Abstract
We used real-time PCR to quantify the denitrifying nitrite reductase gene (nirS), a functional gene of biogeochemical significance. The assay was tested in vitro and applied to environmental samples. The primer-probe set selected was specific for nirS sequences that corresponded approximately to the Pseudomonas stutzeri species. The assay was linear from 1 to 106 gene copies (r2 = 0.999). Variability at low gene concentrations did not allow detection of twofold differences in gene copy number at less than 100 copies. DNA spiking and cell-addition experiments gave predicted results, suggesting that this assay provides an accurate measure of P. stutzeri nirS abundance in environmental samples. Although P. stutzeri abundance was high in lake sediment and groundwater samples, we detected low or no abundance of this species in marine sediment samples from Puget Sound (Wash.) and from the Washington ocean margin. These results suggest that P. stutzeri may not be a dominant marine denitrifier.
Denitrification is one of the important biogeochemical processes in that it is the main sink for fixed nitrogen (28). In agriculture, it accounts for 20 to 30% of fertilizer losses (11), and in marine environments it is thought to account for up to 80% of the loss of the nitrogen load to coastal areas (38). Two of its products, NO and N2O, are involved in global warming and the destruction of the ozone layer. Denitrification is also used in waste treatment facilities to remove excess combined nitrogen (41).
More accurate understanding and modeling of denitrification should be possible if the catalyst can be quantified. We have evaluated the application of real-time PCR to the quantification of the nitrite reductase gene in Pseudomonas stutzeri. P. stutzeri is a denitrifier commonly isolated from both soil (12) and marine environments (44), and it may be of general importance in global denitrification. Nitrite reductase catalyzes a key step in the nitrogen cycle, in that the reduction of nitrite (NO2−) to nitric oxide (NO) converts N to a form no longer available to most of the biota. This enzyme is found as two different variants. One contains copper and is encoded by nirK, while the other contains the hemes c and d1 and is encoded by nirS (45). nirK is found in a wider range of physiological groups, while nirS appears to be more abundant in nature (8). P. stutzeri contains nirS (29).
Several methods have been used to attempt to identify or quantify denitrifiers, including the design of specific PCR primers (6, 17) or probes (39) for genes involved in denitrification or ribosomal DNA genes (26) and immunofluorescence assays using polyclonal antibodies against specific denitrifying bacteria (44) or denitrification enzymes (8). Recently, competitive PCR and most-probable-number PCR were applied to the quantification of nirS-containing denitrifying bacteria (34).
The real-time PCR technique is based on the use of the 5′ nuclease assay, first described by Holland et al. (20) and further improved by the use of fluorescent TaqMan methodology and the ABI Prism 7700 sequence detection system (PE Applied Biosystems, Foster City, Calif.) (13, 19). The system requires the design of a forward and a reverse primer, in addition to a probe that hybridizes between them. The probe is fluorescently labeled at both ends (31). The fluorescent dye at the 5′ end serves as a reporter, and its emission spectra are quenched by the dye at the 3′ end of the probe. During the elongation step of each PCR cycle, the DNA polymerase cleaves the annealed probe with its 5′ nuclease activity. Once separated from the quencher, the reporter fluorescence is detected, resulting in an increase in fluorescence emission. The fluorescence increases logarithmically as the PCR proceeds, until a reagent becomes limiting. A threshold fluorescence intensity is defined within the logarithmic phase. The higher the amount of initial template DNA, the earlier the fluorescence will cross the defined threshold. Copy number of the initial target DNA is thereby determined by comparison to a standard curve.
The advantage of the real-time PCR method over other PCR-based quantification methods is that it focuses on the logarithmic phase of product accumulation rather than on the end product abundance. This technique is therefore more accurate, since it is less affected by amplification efficiency or depletion of a reagent. In addition, real-time PCR measures template abundance over a large dynamic range of around 6 orders of magnitude (19). Finally, this method allows the simultaneous analysis of 96 samples in a short time and reduces the risk of contamination, as no post-PCR manipulation is required. The main disadvantage of real-time PCR is the need for a special thermocycler and reagents that are expensive compared to the equipment utilized by other PCR-based quantification methods.
Real-time PCR has been successfully applied in the medical field, for example in the quantification of various DNA and RNA viruses in patients (16, 23, 27, 33), in the detection of gene amplification (3), mutations, or chromosomal rearrangements (10, 32), and in the quantification of gene expression (2, 43) or detection of various splice variants (25). Recently, real-time PCR was applied to environmental samples in studies that quantified conidia of a human pathogenic mold in airborne samples (18) and for determining the abundance of bacterioplankton in marine samples (40).
In this study, we tested real-time PCR for its use for quantifying an important functional gene in environmental samples.
MATERIALS AND METHODS
Samples.
Soil samples were obtained from an agricultural plot at the Kellogg Biological Station (KBS; Mich.) (1). Groundwater samples were taken from the Schoolcraft (Mich.) and Shiprock uranium mill tailings remedial action (N.Mex.) sites. The uranium recovery process at the latter site used high concentrations of ammonia, some of which entered the groundwater and led to the accumulation of nitrate. The site is located on an elevated terrace, along the south side of the San Juan River. Samples were taken on the terrace (samples 813 and 826), on the floodplain (samples 602, 603, and 619), and from a seep flowing from the base of the escarpment into the floodplain (sample 425). The samples presented increasing concentrations of nitrate in the following order: 602, 826, 619, 425, 603, and 813 (22). P. stutzeri KC, a strain that hydrolyzes carbon tetrachloride (9), was injected into the Schoolcraft aquifer for a bioremediation field test 15 days before the samples were taken from wells 2 m upstream and 1 and 2.5 m downstream from the injection site (M8, M11, and M19, respectively) (21). The freshwater sediment sample was collected from the surface 2 cm of sediment from Wintergreen Lake (Mich.), a small hypereutrophic lake. The marine sediment samples were obtained from the Washington margin of the Pacific Ocean and from Puget Sound (Wash.). Sediment cores were sliced into sections, as described below.
Cultures.
Marine P. stutzeri isolates (strains A3-5, D7-6, D9-1, E4-2, and F9-2) were obtained by L. Wu from the Washington marine sediments (7). nirS clones were obtained by G. Braker by amplification of DNA extracted from Washington margin and Puget Sound sediments, using specific primers (7). All culture collection strains and marine isolates used in this study were grown in nutrient broth (Difco, Detroit, Mich.). Escherichia coli transformants were grown in Luria-Bertani broth (36) amended with kanamycin (50 μg/ml). Shewanella oneidensis MR-1, P. stutzeri KC, and Pseudomonas aeruginosa were grown at 30°C, while all other cultures were grown at 37°C.
DNA extraction and quantitation.
Genomic DNA was extracted from late-exponential-phase cultures. Cells were harvested by centrifugation and resuspended in lysis buffer (50 mM Tris [pH 8], 50 mM EDTA, 100 mM NaCl). The cells were incubated for 15 min at 37°C with lysozyme (3.5 mg/ml), achromopeptidase (70 μg/ml), and RNase A (30 μg/ml). After two freeze-thaw treatments, the cell suspension was incubated for 5 min at 37°C with sodium dodecyl sulfate (1%), followed by incubation with proteinase K (400 μg/ml) (1 h at 60°C). The lysate was extracted twice with phenol-chloroform-isoamyl alcohol. After isopropanol precipitation, the DNA was resuspended in Tris-EDTA buffer (pH 8). Plasmid DNA was extracted from the E. coli transformants with the Wizard Plus SV Miniprep DNA purification system (Promega, Madison, Wis.), according to the manufacturer's instructions. The genomic DNA extractions from the KBS soil, the Shiprock aquifer, and the Wintergreen Lake sediment were performed using the Ultra Clean Soil DNA kit (MO BIO, Solana Beach, Calif.), following the manufacturer's instructions. Genomic DNA was extracted from the Schoolcraft groundwater samples by the method of van Elsas and Smalla (42). This method was also used to extract genomic DNA from the Washington marine sediment samples, with an additional proteinase K treatment (50 μl of a 20-mg/ml solution) after the incubation with sodium dodecyl sulfate. The protocol of Gray and Herwig (15) was used to extract genomic DNA from the Puget Sound samples.
The quality of the extracted DNA was analyzed by electrophoresis on a 0.8% agarose gel. DNA concentrations were measured by absorbance at 260 nm. The P. stutzeri nirS gene copy number was estimated based on the P. stutzeri Zobell genome size (4.29 Mbp) (14) and on the assumption that only 1 copy of nirS is present per genome (24), i.e., 4.4 fg of P. stutzeri DNA = 1 genome copy = 1 nirS copy.
Primers and probe.
Nine P. stutzeri nirS sequences from isolates (7) and 52 non-P. stutzeri nirS sequences from marine isolates, clones, and unrelated species (7) were compared to select conserved regions within the P. stutzeri nirS gene. The primers and probe were designed within conserved regions using the program PrimerExpress (PE Applied Biosystems) (Table 1). The probe was dually labeled with the fluorescent dyes 6-carboxyfluorescein (FAM) and 6-carboxytetramethyl-rhodamine (TAMRA) at the 5′ and 3′ ends, respectively, as recommended by the manufacturers. The primers and probe were synthesized by Integrated DNA Technologies (Coralville, Iowa).
TABLE 1.
Primer and probe sequences compared to example positive and negative control nirS gene sequences
| Example nirS sequence | Forward primerb | Probeb | Reverse primerb |
|---|---|---|---|
| 5′ACAAGGAGCACAACTGGAAGGT3′ | FAM-5′GGCAACCTGTTCGTCAAGACCCA3′-TAMRA | 5′CGCGTCGGCCCAGA3′ | |
| Positive control, P. stutzeri Zobell | 5′ACAAGGAGCACAACTGGAAGGT3′ | 5′GGCAACCTGTTCGTCAAGACCCA3′ | 5′CGCGTCGGCCCAGA3′ |
| Negative control,a P. aeruginosa | 5′ATCCGCAGTACGCCTGGAAGAA3′ | 5′GGCTCGCTGTTCATCAAGACCCA3′ | 5′GGTGTCGACGTAGA3′ |
Underlined bases represent mismatches with the primer-probe set.
Sequences corresponding to P. stutzeri Zobell nirS positions 1260 to 1281, 1310 to 1332, and 1363 to 1350 (forward primer, probe, and reverse primer, respectively).
Real-time PCR.
The increase in fluorescence emission, due to the degradation of the probe by the DNA polymerase in each elongation step, was monitored during PCR amplification using the 7700 Sequence Detector (PE Applied Biosystems). The fluorescence signal was normalized by dividing the emission of the reporter dye (6-carboxyfluorescein) by the emission of the passive reference dye 6-carboxy-X-rhodamine. The parameter CT (threshold cycle) is the fractional cycle number at which the fluorescence emission crosses an arbitrarily defined threshold within the logarithmic increase phase (0.1 in our reactions). The higher the amount of initial template DNA, the earlier the fluorescence will cross the threshold and the smaller will be the CT. The CT values obtained for each sample were compared with a standard curve to determine the initial copy number of the target gene.
The reaction mixture for real-time PCR consisted of 1× TaqMan Universal PCR Master Mix (containing AmpliTaq Gold DNA polymerase, AmpErase uracil-N-glycosylase, which degrades PCR carryover products from previous reactions, deoxynucleoside triphosphates with dUTP, a passive reference [6-carboxy-X-rhodamine], and optimized buffer components) (PE Applied Biosystems), 300 nM forward primer, 900 nM reverse primer, and 525 nM fluorogenic probe. MicroAmp optical caps and tubes were used (PE Applied Biosystems). A total volume of 30 μl was used for the optimization steps and 50 μl was used for the final reactions. PCR conditions were as follows: 2 min at 50°C, 10 min at 95°C, then 40 cycles of 15 s at 95°C and 1 min at 60°C. Negative controls with no template DNA or no probe were run in each reaction.
Specificity.
The DNA extracted from several strains, marine isolates (7), and E. coli transformants (7) was used as positive and negative controls to test the specificity of the primer-probe set. Template DNA (18 ng) was added to each reaction tube.
Sensitivity and detection limit.
Marine isolate E4-2 DNA was chosen as a standard for measuring the sensitivity of the primer-probe set and for generating standard curves in subsequent determinations. This isolate was previously identified as P. stutzeri based on 16S ribosomal DNA sequence identity and physiological characteristics (7). Serial dilutions (10-fold) of P. stutzeri E4-2 DNA were prepared in herring sperm DNA (1 μg · ml−1 in water; Boehringer Mannheim, Indianapolis, Ind.) as a carrier. All determinations were performed in triplicate and 95% confidence intervals were determined (shown as error bars).
Various calibration curves were constructed to determine the lower detection limit of this assay and our ability to discriminate twofold differences in template concentration. A dilution series of marine isolate E4-2 DNA was prepared in a 1-μg · ml−1 solution of herring sperm DNA. Different volumes (2, 4, 10, and 20 μl) of template DNA from this dilution series were added to individual reaction tubes, and the difference was made up with water.
The maximum allowable error (MAE) in order to distinguish a twofold difference in copy number was calculated as follows: ΔCT = m · log(2), where ΔCT is the difference between CT obtained from samples with a twofold difference in target copy number, and m is the slope of the standard curve. The MAE is calculated by this equation: MAE = ΔCT/2. The MAE was calculated for each standard curve generated, and the mean MAE and the 95% confidence interval were determined.
Quantitation of nirS in environmental samples.
Reactions were performed using 100 ng of template DNA. The template copy number was determined from CT values by using a standard curve. Samples that exhibited CT values equal to or higher than the negative controls were considered as below the detection limit. All results were normalized to the quantity of community DNA.
Accuracy.
In order to test for the presence of PCR inhibitors, 106 strain E4-2 genome copies (4.4 ng of DNA) were added to 100 ng of DNA extracted from one environmental sample from each site. Samples with and without the addition of this positive control DNA were compared, and the percent recovery of the added genome copy number was calculated. P. stutzeri abundance was also evaluated in microbial communities constructed from P. stutzeri KC, P. aeruginosa, and E. coli JM109 in different proportions and added to 1 g of sterile quartz sand (Sigma, St. Louis, Mo.). Direct cell counts were obtained before mixing, using a Petroff-Hausser counting chamber. P. stutzeri KC cells were added to the mixtures in various ratios (1, 1/10, 1/102, 1/103, 1/104, 1/105, 1/107, 1/108, and 0). Equal cell numbers of P. aeruginosa and E. coli JM109 were added to achieve 2 × 108 cells in each 300-μl mixture. After cell addition, the mixture was vortexed for 3 s, and DNA was extracted using the Ultra Clean Soil DNA kit (MO BIO), following the manufacturer's instructions. Triplicate samples were analyzed using 100 ng of template DNA. To avoid any problem from different extraction efficiencies, the P. stutzeri KC nirS gene copy number was expressed relative to total DNA extracted. P. stutzeri, P. aeruginosa, and E. coli genome sizes were recently measured and reported as 4.29 Mbp (14), 5.9 Mbp (35), and 4.6 Mbp (4), respectively; their percent G+C content was considered to be 63, 67, and 50%, respectively (30).
To measure the accuracy of real-time estimations of P. stutzeri abundance in environmental samples, different numbers of P. stutzeri cells were added to Wintergreen Lake sediment and KBS soil samples. The number of total cells in the soil or sediment samples was determined by direct counts after staining with 5-(4,6-dichlorotriazine-2-yl) aminofluorescein (5). Serially diluted cells (108, 107, and 106 cells) were added to 0.5 g of sediment or soil samples. After vortexing for 5 s, total DNA was extracted as described above. In order to normalize these values to the total community DNA, the average genome size of soil or sediment bacteria was considered equal to the E. coli genome size, or 4.6 Mbp (4).
RESULTS
Specificity.
DNA extracted from a range of denitrifying isolates was used to test the specificity of the primer-probe combination (Table 2). A logarithmic increase in fluorescence intensity was readily detected by real-time PCR in 17 of 21 P. stutzeri strains. However, we were not able to detect by real-time PCR the P. stutzeri strains corresponding to genomovars 4, 5, and 7 and one strain in genomovar 1. Sequence analysis of nirS of the strains from genomovars 4, 5, and 7 indicated a higher similarity (80, 81, and 82%, respectively) with the nitrite reductase gene of P. aeruginosa than with that of P. stutzeri. We identified several mismatches between these strains and the primer-probe combination used in this study (five to nine mismatches for the primers and four to five for the probe). No non-P. stutzeri strain or clone gave a real-time PCR signal.
TABLE 2.
Specificity of real-time PCR for P. stutzeri DNA
| Species, strain, or clonea (genomovar) | Sourceb | Denitrificationc | nirSc | Real-time PCRd |
|---|---|---|---|---|
| Positive controls | ||||
| P. stutzeri (1) | ATCC 17589 | + | + | + |
| P. stutzeri (1) | ATCC 27951 | + | + | + |
| P. stutzeri (1) | ATCC 17593 | + | + | + |
| P. stutzeri (1) | ATCC 17594 | + | + | + |
| P. stutzeri (1) | CCUG11256 | + | − | |
| P. stutzeri (2) | ATCC 17591 | + | + | + |
| P. stutzeri (2) | ATCC 17587 | + | + | + |
| P. stutzeri Zobell (2) | ATCC 14405 | + | + | + |
| P. stutzeri (2) | ATCC 17592 | + | + | + |
| P. stutzeri (2) | ATCC 17595 | + | + | + |
| P. stutzeri (3) | ATCC 50227 | + | + | + |
| P. stutzeri 19SMN4 (4) | DSM 6084 | + | + | − |
| P. stutzeri DNSP21 (5) | DSM 6082 | + | + | − |
| P. stutzeri (7) | DSM 50238 | + | + | − |
| P. stutzeri JM300 (8) | DSM 10701 | + | + | + |
| P. stutzeri KC | ATCC 55595 | + | + | + |
| P. stutzeri A3-5 | L. Wu | + | + | + |
| P. stutzeri D7-6 | L. Wu | + | + | + |
| P. stutzeri D9-1 | L. Wu | + | + | + |
| P. stutzeri E4-2 | L. Wu | + | + | + |
| P. stutzeri F9-2 | L. Wu | + | + | + |
| Negative controls | ||||
| E. coli K-12 | DSM 498 | − | − | − |
| Paracoccus denitrificans | ATCC 17741 | + | + | − |
| Azospirillum brazilense | DSM 1690 | + | + | − |
| Shewanella oneidensis MR-1 | ATCC 700550 | + | + | − |
| Pseudomonas aeruginosa | ATCC 15692 | + | + | − |
| Pseudomonas balearica | DSM 6083 | + | + | − |
| Marine isolate C10-1s | L. Wu | + | + | − |
| Marine isolate D4-14 | L. Wu | + | + | − |
| nirS clone A4 | G. Braker | + | + | − |
| nirS clone B6 | G. Braker | + | + | − |
| nirS clone A12 | G. Braker | + | + | − |
| nirS clone B76 | G. Braker | + | + | − |
Marine isolates and nirS clones were obtained from the Washington margin and Puget Sound marine sediments (7).
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikroorganismen; CCUG, Culture Collection, University of Göteborg; L. Wu, Oak Ridge National Laboratory, Oak Ridge, Tenn.; G. Braker, Michigan State University, East Lansing, Mich.
Reported presence of denitrification and nirS from literature sources, except for results for the characterized genomovars, which were determined in this study by PCR.
+, logarithmic amplification readily detected; −, no amplification detected.
Sensitivity and detection limit.
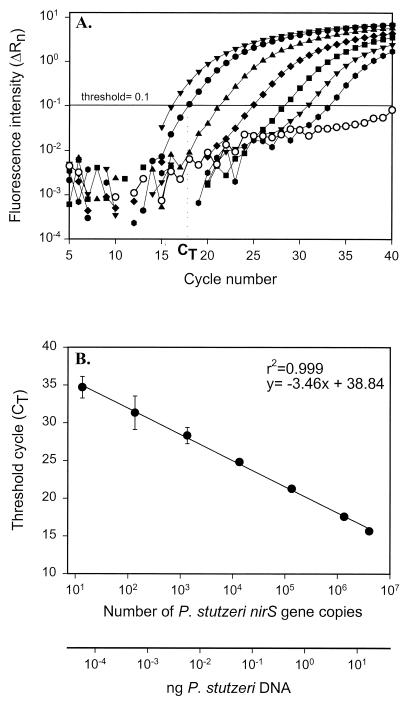
We tested the sensitivity of the real-time detection system using a dilution series of P. stutzeri DNA (Fig. 1A). The threshold value for this and all subsequent analyses was chosen to be 0.1. This value falls within the range of logarithmic fluorescence increase, yet avoids the signal from the no-template control. Nearly all of our no-template controls showed some increase in fluorescence intensity, similar to that shown in Fig. 1A. Since this increase was not logarithmic and similar signals were detected in control reactions without DNA polymerase (data not shown), this fluorescence increase is likely due to probe degradation. The data obtained were used to draw a standard curve relating CT values to the added mass of P. stutzeri DNA and the number of gene copies (Fig. 1B). A linear response was observed over more than 6 orders of magnitude, ranging from 14 to 4.05 × 106 nirS gene copies (r2 = 0.999; Fig. 1B).
FIG. 1.
Generation of standard curve. (A) Increase of fluorescence intensity with cycle number for serially diluted P. stutzeri DNA. Symbols, from left to right: ▾, 17.8 ng; ●, 5.9 ng; ▴, 0.59 ng; ⧫, 59 pg; ■, 5.9 pg; ▾, 0.59 pg;  , 59 fg; ○, no-template control. CT, cycle at which the fluorescence intensity crosses an arbitrary threshold value. (B) Standard curve. Values represent means ± 95% confidence interval (n = 3).
, 59 fg; ○, no-template control. CT, cycle at which the fluorescence intensity crosses an arbitrary threshold value. (B) Standard curve. Values represent means ± 95% confidence interval (n = 3).
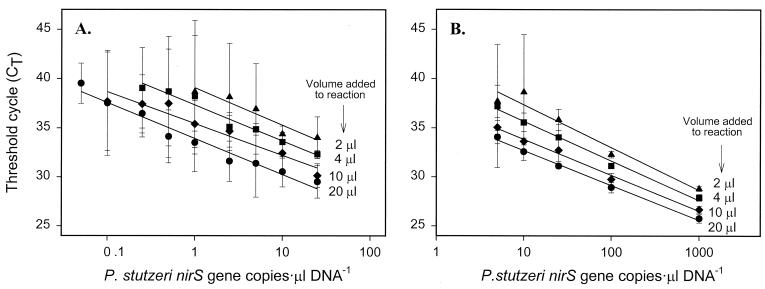
We constructed a series of calibration curves to study the detection limit of the system and our ability to differentiate similar P. stutzeri DNA concentrations. Different volumes (ranging from 2 to 20 μl) of a dilution series of template DNA (ranging from 0.1 to 1,000 nirS gene copies · μl−1) were added to the PCR. Given the 96-well capacity, the analyses were done in a low- and high-concentration set (Fig. 2A and B, respectively). Although the curves remained linear down to 1 nirS copy (20 μl of 0.05 nirS gene copies · μl−1), the variability associated with CT values from low copy numbers precluded our ability to distinguish concentrations in samples with similar amounts. Only when the copy number was 100 or greater were we able to reliably differentiate a twofold difference in P. stutzeri nirS concentration.
FIG. 2.
Limit of detection of the P. stutzeri nirS gene using real-time PCR. The denoted volumes of serially diluted P. stutzeri DNA were added to different reaction mixtures. (A) Low concentrations. (B) High concentrations. Values represent means ± 95% confidence interval (n = 3).
The increase in variability with cycle number is apparent when the upper 95% confidence interval from all determinations is plotted against CT (Fig. 3). Based on the slope of each individual standard curve, we calculated the maximal error in the CT value that would still allow detection of a twofold difference in gene copy number (MAE). The average of the different MAE values is equal to 0.53 (Fig. 3). The corresponding 95% confidence interval is too small to be observed (Fig. 3).
FIG. 3.
Relationship between CT and error. The horizontal line represents the MAE to discriminate between samples containing a twofold difference in P. stutzeri nirS copy number. Values shown represent the means from 14 independent standard curves. The 95% confidence interval is shown as a dashed line.
Accuracy.
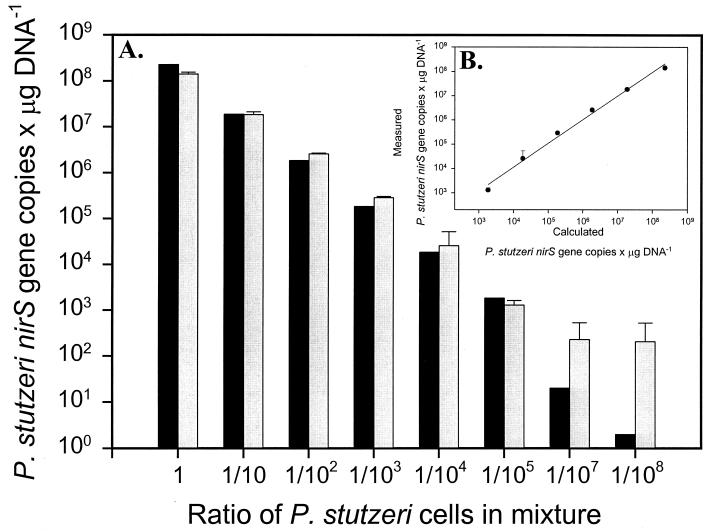
P. stutzeri KC, E. coli JM109, and P. aeruginosa cells were added to sterile quartz sand in various known amounts. P. stutzeri KC contains the targeted nirS gene, while P. aeruginosa has a nirS that is 67% similar in nucleotide sequence. The correlation between the calculated and measured values was extremely high (slope = 0.98, r2 = 0.992) (Fig. 4B). The lowest proportions of P. stutzeri (1/107 and 1/108) overlapped with the background in the real-time PCR measurements.
FIG. 4.
Quantification of nirS DNA in artificial mixtures of P. stutzeri KC, P. aeruginosa, and E. coli cells. (A) Comparison between P. stutzeri nirS gene copies per μg of DNA measured values by real-time PCR (gray bars) and calculated values based on cell counts (black bars). (B) Correlation between calculated and measured values (slope = 0.98, r2 = 0.992). Error bars represent 95% confidence intervals (n = 3) for both panels.
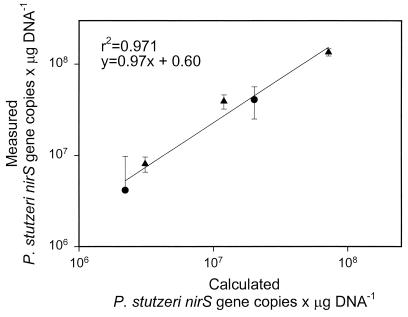
To further test the accuracy of the system, a similar experiment was performed adding known amounts of P. stutzeri cells to different soil and sediment samples. Measured values of nirS correlated well (slope = 0.97, r2 = 0.971) with the values calculated from direct cell counts (Fig. 5).
FIG. 5.
Quantification of nirS in KBS soil (●) and Wintergreen Lake sediment (▴) samples spiked with P. stutzeri cells. The line shows the correlation between P. stutzeri nirS gene copies per microgram of DNA measured by real-time PCR and calculated values. Error bars represent 95% confidence intervals (n = 3).
Analyses of environmental samples.
DNA extracted from environmental samples exhibited a wide range of P. stutzeri nirS gene abundance, as measured by real-time PCR (Table 3). The groundwater and freshwater sediment samples had the highest P. stutzeri nirS copy numbers. Marine sediments consistently displayed low P. stutzeri nirS abundance.
TABLE 3.
P. stutzeri nirS gene abundance in various habitats as measured by real-time PCR
| Habitat | Sample no. or type | No of copies of P. stutzeri nirS DNA/μg of community DNAf
|
P. stutzeri nirS DNA (μg/g of community DNA)f
|
||
|---|---|---|---|---|---|
| Mean | Upper 95% CI | Mean | Upper 95% CI | ||
| Soila | |||||
| KBS (Mich.) | 0× | 1.0 × 102 | 4.4 × 10−4 | ||
| 1× | BDLg | BDL | |||
| 10× | BDL | BDL | |||
| 100× | BDL | BDL | |||
| Groundwater | |||||
| Schoolcraft bioremediation site (Mich.)b | M8 | BDL | BDL | ||
| M11 | 1.1 × 1011 | 4.8 × 105 | |||
| M19 | 2.0 × 107 | 89 | |||
| Shiprock UMTRAc site (N. Mex.) | 425 | 1.9 × 103 | 3.7 × 103 | 8.4 × 10−3 | 1.6 × 10−2 |
| 602 | 3.1 × 105 | 8.8 × 104 | 1.4 | 3.9 × 10−1 | |
| 603 | 2.2 × 105 | 2.1 × 104 | 9.7 × 10−1 | 9.3 × 10−2 | |
| 619 | 2.0 × 107 | 9.4 × 105 | 90 | 4.1 | |
| 813 | 5.0 × 106 | 4.5 × 105 | 22 | 2 | |
| 826 | 5.0 × 105 | 1.6 × 105 | 2.2 | 6.9 × 10−1 | |
| Freshwater sedimentd | |||||
| Wintergreen Lake (Mich.) | 2.2 × 106 | 3.1 × 105 | 9.7 | 1.3 | |
| Marine sedimente | |||||
| Pacific Ocean (Wash.) | 0–0.5 cm | BDL | BDL | ||
| 0.5–1 cm | BDL | BDL | |||
| 1–2 cm | 1.0 × 102 | 2.0 × 102 | 4.4 × 10−4 | 8.8 × 10−4 | |
| 2–3 cm | BDL | BDL | |||
| 3–5 cm | BDL | BDL | |||
| 5–10 cm | BDL | BDL | |||
| Puget Sound (Wash.) | 1–1.5 cm | BDL | BDL | ||
| 1.5–2 cm | 2.1 × 104 | 2.4 × 103 | 9.1 × 10−2 | 1.0 × 10−2 | |
| 6–6.5 cm | 7.0 × 102 | 2.0 × 102 | 3.1 × 10−3 | 8.8 × 10−4 | |
Soils were treated with 0, 1, 10, and 100× the normal field rate (1.1 kg ha−1) of the herbicide 2, 4-dichlorophenoxyacetate (1).
Samples were collected 2 m upstream (M8), 1 m downstream (M11), and 2.5 m downstream (M19) from the well where P. stutzeri KC was injected (21).
UMTRA, uranium mill tailings remedial action.
The surface 2 cm containing the active denitrifiers was analyzed.
Depths within the sediment core are reported.
Results are from triplicate samples when 95% confidence interval (CI) is indicated and from single samples, otherwise.
BDL, below detection limit.
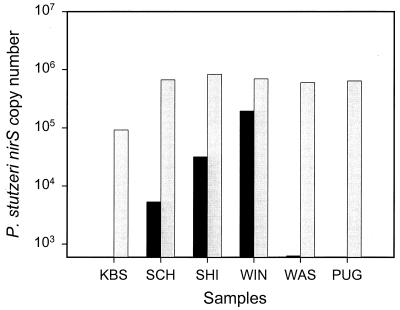
In order to test for the presence of any PCR inhibitors in the environmental samples, 106 genome copies of strain E4-2 were added to each sample. Five of the six samples yielded the expected amount of nirS (Fig. 6). The KBS soil sample showed slightly fewer nirS copies than expected.
FIG. 6.
P. stutzeri nirS copy number measured by real-time PCR without (black bars) and with (gray bars) the addition of 106 P. stutzeri nirS copy numbers to DNA extracted from the following environmental samples: KBS, soil from KBS (Mich.); SCH, groundwater from Schoolcraft bioremediation site (Mich.); SHI, groundwater from Shiprock (N.Mex.); WIN, freshwater sediment from Wintergreen Lake (Mich.); WAS, marine sediment from Washington margin (Wash.); PUG, marine sediment from Puget Sound (Wash.).
DISCUSSION
Good detection methods share four features: specificity, sensitivity, precision, and accuracy. Real-time PCR appears to satisfy these requirements. The primer-probe set we designed for the P. stutzeri nirS gene amplified a group of strains that generally corresponded to the P. stutzeri species (Table 2). Except for one strain, all the representatives of the two more commonly isolated genomovars, 1 and 2, were readily amplified. The sequence similarity of the nirS gene of the P. stutzeri strains in genomovars 4, 5, and 7 to the P. aeruginosa nitrite reductase gene explains the inability to detect these strains by real-time PCR. In addition to cultured strains, we selected a range of isolates and cloned nirS sequences from the Pacific Northwest marine environment for the primer-probe design. This makes our system especially appropriate for application in this environment. Although there may be cross-reactivity with related DNA in the natural microbial communities, none of our habitat-specific negative controls gave a positive reaction. Furthermore, the phylogenetically closest relative, Pseudomonas balearica, was not detected. The probe needed in real-time PCR requires the identification of three specific regions in the DNA sequence, rather than two, which provides for the high specificity. This, however, can make the design of an appropriate primer-probe set more difficult. The 16S rRNA gene would be an alternative target which is more conserved but that would sample a larger organismal group, which may not correlate with function.
The method was linear over more than 6 orders of magnitude and sensitive down to 1 gene copy, similar to the results obtained in other studies (18, 27). However, the high variability associated with low target copies limits precision near the detection limit (Fig. 2). We are able to detect the presence of only 1 copy, but not precisely, e.g., we cannot discriminate a twofold difference at 100 copies or less. The theoretical upper 95% confidence interval to allow the discrimination of a twofold difference in copy number is 0.53 (Fig. 3). Confidence intervals below this value are achieved more frequently at lower CT values, equivalent to higher numbers of target copies. The error values associated with the measurements of P. stutzeri nirS abundance in various environmental samples were consistent with this characterization of precision, i.e., high concentrations have an error value at least 1 order of magnitude smaller than the measured value, while the error was of the same order of magnitude when nirS copy number was small (Table 3).
The method proved to be extremely accurate. When P. stutzeri was mixed in various proportions with E. coli and P. aeruginosa cells, it could be detected when present at 100% to 0.001% in the mixture, presenting a high correlation between expected and measured results over this whole range. Furthermore, spiked samples gave the expected results.
Environmental samples analyzed with real-time PCR displayed a wide range of P. stutzeri nirS abundances. P. stutzeri was highly abundant in freshwater sediment from Wintergreen Lake (Mich.), which agrees with a study by Gamble et al. (12), who identified P. stutzeri as the dominant denitrifier in these sediments. P. stutzeri was either absent or present at extremely low population densities in marine sediments from the Washington margin and Puget Sound (Wash.). Ward and Cockcroft (44) measured the abundance of P. stutzeri in the water column of Monterey Bay, Calif., and found that it represented only 0.02 to 0.08% of the total bacterial community. Our findings indicate that P. stutzeri is also not abundant in marine sediments. This is in agreement with the studies by Braker et al. (7), who observed nirS heterogeneity and the lack of a dominant group in nirS clones isolated from these sites. Furthermore, these results are also consistent with studies done on other denitrification genes. Scala and Kerkhof (37) observed a high diversity among nitrous oxide reductase (nosZ) genes in marine sediments, with no overlap between environmental nosZ sequences and cultured denitrifiers. Groundwater samples from the Schoolcraft bioremediation site (Mich.) followed expected trends, corresponding with the injection of substrates and P. stutzeri strain KC into a contamination plume. We detected no P. stutzeri upgradient from the inoculation site and found the highest P. stutzeri abundance just downgradient from the inoculation site. P. stutzeri was also prevalent at Shiprock, perhaps resulting from the high nitrate contamination at that site.
Real-time PCR is a specific, sensitive, precise, and accurate method for quantifying a gene or organism group in a broad range of environmental sample types. It remains to be seen whether functional gene sequences are conserved enough in a habitat so that a reasonable number of probe-primer sets can provide useful quantitative information for components of a group or process. This same method and primer-probe regions should be effective in quantifying mRNA and hence in determining which group of organisms or gene families are active in denitrification.
ACKNOWLEDGMENTS
This work was supported by Department of Energy grants DE-FG02-98ER62535 and DE-FG02-97ER62469. Oak Ridge National Laboratory is managed by the University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
We kindly thank Jorge Lalucat for providing the P. stutzeri strains with genomovar classifications, Allan Devol for collecting the Puget Sound and Pacific Ocean sediment samples, Phillip Long and the UMTRA personnel for the Shiprock groundwater samples, Liyou Wu for the marine isolates, Gesche Braker for the nirS clones and for providing us DNA extracted from the Pacific Ocean sediments, and Katrina Linning for DNA extracted from the Schoolcraft samples.
REFERENCES
- 1.Asuming-Brempong S. The effect of 2,4-dichlorophenoxyacetate selection on microbial communities in microcosm and field studies and the impact on ecosystem function. Ph.D. thesis. East Lansing: Michigan State University; 1999. [Google Scholar]
- 2.Bièche I, Laurendeau I, Tozlu S, Olivi M, Vidaud D, Lidereau R, Vidaud M. Quantitation of MYC gene expression in sporadic breast tumors with a real-time reverse transcription-PCR assay. Cancer Res. 1999;59:2759–2765. [PubMed] [Google Scholar]
- 3.Bièche I, Olivi M, Champème M-H, Vidaud D, Lidereau R, Vidaud M. Novel approach to quantitative polymerase chain reaction using real-time detection: application to the detection of gene amplification in breast cancer. Int J Cancer. 1998;78:661–666. doi: 10.1002/(sici)1097-0215(19981123)78:5<661::aid-ijc22>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Bloem J. Fluorescent staining of microbes for total direct counts. In: Akkermans A D L, van Elsas J D, de Brujin F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–12. [Google Scholar]
- 6.Braker G, Fesefeldt A, Witzel K-P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol. 1998;64:3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braker G, Zhou J, Wu L, Devol A H, Tiedje J M. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl Environ Microbiol. 2000;66:2096–2104. doi: 10.1128/aem.66.5.2096-2104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne M S, Arunakumari A, Averill B A, Tiedje J M. Immunological identification and distribution of dissimilatory heme cd1 and nonheme copper nitrite reductase in denitrifying bacteria. Appl Environ Microbiol. 1989;55:2924–2931. doi: 10.1128/aem.55.11.2924-2931.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Criddle C S, DeWitt J T, Grbic-Galic D, McCarty P. Transformation of carbon tetrachloride by Pseudomonas sp. strain KC under denitrification conditions. Appl Environ Microbiol. 1990;56:3240–3246. doi: 10.1128/aem.56.11.3240-3246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dölken L, Schüler F, Dölken G. Quantitative detection of t(14; 18)-positive cells by real-time quantitative PCR using fluorogenic probes. BioTechniques. 1998;25:1058–1064. doi: 10.2144/98256cr05. [DOI] [PubMed] [Google Scholar]
- 11.Firestone M K. Biological denitrification. In: Stevenson F J, editor. Nitrogen in agricultural soils. Agronomy monograph 22. Madison, Wis: American Society for Agronomy; 1982. pp. 289–326. [Google Scholar]
- 12.Gamble T N, Betlach M R, Tiedje J M. Numerically dominant denitrifying bacteria from world soils. Appl Environ Microbiol. 1977;33:926–939. doi: 10.1128/aem.33.4.926-939.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson U E M, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 14.Ginard M, Lalucat J, Tümmler B, Römling U. Genome organization of Pseudomonas stutzeri and resulting taxonomic and evolutionary considerations. Int J Syst Bacteriol. 1997;47:132–143. doi: 10.1099/00207713-47-1-132. [DOI] [PubMed] [Google Scholar]
- 15.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gut M, Leutenegger C M, Huder J B, Pedersen N C, Lutz H. One-tube fluorogenic reverse transcription-polymerase chain reaction for the quantitation of feline coronaviruses. J Virol Methods. 1999;77:37–46. doi: 10.1016/S0166-0934(98)00129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallin S, Lindgren P-E. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl Environ Microbiol. 1999;65:1652–1657. doi: 10.1128/aem.65.4.1652-1657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haugland R A, Vesper S J, Wymer L J. Quantitative measurement of Stachybotrys chartarum conidia using real time detection of PCR products with the TaqManTM fluorogenic probe system. Mol Cell Probes. 1999;13:329–340. doi: 10.1006/mcpr.1999.0258. [DOI] [PubMed] [Google Scholar]
- 19.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 20.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′-3′exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyndman D W, Dybas M J, Wiggert D, Zhao X, Wallace R, Voice T, Chan A, Phanikumar M S, Criddle C S. Hydraulic characterization and design of a full-scale biocurtain. Ground Water. 2000;38:462–474. [Google Scholar]
- 22.Ivanova I A, Stephen J R, Chang Y-J, Brüggemann J, Long P E, McKinley J P, Kowalchuk G A, White D C, Macnaughton S J. A survey of 16S rRNA and amoA genes related to autotrophic ammonia-oxidizing bacteria of the β-subdivision of the class proteobacteria in contaminated groundwater. Can J Microbiol. 2000;46:1012–1020. doi: 10.1139/w00-099. [DOI] [PubMed] [Google Scholar]
- 23.Josefsson A, Livak K, Gyllenstein U. Detection and quantitation of human papillomavirus by using the fluorescent 5′ exonuclease assay. J Clin Microbiol. 1999;37:490–496. doi: 10.1128/jcm.37.3.490-496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jüngst A, Wakabayashi S, Matsubara H, Zumft W G. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 1991;279:205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- 25.Kafert S, Krauter J, Ganser A, Eder M. Differential quantitation of alternatively spliced messenger RNAs using isoform-specific real-time RT-PCR. Anal Biochem. 1999;269:210–213. doi: 10.1006/abio.1999.4016. [DOI] [PubMed] [Google Scholar]
- 26.Kerkhof L. A species-specific probe and a PCR assay for the marine bacterium, Pseudomonas stutzeri strain Zobell. Microb Ecol. 1994;27:201–212. doi: 10.1007/BF00182405. [DOI] [PubMed] [Google Scholar]
- 27.Kimura H, Morita M, Yabuta Y, Kuzushima K, Kato K, Kojima S, Matsuyama T, Morishima T. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J Clin Microbiol. 1999;37:132–136. doi: 10.1128/jcm.37.1.132-136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knowles R. Denitrification. Microbiol Rev. 1982;46:43–70. doi: 10.1128/mr.46.1.43-70.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Körner H, Frunzke K, Dohler K, Zumft W G. Immunochemical patterns of distribution of nitrous oxide reductase and nitrite reductase (cytochrome cd1) among denitrifying pseudomonads. Arch Microbiol. 1987;148:20–24. doi: 10.1007/BF00429641. [DOI] [PubMed] [Google Scholar]
- 30.Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Baltimore, Md: Williams & Wilkins; 1984. [Google Scholar]
- 31.Lee L G, Connell C R, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luthra R, McBride J A, Cabanillas F, Sarris A. Novel 5′ exonuclease-based real-time PCR assay for the detection of t(14; 18)(q32; q21) in patients with follicular lymphoma. Am J Pathol. 1998;153:63–68. doi: 10.1016/S0002-9440(10)65546-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martell M, Gómez J, Esteban J I, Sauleda S, Quer J, Cabot B, Esteban R, Guardia J. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J Clin Microbiol. 1999;37:327–332. doi: 10.1128/jcm.37.2.327-332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michotey V, Méjean V, Bonin P. Comparison of methods for quantification of cytochrome cd1-denitrifying bacteria in environmental marine samples. Appl Environ Microbiol. 2000;66:1564–1571. doi: 10.1128/aem.66.4.1564-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratnaningsih E, Dharmsthiti S, Krishnapillai V, Morgan A, Sinclair M, Holloway B W. A combined physical and genetic map of Pseudomonas aeruginosa PAO. J Gen Microbiol. 1990;136:2351–2357. doi: 10.1099/00221287-136-12-2351. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Scala D J, Kerkhof L J. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl Environ Microbiol. 1999;65:1681–1687. doi: 10.1128/aem.65.4.1681-1687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seitzinger S P. Denitrification in aquatic sediments. In: Revsbech N P, Sørensen J, editors. Denitrification in soil and sediment. FEMS Symposium. New York, N.Y: Plenum Press; 1990. pp. 301–322. [Google Scholar]
- 39.Smith G B, Tiedje J M. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl Environ Microbiol. 1992;58:376–384. doi: 10.1128/aem.58.1.376-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki M T, Taylor L T, Delong E F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl Environ Microbiol. 2000;66:4605–4614. doi: 10.1128/aem.66.11.4605-4614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiedje J M. Ecology of denitrification and dissimilatory nitrate reduction to ammonium. In: Zehnder A J B, editor. Biology of anaerobic microorganisms. New York, N.Y: John Wiley & Sons, Inc.; 1988. pp. 179–244. [Google Scholar]
- 42.van Elsas J D, Smalla K. Extraction of microbial community DNA from soils. In: Akkermans A D L, van Elsas J D, de Brujin F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–11. [Google Scholar]
- 43.Wang T, Brown M J. mRNA quantification by real time TaqMan polymerase chain reaction: validation and comparison with RNase protection. Anal Biochem. 1999;269:198–201. doi: 10.1006/abio.1999.4022. [DOI] [PubMed] [Google Scholar]
- 44.Ward B B, Cockcroft A R. Immunofluorescence detection of the denitrifying strain Pseudomonas stutzeri (ATCC 14405) in seawater and intertidal sediment environments. Microb Ecol. 1993;25:233–246. doi: 10.1007/BF00171890. [DOI] [PubMed] [Google Scholar]
- 45.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]