laser Surg, 27(3): 493-498. 10.1089/pho.2008.2343 [DOI] [PubMed] [Google Scholar]
- Lee TJ, Yuan XY, Kerr K, et al. , 2020. Strategies to modulate microRNA functions for the treatment of cancer or organ injury. Pharmacol Rev, 72(3): 639-667. 10.1124/pr.119.019026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DF, Liu J, Guo BS, et al. , 2016. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat Commun, 7: 10872. 10.1038/ncomms10872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DM, Zhang RX, Sun QY, et al. , 2020. Involvement of Bmal1 and circadian clock signaling in chondrogenic differentiation of ATDC5 cells by fluoride. Ecotoxicol Environ Saf, 204: 111058. 10.1016/j.ecoenv.2020.111058 [DOI] [PubMed] [Google Scholar]
- Li J, Xue K, Zheng Y, et al. , 2019. RORA overexpression alleviates nasal mucosal injury and enhances red blood cell immune adhesion function in a mouse model of allergic rhinitis via inactivation of the Wnt/β-catenin signaling pathway. Int Arch Allergy Immunol, 180(2): 79-90. 10.1159/000500637 [DOI] [PubMed] [Google Scholar]
- Li JC, Zou XM, 2019. MiR-652 serves as a prognostic biomarker in gastric cancer and promotes tumor proliferation, migration, and invasion via targeting RORA. Cancer Biomark, 26(3): 323-331. 10.3233/CBM-190361 [DOI] [PubMed] [Google Scholar]
- Li XG, Liu N, Gu B, et al. , 2018. BMAL1 regulates balance of osteogenic-osteoclastic function of bone marrow mesenchymal stem cells in type 2 diabetes mellitus through the NF-κB pathway. Mol Biol Rep, 45(6): 1691-1704. 10.1007/s11033-018-4312-7 [DOI] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, et al. , 2002. A role for casein kinase 2α in the Drosophila circadian clock. Nature, 420(6917): 816-820. 10.1038/nature01235 [DOI] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu BG, et al. , 2011. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med, 17(2): 211-215. 10.1038/nm.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wu YY, Yoshizawa T, et al. , 2016. Basic helix-loop-helix transcription factor DEC2 functions as an anti-apoptotic factor during paclitaxel-induced apoptosis in human prostate cancer cells. Int J Mol Med, 38(6): 1727-1733. 10.3892/ijmm.2016.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wu YY, Seino H, et al. , 2018. Correlation between DEC1/DEC2 and epithelial-mesenchymal transition in human prostate cancer PC3-cells. Mol Med Rep, 18(4): 3859-3865. 10.3892/mmr.2018.9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J, Wang YL, Zhang ZM, et al. , 2017. Activation of MMPs in macrophages by Mycobacterium tuberculosis via the miR-223-BMAL1 signaling pathway. J Cell Biochem, 118(12): 4804-4812. 10.1002/jcb.26150 [DOI] [PubMed] [Google Scholar]
- Luan X, Tian XY, Zhang HX, et al. , 2019. Exercise as a prescription for patients with various diseases. J Sport Health Sci, 8(5): 422-441. 10.1016/j.jshs.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchavova M, Zikan V, Michalska D, et al. , 2011. The effect of timing of teriparatide treatment on the circadian rhythm of bone turnover in postmenopausal osteoporosis. Eur J Endocrinol, 164(4): 643-648. 10.1530/EJE-10-1108 [DOI] [PubMed] [Google Scholar]
- Ma S, Wang DD, Ma CY, et al. , 2019. MicroRNA-96 promotes osteoblast differentiation and bone formation in ankylosing spondylitis mice through activating the Wnt signaling pathway by binding to SOST. J Cell Biochem, 120(9): 15429-15442. 10.1002/jcb.28810 [DOI] [PubMed] [Google Scholar]
- Mao XF, Li XG, Hu W, et al. , 2020. Downregulated brain and muscle aryl hydrocarbon receptor nuclear translocator-like protein-1 inhibits osteogenesis of BMSCs through p53 in type 2 diabetes mellitus. Biol Open, 9(7): bio051482. 10.1242/bio.051482 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mao ZX, Zhu YH, Hao WM, et al. , 2019. MicroRNA-155 inhibition up-regulates LEPR to inhibit osteoclast activation and bone resorption via activation of AMPK in alendronate-treated osteoporotic mice. IUBMB Life, 71(12): 1916-1928. 10.1002/iub.2131 [DOI] [PubMed] [Google Scholar]
- Maronde E, Schilling AF, Seitz S, et al. , 2010. The clock genes Period 2 and Cryptochrome 2 differentially balance bone formation. PLoS ONE, 5(7): e11527. 10.1371/journal.pone.0011527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Pescatore S, Rosbash M, 2014. CLOCK: BMAL1 is a pioneer-like transcription factor. Genes Dev, 28(1): 8-13. 10.1101/gad.228536.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michou L, 2018. Epigenetics of bone diseases. Joint Bone Spine, 85(6): 701-707. 10.1016/j.jbspin.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Min HY, Kim KM, Wee G, et al. , 2016. Bmal1 induces osteoblast differentiation via regulation of BMP2 expression in MC3T3-E1 cells. Life Sci, 162: 41-46. 10.1016/j.lfs.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Mohr AM, Mott JL, 2015. Overview of microRNA biology. Semin Liver Dis, 35(1): 3-11. 10.1055/s-0034-1397344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollazadeh S, Bazzaz BSF, Neshati V, et al. , 2019. Overexpression of microRNA-148b-3p stimulates osteogenesis of human bone marrow-derived mesenchymal stem cells: the role of microRNA-148b-3p in osteogenesis. BMC Med Genet, 20: 117. 10.1186/s12881-019-0854-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao S, Iwata N, Soejima Y, et al. , 2019. Interaction of ovarian steroidogenesis and clock gene expression modulated by bone morphogenetic protein-7 in human granulosa cells. Endocr J, 66(2): 157-164. 10.1507/endocrj.EJ18-0423 [DOI] [PubMed] [Google Scholar]
- Neudecker V, Brodsky KS, Kreth S, et al. , 2016. Emerging roles for microRNAs in perioperative medicine. Anesthesiology, 124(2): 489-506. 10.1097/ALN.0000000000000969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudecker V, Colgan SP, Eltzschig HK, 2017. Novel therapeutic concepts for inflammatory bowel disease—from bench to bedside. J Mol Med (Berl), 95(9): 899-903. 10.1007/s00109-017-1574-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patke A, Murphy PJ, Onat OE, et al. , 2017. Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell, 169(2): 203-215.e13. 10.1016/j.cell.2017.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak D, Domaniewski T, Znorko B, et al. , 2017. The impact of peripheral serotonin on leptin-brain serotonin axis, bone metabolism and strength in growing rats with experimental chronic kidney disease. Bone, 105: 1-10. 10.1016/j.bone.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Quevedo I, Zuniga AM, 2010. Low bone mineral density in rotating-shift workers. J Clin Densitom, 13(4): 467-469. 10.1016/j.jocd.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Rageul J, Park JJ, Zeng PP, et al. , 2020. SDE2 integrates into the TIMELESS-TIPIN complex to protect stalled replication forks. Nat Commun, 11: 5495. 10.1038/s41467-020-19162-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Valekunja UK, Stangherlin A, et al. , 2020. Circadian rhythms in the absence of the clock gene Bmal1 . Science, 367(6479): 800-806. 10.1126/science.aaw7365 [DOI] [PubMed] [Google Scholar]
- Reale ME, Webb IC, Wang X, et al. , 2013. The transcription factor Runx2 is under circadian control in the suprachiasmatic nucleus and functions in the control of rhythmic behavior. PLoS ONE, 8(1): e54317. 10.1371/journal.pone.0054317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan W, Yuan XY, Eltzschig HK, 2021. Circadian rhythm as a therapeutic target. Nat Rev Drug Discov, 20(4): 287-307. 10.1038/s41573-020-00109-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsa WE, Vasanji A, Midura RJ, et al. , 2016. Deficiency of circadian clock protein BMAL1 in mice results in a low bone mass phenotype. Bone, 84: 194-203. 10.1016/j.bone.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F, Kohsaka A, Bhawal UK, et al. , 2018. Potential roles of Dec and BMAL1 genes in interconnecting circadian clock and energy metabolism. Int J Mol Sci, 19(3): 781. 10.3390/ijms19030781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, 2005. The daily rhythms of genes, cells and organs. Biological clocks and circadian timing in cells. EMBO Rep, 6: S9-S13. 10.1038/sj.embor.7400424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Ripperger J, Brown SA, 2003. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms, 18(3): 250-260. 10.1177/0748730403018003007 [DOI] [PubMed] [Google Scholar]
- Schilperoort M, Bravenboer N, Lim J, et al. , 2020. Circadian disruption by shifting the light-dark cycle negatively affects bone health in mice. FASEB J, 34(1): 1052-1064. 10.1096/fj.201901929R [DOI] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, Macrae IJ, 2014. Structural basis for microRNA targeting. Science, 346(6209): 608-613. 10.1126/science.1258040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selfridge JM, Gotoh T, Schiffhauer S, et al. , 2016. Chronotherapy: intuitive, sound, founded.. . but not broadly applied. Drugs, 76(16): 1507-1521. 10.1007/s40265-016-0646-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JF, Tong RY, Zhou M, et al. , 2022. Circadian nuclear receptor Rev-erbα is expressed by platelets and potentiates platelet activation and thrombus formation. Eur Heart J, 43(24): 2317-2334. 10.1093/eurheartj/ehac109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SS, Dole NS, Franceschetti T, et al. , 2016. MicroRNA-433 dampens glucocorticoid receptor signaling, impacting circadian rhythm and osteoblastic gene expression. J Biol Chem, 291(41): 21717-21728. 10.1074/jbc.M116.737890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Tan P, Zhang Z, et al. , 2018. REV-ERB agonism suppresses osteoclastogenesis and prevents ovariectomy-induced bone loss partially via FABP4 upregulation. FASEB J, 32(6): 3215-3228. 10.1096/fj.201600825RRR [DOI] [PubMed] [Google Scholar]
- Sulli G, Manoogian ENC, Taub PR, et al. , 2018. Training the circadian clock, clocking the drugs, and drugging the clock to prevent, manage, and treat chronic diseases. Trends Pharmacol Sci, 39(9): 812-827. 10.1016/j.tips.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun MG, Hu LQ, Wang S, et al. , 2020. Circulating microRNA-19b identified from osteoporotic vertebral compression fracture patients increases bone formation. J Bone Miner Res, 35(2): 306-316. 10.1002/jbmr.3892 [DOI] [PubMed] [Google Scholar]
- Sun X, Guo Q, Wei WH, et al. , 2019. Current progress on microRNA-based gene delivery in the treatment of osteoporosis and osteoporotic fracture. Int J Endocrinol, 2019: 6782653. 10.1155/2019/6782653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XM, Dongol S, Qiu CP, et al. , 2018. miR-652 promotes tumor proliferation and metastasis by targeting RORA in endometrial cancer. Mol Cancer Res, 16(12): 1927-1939. 10.1158/1541-7786.MCR-18-0267 [DOI] [PubMed] [Google Scholar]
- Sun YQ, Kuek V, Liu YH, et al. , 2019. MiR-214 is an important regulator of the musculoskeletal metabolism and disease. J Cell Physiol, 234(1): 231-245. 10.1002/jcp.26856 [DOI] [PubMed] [Google Scholar]
- Swanson CM, Shea SA, Wolfe P, et al. , 2017. Bone turnover markers after sleep restriction and circadian disruption: a mechanism for sleep-related bone loss in humans. J Clin Endocrinol Metab, 102(10): 3722-3730. 10.1210/jc.2017-01147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipaleenmäki H, 2018. Regulation of bone metabolism by microRNAs. Curr Osteoporos Rep, 16(1): 1-12. 10.1007/s11914-018-0417-0 [DOI] [PubMed] [Google Scholar]
- Takarada T, Xu C, Ochi H, et al. , 2017. Bone resorption is regulated by circadian clock in osteoblasts. J Bone Miner Res, 32(4): 872-881. 10.1002/jbmr.3053 [DOI] [PubMed] [Google Scholar]
- Tang ZH, Xu TY, Li YH, et al. , 2020. Inhibition of CRY2 by STAT3/miRNA-7-5p promotes osteoblast differentiation through upregulation of CLOCK/BMAL1/P300 expression. Mol Ther Nucleic Acids, 19: 865-876. 10.1016/j.omtn.2019.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong XY, Chen X, Zhang SH, et al. , 2019. The effect of exercise on the prevention of osteoporosis and bone angiogenesis. Biomed Res Int, 2019: 8171897. 10.1155/2019/8171897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang K, Liu HM, Yang Y, et al. , 2019. Defective circadian control in mesenchymal cells reduces adult bone mass in mice by promoting osteoclast function. Bone, 121: 172-180. 10.1016/j.bone.2019.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbing M, Kirsch AH, Leber B, et al. , 2017. MicroRNAs 223-3p and 93-5p in patients with chronic kidney disease before and after renal transplantation. Bone, 95: 115-123. 10.1016/j.bone.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BX, Yang JK, Fan LJ, et al. , 2021. Osteogenic effects of antihypertensive drug benidipine on mouse MC3T3-E1 cells in vitro. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 22(5): 410-420. 10.1631/jzus.B2000628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wang CH, Meng Y, 2019. MicroRNA-1297 promotes the progression of osteoporosis through regulation of osteogenesis of bone marrow mesenchymal stem cells by targeting WNT5A. Eur Rev Med Pharmacol Sci, 23(11): 4541-4550. 10.26355/eurrev_201906_18029 [DOI] [PubMed] [Google Scholar]
- Wang S, Li F, Lin YK, et al. , 2020. Targeting REV-ERBα for therapeutic purposes: promises and challenges. Theranostics, 10(9): 4168-4182. 10.7150/thno.43834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz PS, Zinna VM, Symeonidi A, et al. , 2019. BMAL1-driven tissue clocks respond independently to light to maintain homeostasis. Cell, 177(6): 1436-1447.e12. 10.1016/j.cell.2019.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu QY, Wang J, Tong X, et al. , 2019. Emerging role of circadian rhythm in bone remodeling. J Mol Med (Berl), 97(1): 19-24. 10.1007/s00109-018-1723-9 [DOI] [PubMed] [Google Scholar]
- Xie Y, Zhang LH, Gao YP, et al. , 2015. The multiple roles of microRNA-223 in regulating bone metabolism. Molecules, 20(10): 19433-19448. 10.3390/molecules201019433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Ochi H, Fukuda T, et al. , 2016. Circadian clock regulates bone resorption in mice. J Bone Miner Res, 31(7): 1344-1355. 10.1002/jbmr.2803 [DOI] [PubMed] [Google Scholar]
- Xue YB, Zhang Y, 2018. Emerging roles for microRNA in the regulation of Drosophila circadian clock. BMC Neurosci, 19: 1. 10.1186/s12868-018-0401-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Smyllie NJ, Morris H, et al. , 2020. Quantitative live imaging of venus: : BMAL1 in a mouse model reveals complex dynamics of the master circadian clock regulator. PLoS Genet, 16(4): e1008729. 10.1371/journal.pgen.1008729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XM, Wood PA, Oh EY, et al. , 2009. Down regulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat, 117(2): 423-431. 10.1007/s10549-008-0133-z [DOI] [PubMed] [Google Scholar]
- Yuan GS, Hua BX, Yang Y, et al. , 2017. The circadian gene Clock regulates bone formation via PDIA3. J Bone Miner Res, 32(4): 861-871. 10.1002/jbmr.3046 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhang LL, Tong XY, et al. , 2017. Mechanical stress regulates bone metabolism through microRNAs. J Cell Physiol, 232(6): 1239-1245. 10.1002/jcp.25688 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Guo JM, Zhang LL, et al. , 2019. MiR-214 attenuates the osteogenic effects of mechanical loading on osteoblasts. Int J Sports Med, 40(14): 931-940. 10.1055/a-1015-0285 [DOI] [PubMed] [Google Scholar]
- Yue DX, Zhao JJ, Chen HZ, et al. , 2020. MicroRNA-7, synergizes with RORα, negatively controls the pathology of brain tissue inflammation. J Neuroinflammation, 17: 28. 10.1186/s12974-020-1710-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, He JJ, Wei YJ, et al. , 2020. Decreased expression of Rev-Erbα in the epileptic foci of temporal lobe epilepsy and activation of Rev-Erbα have anti-inflammatory and neuroprotective effects in the pilocarpine model. J Neuroinflammation, 17: 43. 10.1186/s12974-020-1718-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Li YL, Yu Y, et al. , 2018. MicroRNA-378 promotes osteogenesis-angiogenesis coupling in BMMSCs for potential bone regeneration. Anal Cell Pathol (Amst), 2018: 8402390. 10.1155/2018/8402390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wu YF, Li ZH, et al. , 2021. MiR-144-5p, an exosomal miRNA from bone marrow-derived macrophage in type 2 diabetes, impairs bone fracture healing via targeting Smad1. J Nanobiotechnology, 19: 226. 10.1186/s12951-021-00964-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Li SY, Yuan SJ, et al. , 2019. MicroRNA-23a inhibits osteogenesis of periodontal mesenchymal stem cells by targeting bone morphogenetic protein signaling. Arch Oral Biol, 102: 93-100. 10.1016/j.archoralbio.2019.04.001 [DOI] [PubMed] [Google Scholar]
- Zheng HJ, Liu J, Tycksen E, et al. , 2019. MicroRNA-181a/b-1 over-expression enhances osteogenesis by modulating PTEN/PI3K/AKT signaling and mitochondrial metabolism. Bone, 123: 92-102. 10.1016/j.bone.2019.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, He J, Sun SW, et al. , 2019. Cryptochrome 1 regulates osteoblast differentiation via the AKT kinase and extracellular signal-regulated kinase signaling pathways. Cell Reprogram, 21(3): 141-151. 10.1089/cell.2018.0054 [DOI] [PubMed] [Google Scholar]
- Zhou X, Yu R, Long YL, et al. , 2018. BMAL1 deficiency promotes skeletal mandibular hypoplasia via OPG downregulation. Cell Prolif, 51(5): e12470. 10.1111/cpr.12470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SP, Yao F, Qiu H, et al. , 2018. Coupling factors and exosomal packaging microRNAs involved in the regulation of bone remodelling. Biol Rev, 93(1): 469-480. 10.1111/brv.12353 [DOI] [PubMed] [Google Scholar]
- Zhuo HY, Wang YH, Zhao Q, 2018. The interaction between Bmal1 and Per2 in mouse BMSC osteogenic differentiation. Stem Cells Int, 2018: 3407821. 10.1155/2018/3407821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou YJ, Lin X, Bu JG, et al. , 2020. Timeless-stimulated miR-5188-FOXO1/β-catenin-c-Jun feedback loop promotes stemness via ubiquitination of β-catenin in breast cancer. Mol Ther, 28(1): 313-327. 10.1016/j.ymthe.2019.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Kilroy G, et al. , 2007. Circadian oscillation of gene expression in murine calvarial bone. J Bone Miner Res, 22(3): 357-365. 10.1359/jbmr.061114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.