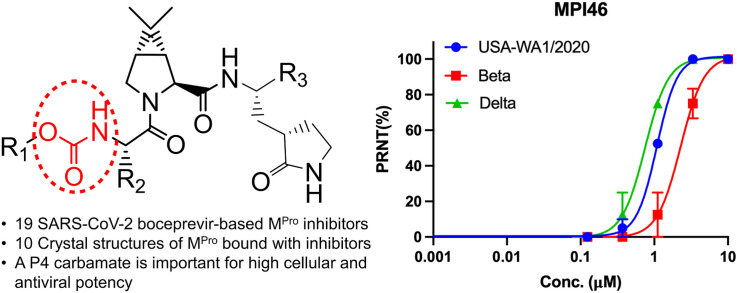
Abstract
Boceprevir is an HCV NSP3 inhibitor that was explored as a repurposed drug for COVID-19. It inhibits the SARS-CoV-2 main protease (MPro) and contains an α-ketoamide warhead, a P1 β-cyclobutylalanyl moiety, a P2 dimethylcyclopropylproline, a P3 tert-butylglycine, and a P4 N-terminal tert-butylcarbamide. By introducing modifications at all four positions, we synthesized 20 boceprevir-based MPro inhibitors including PF-07321332 and characterized their MPro inhibition potency in test tubes (in vitro) and 293T cells (in cellulo). Crystal structures of MPro bound with 10 inhibitors and cytotoxicity and antiviral potency of 4 inhibitors were characterized as well. Replacing the P1 site with a β-(S-2-oxopyrrolidin-3-yl)-alanyl (Opal) residue and the warhead with an aldehyde leads to high in vitro potency. The original moieties at P2, P3 and the P4 N-terminal cap positions in boceprevir are better than other tested chemical moieties for high in vitro potency. In crystal structures, all inhibitors form a covalent adduct with the MPro active site cysteine. The P1 Opal residue, P2 dimethylcyclopropylproline and P4 N-terminal tert-butylcarbamide make strong hydrophobic interactions with MPro, explaining high in vitro potency of inhibitors that contain these moieties. A unique observation was made with an inhibitor that contains a P4 N-terminal isovaleramide. In its MPro complex structure, the P4 N-terminal isovaleramide is tucked deep in a small pocket of MPro that originally recognizes a P4 alanine side chain in a substrate. Although all inhibitors show high in vitro potency, they have drastically different in cellulo potency to inhibit ectopically expressed MPro in human 293T cells. In general, inhibitors with a P4 N-terminal carbamide or amide have low in cellulo potency. This trend is reversed when the P4 N-terminal cap is changed to a carbamate. The installation of a P3 O-tert-butyl-threonine improves in cellulo potency. Three molecules that contain a P4 N-terminal carbamate were advanced to cytotoxicity tests on 293T cells and antiviral potency tests on three SARS-CoV-2 variants. They all have relatively low cytotoxicity and high antiviral potency with EC50 values around 1 μM. A control compound with a nitrile warhead and a P4 N-terminal amide has undetectable antiviral potency. Based on all observations, we conclude that a P4 N-terminal carbamate in a boceprevir derivative is key for high antiviral potency against SARS-CoV-2.
Keywords: COVID-19, SARS-CoV-2, Main protease, Covalent inhibitor, Carbamate
Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MPro, main protease; MPI, main protease inhibitor
Graphical abstract
1. Introduction
COVID-19 is the currently ongoing pandemic that is caused by the coronavirus SARS-CoV-2. To address this emergency, a large variety of drug repurposing research has been conducted to identify approved medications that might be potentially used as COVID-19 treatments [[1], [2], [3], [4], [5]]. Significant part of this research has been targeting the SARS-CoV-2 main protease (MPro) [[6], [7], [8], [9], [10], [11], [12]]. MPro is a peptide fragment of two translation products pp1a and pp1ab of the SARS-CoV-2 RNA genome after the virus infects human cells. Both pp1a and pp1ab are very large polypeptides that need to undergo proteolytic hydrolysis to form 16 nonstructural proteins (nsps). These nsps are essential for the virus to replicate its genome in host cells, evade the host immune system, and package new virions for the infection of new host cells [13]. Intervention of the proteolytic hydrolysis of pp1a and pp1ab is a viable approach to stop SARS-CoV-2 infection. There are two internal peptide fragments from pp1a and pp1b that function as cysteine proteases to hydrolyze all nsps. One is MPro and the other papain-like protease (PLPro). As the major protease, MPro processes the majority of nsps. It is also more conserved than PLPro. MPro genes in SARS-CoV and SARS-CoV-2 share 96% sequence identity [2]. Targeting MPro for drug discovery has been demonstrated as a successful route for the development of SARS-CoV-2 antivirals by the U.S. FDA approval of the emergency use of paxlovid for treating COVID-19 [14]. So far, a number of approved small molecule drugs have been confirmed as potent MPro inhibitors [1,3,5,6,[9], [10], [11],[15], [16], [17]]. One of which is boceprevir [[15], [16], [17]]. Boceprevir is a peptidyl inhibitor of HCV NSP3. HCV NSP3 is a serine protease. Boceprevir contains an α-ketoamide warhead that forms a reversible covalent adduct with its active site serine for high inhibition potency [18]. Besides the α-ketoamide warhead, boceprevir contains a P1 β-cyclobutylalanyl moiety, a P2 dimethylcyclopropylproline, a P3 tert-butylglycine, and a P4 N-terminal tert-butylcarbamide. Boceprevir has been shown as a potent inhibitor of MPro in multiple publications [1,[15], [16], [17]]. Its interactions with MPro have also been structurally characterized using X-ray protein crystallography [[15], [16], [17],19,20]. Although its discovery as a potent MPro inhibitor created excitement about its potential use as a COVID-19 treatment, boceprevir has moderate antiviral activity against SARS-CoV-2 and displays very low potency to inhibit MPro in a human cell host [15,16,21]. In this work, we report a systematic study of boceprevir-based molecules for improved cellular and antiviral potency against the SARS-CoV-2 MPro.
2. Results
2.1. The design and synthesis of MPI29-47 and PF-07321332
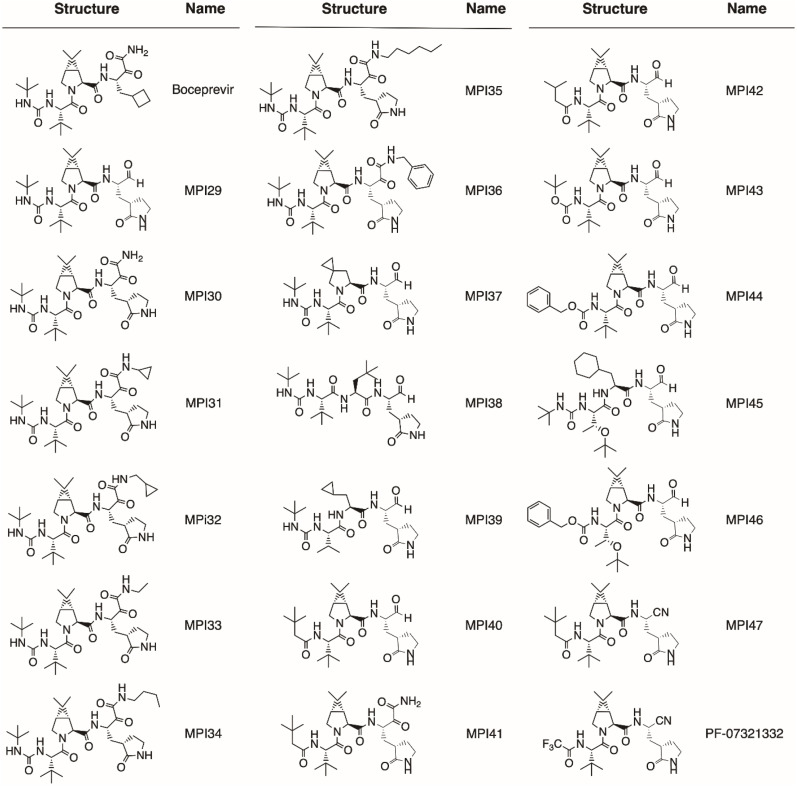
In total, we designed 19 boceprevir-based inhibitors as shown in Fig. 1 [22]. Paxlovid is a combination therapy with two components, ritonavir and nirmatrelvir. Nirmatrelvir is an potent MPro inhibitor and was originally called PF-07321332 [14]. Since it is a boceprevir derivative, we included it in our study as well. MPro contains four binding pockets in its active site to interact with the P1, P2, P4, and P3’ amino acid residues in a substrate [23]. The P1 residue in a substrate is strictly glutamine. Past efforts on the development of MPro inhibitors have been either using a P1 β-(S-2oxopyrrolidin-3-yl)-alanyl (Opal) residue in a peptidyl inhibitor or a pyridine moiety in a nonpeptidyl inhibitor to engage the S1 (P1 binding) pocket of MPro for strong interactions [6,8,16,[24], [25], [26], [27], [28], [29], [30], [31]]. Boceprevir has a P1 β-cyclobutylalanyl moiety that displays loose binding to the MPro S1 pocket in its MPro complex structure [[15], [16], [17],19,20], which explains its relatively moderate MPro inhibition potency. In our designed boceprevir derivatives, this site is replaced by the Opal residue. In MPro-boceprevir complex structures, the P2 dimethylcyclopropylproline binds neatly to the MPro S2 (P2 binding) pocket. Due to its nice fit to the MPro S2 pocket, this site is maintained as dimethylcyclopropylproline in most designed inhibitors. In a small number of inhibitors, other residues such as (S)-5-azaspiro [2,4]heptane-6-carboxylic acid in MPI37, neopentylglycine in MPI38, β-cyclopropylalanine in MPI39, and cyclohexylalanine in MPI45 at the P2 site are introduced. A previous study has indicated that neopentylglycine, β-cyclopropylalanine and cyclohexylalanine at this site lead to either high MPro inhibition potency in test tubes (in vitro) or in 293T cells that express MPro (in cellulo) [32]. The P3 site is either maintained as tert-butylglycine or replaced by O-tert-butylthreonine. tert-Butylglycine at the P3 site in a peptidyl inhibitor is known to generate high in vitro potency and O-tert-butylthreonine is known to cause high in cellulo and antiviral potency [21,32,33]. Boceprevir has a P4 N-terminal tert-butylcarbamide cap. This site is either maintained as a carbamide or replaced by a carbamate or amide.
Fig. 1.
Structures of boceprevir, MPI29-47 and PF-07321332.
Boceprevir has an α-ketoamide warhead that covalently interacts with the MPro active site cysteine C145 to generate a hemithioacetal intermediate. Since the majority of currently known peptidyl inhibitors of MPro have an aldehyde warhead, we replaced the α-ketoamide warhead with an aldehyde in some designed inhibitors. We also tested nitrile at this position since PF-07321332 and some other developed MPro inhibitors have this warhead [14,34]. An advantage of the α-ketoamide warhead is its allowance of appending additional chemical moieties at its α-ketoamide nitrogen. Both aldehyde- and nitrile-containing inhibitors will leave the MPro S3’ (P3’ binding) pocket empty when they bind to MPro. A chemical appendage at the α-ketoamide nitrogen can potentially reach this pocket for additional interactions and therefore lead to high affinity to MPro. MPI31-36 were designed for this purpose. The base molecule of MPI31-36 is MPI30 that is different from boceprevir only at the P1 residue. Please note that MPI30 is a previously developed MPro inhibitor with a name as ML1000, though the structural characterization of its complex with MPro has not been reported [35]. We follow typical peptide coupling chemistry to synthesize all inhibitors including PF-07321332. In general, the synthesis started from the P4 N-terminal cap to the C-terminal warhead. All compounds were strictly characterized to ensure their purity. Synthetic details and characterizations are provided in the supplementary information.
2.2. Kinetic characterizations of MPI29-47 and PF-07321332 on their in vitro enzymatic inhibition potency
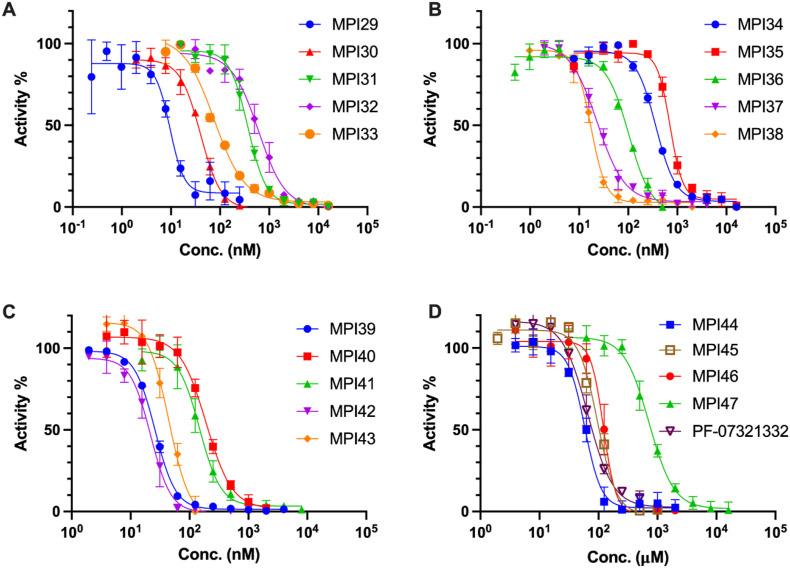
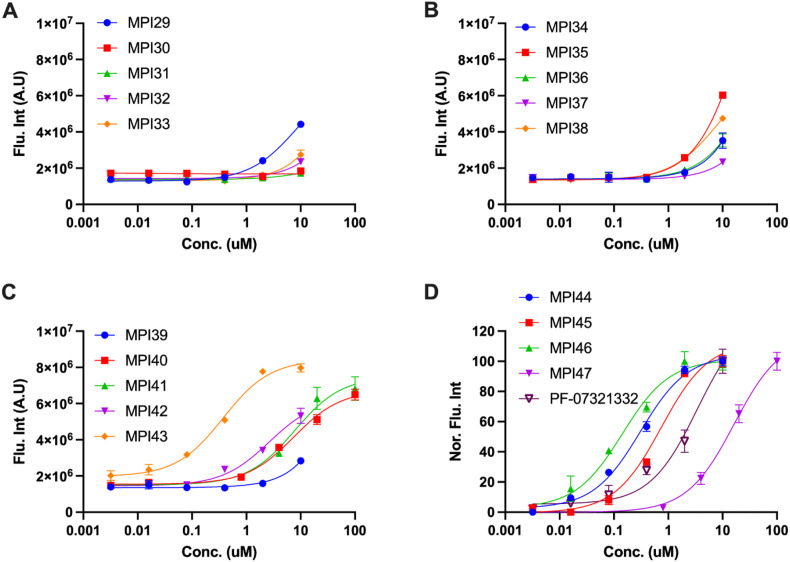
In a previous drug repurposing project, we established a protocol to characterize MPro inhibitors [12]. In this protocol, a fluorogenic substrate Sub3 is used. We followed this protocol to characterize in vitro MPro inhibition potency for all synthesized inhibitors by determining their IC50 values. To conduct the assay, we preincubated 20 nM MPro with varied concentrations of an inhibitor for 30 min before 10 μM Sub3 was added and the fluorescent product formation was recorded in a fluorescence plate reader. 30 min incubation time with MPro is a standard procedure that has been used by multiple labs in the determination of IC50 values for MPro inhibitors [12,16,29,36]. All our designed inhibitors are reversible covalent inhibitors, their incubation times with MPro are not expected to significantly influence their determined IC50 values. As a matter of fact, a previous IC50 analysis in which MPI11 was incubated with MPro for three different times led to very similar characterized IC50 values [32]. For MPI29-47 and PF-07321332, they all exhibited well defined inhibition curves that started from 100% activity without an inhibitor and reached to almost total inhibition when 10 μM of an inhibitor was provided. Data are presented in Fig. 2 . We fit all collected data to a four-parameter variable slope inhibition equation in GraphPad 9.0 to obtain IC50 values for all inhibitors. Determined IC50 values are presented in Table 1 . As shown in Table 1, MPI29 has the highest in vitro potency with an IC50 value as 9.3 nM. Since we used 20 nM MPro for the assay, this IC50 value has reached the detection limit. Real in vitro potency of MPI29 is likely higher than what the determined IC50 value indicates. MPI29 has an aldehyde warhead. MPI30 is different from MPI29 only at the warhead position. It has an α-ketoamide instead. MPI30 has a determined IC50 value as 40 nM. This value is similar to that reported by Westberg et al. [35]. This lower potency than MPI29 is expected since α-ketoamide is less chemically reactive than aldehyde toward a nucleophile. However, MPI30 is 100-fold more potent than boceprevir indicating the essential role of the P1 Opal residue in improving interactions with MPro. MPI31-36 all have much higher in vitro IC50 values than MPI30. Apparently adding a chemical appendage to the α-ketoamide nitrogen leads to less favorable interactions with MPro. Interestingly, MPI33 that has the smallest appendage has an in vitro IC50 value closest to MPI30. Although the benzyl appendage in MPI36 is bigger than that in MPI32-35, it is less detrimental to the in vitro potency indicating that the benzyl group possibly involves some favorable interactions with MPro in comparison to other N-substituents.
Fig. 2.
Inhibition curves of MPI29-47 and PF-07321332 on MPro. Triplicate experiments were performed for each compound. For all experiments, 20 nM MPro was incubated with an inhibitor for 30 min before 10 μM Sub3 was added. The MPro-catalyzed Sub3 hydrolysis rate was determined by measuring linear increase of product fluorescence (Ex: 336 nm/Em: 455 nm) at the initial 5 min reaction time.
Table 1.
Determined enzymatic IC50, cellular EC50, and antiviral EC50 values of MPro inhibitors.
| Compound ID | Enzymatic IC50 (nM) | Cellular EC50 (μM) | Antiviral EC50 (μM) | PDB Entry | Compound ID | Enzymatic IC50 (nM) | Cellular EC50 (μM) | Antiviral EC50 (μM) | CC50 (μM) | PDB Entry |
|---|---|---|---|---|---|---|---|---|---|---|
| Boceprevir | 4200 ± 600 [16]/8000 ± 1500 [15] | >10 | MPI39 | 26 ± 1 | >10 | |||||
| MPI29 | 9.3 ± 0.8 | >5 | 7S6W | MPI40 | 180 ± 20 | 7.4 ± 0.9 | n.d.a/n.d.b/n.d.c | |||
| MPI30 | 40 ± 4 | >10 | 7S6X | MPI41 | 150 ± 20 | 7.7 ± 1.5 | ||||
| MPI31 | 360 ± 50 | >10 | MPI42 | 22 ± 4 | 2.6 ± 0.5 | 7S75 | ||||
| MPI32 | 620 ± 170 | >10 | 7S6Y | MPI43 | 45 ± 5 | 0.37 ± 0.04 | 0.61a/0.36b/1.00c | 34.2 | ||
| MPI33 | 75 ± 9 | >10 | 7S6Z | MPI44 | 59 ± 7 | 0.31 ± 0.03 | 2.94a/0.86b/1.04c | 143.7 | ||
| MPI34 | 370 ± 30 | >10 | 7S70 | MPI45 | 97 ± 11 | 0.74 ± 0.11 | ||||
| MPI35 | 720 ± 80 | >5 | 7S71 | MPI46 | 120 ± 10 | 0.14 ± 0.02 | 1.08a/2.28b/0.75c | 163.4 | ||
| MPI36 | 102 ± 2 | >10 | 7S72 | MPI47 | 720 ± 90 | >10 | ||||
| MPI37 | 23 ± 1 | >10 | 7S73 | PF-07321332 | 66 ± 12 | 3.4 ± 0.8 | 1.30 | |||
| MPI38 | 17 ± 2 | >5 | 7S74 |
n.d.: no detectable value.
Antiviral EC50 value for the USA-WA1/2020 strain.
Antiviral EC50 value for the Beta strain.
Antiviral EC50 value for the Delta strain.
MPI37 is different from MPI29 at the P2 residue. It has a determined in vitro IC50 value as 23 nM. Although replacing the P2 dimethylcyclopropylproline with (S)-5-azaspiro [2,4]heptane-6-carboxylic acid leads to less favorable interactions with MPro, the effect is not dramatic. A similar observation was made with MPI38 that has a P2 neopentylglycine. MPI39 has both P2 and P3 sites different from MPI29. Both residues in MPI39 have been shown in a previous study to favor interactions with MPro [32]. Accordingly, we notice that MPI39 has relatively high in vitro potency. MPI40, MPI42 and MPI43 are different from MPI29 only in their P4 N-terminal cap. MPI40 has a P4 N-terminal amide that is different from MPI29 only at one atom position. However, this slight change leads to about 20-fold loss of in vitro potency. Intriguingly, removing one methyl group from the P4 N-terminal cap of MPI40, which leads to MPI42, improves in vitro potency for about 9 folds. Another interesting observation is on MPI43. MPI43 has a P4 N-terminal carbamate and is different from MPI29 and MPI40 only at the carbamate oxygen position. It has in vitro potency between MPI29 and MPI40. Therefore, switching a P4 N-terminal carbamide nitrogen to oxygen is less detrimental to in vitro potency than switching it to methylene. MPI41 is an α-ketoamide derivative of MPI40. It has in vitro potency surprisingly similar to MPI40 although α-ketoamide is expected to be less reactive than aldehyde. MPI44 is different from MPI43 at its P4 N-terminal O-alkyl group. It has a benzyl group instead of a tert-butyl group as in MPI43. Although two O-alkyl groups are structurally different, MPI43 and MPI44 have similar in vitro potency. In our previous work, we noticed that an O-benzyl group at this position loosely binds MPro [32,33]. Similar in vitro potency between MPI43 and MPI44 indicates that an O-tert-butyl group at this position might involve loose interactions with MPro as well. In our previous work, we discovered that MPI8, a peptidyl aldehyde has the highest in cellulo and antiviral potency among all inhibitors that we synthesized [21]. Due to the high in vitro potency of MPI29, we created chimera inhibitors MPI45 and MPI46 that integrate structural components from both MPI8 and MPI29. MPI45 is a chimera in which the P4 N-terminal cap of MPI29 is fused to the rest of MPI8 and MPI46 is a product made by switching the P2 residue in MPI8 to the one in MPI29. Both MPI45 and MPI46 have in vitro potency similar to MPI8 with an IC50 value about 100 nM [33]. MPI47 is different from MPI40 only at its warhead. It has an in vitro IC50 value as 720 nM. Apparently switching the warhead to nitrile from either aldehyde or α-ketoamide leads to significant loss of in vitro potency. Although PF-07321332 and MPI47 are structurally similar, PF-07321332 has a more than 10-folder lower IC50 value than MPI47. A likely explanation is that the P4 N-terminal trifluoroacetamide cap in PF-07321332 involves unique interactions with MPro that do not exist for MPI47, which is supported by the crystal structure of MPro bound with PF-07321332 [14,37,38].
2.3. X-ray crystallography analysis of MPro bound with 10 inhibitors
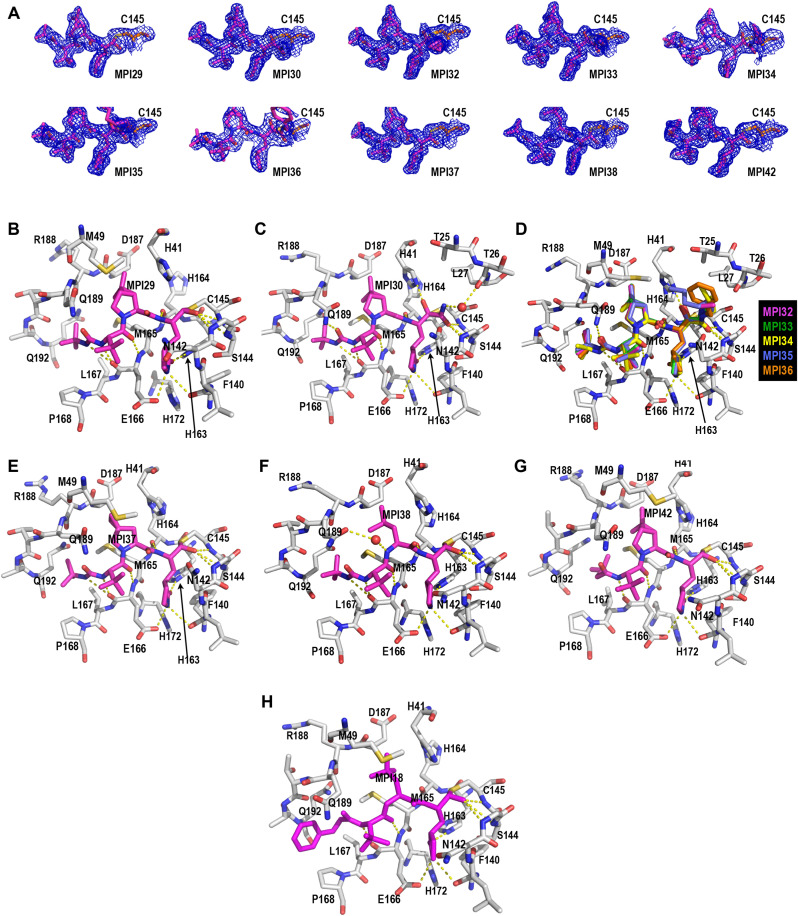
In order to structurally characterize our developed inhibitors in their complexes with MPro. We crystalized MPro in its apo form and then soaked obtained crystals with our designed inhibitors for X-ray crystallography analysis. Using this approach, we successfully determined 10 MPro-inhibitor complex structures to high resolutions. These inhibitors include MPI29, MPI30, MPI32-38, and MPI42. PDB entries for these MPro-inhibitor complex structures are summarized in Table 1. In the active sites of all MPro-inhibitor complexes, electron density is well shaped for unambiguous modeling of inhibitors except for MPI36 at its α-ketoamide N-benzyl group (Fig. 3 A). A covalent bond between C145 of MPro and the warhead of an inhibitor is clearly visible in all structures. MPI29, MPI37, MPI38, and MPI42 have an aldehyde warhead. In the MPro-MPI29 complex as shown in Fig. 3B, MPI29 forms a number of hydrogen bonds with MPro. The amide of the P1 Opal side chain lactam forms three hydrogen bonds with MPro residues including F140 at its backbone oxygen, H163 at one of its imidazole nitrogen atoms and E166 at its side chain carboxylate. These three hydrogen bonds have also been observed in structures of MPro bound with other inhibitors containing a P1 Opal side chain lactam. There are two hydrogen bonds generated between two backbone amides of MPI29 and MPro residues including H164 at its backbone oxygen and E166 at its backbone nitrogen. Similar hydrogen bonds have been observed in structures of MPro bound with other peptidyl inhibitors. The P4 N-terminal carbamide cap of MPI29 uses its two nitrogen atoms to form hydrogen bonds with the backbone oxygen of MPro E166. In previously published structures of MPro bound with peptidyl aldehyde inhibitors that have a P4 N-terminal carbamate, typically only one hydrogen bond is observed between the P4 N-terminal cap and MPro E166. One additional hydrogen bond that is formed between the P4 N-terminal cap and MPro E166 provides an explanation of very high in vitro potency of MPI29. Since the introduction a carbamide nitrogen causes binding energy loss due to desolvation, this energy loss is apparently counteracted possibly by this additional hydrogen bond formed between MPI29 and MPro. In the MPro-MPI29 complex, the hemithioacetal hydroxide is predominantly in an S configuration and poises to interact with three MPro backbone amide NH groups from G143, S144 and C145 that purportedly generate an oxyanion hole to stabilize the transition state of MPro-catalyzed hydrolysis of a substrate. This favorable binding of the hemithioacetal hydroxide at the oxyanion hole likely contributes to strong in vitro potency for peptidyl aldehyde inhibitors. It is debating that whether the hemithioacetal hydroxide is at its deprotonated and therefore charged form. Since a hemithioacetal hydroxide has a much lower pKa value than a regular alcohol and the site prefers a negatively charged hydroxide, it is highly likely that the hemithioacetal hydroxide is negatively charged. If this is the situation, one may generally bear in mind to introduce a negatively charged moiety in a designed inhibitor to bind to this anion hole in MPro for improved affinity. In MPro-MPI29, the P2 dimethylcyclopropylproline is in van der Waals distances to most surrounding MPro residues in the S2 pocket, which indicates favorable hydrophobic interactions. Based on the structure, there is not much space left for adding additional chemical moieties into the inhibitor to improve affinity. However, the plasticity of MPro may allow the change around the S2 pocket to accommodate even large P2 residues in inhibitors. The P3 tert-butylglycine is in the van der Waals distance to the side chain of MPro E166 but apparently doesn't engage other residues in MPro. This is known in most peptidyl inhibitors for MPro. However, the tert-butyl group in the P4 N-terminal cap fits nicely to the MPro P4 binding pocket indicating favorable van der Waals interactions with surrounding MPro residues. All these favorable interactions likely contribute to the exceedingly high in vitro potency of MPI29. In previously published structures of MPro bound with peptidyl inhibitors that contained a P4 N-terminal benzoxycarbonyl (CBZ) group such as the MPro-MPI18 structure shown in Fig. 3H, the CBZ group was typically not structurally defined [32,33,39]. It is apparent that the tert-butyl carbamide fits better than the CBZ group to the S4 pocket of MPro.
Fig. 3.
Crystal structures of MPro bound with 10 inhibitors. (A): Contoured 2Fo-Fc maps at the 1σ level around 10 MPIs and C145 in the active site of MPro. (B–G): The active site structures for MPro bound with (B) MPI29, (C) MPI30, (D) MPI32-MPI36, € MPI37, (F) MPI38, (G) MPI42, and (H) MPI8. The structure of MPI18 is based on the PDB entry 7RVR. It is provided as a comparison. Dashed yellow lines between inhibitors and MPro are hydrogen bonds.
MPI30 is different from MPI29 only at its warhead. It has an α-ketoamide. In its crystal structure as shown in Fig. 3C, MPI30 forms interactions with MPro similar to MPI29 at the P1, P2 and P3 residues and the P4 N-terminal group. We observed one additional hydrogen bond that is formed between the P4 N-terminal carbamide oxygen and the side chain of MPro Q189. Q189 is a residue that adopts different conformations in reported structures. Given that its hydrogen bonding interactions with an inhibitor may improve the inhibitor selectivity over other proteins, one may take advantage of this aspect to design MPro inhibitors. The keto group of the α-ketoamide of MPI30 covalently interacts with MPro C145 to generate a hemithioacetal. Unlike MPI29, the hemithioacetal hydroxide adopts a conformation pointing away from the oxyanion hole. It is in an unambiguously S configuration and interacts with H41 through two water-bridged hydrogen bonds. The amide part of the warhead points toward the oxyanion hole. The amide oxygen is assigned at the oxyanion hole due to its ability to form hydrogen bonds with three backbone NH groups from MPro. The amide nitrogen interacts indirectly with the backbone oxygen of MPro T26 through two water-bridged hydrogen bonds. All these interactions likely contribute to the high in vitro potency of MPI30. As aforementioned, aldehyde-based inhibitors potentially generate a negatively charged hemithioacetal hydroxide to potently bind the MPro anion hole. Give that the hemithioacetal hydroxide points away from the anion hole in the MPro-MPI30 structure, this potential strong interaction is lost. The α-ketoamide is also expected to react with the MPro active site cysteine much weaker than the aldehyde warhead. Apparently both weakening effects are counterbalanced by additional interactions that are generated between MPro and the MPI30 α-ketoamide.
Since MPI32-36 have an MPI30 base structure and different N-appendages on the α-ketoamide, they are presented together in Fig. 3D. Except at the appendage position, all other parts of MPI32-36 interact with MPro similar to MPI30. We designed MPI32-36 with a hope that their α-ketoamide N-appendages may reach the MPro S3′ pocket for favorable interactions. However, all N-appendages have a Z configuration that is energetically favorable and consequently makes them point away from the MPro S3′ pocket. The appendage of MPI35 is long enough to fold back to the MPro S3’ pocket. Since the N-benzyl group of MPI36 has weak electron density around the phenyl ring, it is difficult to assess its interactions with MPro. Its more favorable in vitro potency than MPI34 and MPI35 is likely due to higher structural rigidity of the N-benzyl group than two flexible N-alkyl groups in MPI34 and MPI35. The indirect interaction between the α-ketoamide nitrogen and MPro T26 that was observed in the MPro-MPI30 structure is not observed in MPro complexes with MPI32-36. All N-appendages occupy the original water position and consequently remove interactions involving this water molecule. This might contribute to low in vitro potency of MPI32-36. Other N-aromatic functionalities and modifications to the N-benzyl group may be introduced to MPI36 to improve the binding to MPro. However, delicate tuning will be necessary.
As shown in Fig. 3E and F, MPI37 and MPI38 interact with MPro similar to MPI29. The P2 residue in MPI37 occupies slightly less space than that in MPI29. This may contribute to less in vitro potency of MPI37 than MPI29. In MPro-MPI38, there exists a water molecule that bridges an indirect interaction between the P2 backbone nitrogen and the side chain of MPro Q189. MPI38 displays an MPro binding mode very similar to MPI18 that was previously developed and structurally resolved in its complex with MPro (PDB entry: 7RVR and Fig. 3H) [32]. The only difference is at the P4 N-terminal cap. MPI18 has a P4 N-terminal CBZ group that displays loose binding to MPro and cannot be unambiguously refined in its MPro complex structure. However, MPI38 has an N-tert-butylcarbamide cap whose structure can be clearly refined in the MPro-MPI38 complex. MPI38 is slightly more potent than MPI18. This slightly higher potency might be attributed to more favorable interactions between the P4 N-terminal cap and the MPro P4 binding pocket. Another noticeable difference is at Q189. The side chain of Q189 adopted two different conformations in the two structures. As discussed previously, Q189 is a relatively flexible residue that has shown very different structural modes in MPro-inhibitor complexes.
MPI42 is structurally similar to MPI29 except that it has a P4 N-terminal isovaleramide. As shown in Fig. 3G, MPI42 interacts with MPro similar to MPI29 except at the P4 N-terminal cap. The isovaleramide cap adopts a unique orientation that points deep into the MPro S4 pocket. This is different from most currently known peptidyl aldehyde inhibitors. MPro naturally prefers a P4 valine in its substrates. Isovaleramide is almost structurally identical to valine except that it doesn't have an α-amine. It is apparent that isovaleramide engages favorable interactions with MPro by binding deep to the MPro P4 binding pocket. This explains its much higher in vitro potency than MPI40 that has an additional methyl group at the P4 N-terminal cap. The 3,3-dimethylbutamide cap in MPI40 is too bulky to fit into the relatively small MPro S4 pocket. Due to the high potency induced by this small isovaleramide cap, it is suggested to be incorporated into future inhibitor designs for improved affinity toward MPro.
2.4. Characterizations of in cellulo MPro inhibition potency of MPI29-47 and PF-07321332
When expressed in a human cell host, MPro leads to acute cytotoxicity and drives the host cell to undergo apoptosis. Using this unique feature, we previously developed a cell-based assay to characterize in cellulo potency of MPro inhibitors [21]. In this assay, an inhibitor with in cellulo potency suppresses cytotoxicity from an MPro-eGFP (enhanced green fluorescent protein) fusion protein that is ectopically expressed in 293T cells and consequently leads to host cell survival and enhanced overall expression of MPro-eGFP that can be characterized by flow cytometry. We consider that this assay is more advantageous over a direct antiviral assay in the characterization of MPro inhibitors since an inhibitor may block functions of host proteases such as TMPRSS2, furin, and cathepsin L that are critical for SARS-CoV-2 infection and therefore provides false positive in cellulo potency results for MPro inhibitors [[40], [41], [42]]. False positive results from targeting TMPRSS2, furin, and cathepsin L will lead to wrong structure-activity relationship studies of MPro inhibitors. Using this novel cellular assay, we characterized a number of repurposed drugs for MPro inhibitors. Our results indicated that some of these inhibitors suppress SARS-CoV-2 via mechanisms different from MPro inhibition [21]. Using this novel cellular assay, we characterized all synthesized inhibitors in this work. Inhibitor-driven MPro-eGFP expression data are presented in Fig. 4 and the determined in cellulo EC50 values are summarized in Table 1. MPI29-39 that have a P4 N-terminal carbamide cap all showed low in cellulo potency. Since data for all these inhibitors do not reach a plateau at their highest tested concentrations, their EC50 values can only be estimated as higher than 5 or 10 μM. Although MPI29 has the most in vitro potency among all inhibitors that we synthesized, it has very weak potency in cells. MPI40-42 have a P4 N-terminal amide cap. In comparison to MPI29-39, they showed higher in cellulo potency. Their determined EC50 values are in a single digit μM range. MPI43 has a P4 N-terminal carbamate and is different from MPI29 and MPI40 at only one atom position. It has a determined in cellulo EC50 value as 0.37 μM. This in cellulo potency is 20-fold higher than MPI40 and 160-fold higher than MPI29. Considering that MPI29, MPI40 and MPI43 are different only at one atom position and MPI29 has much higher in vitro potency than MPI43, their drastically reversed potency in cells is intriguing. These three inhibitors must have very different plasma/cellular stability, cellular permeability, or both. MPI44 has a P4 N-terminal CBZ cap. It has a determined in cellulo EC50 value similar to that of MPI43. Since MPI43 and MPI44 have similar in vitro potency as well, the identity of a P4 N-terminal carbamate seems to have little effect on an inhibitor's in vitro and in cellulo potency. MPI45 and MPI46 each have chemical moieties at two positions that are switched from that in MPI29 to that in MPI8. Both have high in cellulo potency. Their determined EC50 values are 0.74 and 0.14 μM respectively. It is evident that chemical moieties in MPI8 are optimal for high in cellulo potency. The in cellulo potency of MPI45 is much lower than that of MPI46 likely due to its P4 N-terminal carbamide cap. MPI47 have a P4 N-terminal amide. Its determined in cellulo EC50 value is similar to that for MPI40-42 that also contains a P4 N-terminal amide. All four compounds have relatively low cellular potency. PF-07321332 has a determined in cellulo EC50 value as 3.4 μM. Its better in cellulo potency than other inhibitors with an N-terminal amide is likely due to its N-terminal trifluoro group that may protect the N-terminal amide from hydrolysis and/or facilitate the permeability into cells. PF-07321332 was previously shown with an antiviral EC50 value as 78 nM to inhibit SARS-CoV-2 in infecting ACE2+ A549 cells [14]. Please note that both different cells and different systems were used in this analysis. Direct comparison of the antiviral EC50 value from this study to the in cellulo EC50 value determined in our study is not meaningful. Multiple reasons likely contribute to the discrepancy.
Fig. 4.
Cellular potency of MPI29-47 in their inhibition of MPro to drive host 293T cell survival and overall MPro-eGFP expression. In D, fluorescent intensity is normalized due to that data for PF-07321332 was collected at a different time using different setups.
2.5. Characterizations of antiviral potency and cytotoxicity of four selected inhibitors on three SARS-CoV-2 variants
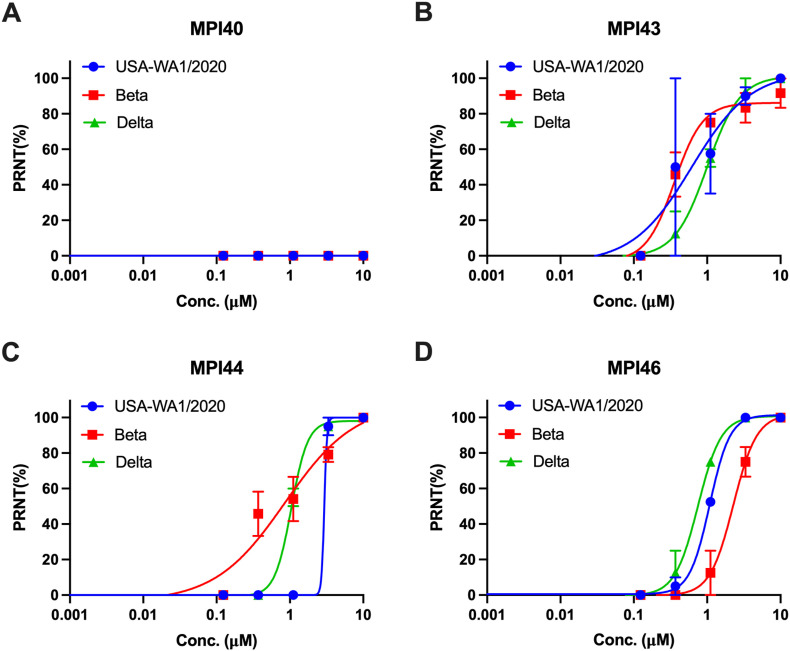
Three most cellularly potent inhibitors MPI43, MPI44 and MPI46 were advanced to antiviral assays. One inhibitor MPI40 that has low cellular potency was characterized as well as a negative control. To quantify their antiviral EC50 values, we conducted plaque reduction neutralization tests of three SARS-CoV-2 variants including USA-WA1/2020, Beta and Delta in Vero E6 cells for all four inhibitors. We infected Vero E6 cells by virus in the presence of an inhibitor at various concentrations for three days and then quantified viral plaque reduction. Based on viral plaque reduction data, we determined antiviral EC50 values for all inhibitors. MPI40 showed no inhibition at all tested concentrations, which matches its detected low in cellulo potency. All other three inhibitors have high antiviral potency. Their determined antiviral EC50 values are similar and around 1 μM (Fig. 5 and Table 1). MPI46 shows the most well shaped antiviral data. It inhibits the Delta variants slightly better than the other two SARS-CoV-2 variants. A similar assay of PF-07321332 in its inhibition of USA-WA1/2020 in Vero R6 cells led to a determined antiviral EC50 value as 1.3 μM (Supplementary Fig. S1). Please note that the determined antiviral EC50 value for PF-07321332 as 75 nM in Vero E6 cells was obtained by the combination use of 2 mM CP-100356 that is an inhibitor of the prototypical ABC transporter [14]. In our study, CP-100356 was not used in all the compounds tested.
Fig. 5.
Plaque reduction neutralization tests (PRNTs) of MPI40, MPI43, MPI44, and MPI46 on their inhibition of three SARS-CoV-2 strains USA-WA1/2020, Beta and Delta in Vero E6 cells. Two repeats were conducted for each concentration.
We have also characterized cytotoxicity for MPI43, MPI44 and MPI46 in 293T cells using the MTT assay [43]. Cytotoxicity curves of these inhibitors are shown in Supplementary Fig. S2. Determined CC50 values are 34.2, 143.7 and 163.4 μM for MPI43, MPI44 and MPI46, respectively. In term of selectivity index (CC50/antiviral EC50), MPI43, MPI44 and MPI46 range from 56 to 233.
3. Discussion
As a potential repurposed drug for COVID-19, boceprevir provided a high hope during the early phase of the COVID-19 pandemic. However, it is only moderate against SARS-CoV-2. Although it shows inhibition of MPro, it has very weak potency to inhibit MPro in a human cell host [21]. Lin et al. made ML1000 (MPI30 in our series) based on boceprevir. Although it showed high in vitro potency, its antiviral potency was minimal [35]. In our study, both MPI29 and MPI30 are highly potent MPro inhibitors in vitro. The in vitro potency of MPI29 has even reached the detection limit of our kinetic assay. However, both inhibitors show very low in cellulo potency. Except MPI45 that has some MPI8 moieties, all other inhibitors that have a P4 N-terminal carbamide cap all display low in cellulo potency. Based on these data, we can conclude that MPro inhibitors that have a P4 N-terminal carbamide will have low cellular and antiviral potency. Although reasons for this low cellular and antiviral potency need to be further investigated, the emergency of COVID-19 demands to focus on other routes to develop MPro inhibitors as potential SARS-CoV-2 therapeutics.
An alternative to the P4 N-terminal carbamide is amide. We synthesized several inhibitors that contain a P4 N-terminal amide. In comparison to inhibitors that have a P4 N-terminal carbamide, P4 amide-containing inhibitors are relatively more potent in cells but their cellular potency is moderate. In comparison to P4 amide-containing inhibitors that we designed, PF-07321332 that has an N-terminal trifluoroacetamide is much more potent in cells. It is highly possible that the bulky trifluoroacetyl group protects the N-terminal trifluoroacetamide of PF-07321332 from degradation, a situation that does not exist for other P4 amide-containing inhibitors that we generated. It is also possible the trifluoroacetyl group assists cellular permeability of PF-07321332. Unlike inhibitors with a P4 N-terminal carbamide, all three inhibitors with a P4 N-terminal carbamate show high cellular potency and also high antiviral potency. These three inhibitors have also low cytotoxicity. Our collected data support a conclusion that a P4 N-terminal carbamate in a boceprevir-based MPro inhibitor is key for high cellular and antiviral potency.
In other positions, a P1 Opal residue, a P2 dimethylcyclopropylproline, and a P3 tert-butylglyine or O-tert-butylthreonine are optimal for high cellular and antiviral potency. In previous work, we discovered that a P3 O-tert-butylthreonine is critical for an MPro inhibitor to have high cellular and antiviral potency [32]. This observation is supported by the high cellular and antiviral potency of MPI46. MPI8 was previously discovered as a highly potent SARS-CoV-2 inhibitor with an EC50 value as 30 nM [21,33,39]. All boceprevir-based inhibitors that we developed in this work do not reach the potency of MPI8. To improve cellular and antiviral potency of inhibitors such as MPI43, MPI44 and MPI46, a possible route is to search for alternative O-alkyl groups in the P4 N-terminal carbamate cap for improved binding to the MPro P4 binding pocket.
For the warhead, all current data indicate that aldehyde has the best activity. Changing it to either α-ketoamide or nitrile significantly decreases both in vitro and in cellulo potency of an inhibitor. Compounds with an aldehyde will lead to a concern of cytotoxicity. Cytotoxicity and potency of an aldehyde inhibitor for MPro need to be carefully balanced when it is finally applied in animals or human patients. Pharmacokinetic (PK)/pharmacodynamic (PD) features will also need to be considered. Most currently reported MPro inhibitors do not have PK/PD results in animals available. We recently characterized MPI8 PK results in rats [44]. MPI8 displayed a relatively quick clearance rate in rats with a half-life as 1.22 h by oral administration. However, PK results for PF-07321322 have shown that it is stable in animals, although it is metabolized by CYP3A4 in humans [14]. The nitrile warhead and other structural moieties likely contribute to this high stability. Since PF-07321332 is a component of an approved drug for COVID-19 already, combining characteristics within PF-07321332 such as the nitrile warhead that has low cytotoxicity concern and that from other boceprevir and/or MPI8-based inhibitors including MPI43, MPI44 and MPI46 for improved PK/PD features need to be explored. PF-07321332 requires to be used together with ritonavir due to its metabolic instability. Strong side effects from ritonavir and serious and sometimes fatal drug interactions between ritonavir and other medications have been reported [[45], [46], [47], [48], [49]]. An obvious drug development goal is to develop a PF-07321332-based SARS-CoV-2 antiviral that will not require the combination use with ritonavir. The metabolization weak point of PF-07321332 is at the P1 Opal side chain lactam. Changing this site to other chemical moieties for improved PK characteristics is likely but has not yet been reported.
We also synthesized a number of α-ketoamide inhibitors with different alkyl substituents at the α-ketoamide nitrogen. The purpose was to make these N-appendages reach the MPro P3′ binding pocket. Unfortunately, all these inhibitors have low in vitro and in cellulo potency. Structures of their complexes with MPro all show a Z configuration at the α-ketoamide. The Z configuration is more energetically favored than the E form. It is evident that steric hindrance between the hemithioacetal hydroxide and an N-appendage prevents the N-appendage from adopting a favorable position to interact with the MPro P3′ binding pocket. Although further optimization in related MPro inhibitors may likely generate interactions with the MPro P3′ binding pocket, we suggest other routes to take advantage of the MPro P3′ binding pocket. In our characterized structures, the P4 N-terminal isovaleramide of MPI42 shows a unique conformation that binds deep into the MPro P4 binding pocket. The N-terminal trifluoroacetamide of PF-07321332 binds at the same MPro pocket [14]. This unique binding explains MPI42's much higher in vitro potency than its close relative MPI40. We think MPI42 points to a unique way to develop potent MPro inhibitors by exploring P4 N-terminal chemical moieties with a similar size as isovaleramide.
4. Conclusion
Based on our systematic analysis of boceprevir-based MPro inhibitors, we conclude that a P4 N-terminal carbamate in an inhibitor is critical to yield high cellular and antiviral potency. However, chemical moieties at all sites including the P4 N-terminal carbamate and the warhead might be continuously optimized. Combined features from PF-07321332 (e.g. nitrile warhead) and newly developed inhibitors such as MPI43, MPI44 and MPI46 can be considered. In vitro DMPK and ADME assays will provide insights into how to further optimize the boceprevir-based MPro inhibitors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by Welch Foundation (grant A-1715), DHHS–NIH (R21AI164088 and R21EB032983), TAMU COS Strategic Transformative Research Program, and Texas A&M X Grants. The ALS-ENABLE beam-lines are supported in part by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169-01 and the Howard Hughes Medical Institute. The Advanced Light Source is a Department of Energy Office of Science User Facility under Contract No. DE-AC02-05CH11231.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2022.114596.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Baker J.D., Uhrich R.L., Kraemer G.C., Love J.E., Kraemer B.C. A drug repurposing screen identifies hepatitis C antivirals as inhibitors of the SARS-CoV2 main protease. PLoS One. 2021;16 doi: 10.1371/journal.pone.0245962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Abranyi-Balogh P., Brandao-Neto J., Carbery A., Davison G., Dias A., Downes T.D., Dunnett L., Fairhead M., Firth J.D., Jones S.P., Keeley A., Keseru G.M., Klein H.F., Martin M.P., Noble M.E.M., O'Brien P., Powell A., Reddi R.N., Skyner R., Snee M., Waring M.J., Wild C., London N., von Delft F., Walsh M.A. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., Chang M.W., Chan J.F., Cao J., Poon V.K., Herbert K.M., Cheng K., Nguyen T.H., Rubanov A., Pu Y., Nguyen C., Choi A., Rathnasinghe R., Schotsaert M., Miorin L., Dejosez M., Zwaka T.P., Sit K.Y., Martinez-Sobrido L., Liu W.C., White K.M., Chapman M.E., Lendy E.K., Glynne R.J., Albrecht R., Ruppin E., Mesecar A.D., Johnson J.R., Benner C., Sun R., Schultz P.G., Su A.I., Garcia-Sastre A., Chatterjee A.K., Yuen K.Y., Chanda S.K. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunther S., Reinke P.Y.A., Fernandez-Garcia Y., Lieske J., Lane T.J., Ginn H.M., Koua F.H.M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J.D., Gelisio L., Brehm W., Dunkel I., Seychell B., Gieseler H., Norton-Baker B., Escudero-Perez B., Domaracky M., Saouane S., Tolstikova A., White T.A., Hanle A., Groessler M., Fleckenstein H., Trost F., Galchenkova M., Gevorkov Y., Li C., Awel S., Peck A., Barthelmess M., Schlunzen F., Lourdu Xavier P., Werner N., Andaleeb H., Ullah N., Falke S., Srinivasan V., Franca B.A., Schwinzer M., Brognaro H., Rogers C., Melo D., Zaitseva-Doyle J.J., Knoska J., Pena-Murillo G.E., Mashhour A.R., Hennicke V., Fischer P., Hakanpaa J., Meyer J., Gribbon P., Ellinger B., Kuzikov M., Wolf M., Beccari A.R., Bourenkov G., von Stetten D., Pompidor G., Bento I., Panneerselvam S., Karpics I., Schneider T.R., Garcia-Alai M.M., Niebling S., Gunther C., Schmidt C., Schubert R., Han H., Boger J., Monteiro D.C.F., Zhang L., Sun X., Pletzer-Zelgert J., Wollenhaupt J., Feiler C.G., Weiss M.S., Schulz E.C., Mehrabi P., Karnicar K., Usenik A., Loboda J., Tidow H., Chari A., Hilgenfeld R., Uetrecht C., Cox R., Zaliani A., Beck T., Rarey M., Gunther S., Turk D., Hinrichs W., Chapman H.N., Pearson A.R., Betzel C., Meents A. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372:642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghahremanpour M.M., Tirado-Rives J., Deshmukh M., Ippolito J.A., Zhang C.H., Cabeza de Vaca I., Liosi M.E., Anderson K.S., Jorgensen W.L. Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. ACS Med. Chem. Lett. 2020;11:2526–2533. doi: 10.1021/acsmedchemlett.0c00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 9.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X., Duan Y., Yu J., Yang X., Yang X., Yang K., Liu X., Guddat L.W., Xiao G., Zhang L., Yang H., Rao Z. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 10.Li Z., Li X., Huang Y.Y., Wu Y., Liu R., Zhou L., Lin Y., Wu D., Zhang L., Liu H., Xu X., Yu K., Zhang Y., Cui J., Zhan C.G., Wang X., Luo H.B. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc. Natl. Acad. Sci. U.S.A. 2020;117:27381–27387. doi: 10.1073/pnas.2010470117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma C., Hu Y., Townsend J.A., Lagarias P.I., Marty M.T., Kolocouris A., Wang J., Ebselen, Disulfiram Carmofur. PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol. Transl. Sci. 2020;3:1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vatansever E.C., Yang K.S., Drelich A.K., Kratch K.C., Cho C.C., Kempaiah K.R., Hsu J.C., Mellott D.M., Xu S., Tseng C.K., Liu W.R. Bepridil is potent against SARS-CoV-2 in vitro. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2012201118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cevik M., Kuppalli K., Kindrachuk J., Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. 2020;371:m3862. doi: 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- 14.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 15.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oerlemans R., Ruiz-Moreno A.J., Cong Y., Dinesh Kumar N., Velasco-Velazquez M.A., Neochoritis C.G., Smith J., Reggiori F., Groves M.R., Domling A. Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics. RSC Med. Chem. 2020;12:370–379. doi: 10.1039/d0md00367k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howe A.Y., Venkatraman S. The discovery and development of boceprevir: a novel, first-generation inhibitor of the hepatitis C virus NS3/4A serine protease. J. Clin. Transl. Hepatol. 2013;1:22–32. doi: 10.14218/JCTH.2013.002XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kneller D.W., Galanie S., Phillips G., O'Neill H.M., Coates L., Kovalevsky A. Malleability of the SARS-CoV-2 3CL M(pro) active-site cavity facilitates binding of clinical antivirals. Structure. 2020;28:1313–1320 e1313. doi: 10.1016/j.str.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao J., Li Y.S., Zeng R., Liu F.L., Luo R.H., Huang C., Wang Y.F., Zhang J., Quan B., Shen C., Mao X., Liu X., Sun W., Yang W., Ni X., Wang K., Xu L., Duan Z.L., Zou Q.C., Zhang H.L., Qu W., Long Y.H., Li M.H., Yang R.C., Liu X., You J., Zhou Y., Yao R., Li W.P., Liu J.M., Chen P., Liu Y., Lin G.F., Yang X., Zou J., Li L., Hu Y., Lu G.W., Li W.M., Wei Y.Q., Zheng Y.T., Lei J., Yang S. SARS-CoV-2 M(pro) inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371:1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao W., Cho C.D., Geng Z.Z., Shaabani N., Ma X.R., Vatansever E.C., Alugubelli Y.R., Ma Y., Chaki S.P., Ellenburg W.H., Yang K.S., Qiao Y., Allen R., Neuman B.W., Ji H., Xu S., Liu W.R. Evaluation of SARS-CoV-2 main protease inhibitors using a novel cell-based assay. ACS Cent. Sci. 2022;8:192–204. doi: 10.1021/acscentsci.1c00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W.R., Fierke C.A., Xu S., Yang K.S., Ma X.R., Ma Y., Alugubelli Y.R., Vatansever E.C., Cho C.-C., Geng Z.Z., Khatua K. World Intellectual Property Organization; 2021. SARS-CoV-2 Main Protease Inhibitors. [Google Scholar]
- 23.Yang K.S., Ma X.R., Ma Y., Alugubelli Y.R., Scott D.A., Vatansever E.C., Drelich A.K., Sankaran B., Geng Z.Z., Blankenship L.R., Ward H.E., Sheng Y.J., Hsu J.C., Kratch K.C., Zhao B., Hayatshahi H.S., Liu J., Li P., Fierke C.A., Tseng C.K., Xu S., Liu W.R. A quick route to multiple highly potent SARS-CoV-2 main protease inhibitors. ChemMedChem. 2021;16:942–948. doi: 10.1002/cmdc.202000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai W., Jochmans D., Xie H., Yang H., Li J., Su H., Chang D., Wang J., Peng J., Zhu L., Nian Y., Hilgenfeld R., Jiang H., Chen K., Zhang L., Xu Y., Neyts J., Liu H. Design, synthesis, and biological evaluation of peptidomimetic aldehydes as broad-spectrum inhibitors against enterovirus and SARS-CoV-2. J. Med. Chem. 2022;65:2794–2808. doi: 10.1021/acs.jmedchem.0c02258. [DOI] [PubMed] [Google Scholar]
- 27.Vankadara S., Wong Y.X., Liu B., See Y.Y., Tan L.H., Tan Q.W., Wang G., Karuna R., Guo X., Tan S.T., Fong J.Y., Joy J., Chia C.S.B. A head-to-head comparison of the inhibitory activities of 15 peptidomimetic SARS-CoV-2 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2021;48 doi: 10.1016/j.bmcl.2021.128263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vuong W., Fischer C., Khan M.B., van Belkum M.J., Lamer T., Willoughby K.D., Lu J., Arutyunova E., Joyce M.A., Saffran H.A., Shields J.A., Young H.S., Nieman J.A., Tyrrell D.L., Lemieux M.J., Vederas J.C. Improved SARS-CoV-2 M(pro) inhibitors based on feline antiviral drug GC376: structural enhancements, increased solubility, and micellar studies. Eur. J. Med. Chem. 2021;222 doi: 10.1016/j.ejmech.2021.113584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C.H., Stone E.A., Deshmukh M., Ippolito J.A., Ghahremanpour M.M., Tirado-Rives J., Spasov K.A., Zhang S., Takeo Y., Kudalkar S.N., Liang Z., Isaacs F., Lindenbach B., Miller S.J., Anderson K.S., Jorgensen W.L. Potent noncovalent inhibitors of the main protease of SARS-CoV-2 from molecular sculpting of the drug perampanel guided by free energy perturbation calculations. ACS Cent. Sci. 2021;7:467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konno S., Kobayashi K., Senda M., Funai Y., Seki Y., Tamai I., Schakel L., Sakata K., Pillaiyar T., Taguchi A., Taniguchi A., Gutschow M., Muller C.E., Takeuchi K., Hirohama M., Kawaguchi A., Kojima M., Senda T., Shirasaka Y., Kamitani W., Hayashi Y. 3CL protease inhibitors with an electrophilic arylketone moiety as anti-SARS-CoV-2 agents. J. Med. Chem. 2022;65:2926–2939. doi: 10.1021/acs.jmedchem.1c00665. [DOI] [PubMed] [Google Scholar]
- 31.Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M(pro) and cathepsin L. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y., Yang K.S., Geng Z.Z., Alugubelli Y.R., Shaabani N., Vatansever E.C., Xinyu R.M., Cho C.-C., Khatua K., Xiao J., Blankenship L.R., Yu G., Sankaran B., Li P., Allen R., Ji H., Xu S., Liu W.R. A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals. Eur. J. Med. Chem. 2022 doi: 10.1016/j.ejmech.2022.114570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang K.S., Ma X.R., Ma Y., Alugubelli Y.R., Scott D.A., Vatansever E.C., Drelich A.K., Sankaran B., Geng Z.Z., Blankenship L.R., Ward H.E., Sheng Y.J., Hsu J.C., Kratch K.C., Zhao B., Hayatshahi H.S., Liu J., Li P., Fierke C.A., Tseng C.K., Xu S., Liu W.R. A quick route to multiple highly potent SARS-CoV-2 main protease inhibitors. ChemMedChem. 2021;16:942–948. doi: 10.1002/cmdc.202000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breidenbach J., Lemke C., Pillaiyar T., Schakel L., Al Hamwi G., Diett M., Gedschold R., Geiger N., Lopez V., Mirza S., Namasivayam V., Schiedel A.C., Sylvester K., Thimm D., Vielmuth C., Phuong Vu L., Zyulina M., Bodem J., Gutschow M., Muller C.E. Targeting the main protease of SARS-CoV-2: from the establishment of high throughput screening to the design of tailored inhibitors. Angew. Chem. Int. Ed. 2021;60:10423–10429. doi: 10.1002/anie.202016961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westberg M., Su Y., Zou X., Ning L., Hurst B., Tarbet B., Lin M.Z. Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases. bioRxiv. 2020;2009:2020. 2015.275891. [Google Scholar]
- 36.Ma C., Xia Z., Sacco M.D., Hu Y., Townsend J.A., Meng X., Choza J., Tan H., Jang J., Gongora M.V., Zhang X., Zhang F., Xiang Y., Marty M.T., Chen Y., Wang J. Discovery of di- and trihaloacetamides as covalent SARS-CoV-2 main protease inhibitors with high target specificity. J. Am. Chem. Soc. 2021;143:20697–20709. doi: 10.1021/jacs.1c08060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y., Fang C., Zhang Q., Zhang R., Zhao X., Duan Y., Wang H., Zhu Y., Feng L., Zhao J., Shao M., Yang X., Zhang L., Peng C., Yang K., Ma D., Rao Z., Yang H. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell. 2022;13:689–693. doi: 10.1007/s13238-021-00883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang K.S., Leeuwon S.Z., Xu S., Liu W.R. Evolutionary and structural insights about potential SARS-CoV-2 evasion of nirmatrelvir. J. Med. Chem. 2022 doi: 10.1021/acs.jmedchem.2c00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma X.R., Alugubelli Y.R., Ma Y., Vatansever E.C., Scott D.A., Qiao Y., Yu G., Xu S., Liu W.R. MPI8 is potent against SARS-CoV-2 by inhibiting dually and selectively the SARS-CoV-2 main protease and the host cathepsin L. ChemMedChem. 2022;17 doi: 10.1002/cmdc.202100456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao M.M., Yang W.L., Yang F.Y., Zhang L., Huang W.J., Hou W., Fan C.F., Jin R.H., Feng Y.M., Wang Y.C., Yang J.K. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Targeted Ther. 2021;6:134. doi: 10.1038/s41392-021-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng Y.W., Chao T.L., Li C.L., Chiu M.F., Kao H.C., Wang S.H., Pang Y.H., Lin C.H., Tsai Y.M., Lee W.H., Tao M.H., Ho T.C., Wu P.Y., Jang L.T., Chen P.J., Chang S.Y., Yeh S.H. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar P., Nagarajan A., Uchil P.D. Analysis of cell viability by the MTT assay. Cold Spring Harb. Protoc. 2018;2018:469–471. doi: 10.1101/pdb.prot095505. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Xie H., Alugubelli Y.R., Ma Y., Xu S., Ma J., Liu W.R., Liang D. vol. 15. 2022. Accurate mass identification of an interfering water adduct and strategies in development and validation of an LC-MS/MS method for quantification of MPI8, a potent SARS-CoV-2 main protease inhibitor; p. 676. (Rat Plasma in Pharmacokinetic Studies, Pharmaceuticals (Basel)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sulkowski M.S., Thomas D.L., Chaisson R.E., Moore R.D. Hepatotoxicity associated with antiretroviral therapy in adults infected with human immunodeficiency virus and the role of hepatitis C or B virus infection. JAMA. 2000;283:74–80. doi: 10.1001/jama.283.1.74. [DOI] [PubMed] [Google Scholar]
- 46.Tebas P., Powderly W.G., Claxton S., Marin D., Tantisiriwat W., Teitelbaum S.L., Yarasheski K.E. Accelerated bone mineral loss in HIV-infected patients receiving potent antiretroviral therapy. AIDS. 2000;14:F63–F67. doi: 10.1097/00002030-200003100-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nolan D., Reiss P., Mallal S. Adverse effects of antiretroviral therapy for HIV infection: a review of selected topics. Expet Opin. Drug Saf. 2005;4:201–218. doi: 10.1517/14740338.4.2.201. [DOI] [PubMed] [Google Scholar]
- 48.Henry J.A., Hill I.R. Fatal interaction between ritonavir and MDMA. Lancet. 1998;352:1751–1752. doi: 10.1016/s0140-6736(05)79824-x. [DOI] [PubMed] [Google Scholar]
- 49.Mansuri Z., Shah B., Adnan M., Chaudhari G., Jolly T. vol. 22. Care Companion CNS Disord.; 2020. (Ritonavir/Lopinavir and its Potential Interactions with Psychiatric Medications: A COVID-19 Perspective, Prim). 20com02677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.