Abstract
Outbreaks of food- and waterborne gastroenteritis are being increasingly reported throughout the world. The analysis of environmental samples by newer diagnostic techniques such as reverse transcription-PCR (RT-PCR) amplification of nucleic acid has begun to identify human enteric viruses (predominantly “Norwalk-like” viruses [NLVs]) as the cause of many of these outbreaks. To streamline NLV detection from environmental samples such as shellfish, we have developed an RT-PCR–oligoprobe amplification and detection method using several new procedures that enable confirmed RT-PCR amplification and product detection in 1 day. The new steps include replacing reverse transcriptase and Taq polymerase with rTth polymerase, a heat-stable enzyme that functions as both a reverse transcriptase and DNA polymerase, in a single-tube, single-buffer, elevated temperature reaction. An internal standard Norwalk virus (NV) RNA control is added to each RT-PCR to identify sample inhibition, and thermolabile uracil N-glycosylase is incorporated into the reaction to prevent PCR product carryover contamination. Finally, RT-PCR-generated amplicons are detected in microtiter wells using virus-specific biotinylated oligoprobes in an enzyme-linked immunosorbent assay-based format. The DNA enzyme immunoassay is based on the capture of PCR product by biotinylated probes fixed onto individual streptavidin-coated wells. Using this method, low levels of NV were detected in stool and both NLV and hepatitis A virus were detected in bivalve mollusks following bioaccumulation. The method also successfully detected NLV in oysters implicated in an outbreak of NLV gastroenteritis. This method dramatically decreases the time needed for analysis and is amenable to automation.
“Norwalk-like” viruses (NLVs), previously described as small round structured viruses, are the most common cause of nonbacterial gastroenteritis outbreaks in the United States and other countries (12, 39). These small, nonenveloped RNA viruses are resistant to environmental degradation and chemical treatment processes and have been documented to be the most common agents of food- and waterborne outbreaks of nonbacterial gastroenteritis (7, 10, 11, 18, 23, 24). No animal model or cell culture system has been developed for the isolation or propagation of these viruses. Methods incorporating reverse transcription-PCR (RT-PCR) amplification that target short segments of the viral genome have been developed for NLV detection from clinical and environmental samples, including water concentrates, shellfish, and stools (2, 16, 17, 20, 36). Although each environmental sample type is unique, they all have the common feature of containing a wide variety of potential inhibitors of the RT-PCR. Several methods have been developed in attempts to overcome these inhibitors (1, 8, 9, 34, 35). However, most studies have not documented if inhibitors have been successfully removed from each sample prior to PCR amplification. We and others have developed internal standard RNA controls for Norwalk virus (NV) and hepatitis A virus (HAV), which when incorporated into each sample can determine if a negative RT-PCR result is due to sample inhibition (4, 5, 36).
RT-PCR is very specific and sensitive, enabling the detection of as few as 10 to 40 genomic copies of viral nucleic acid (5, 36). This sensitivity can also contribute to potential sample contamination by PCR product carryover into subsequent experiments. In addition, when assaying with RT-PCR for NLVs from processed environmental samples, production of nonspecific amplicons in conjunction with virus-specific amplicons can prevent successful interpretation of results by gel electrophoresis (2, 5, 37). In many cases, the specificity of PCR amplicons is determined either by direct sequencing or internal oligoprobing. Direct sequencing can rapidly and unequivocally confirm that appropriate amplification occurred but requires amplicons to be relatively pure (i.e., free of nonspecific products) and concentrated and can become expensive for routine screening of all samples. Most researchers only sequence samples that are positive by gel electrophoresis and thus may miss samples that are positive only after amplicon detection using more sensitive methods. In addition, direct sequencing of amplicons generated from environmental samples containing very few target organisms is very difficult and rarely successful. Environmental samples can be sequenced after nested PCR, but this technique greatly increases the potential for sample contamination (27, 29, 41). Internal oligoprobing, using radioactive or nonradioactive probes, can greatly enhance amplicon sensitivity and specificity compared to gel electrophoresis, with a 10- to 100-fold improvement in detection sensitivity (6). However, internal oligoprobing of NLVs can suffer from variability occurring during Southern or slot blot transfer of amplicons onto membranes and is time-consuming and cumbersome, requiring 1 to 2 days before results are obtained.
To streamline NLV detection from environmental samples such as shellfish, we have developed an RT-PCR–oligoprobe amplification and detection method incorporating several new procedures, enabling confirmed RT-PCR amplification and product detection in 1 day. The new steps include replacing reverse transcriptase and Taq polymerase with rTth polymerase, a heat-stable enzyme that functions as both a reverse transcriptase and DNA polymerase, in a single-tube, single-buffer, elevated temperature reaction. An internal standard NV RNA control is added to each RT-PCR to identify sample inhibition, and thermolabile uracil N-glycosylase (HK-UNG) is incorporated into the reaction to prevent PCR product carryover contamination. Finally, RT-PCR-generated amplicons are detected in microtiter wells using virus-specific biotinylated oligoprobes in an enzyme-linked immunosorbent assay (ELISA)-based format. The DNA enzyme immunoassay (DEIA) is based on the capture of PCR product by biotinylated probes fixed onto individual streptavidin-coated wells. DEIA permits rapid, potentially automated PCR product detection. The sensitivity and specificity of the developed method were determined, and the method was tested on bivalve mollusks that had bioaccumulated NV and HAV. The method was then tested on oysters implicated in an outbreak of NLV gastroenteritis.
MATERIALS AND METHODS
Virus preparations.
Stools containing NV collected from human volunteers following challenge with NV 8fIIa (14, 17) were diluted to 10% suspensions in phosphate-buffered saline (PBS) (pH 7.4). NV suspensions were titered by RT-PCR unit (PCRU) end-point dilution using heat to release the viral genome from the capsid protein (36). HAV (HM-175 strain) was propagated in FRhK4 monolayers, and monodispersed preparations were prepared and stored at −70°C as described previously (5).
Shellfish.
Two commercially important shellfish groups were studied: the oyster, Crassostrea virginica, and a hardshell clam, Mercenaria mercenaria. Live oysters harvested from Gulf coast estuaries were obtained from a commercial wholesaler (The Dutchman's Seafood, Houston, Tex.) 24 to 48 h after harvest. Live clams were obtained from the South Carolina Department of Natural Resources, Charleston. All shellfish were stored at 4°C and used within 6 days. Shellfish were scrubbed, rinsed, and allowed to acclimatize in synthetic seawater (Instant Ocean; Aquarium Systems, Mentor, Ohio) before experiments.
Bioaccumulation.
Shellfish were allowed to bioaccumulate NV as previously described (37). Briefly, shellfish were placed in separate polycarbonate trays (control and test tanks) containing 5 liters of synthetic seawater. A virus input of 105 PCRU (20 PCRU/ml) of NV and 105 PFU of HAV was mixed with 0.5 g of sterile (autoclaved) stool in 10 ml of PBS (to increase uptake in shellfish due to pathogen-particle association) (28, 32) for 10 min before addition to 5 liters of seawater in the test tank. Algae flakes (Kyorin Co., Ltd., Himeji, Japan) were added to stimulate shellfish feeding during bioaccumulation. A control tank was prepared in an identical manner to the test tank except with no addition of virus. Seawater was continuously aerated to maintain adequate oxygen levels. Tanks were covered during bioaccumulation to reduce the potential for cross-contamination by aerosolization of pathogens caused by aeration and mixing of the seawater. Bioaccumulation lasted 24 h, with a complete change of seawater and components (algae and stool-associated virus in the test tank or algae and sterile stool in the control tank) at 8 h.
Processing of tissues for detection by RT-PCR.
Oysters with bioaccumulated virus were cleaned and shucked, and the digestive diverticula were recovered. Individual shellfish tissue samples were processed for virus recovery by a previously described method (5). Briefly, digestive diverticula were homogenized, extracted with chloroform-butanol, and precipitated with Cat-floc (Calgon, Ellwood City, Pa.), followed by polyethylene glycol 6000 (BDH Laboratory Supplies, Poole, England) precipitation. Viral nucleic acid was extracted and purified from the suspended polyethylene glycol pellet by digestion with 0.2 mg of proteinase K per ml, phenol-chloroform extraction, ethanol precipitation, 1.4% (wt/vol) cetyltrimethylammonium bromide (Sigma, St. Louis, Mo.) precipitation, and a final ethanol precipitation. Viral nucleic acid was suspended in 100 μl of RNase-free H2O. Twenty microliters of extracted viral nucleic acid was used in an individual RT-PCR amplification.
Two-enzyme RT-PCR.
Conventional two-enzyme RT-PCRs were performed as described previously using NV-specific p35 and p36 primers (5). Briefly, 20 μl of sample was reverse transcribed in a total volume of 30 μl. The RT mix contained 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 3.3 μM downstream primer, 667 μM deoxynucleoside triphosphates, 20 U of RNasin (Promega), and 5 U of avian myeloblastosis virus reverse transcriptase (Life Sciences, Inc., St. Petersburg, Fla.). In addition, the NLV RT mix contained approximately 50 copies of NV RNA internal standard control (36). The RT reaction was incubated for 1 h at 43°C, heated for 5 min at 95°C, and placed on ice. Seventy microliters of PCR reagents was added to the RT mix to yield a solution containing 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 1 μM concentration of each primer, 200 μM deoxynucleoside triphosphates, and 5 U of Taq polymerase (Perkin-Elmer, Foster City, Calif.). cDNA was amplified using either a PTC-100 or PTC-200 thermal cycler (MJ Research, Inc., Cambridge, Mass.), with an initial 2-min denaturation at 94°C followed by 40 cycles of 92°C denaturation for 15 s, 55 or 57°C annealing for 30 s, and extension for 30 s at 72°C, with a final 7-min extension at 72°C. The expected product sizes using the NVp35 and p36 primers were 470 bp for NV and 347 bp for the internal standard control.
RT-PCR internal standard control.
An NV RNA internal standard control consisting of NV RNA with a 123-bp deletion was generated by transcription from an NV plasmid constructed as previously described (36). The internal control transcript (50 to 100 copies per reaction) was added to NV-specific amplification reactions that were amplified with NVp35 and p36 primers.
Single-enzyme RT-PCR.
Single-enzyme RT-PCR using rTth (Perkin-Elmer) was performed in 50-μl reactions with the incorporation of HK-UNG (Epicentre Technologies, Madison, Wis.). An MJR PTC-200 thermal cycler using 200-μl thin-wall tubes and no oil overlay was used. The RT-PCR mixture consisted of 1× EZ buffer; 300 μM (each) dATP, dCTP, and dGTP; 600 μM dUTP; 1 μM upstream and downstream primers; 2.5 mM Mn(OAc)2; 20 U of RNasin; 5U of rTth; 0.5 U of HK-UNG; approximately 50 copies of internal standard; and 20 μl of sample. Samples were incubated for 5 min at 37°C to degrade any contaminating dUTP-containing PCR products, 10 min at 68°C to inactivate HK-UNG, 10 min at 55 or 57°C for downstream primer annealing, and 50 min at 60°C for RT. cDNA was amplified using the following cycling conditions: initial denaturation at 94°C for 2 min followed by 40 cycles of template denaturation at 92°C for 15 s, primer annealing at 55 or 57°C for 30 s, and primer extension at 60°C for 30 s, followed by a final extension at 60°C for 5 min. For HAV studies, the HAV-specific primers HAVp3 and HAVp4 (5, 38) were used for amplification using the above amplification conditions, generating a 248-bp product.
Conventional oligoprobe hybridization and detection.
For Southern hybridization oligoprobing, the NV-specific oligonucleotide NVp69 (22) was 5′ end labeled with digoxigenin using terminal transferase according to the manufacturer's instructions (Boehringer-Mannheim, Indianapolis, Ind.). Southern blot hybridization assays were performed as described previously (37). Briefly, DNA was transferred to a positively charged nylon membrane by vacuum transfer for 2 h and fixed to the membrane by UV cross-linking. The membrane was prehybridized for 30 min at 55°C in prehybridization buffer (Express Hyb; ClonTech, Palo Alto, Calif.) and then placed into a hybridization solution (Express Hyb plus 25 nM digoxigenin-labeled NVp69). After hybridization for 1 h at 55°C, the membrane was washed twice at room temperature for 5 min in 2× SSC–0.1% sodium dodecyl sulfate (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and twice at 45°C for 15 min in 0.5× SSC–0.1% sodium dodecyl sulfate. The hybridized probe was detected by use of the Genius 3 nucleic acid detection kit (Boehringer-Mannheim) according to the manufacturer's protocol. Digoxigenin-labeled markers (Boehringer-Mannheim) were used for molecular weight size determination following gel electrophoresis and Southern blot hybridization. For HAV studies, the HAV-specific probe HAVp8 (38) was used in Southern hybridization studies as described previously.
DEIA PCR product detection.
DEIAs (GEN-ETI-K DEIA; DiaSorin Inc., Stillwater, Minn.) used in conjunction with the NLV and HAV RT-PCR assays were developed. Manufacturer instructions were followed, with slight modifications. Briefly, streptavidin-coated microtiter eight-well strips were incubated with 100 μl of biotinylated probe overnight at 4°C. For the NLV DEIA, biotinylated and high-pressure liquid chromatography (HPLC)-purified oligoprobes NVp69 and SR65 (2) were purchased (Integrated DNA Technologies, Inc., Coralville, Iowa). Coated strips were washed five times with 200 μl of 1× wash buffer (PBS, Tween 20, and preservatives) provided with the kit, with a 30-s incubation per wash. Washed strips were air dried and stored for up to 1 month at 4°C. Amplified PCR products were denatured for 10 min at 95°C and placed on ice until use. Sufficient probe-coated wells were prepared for one blank well (only chromogen and substrate), one negative DEIA control well, and all sample wells including amplification controls, by adding 100 μl of the hybridization buffer (Denhardt's solution, SSC, Tris-HCl buffer, EDTA, and preservatives) provided with the kit into each well. Ten microliters of the DEIA negative control or denatured amplified PCR products was dispensed into the appropriate wells. Samples were incubated for 1 h at 50°C, washed five times with wash buffer, incubated for 30 min with 100 μl of anti-double-stranded DNA antibody (1:50 dilution), washed five times with wash buffer, incubated for 30 min with 100 μl of a 1:50 dilution of enzyme tracer (protein A conjugated to horseradish peroxidase, MES buffer, proteins, stabilizers, and preservatives), washed five times with wash buffer, and incubated for 30 min in the dark after the addition of 100 μl of a 1:1 dilution of chromogen (3,3′,5,5′ tetramethylbenzidine derivative in citrate buffer) and substrate (hydrogen peroxide and citrate buffer). The reaction was stopped by the addition of 200 μl of 1 N sulfuric acid to each well, and the absorbance was determined spectrophotometrically (MR5000; Dynatech Laboratories, Chantilly, Va.) within 10 min of addition of sulfuric acid. Wells were read at 450 nm and 630 nm, with subtraction of the optical density at 630 nm (OD630) from the OD450. A probe-coated well, to which no reagents were added until the chromogen and substrate step, was used to blank the spectrophotometer. For HAV studies, the HAV-specific biotinylated and HPLC-purified oligoprobe HAVp8 (38) was purchased from Genosys (Woodlands, Tex.) and used at a concentration of 25 pg/μl and a hybridization temperature of 50°C.
Detection of an NLV from oysters implicated in an outbreak of gastroenteritis.
The digestive diverticula from oysters collected during an outbreak of gastroenteritis in France were processed by the described method. The primers CDR 1U (GCC CAR GCT GAA ATG AC) and CDR L1B (GCR TTG GTG GTG ATG ACT AT) were used to amplify a 347-bp region of the polymerase region. PCR product was detected using the biotinylated and HPLC-purified probe CDR U2D (TGG GAC GAC TAT GGA ATG AC) (42) at a concentration of 100 pg/μl and a hybridization temperature of 50°C using the DEIA method described above.
RESULTS
Single tube-single enzyme rTth assay. (i) Determination of digestion time required to eliminate dUTP-containing contaminants.
In order to estimate the effectiveness of UNG removal of contaminating PCR products, experiments were conducted to determine at what dilution PCR product could be reamplified. PCR products from previous experiments amplified by rTth without UNG, using either dTTP or dUTP in the reaction mix, were serially diluted 10-fold and amplified with rTth without UNG. Following amplification and agarose gel electrophoresis, the bands for these products were visible at a 10−6 dilution of template. Amplicons were not detectable by agarose gel electrophoresis beyond a 10−2 dilution prior to reamplification. Therefore, 10−3 dilutions of uracil-containing amplicons seeded into RT-PCRs were used to test the effectiveness of UNG. This represented at least 1,000 PCRU of amplicons to be eliminated, and the input amplicons would not be visible on a gel without amplification having taken place. The time required for UNG digestion of amplicon contaminants was examined. Digestion times of 30, 15, and 0 min were tested, followed by a 10-min heat inactivation of UNG. Thymidine-containing amplicons, used as controls, were amplified under all conditions tested. Uracil-containing amplicons were not amplified under any of these conditions. Although significant digestion occurred in less than 1 min, 5 min of UNG digestion was chosen as a conservative time to ensure complete digestion of PCR product contamination.
(ii) Effect of digested amplicons on detection of viral RNA.
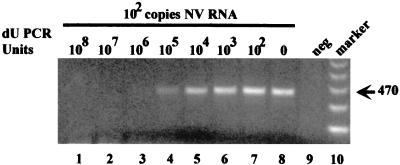
When the efficacy of the UNG system was evaluated in the presence of contaminating uracil-containing amplicons generated from internal standard and viral RNA amplification, not only was the amplification of uracil-containing amplicons prevented as expected, but viral RNA also were not amplified. We found that when ≥107 PCRU of amplicons were present in the reaction mixture, amplification of these amplicons and a low copy number of viral RNA (100 PCRU) were not detectable (Fig. 1, lanes 1 and 2). When 106 to 104 PCRU of dUTP-containing amplicons was present, the magnitude of viral RNA amplification increased (Fig. 1, lanes 3 to 5). At levels of 103 or fewer PCRU there was no observable inhibition of viral RNA amplification (Fig. 1, lanes 6 and 7). The dUTP-containing amplicons were not amplified at any concentration.
FIG. 1.
RT-PCR coamplification of internal standard amplicons with NV RNA using rTth, NV p35 and p36 primers, and UNG. Lanes 1 to 8 contain 100 copies of NV (10 μl of a 10−3 dilution of NV RNA) and 10 μl of a 10-fold dilution series of PCR product amplified with dUTP (lane 1, 10−2 dilution of PCR product [108 PCRU], through lane 7, 10−8 dilution of PCR product [102 PCRU]). Lane 9, negative reagent control; lane 10, 123-bp molecular weight marker.
Detection of PCR product by DEIA. (i) Optimization of DEIA probe concentration and hybridization temperature.
Two biotinylated probes, NVp69 and SR65, were used to detect amplicons generated from internal standard and NV RNA, respectively (NVp69 will detect both internal standard and NV amplicons and SR65 will only detect NV amplicons), for both rTth and two-enzyme RT-PCR. NVp69 is used to determine if inhibitors are present, and SR65 is used to detect the virus of interest (i.e., NV). Conditions were varied from 0.025 to 0.5 ng of NVp69 per μl and from 0.05 to 0.5 ng of SR65 per μl to determine probe concentrations that generated the lowest backgrounds without loss of product detection sensitivity. Concurrently, probe hybridization temperature was optimized between 50 and 55°C for each probe concentration. Following quadruplicate end-point dilution trials, optimum specificity and sensitivity were obtained using a hybridization temperature of 50°C and probe concentrations of 0.05 and 0.025 ng/μl for SR65 and NVp69, respectively. There was no difference in product detection when either rTth or the two-enzyme method was used to generate amplicons.
(ii) Determination of DEIA background levels.
To obtain a background DEIA OD level, 18 virus-free negative-control volunteer stools were processed and RT-PCR amplified with NVp35 and p36 primers, using either the rTth or two-enzyme method with and without the addition of the NV internal standard control. Amplicons were assayed by both Southern blot hybridization and DEIA. For each probe, OD results were combined and a standard deviation (SD) was calculated. For SR65 with and without the internal standard and NVp69 without the internal standard (NVp69 with the internal standard is used to detect inhibition and thus always results in a strong signal when no inhibition is present), the sample OD results (mean ± SD) for negative-control stools were 0.056 ± 0.004 (n = 16), 0.059 ± 0.018 (n = 17), and 0.038 ± 0.010 (n = 17), respectively. A subset (n = 8) of negative-control stool samples was assayed in three separate experiments to determine interexperimental variability for both probes. The sample mean OD readings were less than or equal to 0.050. The SD for any of the samples did not exceed 0.010. The means (SD) of probes SR65 and NVp69 for all eight samples over the three experiments were 0.039 (0.002) and 0.025 (0.002), respectively. The cutoff OD value for positive samples for each subsequent experiment was calculated by averaging the DEIA negative control and RT-PCR negative control from that experiment and adding 0.05 (greater than 2 SD for any probe condition tested). If this cutoff value was less than 0.1, a more conservative value of 0.1 was used as the cutoff OD.
(iii) Comparison between DEIA and Southern blot sensitivity.
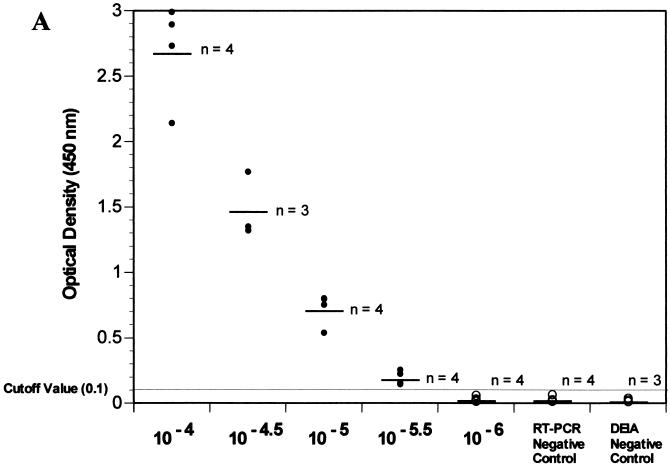
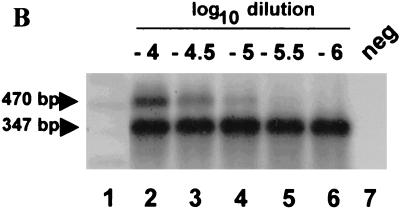
The DEIA assay detected NV RNA RT-PCR amplicons at similar or in some cases greater end points compared to Southern blot hybridization. Replicate samples of an RT-PCR-amplified NV RNA half-log10 dilution series pool using rTth, NVp35 and p36 primers, UNG, and 50 copies of internal control were analyzed by DEIA (probe SR65) and Southern blot hybridization (probe NVp69) (Fig. 2). Both assay methods produced an NV end point of 20 μl of a 10−5.5 dilution. However, the Southern blot end point was quite faint, while the numerical DEIA results provided a clear determination of the viral RT-PCR end point (Fig. 2).
FIG. 2.
DEIA using probe SR65 (A) and Southern blot using probe NVp69 (B) detection of pooled RT-PCR amplification of NV half-log10 dilution series end-point determination. RT-PCR conditions included use of rTth, UNG, NV p35 and p36 primers, and addition of 50 copies of internal standard (except lane 7).
(iv) Determination of DEIA specificity.
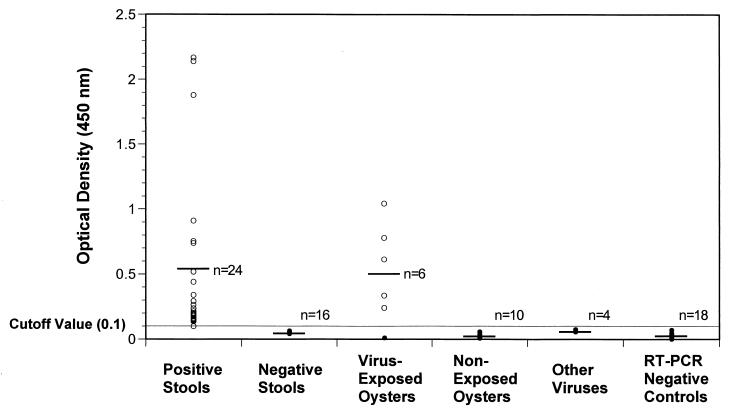
NV-positive stools (n = 24), virus-negative stools (n = 16), shellfish with bioaccumulated NV (n = 6), and negative-control shellfish with bioaccumulation (n = 10) were assayed by the developed rTth RT-PCR method and detected by both Southern blot hybridization and DEIA. There was 100% concordance between both methods. Figure 3 shows SR65 probe DEIA results for all the samples. In addition, RT-PCR or PCR products from poliovirus type 1, adenovirus serotype 4 strain 75680, human astrovirus type 2, and HAV, amplified with virus-specific primers and confirmed with virus-specific probes, were tested by the DEIA using SR65 and NVp69 probes. None of the non-NV products were positive using SR65 and NVp69 probes, with all of the OD readings being less than 0.1 (Fig. 3).
FIG. 3.
SR65 probe DEIA results of RT-PCR amplicons generated by RT-PCR using rTth, UNG, NV p35 and p36 primers, and 50 to 100 copies of internal standard. Viral RNA was recovered from stools and oysters using described methods prior to amplification.
(v) Detection of NLV and HAV from mollusks with bioaccumulation.
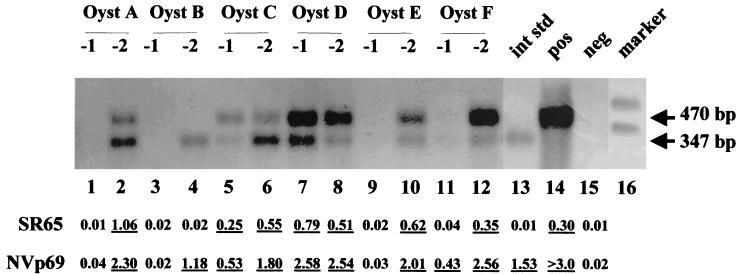
Digestive diverticula from oysters and clams with bioaccumulated NV and HAV were processed by the described virus recovery method and assayed by the developed single-tube single-enzyme rTth method. Resulting PCR products were subsequently detected by both conventional gel and Southern blot analysis as well as by the DEIA method. Nine of 12 oysters and 5 of 8 clam samples were positive (one inhibited clam sample retested at a 10-fold lower dilution was positive) for NV following RT-PCR amplification and either Southern blot or DEIA oligoprobe detection. Both detection methods gave concordant results. Ten negative-control shellfish were processed, assayed by RT-PCR, and examined by DEIA. None of the control sample background OD levels were above 0.07, indicating that even after complex environmental samples such as shellfish are processed, amplified, and analyzed by DEIA, background OD readings are as low as reagent controls. Figure 4 shows NV recovery from a representative subset of oysters by both Southern transfer hybridization using the NVp69 probe and DEIA using both the NVp69 and SR65 probes.
FIG. 4.
Southern transfer oligoprobe and DEIA detection following rTth RT-PCR amplification of internal standard and NV bioaccumulated in oysters. RT-PCR of 10-fold (lanes 1, 3, 5, 7, 9, and 11) and 100-fold (lanes 2, 4, 6, 8, 10, and 12) dilutions of RNA of individual oyster (Oyst) extracts obtained following dissection of the digestive diverticula from oysters after 24 h of bioaccumulation with 105 NV PCRU/5 liters. Lanes 1 to 14 contain 100 copies of internal standard. Other lanes include 100 copies of an internal standard (int std) control (lane 13), an NV RT-PCR-positive control (lane 14), a negative reagent control (lane 15), and a digoxigenin-labeled molecular size marker (Marker VIII; Boehringer-Mannheim) (lane 16). Numbers at the right are molecular sizes in base pairs. Below the Southern blot are the DEIA OD readings, with underlined numbers corresponding to positive DEIA samples.
Nine of the oysters with bioaccumulation assayed for NV were also assayed for the presence of HAV RNA using the single-enzyme RT-PCR–DEIA method for HAV (there was insufficient sample to assay the other oysters or clams). HAV was detected in six of the seven oysters from which NV was detected, and HAV was not detected in the two oysters that were negative for NV. HAV was also not detected in any of the seven control shellfish that were not exposed to virus. Concordant results were obtained using Southern hybridization.
(vi) Detection of NLV genogroup I virus from oysters implicated in an NLV outbreak.
A pool of oyster diverticula made from oysters collected in association with an outbreak of gastroenteritis following shellfish consumption was examined. The outbreak strain could not be amplified using polymerase-specific primers. However, a genogroup I NLV was amplified from the shellfish using both capsid-specific (15) and helicase-specific primers. The single-enzyme RT-PCR–DEIA method was modified to use the helicase-specific primers, and virus-specific amplicons were detected in the outbreak shellfish (OD450 = 0.89, negative control OD450 = 0.008). The results were concordant with Southern hybridization results.
DISCUSSION
The use of molecular techniques has dramatically improved our ability to identify human pathogens from clinical and environmental samples. Our laboratory and others have developed methods to recover low levels of human enteric viruses from such samples for subsequent analysis by RT-PCR (2, 19, 35, 36, 38, 40).
This paper describes improvements to the conventional two-enzyme RT-PCR amplification and subsequent Southern transfer and oligoprobe detection of NLVs. The rTth single-enzyme method uses a Perkin-Elmer recombinant thermostable DNA polymerase from Thermus thermophilus (rTth) which has both reverse transcriptase and polymerase activity. A key component is the use of a unique buffer in which Tris is replaced with bicine. This enables both the Mn2+ concentration and the pH to be controlled precisely enough to allow both RT and PCR amplification without opening the tube and changing the buffer between reactions. The advantages of this system include an easier and quicker setup and the reduced likelihood of cross-contamination.
A screening system for environmental samples must address the possibility of carryover contamination of samples by previously amplified PCR products (resulting in false positives). Carryover contamination is a particular concern at facilities where personnel are less experienced in molecular biology techniques. One carryover prevention strategy consists of utilizing dUTP in place of dTTP in the RT-PCR reaction mix (21, 30, 31). Using this strategy, all samples are treated with UNG before RT-PCR to destroy any dUTP-containing amplicons that may have contaminated the sample during preparation. This prevents contaminating amplicons from being amplified. We have developed a carryover prevention method that uses a heat-labile UNG (HK-UNG). Following the elimination of dUTP-containing amplicons by HK-UNG, the HK-UNG enzyme is inactivated by heat prior to RT. There was no loss in the sensitivity of viral RNA detection when HK-UNG was added to the RT-PCR mixture (data not shown). Interestingly, when greater than 106 contaminating amplicons were seeded into a reaction tube containing low levels of test RNA, RT-PCR of the target RNA was completely ablated (Fig. 1, lanes 1 and 2), suggesting that while gross PCR product contamination will be eliminated by the UNG, the degraded contaminants may also inhibit PCR. Fortunately, such inhibition will be detected by the internal standard control.
Although the manufacturer claimed that the HK-UNG was completely inactivated following heat denaturation, we noticed at least a 10-fold loss in sensitivity (level of detection) when amplified samples were stored for more than 24 h at 4°C following RT-PCR amplification. We did not observe any loss of product when samples were stored at −20°C for up to 7 days postamplification (data not shown).
Southern blot hybridization is a labor-intensive detection system that requires 1 to 2 days to complete. Newer detection assays have been developed that allow more rapid evaluation of PCR results (3, 4, 13, 25, 26, 33, 43). We developed a DEIA to be used in conjunction with the NLV and HAV RT-PCRs. This ELISA-based format enables very rapid analysis of large numbers of PCR samples and is amenable to automation. As with other ELISA-based assays, it is important to determine and optimize background absorbance for each probe. Probe concentration and hybridization temperature are key factors during optimization. For the probes used in these experiments, parameters were selected that provided good sensitivity and low background. The DEIA detected NV and internal standard RNA with similar or, in some experiments, greater sensitivity than was achieved by Southern blot hybridization (Fig. 2).
A distinct advantage of the DEIA is the elimination of the subjective interpretation of the presence or absence of bands on a gel or Southern blot. DEIA provides a numerical result, in contrast to a band with varying levels of intensity present on a gel or a Southern or slot blot. In a DEIA, a numerical cutoff value is generated by the negative-control samples. Samples are either above or below this cutoff value. Results obtained following more conventional detection (e.g., Southern blot hybridization) are more subjective, and interpretations of results can vary, especially when signal intensity is weak. The DEIA detection assay achieves a significant savings in time, with results now available in less than 4 h instead of 1 to 2 days.
Although many viral extraction procedures for bivalve mollusks have been described, no method is 100% efficient. By incorporating an internal standard control into the RT-PCR, we can identify samples that contain inhibitors. Many other detection methods do not allow the researcher to distinguish between samples absent of virus or false-negative samples due to sample inhibition. This point is exemplified by the difference between oysters A, B, and F in Fig. 4. All three oysters contained sample inhibition at the 10−1 dilution. Oysters A and F are positive for both the internal standard and NV at the 10−2 dilution, indicating that these two oysters contained virus. Oyster B, however, is only positive for the internal standard at the 10−2 dilution. Thus, oyster B does not contain virus and is a true negative sample. Bioaccumulation experiments use live animals, and we speculate that shellfish from which virus was not detected may not have bioaccumulated virus during the bioaccumulation period. Alternatively, bioaccumulated virus may have been present at a level below the level of virus detection.
The general applicability of the developed method for the detection of viruses in shellfish was confirmed by the results for HAV and by using oysters implicated in an outbreak of NLV gastroenteritis in France. In the latter group, a genogroup I NLV was detected in a pool of tested oysters. Application of this method to an outbreak investigation suggests the broader applicability of the method for the detection of NLVs, although the inability of polymerase-specific primers to detect the outbreak strain highlights the limitation of a single set of primers to detect all NLVs. Current research is examining the sensitivity and specificity of PCR product detection using multiple biotinylated probes present in a single streptavidin-coated well. This could further improve the applicability of the method in outbreak investigations.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Oceanic and Atmospheric Administration (NA77FD0080), the Environmental Protection Agency (CX 827430-01-0), and the European Union (QLK1-1999-00634).
REFERENCES
- 1.Abbaszadegan M, Huber M S, Gerba C, Pepper I L. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl Environ Microbiol. 1993;59:1318–1324. doi: 10.1128/aem.59.5.1318-1324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando T, Monroe S S, Gentsch J R, Jin Q, Lewis D C, Glass R I. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J Clin Microbiol. 1995;33:64–71. doi: 10.1128/jcm.33.1.64-71.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnal C, Ferre-Aubineau V, Besse B, Billaudel S. Simplified reverse transcription polymerase chain reaction procedure with detection by microplate hybridization for routine screening of hepatitis A virus. Can J Microbiol. 1998;44:298–302. [PubMed] [Google Scholar]
- 4.Arnal C, Ferre-Aubineau V, Mignotte B, Imbert-Marcille B M, Billaudel S. Quantification of hepatitis A virus in shellfish by competitive reverse transcription-PCR with coextraction of standard RNA. Appl Environ Microbiol. 1999;65:322–326. doi: 10.1128/aem.65.1.322-326.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atmar R L, Neill F H, Romalde J L, Le Guyader F, Woodley C M, Metcalf T G, Estes M K. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl Environ Microbiol. 1995;61:3014–3018. doi: 10.1128/aem.61.8.3014-3018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atmar R L, Neill F H, Woodley C M, Manger R, Fout G S, Burkhardt W, Leja L, McGovern E R, Le Guyader F, Metcalf T G, Estes M K. Collaborative evaluation of a method for the detection of Norwalk virus in shellfish tissues by PCR. Appl Environ Microbiol. 1996;62:254–258. doi: 10.1128/aem.62.1.254-258.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beller M, Ellis A, Lee S H, Drebot M A, Jenkerson S A, Funk E, Sobsey M D, Simmons O D I, Monroe S S, Ando T, Noel J, Petric M, Middaugh J P, Spika J S. Outbreak of viral gastroenteritis due to a contaminated well. International consequences. JAMA. 1997;278:563–568. [PubMed] [Google Scholar]
- 8.Boom R, Sol C, Beld M, Weel J, Goudsmit J, Wertheim-van Dillen P. Improved silica-guanidiniumthiocyanate DNA isolation procedure based on selective binding of bovine alpha-casein to silica particles. J Clin Microbiol. 1999;37:615–619. doi: 10.1128/jcm.37.3.615-619.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boom R, Sol C J, Salimans M M, Jansen C L, Wertheim-van Dillen P M, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniels N A, Bergmire-Sweat D, Schwab K J, Hendricks K A, Reddy S, Rowe S M, Fankhauser R L, Monroe S S, Atmar R L, Glass R I, Mead P. A foodborne outbreak of gastroenteritis associated with Norwalk-like viruses: first molecular traceback to deli sandwiches contaminated during preparation. J Infect Dis. 2000;181:1467–1470. doi: 10.1086/315365. [DOI] [PubMed] [Google Scholar]
- 11.Dowell S F, Groves C, Kirkland K B, Cicirello H G, Ando T, Jin Q, Gentsch J R, Monroe S S, Humphrey C D, Slemp C, Dwyer D M, Meriwether R A, Glass R I. A multistate outbreak of oyster-associated gastroenteritis: implications for interstate tracing of contaminated seafood. J Infect Dis. 1995;171:1497–1503. doi: 10.1093/infdis/171.6.1497. [DOI] [PubMed] [Google Scholar]
- 12.Fankhauser R L, Noel J S, Monroe S S, Ando T, Glass R I. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J Infect Dis. 1998;178:1571–1578. doi: 10.1086/314525. [DOI] [PubMed] [Google Scholar]
- 13.Ferre-Aubineau V, Imbert-Marcille B M, Raffi F, Besse B, Loirat R, Billaudel S. Colorimetric microtiter plate hybridization assay using monoclonal antibody for detection of an amplified human immunodeficiency virus target. J Virol Methods. 1995;55:145–151. doi: 10.1016/0166-0934(95)00058-3. [DOI] [PubMed] [Google Scholar]
- 14.Graham D Y, Jiang X, Tanaka T, Opekun A R, Madore H P, Estes M K. Norwalk virus infection of volunteers: new insights based on improved assays. J Infect Dis. 1994;170:34–43. doi: 10.1093/infdis/170.1.34. [DOI] [PubMed] [Google Scholar]
- 15.Hafliger D, Gilgen M, Luthy J, Hubner P. Seminested RT-PCR systems for small round structured viruses and detection of enteric viruses in seafood. Int J Food Microbiol. 1997;37:27–36. doi: 10.1016/s0168-1605(97)00041-x. [DOI] [PubMed] [Google Scholar]
- 16.Jaykus L A, De Leon R, Sobsey M D. A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization. Appl Environ Microbiol. 1996;62:2074–2080. doi: 10.1128/aem.62.6.2074-2080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X, Wang J, Graham D Y, Estes M K. Detection of Norwalk virus in stool by polymerase chain reaction. J Clin Microbiol. 1992;30:2529–2534. doi: 10.1128/jcm.30.10.2529-2534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilgore P E, Belay E D, Hamlin D M, Noel J S, Humphrey C D, Gary H E, Jr, Ando T, Monroe S S, Kludt P E, Rosenthal D S, Freeman J, Glass R I. A university outbreak of gastroenteritis due to a small round-structured virus. Application of molecular diagnostics to identify the etiologic agent and patterns of transmission. J Infect Dis. 1996;173:787–793. doi: 10.1093/infdis/173.4.787. [DOI] [PubMed] [Google Scholar]
- 19.Kukkula M, Maunula L, Silvennoinen E, von Bonsdorff C H. Outbreak of viral gastroenteritis due to drinking water contaminated by Norwalk-like viruses. J Infect Dis. 1999;180:1771–1776. doi: 10.1086/315145. [DOI] [PubMed] [Google Scholar]
- 20.Lees D N, Henshilwood K, Green J, Gallimore C I, Brown D W G. Detection of small round structured viruses in shellfish by reverse transcription-PCR. Appl Environ Microbiol. 1995;61:4418–4424. doi: 10.1128/aem.61.12.4418-4424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legeay O, Bounaix S, Denis M, Arnauld C, Hutet E, Cariolet R, Albina E, Jestin A. Development of a RT-PCR test coupled with a microplate colorimetric assay for the detection of a swine Arterivirus (PRRSV) in boar semen. J Virol Methods. 1997;68:65–80. doi: 10.1016/s0166-0934(97)00110-9. [DOI] [PubMed] [Google Scholar]
- 22.Le Guyader F, Estes M K, Hardy M E, Neill F H, Green J, Brown D, Atmar R L. Evaluation of a degenerate primer for the PCR detection of human caliciviruses. Arch Virol. 1996;141:2225–2235. doi: 10.1007/BF01718228. [DOI] [PubMed] [Google Scholar]
- 23.Le Guyader F, Neill F H, Estes M K, Monroe S S, Ando T, Atmar R L. Detection and analysis of a small round-structured virus strain in oysters implicated in an outbreak of acute gastroenteritis. Appl Environ Microbiol. 1996;62:4268–4272. doi: 10.1128/aem.62.11.4268-4272.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maguire A J, Green J, Brown D W G, Desselberger U, Gray J J. Molecular epidemiology of outbreaks of gastroenteritis associated with small round-structured viruses in East Anglia, United Kingdom, during the 1996–1997 season. J Clin Microbiol. 1999;37:81–89. doi: 10.1128/jcm.37.1.81-89.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantero G, Zonaro A, Albertini A, Bertolo P, Primi D. DNA enzyme immunoassay: general method for detecting products of polymerase chain reaction. Clin Chem. 1991;37:422–429. [PubMed] [Google Scholar]
- 26.Maunula L, Piiparinen H, von Bonsdorff C H. Confirmation of Norwalk-like virus amplicons after RT-PCR by microplate hybridization and direct sequencing. J Virol Methods. 1999;83:125–134. doi: 10.1016/s0166-0934(99)00115-9. [DOI] [PubMed] [Google Scholar]
- 27.McGoldrick A, Bensaude E, Ibata G, Sharp G, Paton D J. Closed one-tube reverse transcription nested polymerase chain reaction for the detection of pestiviral RNA with fluorescent probes. J Virol Methods. 1999;79:85–95. doi: 10.1016/s0166-0934(99)00010-5. [DOI] [PubMed] [Google Scholar]
- 28.Metcalf T G, Mullin B, Eckerson D, Moulton E, Larkin E P. Bioaccumulation and depuration of enteroviruses by the soft-shelled clam, Mya arenaria. Appl Environ Microbiol. 1979;38:275–282. doi: 10.1128/aem.38.2.275-282.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellett P E, Spira T J, Bagasra O, Boshoff C, Corey L, de Lellis L, Huang M L, Lin J C, Matthews S, Monini P, Rimessi P, Sosa C, Wood C, Stewart J A. Multicenter comparison of PCR assays for detection of human herpesvirus 8 DNA in semen. J Clin Microbiol. 1999;37:1298–1301. doi: 10.1128/jcm.37.5.1298-1301.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poddar S K, Sawyer M H, Connor J D. Optimized PCR amplification of influenza A virus RNA using Tth DNA polymerase, incorporating uracil N glycosylase (UNG) in a single tube reaction. J Clin Lab Anal. 1997;11:323–327. doi: 10.1002/(SICI)1098-2825(1997)11:6<323::AID-JCLA2>3.0.CO;2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poljak M, Seme K. Simple one-tube reverse transcription-polymerase chain reaction protocol containing anticontamination procedure for detection of GB virus C/hepatitis G virus RNA. J Virol Methods. 1998;71:1–6. doi: 10.1016/s0166-0934(97)00205-x. [DOI] [PubMed] [Google Scholar]
- 32.Rao V C, Seidel K M, Goyal S M, Metcalf T G, Melnick J L. Isolation of enteroviruses from water, suspended solids, and sediments from Galveston Bay: survival of poliovirus and rotavirus adsorbed to sediments. Appl Environ Microbiol. 1984;48:404–409. doi: 10.1128/aem.48.2.404-409.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabouraud A, Smith J S, Orciari L A, de Mattos C, de Mattos C, Rohde R. Typing of rabies virus isolates by DNA enzyme immunoassay. J Clin Virol. 1999;12:9–19. doi: 10.1016/s1386-6532(98)00006-7. [DOI] [PubMed] [Google Scholar]
- 34.Schwab K J, De Leon R, Sobsey M D. Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription-PCR. Appl Environ Microbiol. 1995;61:531–537. doi: 10.1128/aem.61.2.531-537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwab K J, De Leon R, Sobsey M D. Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcriptase PCR. Appl Environ Microbiol. 1996;62:2086–2094. doi: 10.1128/aem.62.6.2086-2094.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwab K J, Estes M K, Neill F H, Atmar R L. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J Clin Microbiol. 1997;35:511–514. doi: 10.1128/jcm.35.2.511-514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwab K J, Neill F H, Estes M K, Metcalf T G, Atmar R L. Distribution of Norwalk virus within shellfish following bioaccumulation and subsequent depuration by detection using RT-PCR. J Food Prot. 1998;61:1674–1680. doi: 10.4315/0362-028x-61.12.1674. [DOI] [PubMed] [Google Scholar]
- 38.Schwab K J, Neill F H, Fankhauser R L, Daniels N A, Monroe S S, Bergmire-Sweat D A, Estes M K, Atmar R L. Development of methods to detect “Norwalk-like viruses” (NLVs) and hepatitis A virus in delicatessen foods: application to a food-borne NLV outbreak. Appl Environ Microbiol. 2000;66:213–218. doi: 10.1128/aem.66.1.213-218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinje J, Altena S A, Koopmans M. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J Infect Dis. 1997;176:1374–1378. doi: 10.1086/517325. [DOI] [PubMed] [Google Scholar]
- 40.Vinje J, Deijl H, van Der H R, Lewis D, Hedlund K O, Svensson L, Koopmans M P. Molecular detection and epidemiology of Sapporo-like viruses. J Clin Microbiol. 2000;38:530–536. doi: 10.1128/jcm.38.2.530-536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whelen A C, Felmlee T A, Hunt J M, Williams D L, Roberts G D, Stockman L, Persing D H. Direct genotypic detection of Mycobacterium tuberculosis rifampin resistance in clinical specimens by using single-tube heminested PCR. J Clin Microbiol. 1995;33:556–561. doi: 10.1128/jcm.33.3.556-561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyn-Jones A P, Pallin R, Dedoussis D, Shore J, Sellwood J. The detection of small round-structured viruses in water and environmental materials. J Virol Methods. 2000;87:99–107. doi: 10.1016/s0166-0934(00)00157-9. [DOI] [PubMed] [Google Scholar]
- 43.Zerbini M, Gallinella G, Manaresi E, Musiani M, Gentilomi G, Venturoli S. Standardization of a PCR-ELISA in serum samples: diagnosis of active parvovirus B19 infection. J Med Virol. 1999;59:239–244. doi: 10.1002/(sici)1096-9071(199910)59:2<239::aid-jmv19>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]