Abstract
Simple Summary
Commercial routes are reported as the main cause of biological invasions. Particularly, naval trade may accidentally bring several species to new areas where they are not native. This is particularly evident for coastal areas, where most biological invasions occur. In our work, we reported, for the first time, the presence of the ocellated skink, native to the largest Italian islands (Sardinia, Sicily and surrounding islets in a port area of continental Central Italy). We collected several individuals of this alien population and we sampled them for molecular analyses, comparing them with those naturally occurring in Sardinia, Sicily and the Mediterranean basin, including individuals accidentally introduced to peninsular Southern Italy. Differently from what previously suggested, the nucleus in Portici (Southern Italy) may have originated from Sardinia. The intense cork trade and touristic traffic between Sardinia and Southern Tuscany may have been responsible for the introduction of this lizard also to Central Italy.
Abstract
The ocellated skink (Chalcides ocellatus) is a widespread lizard, naturally distributed between the Maghreb and coastal Pakistan, with few insular populations in the Mediterranean coastal area. Some populations of this species have also been recorded in peninsular Italy, Campania and Southern Tuscany due to accidental introductions via touristic and commercial routes. In this work, we conducted genetic analyses on mitochondrial DNA COXI, cytb and 16S mtDNA genes on a sample of Italian insular and peninsular populations. Differently from what previously suggested, the nucleus in Portici (Southern Italy) may have originated from Sardinia. The intense trade and touristic traffic between Sardinia and Southern Tuscany may have been responsible for the introduction of this lizard also to Central Italy.
Keywords: Chalcides ocellatus, mitochondrial DNA, port areas, species introduction, Reptilia
1. Introduction
Maritime transport supports 80% of the current global trade, a quarter of which crosses the Mediterranean [1,2,3]. The increasingly heavy commercial traffic has required considerable human movements and trade in goods, which have in turn promoted and encouraged the alteration of the native distribution of many animal and plant species [1,2,4]. Ports and naval trade represent one of the main pathways of introduction and hubs of alien and invasive species [2,3,5,6,7], being a strategic gate for access to the main tourist areas and routes across coastal areas. In this context, over 600 municipalities in Italy locate along the coastline, hosting 27% of the human population of Italy and constituting a nerve center for both tourism and trade. Furthermore, Italy also holds the European record for the number of alien species, including 30% of its terrestrial and freshwater vertebrates [8]. Among those, the detection of alien reptiles has increased in recent decades in coastal areas following the increased importance of reptiles as pets and, mostly, accidental introductions with the brick, plant, and lumber trade [9,10]. Although most releases fail to establish a reproductive population, as regarding only single individuals, many reptile species have successfully colonized areas where there were historically absent. Human-mediated dispersal has allowed several gecko species to reach most Italian regions even far from the coastline (e.g., Hemidactylus turcicus, Mediodactylus kotschyi, and Tarentola mauritanica) [11,12,13,14]). Two populations of Mediterranean chameleon (Chamaeleo chamaeleon) occur in Southern Italy, due to multiple introduction events [15,16]. Similarly, the Brahminy blindsnake Indotyphlops braminus is a semi-fossorial snake native to the Indo-Malayan region, able to survive also inside pots [17]. This adaptability has allowed it to disperse outside its native range through the international plant trade, with at least two populations also in Italy, one in Sicily and one on Ischia island, Campania [17,18].
The ocellated skink (Chalcides ocellatus) is a widespread lizard species naturally occurring throughout Northern Africa, Western Asia, and South-Eastern Europe [19,20]. Traditionally, the subspecies C. o. tiligugu is distributed throughout Italy, i.e., in Sicily and Sardinia [10] and Tunisia [10,21,22]. Two other subspecies from Linosa (C. o. linosae) and Lampedusa islands (C. o. zavattarii) are not genetically supported [23] and, thus, they should be considered as invalid [10]. A small population is also present in the surroundings of Portici (province of Naples), most likely as the result of an accidental introduction [24]. However, no information is available to confirm its actual origin. In spring 2021, a photo showing two individuals of ocellated skink in Tuscany, Central Italy, was posted on Facebook (Figure 1A), and solicited our attention to the origin of these animals. Other photos followed and confirmed the species identification (Figure 1B,C).
Figure 1.
(A) the first photograph documenting the ocellated skink in Tuscany (© Ugo Preziosi); (B,C) individuals of this species found in the surroundings of Piombino port (province of Livorno, Central Italy, © Matteo Riccardo Di Nicola).
Previous molecular phylogenetic inferences [19,20,21] include south-eastern Mediterranean and northern African populations, with a few samples from the two Italian major islands, Sicily and Sardinia. The recent findings of some samples from Peninsular Italy (Piombino, Livorno and Portici, Naples) allowed us to investigate their genetic relationships and to infer their origin.
2. Materials and Methods
2.1. Sample Collection
In Tuscany, samples were collected in a crop area with small vegetable gardens in the Piombino municipality, i.e., in the immediate surroundings of the port and in Torre Mozza (Environment Ministry MATTM permits: 0065971, 18 June 2021; ISPRA: Prot. 31076, 11 June 2021: Figure 2).
Figure 2.
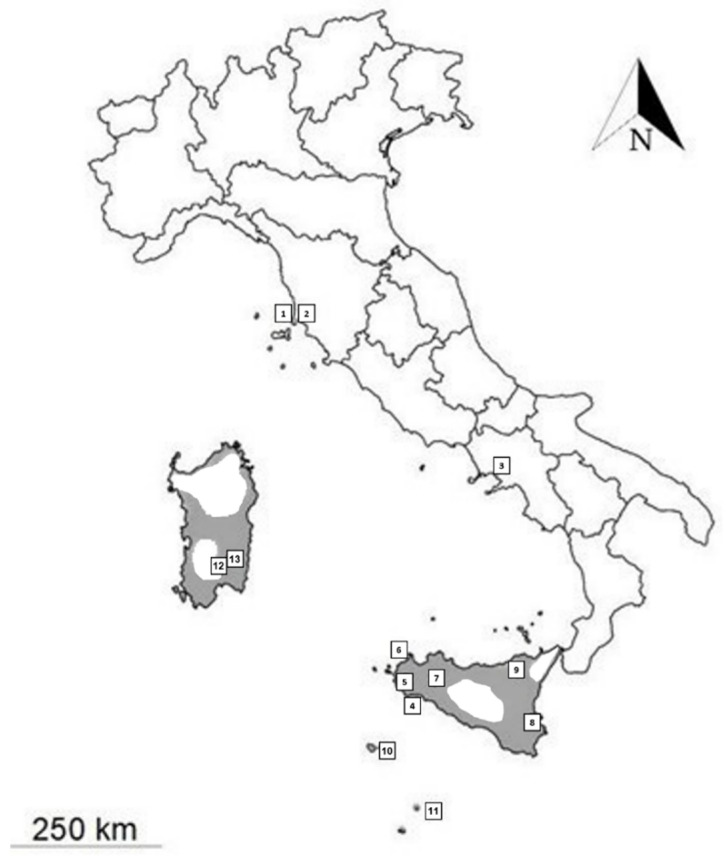
Updated distribution (gray areas) of the ocellated skink in Italy and sites of sample origin (from [10], modified): (1) Piombino; (2) Torre Mozza; (3) Portici; (4) Triscina di Selinunte; (5) Mazara del Vallo; (6) Riserva dello Zingaro; (7) Ficuzza; (8) Siracusa; (9) Rocche del Crasto; (10) Pantelleria island; (11) Linosa Island; (12) Serdiana; (13) Dolianova. One sample was analyzed from each place.
Skinks were searched in June to September 2021, following the monitoring program [25]. Four pitfall traps baited with insect larvae were also employed. They were checked twice a day to verify the presence of any animal fallen inside. Moreover, we also provided a shelter inside and a small container with water to assure survivorship. Each individual was sampled by cutting a small piece (1–2 mm) of the tail. This action does not affect skink health, since this species, like others of the same family, is particularly prone to autotomy, i.e., voluntary tail-loss allowing escaping for defense. Tissue samples were preserved in absolute ethanol before genetic analyses. The sample from Portici was derived from an individual found dead on a sidewalk. Further samples were previously collected in other localities included in the skink range by some authors (Sicily and Sicilian Islands: Giuseppe Mazza, Matteo Riccardo Di Nicola and Francesco Paolo Faraone; Sardinia: Matteo Riccardo Di Nicola and Sergio Mezzadri), and stored in absolute ethanol in museums (sample MZUT R203, Linosa) or private collections (Table 1, Figure 2).
Table 1.
Samples of Chalcides ocellatus and outgroups (C. chalcides, C. viridanus) included in our analyses, locality of collection, coordinates, amplified genes and GenBank accession numbers (COXI, 12S, cytb). *, sample MZUT R203 (Turin, Piedmont, NW Italy).
| Dataset 1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Dataset 2 | |||||||||
| Species | Region/District | Locality (Point in Figure 2) | Latitude | Longitude | Sample Label | Accession Numbers | COXI | 12S | CYTB |
| C. ocellatus | Saudi Arabia | Ash Shihyah | 26.269 | 43.597 | CH01_ARABIA | ON534012, ON534203, ON551370 | X | X | X |
| C. ocellatus | Tuscany/Livorno | Torre Mozza (2) | 42.946 | 10.693 | CH02_TORRE_MOZZA | ON534009, ON534204, ON551371 | X | X | X |
| C. ocellatus | Sicily/Trapani | Pantelleria island (10) | 36.817 | 12.003 | CH05_PANTELLERIA | ON534007, ON534202, ON551381 | X | X | X |
| C. ocellatus | Sicily/Trapani | Triscina di Selinunte (4) | 37.584 | 12.785 | CH06_SELINUNTE | ON534013, ON534206, ON551377 | X | X | X |
| C. ocellatus | Sicily/Trapani | Ficuzza (7) | 37.883 | 13.373 | CH07_FICUZZA | ON534014,ON534207, ON551375 | X | X | X |
| C. ocellatus | Sicily/Trapani | Mazara del Vallo (5) | 37.650 | 12.599 | CH08_MAZARA | ON534015, ON534208, ON551378 | X | X | X |
| C. ocellatus | Sicily/Messina | Rocche del Crasto (9) | 38.023 | 14.746 | CH09_MESSINA | ON534019, ON53420, ON551376 | X | X | X |
| C. ocellatus | Sicily/Trapani | Riserva dello Zingaro (6) | 38.081 | 12.808 | CH010_ZINGARO | ON534016, ON534209, ON551379 | X | X | X |
| C. ocellatus | Sardegna/Sud Sardegna | Dolianova (13) | 39.374 | 9.180 | CH011_SARDINIA | ON551373 | X | ||
| C. ocellatus | Tuscany/Livorno | Piombino (1) | 42.924 | 10.544 | CH015_PIOMBINO | ON534010, ON534205, ON551372 | X | X | X |
| C. ocellatus | Sicily/Agrigento | Linosa island (11) * | 35.870 | 12.862 | CH016_LINOSA | ON534017, ON534210, ON551382 | X | X | X |
| C. ocellatus | Campania/Naples | Portici (3) | 40.804 | 14.349 | CH017_PORTICI | ON534011, ON534211, ON551380 | X | X | X |
| C. ocellatus | Sicily/Siracusa | Siracusa (8) | 37.080 | 15.286 | CH018_SIRACUSA | ON534018, ON534212, ON551374 | X | X | X |
| C. ocellatus | Sardegna/Sud Sardegna | Serdiana (12) | 39.372 | 9.159 | CH019_SARDINIA | ON534008,ON534213, ON551383 | X | X | X |
| C. ocellatus | Algeria | EU278169_ALGERIA | EU278169 | X | |||||
| C. ocellatus | Morocco | EU278171_MOROCCO | EU278171 | X | |||||
| C. ocellatus | Turkey | EU278180_TURKEY | EU278180 | X | |||||
| C. ocellatus | Syria | FJ980143_SYRIA | FJ980143 | X | |||||
| C. ocellatus | Greece | FJ980268_GREECE | FJ980268 | X | |||||
| C. ocellatus | Egypt | EU278181_EGYPT | EU278181 | X | |||||
| C. ocellatus | Israel | EU278184_ISRAEL | EU278184 | X | |||||
| C. ocellatus | Tunisia | EU278194_TUNISIA | EU278194 | X | |||||
| C. ocellatus | Sardinia | EU278194_SARTIL | EU278186 | X | |||||
| C. ocellatus | Tunisia | EU278188_TUNTIL | EU278188 | X | |||||
| C. chalcides | Italy | Giglio island (Grosseto) | EU278211_CCGIGLIO | EU278211 | X | ||||
| C. chalcides | Italy | Piombino (Livorno) | EU278212_CCPIOMBINO | EU278212 | X | ||||
| C. viridanus | Spain | Canary Islands | EU278116_CVCANARY | EU278211 | X | ||||
2.2. Molecular Analyses
Samples of C. ocellatus collected in Italy were sequenced and analyzed, whereas other sequences of congeneric species (C. chalcides and C. viridanus) were employed as outgroups (Table 1). All samples were preserved in 96% ethanol and genomic DNA was extracted using QIAGEN Blood and Tissue kit (Qiagen Inc., Germantown, MD, USA), following the manufacturer’s protocol. Mitochondrial DNA sequencing PCR amplification products were obtained from three genes: the Cytochrome Oxidase I (hereafter, COXI), 12S mitochondrial gene (hereafter, 12S) and Cytochrome b (hereafter, cytb). The primers used are described in Table 2.
Table 2.
Primers used for the molecular analyses of Chalcides ocellatus.
| Target Gene | Label | Sequence 5′-3′ | Reference | Fragment Length (bp) |
|---|---|---|---|---|
| cytb | L14841 | AAAAAGCTTCCATCCAACATCTCACATGATGAAA | [26] | 325 |
| H15149 | AAACTGCAGCCCCTCAGAATGATATTTGTCCTCA | |||
| 12S | 12StPhe | AAAGCACRGCACTGAAGATGC | [27] | 350 |
| 12 g | TATCGATTATAGGACAGGCTCCTCTA | [28] | ||
| COXI | LCO1490 | GGTCAACAAATCATAAAGATATTGG | [29] | 670 |
| HCO 2198 | TAAACTTCAGGGTGACCAAAAAATCA |
The PCR profiles were the same reported by Kornilios et al. [19] for 12S and cytb; for COXI, we followed the protocol described by Baratti et al. [30]. PCR products were run on a 1.5% agarose gel purified (ExoSAP-IT, Amersham Biosciences, Amersham, UK) and sequenced with a sequencing kit (ABI Big Dye Terminator Cycle Sequencing v. 2.0- ABI PRISM, Applied Biosystems, Foster City, CA, USA). GenBank accession numbers are reported in Table 1. Electropherograms were visualized with CHROMAS 1.45 (http://www.technelysium.com.au (accessed on 9 June 2022)). The sequences were manually corrected and then aligned using CLUSTALX 1.81 [31]. In order to determine if sequences were nuclear (NUMTs [32,33]) or mitochondrial copies, we followed three steps. First, sequence chromatograms were checked for double signals. Next, coding sequence alignments were inspected for frameshift mutations and/or stop codons. Finally, the corrected sequences were compared to those in GenBank: we compared our sequences to the ones deposited in the NCBI database using BLASTx and BLASTn. We analyzed two datasets: one with sequences by cytb (Dataset 2), only because it was the gene with more sequences in GenBank and it allowed us to compare our sequences with a higher number of sequences (Table 1); all three genes constituted the other dataset (Dataset 1).
For both datasets, we carried out a phylogenetic reconstruction by Neighbor-Joining (NJ), Bayesian (BI) and a Maximum Likelihood (ML) phylogenetic analysis. Analyses were carried out using genetic models selected by jModelTest [34], with the Akaike Information Criterion (AIC). The HKY (Hasegawa–Kishino–Yano) nucleotide substitution model was selected for cytb, whilst GTR (General Time Reversible) was used for the combined dataset. Models were corrected for rate heterogeneity among sites with a Gamma (G) distribution [35]. The NJ was performed by MEGA software and 1000 bootstrap replicates. The BI analysis was conducted with MrBayes v.3.12 [36], using the best model selected by Modeltest. Four chains of Markov chain Monte Carlo (MCMC) were run simultaneously and sampled every 1000 generations for 4 million generations. The first 1000 sampled trees from each run were discarded as burn-in. The proportion of trees that contained the clade was given as the posterior probability (PP) to estimate the robustness of each clade. Branch supports were assessed by 100 non-parametric bootstrap replicates. A strict consensus tree was calculated when there was more than one tree. ML phylogenetic analysis was conducted through the SeaView software [37]. We selected the optimized choices and we obtained the tree-searching operations by Nearest-Neighbor Interchange (NNI) and Subtree Pruning–Regrafting (SPR).
3. Results
In our work, we confirmed, for the first time, the presence of a population of ocellated skink on the Tuscan coast, between the ports of Piombino and Torre Mozza. The population was confirmed to be reproductive, as two juveniles were also observed. In this area, seven adult individuals of ocellated skink (N = 4 males, 3 females) were sampled and immediately released.
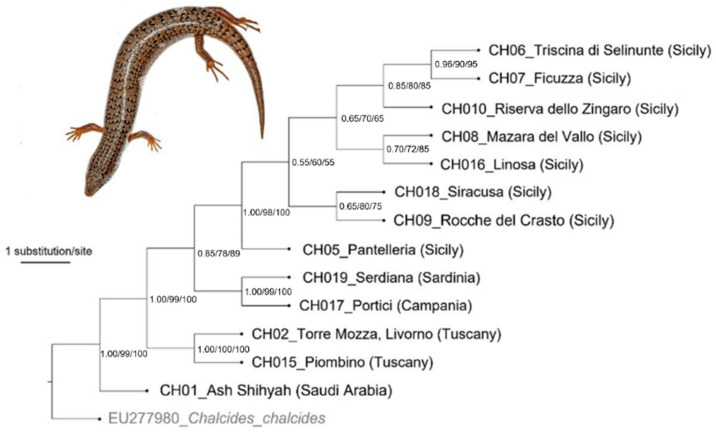
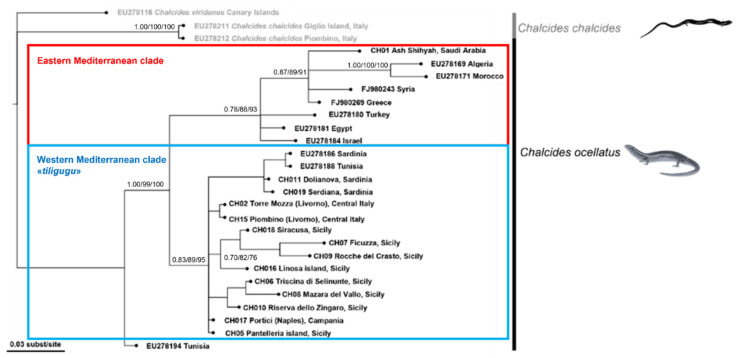
After alignment and trimming, we examined 585 base pairs for COXI, 366 base pairs for 12S and 287 base pairs for cytb. The sequences were analyzed in two datasets: the former (Figure 3), named Dataset 1, included all three genes (1238 bp), including the first COXI barcode sequences for this species, and the latter, named Dataset 2, included a higher number of sequences, but only the cytb gene portion (Figure 4).
Figure 3.
Maximum-likelihood phylogenetic tree of Chalcides ocellatus mtDNA sequences (three genes, Dataset 1). The numbers at nodes indicate Bayesian posterior probability, Maximum-Likelihood and Neighbor-Joining bootstrap values.
Figure 4.
Maximum-likelihood phylogenetic tree of Chalcides ocellatus mtDNA sequences (cytb, Dataset 2). The numbers at nodes indicate Bayesian posterior probability, Maximum-Likelihood and Neighbor-Joining bootstrap values. Main clades in the C. ocellatus group are shown.
The different phylogenetic reconstructions applied to both datasets gave similar results (Figure 3 and Figure 4). Within C. ocellatus sequences, two main clades occurred. The former included southern Mediterranean populations (Greece, Syria, Egypt, Algeria, Morocco and Saudi Arabia), except for Tunisia, which resulted as the sister clade of all the other C. ocellatus groups. The second clade was constituted by the group of Tunisian and Italian populations. However, the last group looked quite close to Sardinian populations but not to the Sicilian or other peninsular populations, even though the tree is largely not solved at the interspecies level. The Linosa sample was included in the Sicilian clade, even if the relationships among Sicilian populations were not highly supported. However, the phylogenetic relationships within different Italian populations were not well resolved. The Dataset 1 tree showed two sister groups: populations from Tuscany (Piombino and Torre Mozza, Livorno) in one group and Sicilian, Sardinian and Portici populations in the other group. Sicilian samples are grouped together and they have the Pantelleria sample as a sister group. The Portici sample looked quite close to Sardinia, whereas Linosa confirmed its position inside the Sicilian group even if the support values at nodes of this group were not high.
Sardinian populations were very close to one of the Tunisian sequences, and appeared as the only one with such high genetic affinity, as enlightened also by genetic distances (Table 3).
Table 3.
Genetic distances calculated among the studied populations of Chalcides ocellatus with cytb distances above diagonal and 12S + COXI below the diagonal.
| Torre Mozza | Piombino | Serdiana | Siracusa | Ficuzza | Rocche Crasto | Triscina Selinunte | Mazara Vallo | Riserva Zingaro | Portici | Pantelleria | Linosa | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Torre Mozza | 0.5% | 2% | 2% | 2% | 2% | 2% | 2% | 2% | 2% | 2% | 2% | |
| Piombino | 1.4% | 2% | 2% | 2% | 2% | 2% | 2% | 2% | 2% | 2% | 2% | |
| Serdiana | 4.3% | 3.6% | 0% | 0% | 0% | 0% | 0% | 0% | 2% | 0% | 0% | |
| Siracusa | 2.5% | 3.5% | 1.4% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
| Ficuzza | 4% | 4% | 2.6% | 1.6% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
| Rocche Crasto | 2.5% | 3.2% | 1.4% | 0.1% | 1.6% | 0% | 0% | 0% | 0% | 0% | 0% | |
| Triscina Selinunte | 3.2% | 2.5% | 2.1% | 0.1% | 0.5% | 1% | 0% | 0% | 0% | 0% | 0% | |
| Mazara Vallo | 2.3% | 4% | 1.2% | 0.2% | 1.4% | 1.4% | 2% | 0% | 0% | 0% | 0% | |
| Riserva Zingaro | 2% | 2% | 1.7% | 0.7% | 1% | 1% | 1% | 3% | 0% | 0% | 0% | |
| Portici | 4.3% | 3.6% | 0.2% | 1% | 2.6% | 1.8% | 2% | 4% | 1% | 0% | 0% | |
| Pantelleria | 2.5% | 2.5% | 1.7% | 1% | 1.6% | 0.7% | 2% | 0.5% | 1% | 0% | 0% | |
| Linosa | 2.5% | 2.5% | 1.2% | 0.2% | 0.9% | 1.4% | 1.4% | 1.5% | 1.3% | 2% | 2% |
4. Discussion
The ocellated skink is a widespread lizard species, with populations throughout the Mediterranean basin, up to the Middle East. The species may have evolved in Northern Africa and, afterward, expanded its range until coastal Pakistan [19]. In our work, we reported the presence of this species in a coastal area of Central Italy (Tuscany) for the first time, and we reconfirmed the presence of this species in the surroundings of Naples (Southern Italy). We also performed the first molecular analyses on both peninsular populations. The occurrence of the ocellated skink in Mediterranean islands and the lack of its presence in Corsica and the European mainland strongly suggest recent introduction events, i.e., after the Last Glacial Maximum (i.e., 18,000 years ago [38]), as is currently assumed for other Sardinian reptiles such as Testudo graeca [39], Chalcides chalcides [21], Natrix maura [40] and Hemorrhois hippocrepis [41]. Particularly, ocellated skinks may have reached Mediterranean islands through commercial routes between Europe and North Africa [19]. Carranza et al. [21] sustained that almost all the populations of C. ocellatus clade can be assigned to a single species, although they exhibit intraspecific genetic divergence with 6–8% values in cytb + 12S. Consequently, a split in different taxa must be evaluated.
We are aware of the limitation due to our small sample size, as our analysis was based on few mitochondrial DNA-only loci from a limited number of individual samples. However, we provided the first COXI barcoding sequences for this species and the first available information on the origins of the only two populations occurring in peninsular Italy; the one in Piombino is still unreported in the scientific literature.
In our analyses, the Italian populations showed very low genetic distance values, with the highest between the Tuscany coast (Torre Mozza and Piombino) and the other populations (Table 3). Sicilian and Sardinian samples always showed low values for both the COXI and 12S + cytb divergences. Maio et al. [24] suggested that the small population present in the surroundings of Portici (province of Naples) is most likely the result of an accidental introduction from Sicily. Conversely, molecular analyses suggested that it may actually represent an introduction from Sardinia. The first record of the ocellated skink in Campania dates back to 1863 in the former Royal Park of Portici (Naples). However, in this area, the population of this lizard underwent several fluctuations, with no individual intercepted between 1990 and 2014. Afterwards, at least two observations of individuals morphologically ascribed to the Mediterranean subspecies C. o. tiligugu were detected (the first one reported by [24], the second one reported in this work), suggesting that the population is not extinct. As for Tuscany, at least seven individuals, morphologically ascribed to C. o. tiligugu (i.e., showing light dorso-lateral bands running from the head to the tail root [10]) were detected through addressed surveys in 2021; the presence of juveniles provided support to local reproduction events. The presence of the ocellated skink in the immediate surroundings of the Piombino port area suggests that these individuals have been introduced from Sardinia, possibly via the cork trade between Sardinia and the Italian mainland [42]. This hypothesis is also confirmed by the fact that the vegetable gardens where ocellated skinks were detected in Piombino are located near a parking area for trucks of cork from Sardinia.
Other Sardinian species followed the same colonization pattern, e.g., the Mediterranean snakefly (Fibla maclachlani), recorded since 2005 also in the coastal part of Southern Tuscany, where it is currently expanding [43]. This species is endemic to the Mediterranean largest islands and can survive at its larval stages in cork bark [43]. The ocellated skink is quite common in Mediterranean habitats characterized by the presence of Quercus suber L. [44]. The intense trade and touristic routes between Sardinia and Piombino may thus have promoted the colonization of the Italian mainland by several Sardinian species [43,45,46]. However, despite representing an actual introduction, we suggest that impacts of C. ocellatus on the Italian mainland would not represent a threat to native biodiversity, given the low population densities [24]. If populations were to increase in size, some competition with native lizards may occur [44]. Further monitoring is necessary to determine the future increase in population size and geographic range in introduction areas.
Our molecular phylogeny did not confirm previous hypotheses on the existence of a different subspecies from Linosa island (C. o. linosae [10]), which may thus represent a different insular form of the same subspecies occurring in Sicily, thus confirming the conclusions by Stöck et al. [23]. However, sequences shown by Stöck et al. [23] were not retrieved from online genetic databases. In our analyses, the Linosa population was always inside the Sicilian group, even though the low support node values did not suggest an exact position.
5. Conclusions
Despite the wide distribution range of the ocellated skink, its scattered presence in Europe makes this species a conservation concern [25]. Thus, although representing introduced populations, the monitoring of ocellated skinks in peninsular Italy deserves future attention also in port areas.
Port areas constitute a typically perturbed anthropogenic ecosystem, widespread and highly globally interconnected. Therefore, they have considerable potential to be hubs for the diffusion of aquatic and terrestrial alien species [3,8]. The stabilization processes of new species are often reported starting from the port or circum-port areas [3]. In this context, our findings of a population of ocellated skink in a Tuscan coastal area and the confirmation of the presence of the species near an important port area in Campania provide further confirmation of the importance of ports as pathways of alien species introduction [3].
Acknowledgments
The authors would like to thank Ugo Preziosi, who recorded the ocellated skink in Tuscany for the first time and who allowed us to trap ocellated skinks on his private property. We also thank Alberto Nocciolini, who helped us in data collection. We thank three anonymous referees, who kindly improved our MS with useful comments. E. Bassett (University of Birmingham) kindly took the time to revise English language of our MS.
Author Contributions
Conceptualization, E.M., M.B. and G.B.; sample collection, B.B., F.P.F., F.A., R.B., G.M., M.R.D.N., A.V. and S.M.; methodology, M.B.; software, E.M. and M.B.; validation, M.B., F.P.F. and F.A.; formal analysis, E.M.; investigation, E.M.; resources, M.B. and M.Z.; data curation, A.V.; writing—original draft preparation, E.M., A.V. and M.B.; writing—review and editing, M.B.; visualization, F.A. and M.B.; supervision, M.B.; project administration, E.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
In Italy there are not official ethical approvals for fieldworks. We provided indications of our methods when we asked for the research and capture permits (Environment Ministry MATTM permits: 0065971, 18.06.2021; ISPRA: Prot. 31076, 11.06.202). Moreover, the actions that we made (tail cutting and pitfall trapping) did not cause particular problems to the captured animals. The tail cutting is the least invasive procedure for a skink, which is used to autotomy (we found and observed several animals without tail, i.e., likely lost in the attempt to avoid cat-predation). This is far less problematic than other methods i.e., toe-clipping. The use of pitfall trapping is a classical method for skin and other reptile study. But we added some more information, i.e the daily check and the fact that the pitfalls had shelters inside.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are included in the MS or deposited on GenBank.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Fieldwork of E.M. and A.V. was supported by Mur-FOE-Project Capitale Naturale-Task “Biodiversità”.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Seebens H. Invasion ecology: Expanding trade and the dispersal of alien species. Curr. Biol. 2019;29:120–122. doi: 10.1016/j.cub.2018.12.047. [DOI] [PubMed] [Google Scholar]
- 2.Tingley R., García-Díaz P., Arantes C.R.R., Cassey P. Integrating transport pressure data and species distribution models to estimate invasion risk for alien stowaways. Ecography. 2018;41:635–646. doi: 10.1111/ecog.02841. [DOI] [Google Scholar]
- 3.Inghilesi A.F., Mazza G., De Silva J., Francardi V., Nuccitelli L., Pennacchio F., Sinatra G., Sposimo P., Gherardi F. Valutazione dell’incidenza degli aeroporti di Fiumicino e Ciampino e del porto di Civitavecchia sull’introduzione di fauna alloctona. In: Monaco A., editor. Alieni, la Minaccia Delle Specie Alloctone per la Biodiversità del Lazio. Palombi Editori; Roma, Italia: 2014. pp. 98–115. [Google Scholar]
- 4.Turbelin A.J., Malamud B.D., Francis R.A. Mapping the global state of invasive alien species: Patterns of invasion and policy responses. Glob. Ecol. Biogeogr. 2017;26:78–92. doi: 10.1111/geb.12517. [DOI] [Google Scholar]
- 5.Hulme P.E. Trade, transport and trouble: Managing invasive species in an era of globalization. J. Anim. Ecol. 2009;46:10–18. doi: 10.1111/j.1365-2664.2008.01600.x. [DOI] [Google Scholar]
- 6.Hulme P.E. Invasion pathways at a crossroad: Policy and research challenges for managing alien species introductions. J. Anim. Ecol. 2015;52:1418–1424. doi: 10.1111/1365-2664.12470. [DOI] [Google Scholar]
- 7.van den Burg M.P., Brisbane J.L., Knapp C.R. Post-hurricane relief facilitates invasion and establishment of two invasive alien vertebrate species in the Commonwealth of Dominica, West Indies. Biol. Invasions. 2020;22:195–203. doi: 10.1007/s10530-019-02107-5. [DOI] [Google Scholar]
- 8.Bertolino S., Ancillotto L., Bartolommei P., Benassi G., Capizzi D., Gasperini S., Lucchesi M., Mori E., Scillitani L., Sozio G., et al. A framework for prioritising present and potentially invasive mammal species for a national list. NeoBiota. 2020;62:31–54. doi: 10.3897/neobiota.62.52934. [DOI] [Google Scholar]
- 9.Karnakowski W. Quarantine pests intercepted in consignments of ornamental plants imported into Poland in 1993/1998. OEPP/EPPO Bull. 1999;29:45–49. doi: 10.1111/j.1365-2338.1999.tb00792.x. [DOI] [Google Scholar]
- 10.Di Nicola M., Cavigioli L., Luiselli L., Andreone F. Anfibi & Rettili d’Italia. Edizione Aggiornata. Edizioni Belvedere; Latina, Italy: 2021. p. 576. Historia Naturae. [Google Scholar]
- 11.Mares G., Novarini N. A likely population of the alien gecko Mediodactylus kotschyi (Steindachner, 1870) in the province of Belluno (Northeastern Italian Alps) Boll. Mus. Stor. Nat. Venezia. 2020;71:83–88. [Google Scholar]
- 12.Mezzasalma M., Odierna G., Maio N., Petraccioli A., Picariello O., Guarino F.M. Habitat features and distribution of Hemidactylus turcicus and Tarentola mauritanica in Campania (southern Italy) In: Di Tizio L., Di Cerbo A.R., Di Francesco N., Cameli A., editors. Atti VIII Congresso Nazionale Societas Herpetologica Italica. Ianieri Edizioni; Pescara, Italy: 2010. pp. 129–133. [Google Scholar]
- 13.Mori E., Plebanim M. First records of Moorish gecko Tarentola mauritanica and Turkish gecko Hemidactylus turcicus (Squamata, Gekkonidae) in the Southern Metalliferous Hills, Tuscany, Italy. Atti Soc. Tosc. Sci. Nat. 2012;119:51–54. [Google Scholar]
- 14.De Fatis K.T., Di Nicola M.R., Lebech Nassling Iversen D. Prima segnalazione di Mediodactlylus kotschyi (Steindachner, 1870) per il Trentino-Alto Adige/Südtirol (Italia) (Squamata: Gekkonidae) Studi Trentini Sci. Nat. 2020;99:25–27. [Google Scholar]
- 15.Andreone F., Angelici F.M., Carlino P., Tripepi S., Crottini A. The common chameleon Chamaeleo chamaeleon in southern Italy: Evidence for allochthony of populations in Apulia and Calabria (Reptilia: Squamata: Chamaeleonidae) Ital. J. Zool. 2016;83:372–381. doi: 10.1080/11250003.2016.1186236. [DOI] [Google Scholar]
- 16.Basso R., Vannuccini M.L., Nerva L., Mazza G., Seno M., Mori E. Multiple origins of the common chameleon in southern Italy. Herpetozoa. 2019;32:11–19. doi: 10.3897/herpetozoa.32.e35611. [DOI] [Google Scholar]
- 17.Faraone F.P., Barraco L., Giacalone G., Muscarella C., Schifani E., Vecchioni L. First records of the Brahminy blind snake, Indotyphlops braminus (Daudin, 1803) (Squamata: Typhlopidae), in Italy. Herpetol. Notes. 2019;12:1225–1229. [Google Scholar]
- 18.Paolino G., Scotti R., Grano M. First detection of the “flowerpot snake” Indotyphlops braminus (Daudin, 1803) (Serpentes Typhlopidae) in Ischia (Italy): A new possible invasive species. Biodiv. J. 2019;10:321–324. doi: 10.31396/Biodiv.Jour.2019.10.4.321.324. [DOI] [Google Scholar]
- 19.Kornilios P., Kyriazi P., Poulakakis N., Kumlutaş Y., Ilgaz Ç., Mylonas M., Lymberakis P. Phylogeography of the ocellated skink Chalcides ocellatus (Squamata, Scincidae), with the use of mtDNA sequences: A hitch-hiker’s guide to the Mediterranean. Mol. Phylogenet. Evol. 2010;54:445–456. doi: 10.1016/j.ympev.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Lavin B.R., Papenfuss T.J. The phylogenetic position of Chalcides ocellatus (Squamata: Scincidae) from Yemen and Somalia. Zootaxa. 2012;3221:26–36. doi: 10.11646/zootaxa.3221.1.2. [DOI] [Google Scholar]
- 21.Carranza S., Arnold E.N., Geniez P., Roca J., Mateo J.A. Radiation, multipledispersal and parallelism in the skinks, Chalcides and Sphenops (Squamata: Scincidae), with comments on Scincus and Scincopus the age of the Sahara Desert. Mol. Phylogenet. Evol. 2008;46:1071–1094. doi: 10.1016/j.ympev.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 22.Sindaco R., Jeremčenko V.K. The Reptiles of the Western Palearctic. Volume 1 Edizioni Belvedere; Latina, Italy: 2008. [Google Scholar]
- 23.Stöck M., Grifoni G., Armor N., Scheidt U., Sicilia A., Novarini N. On the origin of the recent herpetofauna of Sicily: Comparative phylogeography using homologous mitochondrial and nuclear genes. Zool. Anz. 2016;261:70–81. doi: 10.1016/j.jcz.2015.10.005. [DOI] [Google Scholar]
- 24.Maio N., Mancini D., Mezzasalma M., Odierna G., Petraccioli A., Nugnes F., Picariello O.L.A., Guarino F.M. Extinct or not extinct: The case of the Chalcides ocellatus (Squamata: Scincidae) population from the Park of the ex Bourbonic Royal Palace of Portici (Naples, Italy) Nat. Hist. Sci. 2015;2:43–54. [Google Scholar]
- 25.Stoch F., Genovesi P. Manuali per il Monitoraggio di Specie e Habitat di Interesse Comunitario (Direttiva 92/43/CEE) in Italia: Specie Animali. ISPRA Publisher; Roma, Italy: 2016. Serie Manuali e Linee Guida, 141/2016. [Google Scholar]
- 26.Kocher T.D., Thomas W.K., Meyer A., Edwards S.V., Pääbo S., Villablanca F.X., Wilson A.C. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Nat. Acad. Sci. USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiens J.J., Reeder T.W. Phylogeny of the spiny lizards (Sceloporus) based on molecular and morphological evidence. Herpetol. Monogr. 1997;11:1–101. doi: 10.2307/1467007. [DOI] [Google Scholar]
- 28.Leaché A.D., Reeder T.W. Molecular systematics of the eastern fence lizard (Sceloporus undulatus): A comparison of parsimony, likelihood, and Bayesian approaches. Syst. Biol. 2002;51:44–68. doi: 10.1080/106351502753475871. [DOI] [PubMed] [Google Scholar]
- 29.Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 30.Baratti M., Goti E., Messana G. High level of genetic differentiation in the marine isopod Sphaeroma terebrans (Crustacea Isopoda Sphaeromatidae) as inferred by mitochondrial DNA analysis. J. Exp. Mar. Biol. Ecol. 2005;315:225–234. doi: 10.1016/j.jembe.2004.09.020. [DOI] [Google Scholar]
- 31.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song H., Buhay J.E., Whiting M.F., Crandall K.A. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Nat. Acad. Sci. USA. 2008;105:13486–13491. doi: 10.1073/pnas.0803076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buhay J.E. COI-like sequences are becoming problematic in molecular systematic and DNA barcoding studies. J. Crustacean Biol. 2009;29:96–110. doi: 10.1651/08-3020.1. [DOI] [Google Scholar]
- 34.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Meth. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 36.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 37.Gouy M., Guindon S., Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 38.Shackleton J.C., Van Andel T.H., Runnels C.N. Coastal paleogeography of the central and western Mediterranean during the last 125,000 years and its archaeological implications. J. Field Archaeol. 1984;11:307–314. [Google Scholar]
- 39.Fritz U., Harris D.J., Fahd S., Rouag R., Martínez E.G., Casalduero A.G., Široký P., Kalboussi M., Jdeidi T.B., Hundsdörfer A.K. Mitochondrial phylogeography of Testudo graeca in the Western Mediterranean: Old complex divergence in North Africa and recent arrival in Europe. Amphibia-Reptilia. 2009;30:63–80. doi: 10.1163/156853809787392702. [DOI] [Google Scholar]
- 40.Guicking D., Joger U., Wink M. Molecular phylogeography of the viperine snake Natrix maura (Serpentes: Colubridae): Evidence for strong intraspecific differentiation. Org. Divers. Evol. 2008;8:130–145. doi: 10.1016/j.ode.2007.05.001. [DOI] [Google Scholar]
- 41.Faraone F.P., Melfi R., Di Nicola M.R., Giacalone G., Lo Valvo M. Phylogenetic relationships of the Italian populations of Horseshoe Whip Snake Hemorrhois hippocrepis (Serpentes, Colubridae) Acta Herpetol. 2020;15:129–135. [Google Scholar]
- 42.Corona P., Quatrini V., Schirru M., Dettori S., Puletti N. Towards the economic valuation of ecosystem production from cork oak forests in Sardinia (Italy) iForest-Biogeosci. For. 2018;11:660. doi: 10.3832/ifor2558-011. [DOI] [Google Scholar]
- 43.Pantaleoni R.A., Cocco A., Floris I., Letardi A., Loru L. Going overseas: From island to continent colonization in the Mediterranean snakefly Fibla maclachlani (Albarda, 1891) BioInvasions Rec. 2019;8:442–451. doi: 10.3391/bir.2019.8.2.27. [DOI] [Google Scholar]
- 44.Capula M., Luiselli L. Resource partitioning in a Mediterranean lizard community. Ital. J. Zool. 1994;61:173–177. doi: 10.1080/11250009409355879. [DOI] [Google Scholar]
- 45.Casale A., Bastianini M., Minniti M. Sulla presenza in Toscana di Carabus Macrothorax morbillosus Fabricius (Coleoptera, Carabidae, Carabini) e sul suo significato zoogeografico. Frustula Entomol. 1987;10:67–72. [Google Scholar]
- 46.Trozzi C., Vaccaro R., Nicolo L. Air pollutants emissions estimate from maritime traffic in the Italian harbours of Venice and Piombino. Sci. Total Environ. 1995;169:257–263. doi: 10.1016/0048-9697(95)04656-L. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are included in the MS or deposited on GenBank.