Abstract
Activation of integrins by Mn2+ is a benchmark in the integrin field, but how Mn2+ works and whether it reproduces physiological activation is unknown. We show that Mn2+ and high Mg2+ concentrations compete with Ca2+ at the ADMIDAS and shift the conformational equilibrium toward the open state, but the shift is far from complete. Additionally, replacement of Mg2+ by Mn2+ at the MIDAS increases the intrinsic affinities of both the high-affinity open and low-affinity closed states of integrins, in agreement with stronger binding of Mn2+ than Mg2+ to oxygen atoms. Mutation of the ADMIDAS increases the affinity of closed states and decreases the affinity of the open state and thus reduces the difference in affinity between the open and closed states. An important biological function of the ADMIDAS may be to stabilize integrins in highly discrete states, so that when integrins support cell adhesion and migration, their high and low affinity correspond to discrete on and off states, respectively.
INTRODUCTION
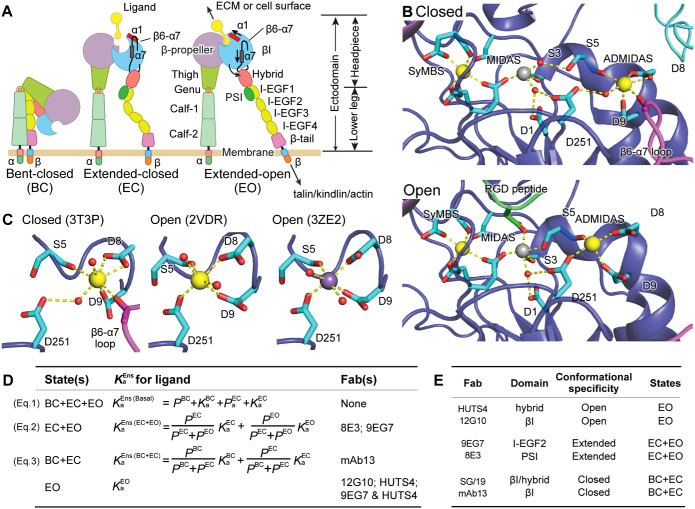
Integrins are large, αβ heterodimeric membrane receptor glycoproteins that bind to the extracellular matrix (ECM) and the surfaces of other cells and transmit signals and forces between the extracellular environment and the intracellular cytoskeleton. Their α- and β-subunits associate in a large, ligand-binding head and connect to transmembrane domains through multiple leg domains (Figure 1A). Most integrins, including α5β1, exist as ensembles with three distinct conformations: bent-closed (BC), extended-closed (EC), and extended-open (EO; Springer and Dustin, 2012). Integrin affinity for extracellular ligands is tightly controlled by association with the cytoskeleton, which applies tensile force through ligand-bound integrins that stabilizes the high-affinity EO conformation (Li and Springer, 2017; Kechagia et al., 2019; Sun et al., 2019). The three integrin conformations interconvert by two distinct motions. (1) Change between the bent and extended conformations involves bending between the upper and lower legs in each subunit, that is, between thigh and calf-1 in the α-subunit and between I-EGF1 and I-EGF2 in the β-subunit. (2) “Opening” of the ligand-binding site, that is, activation to its high-affinity state, is achieved by conformational change within the β-subunit βI domain that is linked to a change in orientation at the βI domain–hybrid domain interface. The βI domain α1 and α7 helices each shift, with movement of the β6–α7 loop away from the active site and the β1–α1 loop toward the ligand-binding site (Figure 1, A and B). These movements are linked to a change in orientation at the interface between the βI and hybrid domains (Takagi et al., 2002; Xiao et al., 2004; Luo et al., 2007; Springer and Dustin, 2012; Su et al., 2016; Dong et al., 2017; Arimori et al., 2021; Schumacher et al., 2021).
FIGURE 1:
Integrin conformational states and their equilibria. (A) Domain organization and conformational states of integrin α5β1. The α1 helix, α7 helix, and β6–α7 loop that move during opening of the βI domain are labeled. ECM: extracellular matrix. (B) Details around the βI domain metal ion binding sites of closed and liganded, open conformations. (C) The coordination number of the ADMIDAS in the closed state is 7 and in the open state is 6. B and C use integrin αIIbβ3, for which high-resolution closed and open structures are available. Closed: PDB 3T3P (Zhu et al., 2012), open: PDB 2VDR (Springer et al., 2008) and PDB 3ZE2, chain D with a soaked-in Mn2+ ion (Zhu et al., 2013). Physiologically, the SyMBS and ADMIDAS are occupied by Ca2+ (gold spheres), while the MIDAS is occupied by Mg2+ (silver sphere). Mn2+ is shown as a violet sphere. β-MIDAS residues (D1-S3-S5-D8-D9) and other coordinating residues are shown with cyan carbons in stick, except that the β6–α7 loop and its carbonyl are shown in magenta. The RGD Asp is shown as green. Oxygens are shown in red. Small red spheres are water molecules, except that one oxygen in 3ZE2 is from a glycerol molecule used as a cryoprotectant. (D) Equation 1 relates the basal ensemble affinity 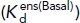 to the affinities of the BC
to the affinities of the BC 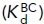 , EC
, EC 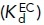 , and EO
, and EO 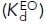 states with the populations of these states (PBC, PEC, and PEO, respectively). Equations 2 and 3 show how the affinity of the extended
states with the populations of these states (PBC, PEC, and PEO, respectively). Equations 2 and 3 show how the affinity of the extended 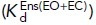 and closed
and closed 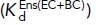 ensemble is described (Li et al., 2017). Listed to the right are the Fabs that are specific for and stabilize these states or ensembles. (E) The domain and conformational specificities of the Fabs used here.
ensemble is described (Li et al., 2017). Listed to the right are the Fabs that are specific for and stabilize these states or ensembles. (E) The domain and conformational specificities of the Fabs used here.
The integrin βI domain has a unique arrangement of three closely spaced metal-ion binding sites, the metal ion–dependent adhesion site (MIDAS), the site adjacent to MIDAS (ADMIDAS), and the synergistic metal-ion binding site (SyMBS) (Figure 1B). Under physiological conditions (∼1 mM Mg2+/∼1 mM Ca2+), the central MIDAS is occupied by Mg2+, while the neighboring SyMBS and ADMIDAS bind Ca2+ ions (Xiong et al., 2001; Xiao et al., 2004). The MIDAS metal ion coordinates the Asp or Glu sidechain, the only invariant feature shared by all integrin ligands. The integrin α5β1 ligand fibronectin contains an Arg–Gly–Asp (RGD) motif that binds through its Asp sidechain to the MIDAS Mg2+ ion. The Mg2+ ion and Asp oxygen electron orbitals overlap, and thus their bond is partially covalent, particularly strong, and more important than the Arg residue of RGD for affinity (Xiao et al., 2004; Zhu et al., 2013; Lin et al., 2016). The nearby Ca2+ ions in the ADMIDAS and SyMBS regulate ligand-binding affinity. Studies with multiple integrins have shown that low, ∼50 µM concentrations of Ca2+ enhance binding and higher (1–10)-mM concentrations of Ca2+ inhibit binding (Marlin and Springer, 1987; Staatz et al., 1989; Dransfield et al., 1992; Mould et al., 1995a; Hu et al., 1996; Labadia et al., 1998; Chen et al., 2003, 2006; Valdramidou et al., 2008). Mutational studies show that the SyMBS is the site at which low concentrations of Ca2+ enhance binding (Chen et al., 2003; Chen et al., 2006; Valdramidou et al., 2008). Mn2+ has the remarkable ability to substantially enhance integrin binding (Gailit and Ruoslahti, 1988) and has been shown to replace Ca2+ and Mg2+ at the SyMBS, MIDAS, and ADMIDAS in crystal structures (Xiong et al., 2002; Zhu et al., 2013). Mn2+ competes with the inhibitory effect of Ca2+ and thus part of its effect may occur at the ADMIDAS (Chen et al., 2003; Mould et al., 2003a). α4β7 is an unusual integrin that mediates rolling adhesion as well as firm adhesion of leukocytes on vessel walls in vascular flow in Ca2+ and Mg2+. In Ca2+, α4β7 mediates rolling adhesion, whereas in Mg2+ and Mn2+, it mediates firm adhesion. When ADMIDAS-coordinating residues are mutated, firm adhesion is abolished, but vigorous rolling adhesion is seen in Ca2+, Mg2+, and Mn2+ (Chen et al., 2003).
Mould and Humphries have highlighted regulatory roles for the ADMIDAS and Mn2+ in integrin α5β1 (Mould et al., 2002, 2003a, 2003b). Mutational removal of either of the two Asp sidechains that coordinate the ADMIDAS metal ion reduced ligand binding affinity by 7- and 2.5-fold for Asp-137 and Asp-138, respectively, and decreased the exposure of two epitopes in the hybrid domain that report headpiece opening, 15/7 and HUTS4, suggesting that the ADMIDAS was important in regulating or relaying conformational change between the βI and hybrid domains. Consistent with crystal structures (Xiong et al., 2002), Mould and Humphries also inferred binding of Mn2+ at the MIDAS from the effect of MIDAS mutations on the binding of the 12G10 mAb to a nearby epitope that reports the open, high-affinity conformation of the βI domain.
Since the discovery of its enhancing properties (Gailit and Ruoslahti, 1988), Mn2+ has become a mainstay in integrin publications, particularly because of the relatively low affinity of integrins for ligands. However, how Mn2+ works is largely unknown. Mn2+ has been viewed on one hand as a positive control for a fully activated integrin, as fully opening integrins and maintaining integrins in a high-affinity conformation, or on the other hand as shifting integrins toward the high-affinity conformation.
Here, we address many important remaining questions about how Mn2+ and the ADMIDAS regulate integrin affinity, using well-studied integrin α5β1 as our model. We have defined the nature of Mn2+-induced integrin activation using recently developed methods that allow integrins to be stabilized in specific conformational states using conformation-specific antibody Fab fragments (Li et al., 2017; Li and Springer, 2018; Figure 1, D and E). These methods allow the affinity of any ensemble of integrin states to be deconvolved into two components: 1) the intrinsic affinity of each integrin conformational state for the particular integrin ligand, and 2) the population in the ensemble of each conformational state (Figure 1D). Surprisingly, we find that Mn2+ both markedly increases the intrinsic affinity of the α5β1 EO state for the ligand and also increases the population of the EO state within integrin ensembles on the cell surface and integrin ectodomain fragment ensembles in solution; however, the EO state is not completely populated. Thus Mn2+ shifts conformational ensembles but does not do so to completion. Our study also reveals an important contribution of the ADMIDAS to enhancing the large separation in affinity between the low-affinity BC and EC states and the high-affinity EO state and enhancing the fidelity of these states.
RESULTS
Influence of cations on intrinsic-ligand binding affinity and conformational equilibrium of the soluble α5β1 ectodomain
Most experiments in this investigation used highly purified wild-type or mutant α5β1 ectodomain fragments with high-mannose N-glycans (Figure 2A). Integrin stability was adversely affected by removal of metal ions, and therefore we stored α5β1 at 20- to 100-µM concentrations (4 to 20 mg/ml) flash frozen in Tris-buffered saline (TBS) containing 1 mM CaCl2 and 1 mM MgCl2 and typically diluted it more than 1000-fold in assays so that residual Mg2+ and Ca2+ concentrations were <1 μM (Materials and Methods); however, Ca2+ is present in laboratory deionized water at concentrations of ∼5–10 μM. Fluorescence polarization (FP) of FITC-labeled fibronectin-mimetic peptides was used with integrin ectodomains to titrate metal ions (Figure 2B) or conformation-specific Fab fragments or measure ligand-binding affinities. In FP assays, when a small fluorescent ligand binds to integrin, the FP signal increases because the integrin–ligand complex rotates more slowly than the free ligand and fluorescence emission retains a polarization closer to its excitation polarization. Thus, the FP value is directly related to the fraction of ligand that is bound to integrin. In an ensemble, FP is a function of 1) the intrinsic affinity of each conformational state for the ligand and 2) the population of each state in the ensemble (Figure 1D).
FIGURE 2:
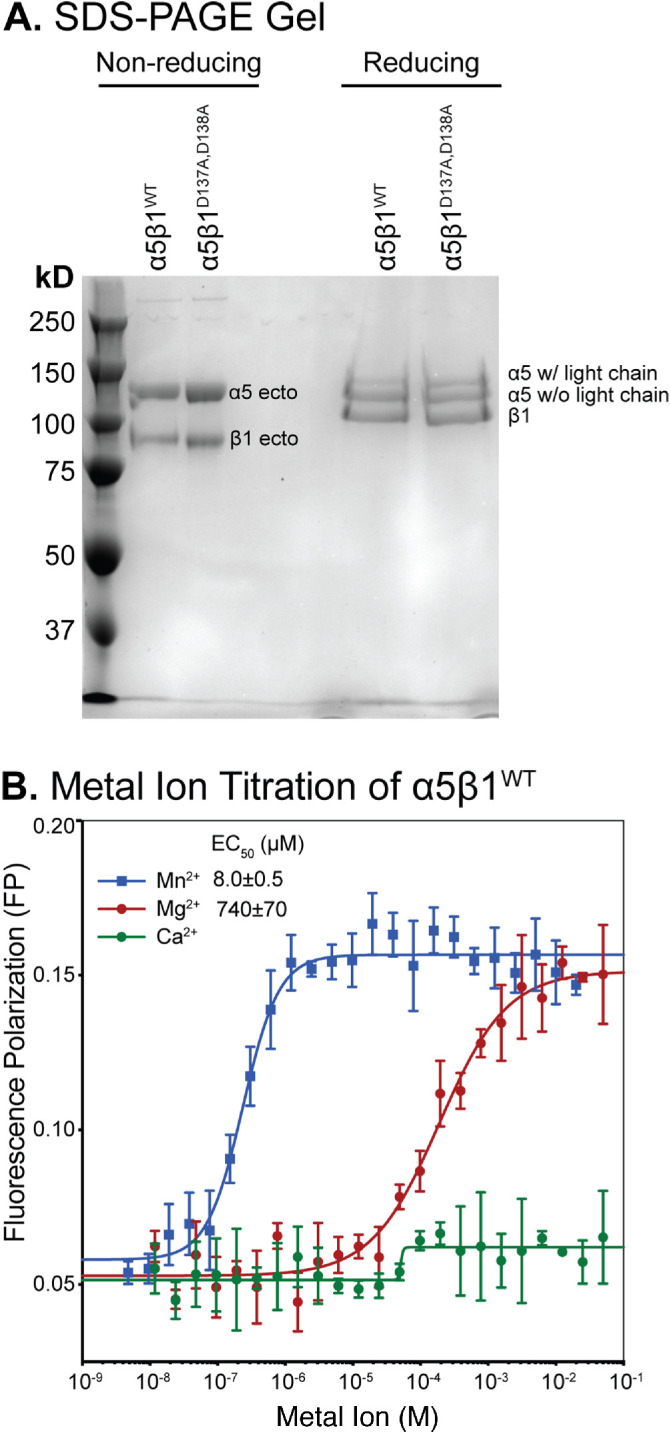
Integrin preparations and metal ion effects on α5β1WT. (A) SDS–PAGE (7.5% polyacrylamide) of α5β1WT and α5β1D137A, D138A under nonreducing and reducing conditions. The α5 chain is divided into two disulfide-linked fragments by furin cleavage. (B) FP assay showing the effect of metal ions on the binding of α5β1WT (20 nM) to FITC-cRGD peptide (5 nM) in pH 7.4 Tris-buffered saline. Titrations were done in triplicate.
Accuracy of FP is dependent on the affinity of the ligand for the integrin or mutant under the particular metal-ion conditions used and on binding sufficient ligand to have a good FP signal change while avoiding depleting free integrin at low integrin concentrations where integrin affinity is high. Therefore, we have used FITC-labeled fibronectin-mimetic peptides that bind to α5β1 with lower affinity, GRGDSPK (RGD); or higher affinity, disulfide-cyclized ACRGDGWCG (cRGD; Koivunen et al., 1995).
Metal-ion titrations with constant concentrations of ligand and α5β1 ectodomain revealed that Mn2+ and Mg2+ supported FITC-cRGD binding to α5β1 with EC50 values of 8 and 740 µM, respectively (Figure 2B). Above the EC50 values, the FP signal for Mn2+ was slightly higher than that for Mg2+, consistent with the higher ligand-binding affinity of α5β1 in Mn2+ than in Mg2+, as shown below. In contrast, the FP value in Ca2+ remained low up to 100 mM, showing that Ca2+ is insufficient for ligand binding. However, we saw a slight increase in FP in Ca2+ titrations above ∼50 µM, consistent with the low residual Mg2+ concentration from the purified integrin preparation and synergy between low concentrations of Ca2+ and Mg2+ for supporting integrin–ligand binding (Marlin and Springer, 1987; Mould et al., 1995a; Labadia et al., 1998; Chen et al., 2003; Valdramidou et al., 2008). In further experiments, we used 2 mM Mn2+ and 50 mM Mg2+, which were ∼100-fold above their EC50 values and thus sufficient to saturate the affinity-responsive metal ion–binding site(s) of α5β1.
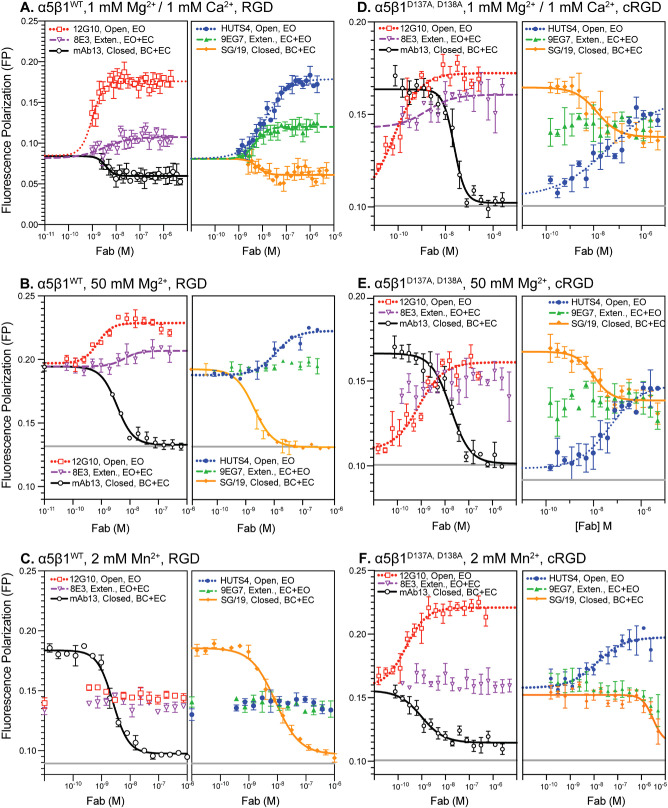
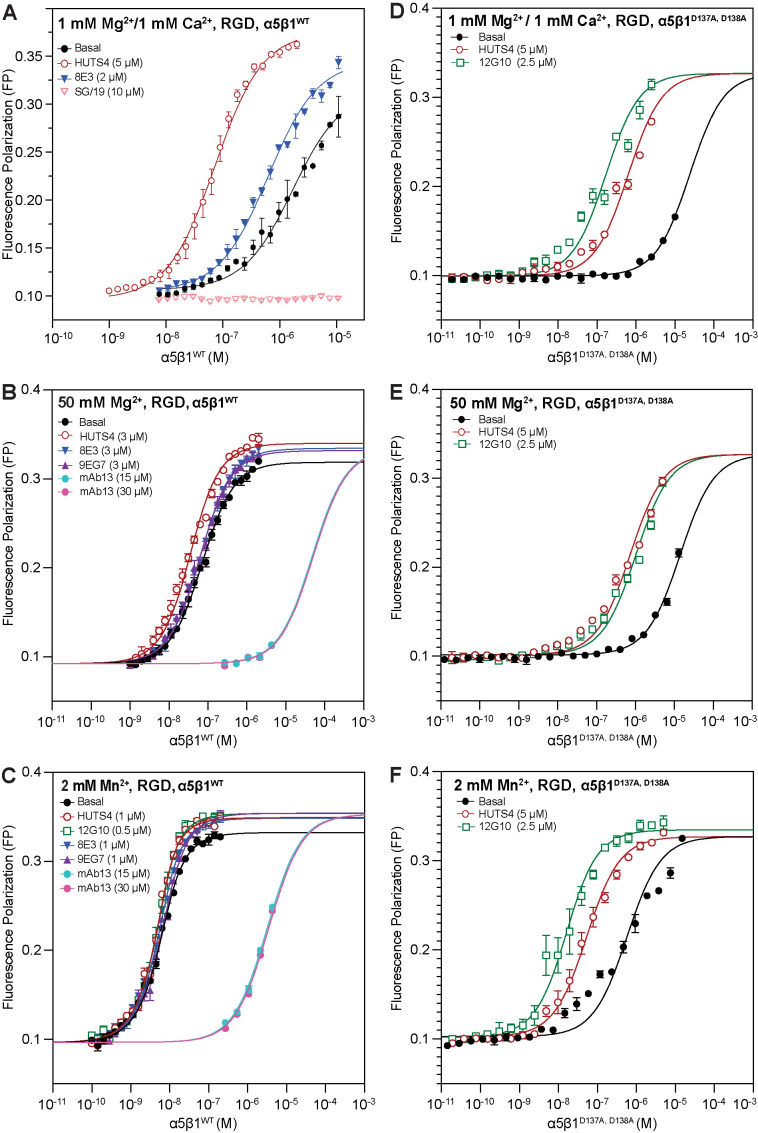
The concentration of a conformation-specific Fab required to stabilize saturably an integrin in a specific conformational state is directly related to the free energy of that state relative to the other states; higher-energy states are less populated and require higher Fab concentrations to stabilize. Therefore, we determined EC50 values of Fab required to alter ligand binding affinity in 50 mM Mg2+ and 2 mM Mn2+ (Figure 3, A–C, and Supplemental Table S1) to supplement values previously determined in 1 mM Mg2+/1 mM Ca2+ (Li et al., 2017). Based on these results, concentrations of conformation-specific Fabs at least 40- to 100-fold higher than their EC50 values were used in subsequent experiments to stabilize specific conformation(s) saturably.
FIGURE 3:
Impact of increasing Fab concentration on the binding of FITC-RGD peptide for α5β1WT and α5β1D137A, D138A ectodomains. To determine saturating concentrations of Fabs for each metal ion condition, used later in experiments to measure the intrinsic affinities of the extended open state and extended and closed ensembles, Fabs were titrated in the presence of a constant concentration of (A–C) α5β1WT or (D–F) α5β1D137A, D138A ectodomain and RGD ligand (FITC-RGD for α5β1WT and FITC-cRGD for α5β1D137A, D138A). EC50 values for each titration are summarized in Supplemental Table S1. Concentrations of integrin and ligand used in each titration are summarized in Supplemental Table S2. Fab titrations of α5β1WT ectodomain under physiological conditions (A, 1 mM Mg2+/1 mM Ca2+) were previously reported (Li et al., 2017). Fitting curves are not shown for datasets where the fitting error (SE) is comparable to or greater than the fitted value of the parameter. Titrations were done in triplicate.
We next used FP to measure the affinities of different conformational states of the wild-type α5β1 ectodomain for FITC-RGD in 50 mM Mg2+ and 2 mM Mn2+ and compared the results with previous measurements in 1 mM Mg2+/1 mM Ca2+ (Li et al., 2017). In 50 mM Mg2+ and 2 mM Mn2+, basal affinity for RGD was much higher than in 1 mM Mg2+/Ca2+ (Figure 4, A–C, and Table 1). Furthermore, whereas Fabs that stabilized the extended and open states greatly increased the affinity of α5β1 ectodomain in 1 mM Mg2+/1 mM Ca2+ (Figure 4A), they increased affinity much less in 50 mM Mg2+ and 2 mM Mn2+ (Figure 4, B and C). These results suggested that the EO state was much more populated in 50 mM Mg2+ and 2 mM Mn2+ than in 1 mM Mg2+/1 mM Ca2+. The affinity of the closed states, BC+EC, was much lower, as shown with mAb13 (Figure 4, A–C). To ensure that the concentration of mAb13 was high enough to saturably stabilize the closed states, two different mAb13 Fab concentrations were tested (Figure 4, B and C). The very similar results confirmed that saturation was achieved. We were unable to express enough α5β1 ectodomain to use more than 10-µM concentrations (2 mg/ml) and therefore could not reach maximal ligand binding for all states in Figure 4. However, in each divalent cation condition, some integrin/Fab combinations gave maximal binding. By sharing the maximum value for all conditions in each panel in Figure 4, we achieved fit errors that were well below fit values (Table 1).
FIGURE 4:
Ensemble and intrinsic affinities of α5β1WT and α5β1D137A, D138A ectodomains for FITC-RGD. Basal ensemble and intrinsic affinities of both α5β1WT (A–C) and α5β1D137A, D138A ectodomains (D–F) for FITC-RGD were determined in the presence of 1 mM Mg2+/1 mM Ca2+, 50 mM Mg2+, and 2 mM Mn2+. Intrinsic affinities of the open (EO, 12G10 and HUTS4), extended (EO+EC, 9EG7 and 8E3), and closed (EC+BC, mAb13) ensembles were determined by titrating integrin in the presence of FITC-RGD (5 nM) and a saturating concentration of Fab. The concentration of Fab used in each titration are shown in each panel. The titrations of α5β1WT ectodomain in 1 mM Mg2+/1 mM Ca2+ (panel A) were previously reported in Figure 4A in Li et al. (2017). All assays also contain pH 7.4, 20 mM Tris, and 150 mM NaCl. Fitted affinities (Kd) are reported in Supplemental Table S3. The titrations shown were done in triplicate, and basal ensemble affinities (panels of B and C) were measured three times (averaged in Table 1).
TABLE 1:
α5β1 ectodomain ensemble affinities and populations.a
| Ligand | α5β1 WT | α5β1 D137A, D138A | |||||
|---|---|---|---|---|---|---|---|
| 1 mM Mg2+, 1 mM Ca2+ | 50 mM Mg2+ | 2 mM Mn2+ | 1 mM Mg2+,1 mM Ca2+ | 50 mM Mg2+ | 2 mM Mn2+ | ||
| RGD |
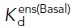 (nM) (nM) |
2,300 ± 800b | 63 ± 4c | 3.9 ± 0.6c | 24000 ± 3600f | 14000 ± 1300f | 580 ± 62f |
| RGD |
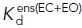 (nM) (nM) |
620 ± 50b | 54 ± 3f,i | 2.8 ± 0.3f,i | — | — | — |
| RGD |
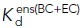 (nM) (nM) |
≥220,000b,e | 49,000 ± 9000f,j | 3,300 ± 130f,j | — | — | — |
| RGD |
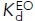 (nM) (nM) |
71 ± 5b | 26 ± 2f,k | 1.6 ± 0.2f,k | 170 ± 14f,l | 1000 ± 76f,l | 14 ± 2f,l |
| RGD |
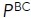 (%) (%) |
73.2 ± 9.6b | 14.3 ± 7.2 | 28.2 ± 13.5 | — | — | — |
| RGD |
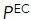 (%) (%) |
23.7 ± 8.7b | 44.5 ± 6.4 | 30.8 ± 10.4 | — | — | — |
| RGD |
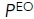 (%) (%) |
3.1 ± 1.1b | 41.2 ± 4.1 | 41.0 ± 8.1 | — | — | — |
| RGD |
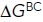 (kcal/mol) (kcal/mol) |
–1.8 ± 0.3b | 0.62 ± 0.3 | 0.22 ± 0.4 | — | — | — |
| RGD |
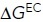 (kcal/mol) (kcal/mol) |
–1.2 ± 0.1b | –0.04 ± 0.1 | 0.17 ± 0.2 | — | — | — |
| cRGD |
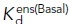 (nM) (nM) |
47 ± 3b | 1.6 ± 0.1g | 0.20 ± 0.05g | 65 ± 49d | 76 ± 4c | 2.5 ± 2c |
| cRGD |
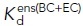 (nM) (nM) |
7000 ± 3400b | 1500 ± 200g | 46 ± 5g | 770 ± 150 h | 1200 ± 13 h,j | 14 ± 0.7h,j |
aValues are ± fitting errors, unless footnotedc,d.
bPreviously reported in Figure 4A of (Li et al., 2017).
cAverage of three experiments. Errors (±) are standard deviations between independent experiments.
dAverage of five experiments. Errors (±) are standard deviations between independent experiments.
eThe value for 220,000 nM was estimated from the intrinsic affinity fold difference between the BC+EC and the EO states for multiple RGD ligands. The value for 600,000 nM was estimated from the sensitivity of the RGD binding assay in Figure 4A of (Li et al., 2017).
fTitrations in Figure 4.
gTitrations in Supplemental Figure S1.
hTitrations in Supplemental Figure S5.
iAffinity averaged from titrations in the presence of 9EG7 and 8E3.
jAffinity averaged from titrations in the presence of mAb13.
kAffinity averaged from titrations in the presence of 12G10 and HUTS4.
lAffinity from titrations in the presence of 12G10.
The calculated results of these measurements of the affinity of wild-type α5β1 ectodomain for RGD under different metal-ion conditions showed that Mn2+ did not completely stabilize the high-affinity integrin state (Table 1, top left). The affinity for RGD of the basal ensemble (with all three conformational states) increased 600-fold from 2300 nM in 1-mM Mg2+/1 mM Ca2+ to 3.9 nM in 2-mM Mn2+. However, affinity was increased by two distinct mechanisms: an increase in the population of the EO state and an increase in its intrinsic affinity. The population of the EO state of the α5β1 ectodomain increased from 3.1% in 1 mM Mg2+/1 mM Ca2+ to 41% in 2 mM Mn2+. Furthermore, the EO state intrinsic affinity increased from 71 nM in 1 mM Mg2+/1 mM Ca2+ to 1.6 nM in 2 mM Mn2+. Although the results with 50 mM Mg2+ were largely intermediate between those with 1 mM Mg2+/1 mM Ca2+ and 2 mM Mn2+, there was an interesting difference between the effects on population and intrinsic affinity. The population of the EO state was almost identical in 50 mM Mg2+ and 2 mM Mn2+; however, the intrinsic affinity in 50 mM Mg2+ differed less from that in 1 mM Mg2+/1 mM Ca2+ (2.7-fold) than that in 2 mM Mn2+ (16-fold). Thus, while Mn2+ shifted only a portion of α5β1 ectodomain molecules to the EO state, the large increase in intrinsic affinity resulted in supraphysiologic activation. While shifting the entire integrin population in 1 mM Mg2+/1 mM Ca2+ to the EO state would result in an affinity of 71 nM, the basal integrin ectodomain ensemble in 2 mM Mn2+ has a substantially higher affinity of 3.9 nM.
Previous results on the α5β1 ectodomain showed that its BC and EC states have identical ligand-binding affinities (Li et al., 2017; Li and Springer, 2018), and therefore we measured affinities for the combined BC and EC states here (Table 1, top left). The intrinsic affinity of the BC+EC states also increased in 50 mM Mg2+ and further increased in 2 mM Mn2+. To buttress these results, and for direct comparisons to measurements on the ADMIDAS mutant, α5β1D137A, D138A, later in this manuscript, we also measured affinities of wild-type α5β1 for FITC-cyclic RGD (cRGD; Table 1, bottom left, Supplemental Figure S1). The results confirmed that 50 mM Mg2+ and 2 mM Mn2+ increased the affinities for cRGD of the basal ensemble and the closed BC+EC states (Supplemental Figure S1). Affinities were well determined for low-affinity measurements on the basal ensemble and on the BC+EC states and less well determined for the high-affinity EO state, for which ligand depletion was significant and values have been omitted from Table 1. Mn2+ increased BC+EC affinity 150-fold compared with 1 mM Mg2+/1 mM Ca2+. The BC+EC affinity of 1500 nM in 50 mM Mg2+ differed less from the affinity in 1 mM Mg2+/1 mM Ca2+ (4.7-fold) than from the affinity in 2 mM Mn2+ (33-fold; Table 1), similar to the trend found for the EO state described in the previous paragraph.
Influence of cations on the conformational equilibrium of intact α5β1 on the cell surface
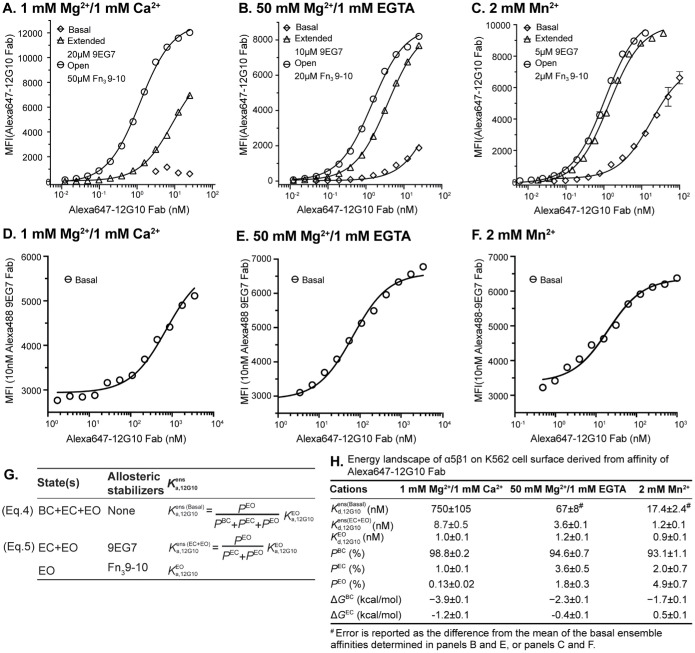
K562 cells express α5β1 as the only RGD-binding integrin. We were unable to measure fibronectin binding to cells, as we had done previously in 1 mM Mg2+/1 mM Ca2+ (Li et al., 2017; Li and Springer, 2018), because its affinity for the EO state was too high to be quantified accurately in 2 mM Mn2+. Furthermore, the fluorescence polarization experiments described above required 4–6 h for RGD peptide to equilibrate with the EO conformation. Because cell viability declined during incubations in binding buffer on this time scale, we determined the conformational equilibrium of α5β1 on K562 cells by measuring the affinity of EO-specific Fab 12G10 for α5β1 ensembles on K562 cells as previously described (Li et al., 2017). The affinity of these ensembles was proportional to their content (the population) of the EO state. As we were working with cells, which might release Ca2+, we used 1 mM EGTA with the 50 mM–Mg2+ condition.
We made two types of measurements. First, we measured binding of Alexa647-labeled 12G10 Fab after 1 h at 21°C by flow cytometry without washing and subtracted background with the same concentration of Alexa647-12G10 Fab with a 500-fold excess of unlabeled 12G10 Fab. In these experiments, we measured Alexa647-12G10 Fab affinity for α5β1 on K562 cells in basal ensembles, the EC+EO ensemble in the presence of 9EG7 Fab and the EO state in the presence of the fibronectin Fn3 domain 9–10 fragment (Figure 5, A–C and G). However, we could not saturably populate the EO state in 1 mM Mg2+/1 mM Ca2+ without a large signal from unbound Alexa647–12G10 Fab. Therefore, in a second type of experiment, we measured binding of a fixed concentration (10 nM) of Alexa488-labeled 9EG7 Fab as a reporter and were able to use 100-fold higher concentrations of Alexa647–12G10 Fab than in the first type of experiment to determine its affinity for α5β1 on K562 cells in basal ensembles under all three metal-ion conditions (Figure 5, D–F).
FIGURE 5:
Influence of cations on the conformational equilibrium of intact α5β1WT. (A–C) Binding of Alexa647-labeled 12G10 Fab for defined intact α5β1WT ensembles under 1 mM Mg2+/1 mM Ca2+, A, 50 mM Mg2+/1 mM EGTA, B, and 2 mM Mn2+, C by flow cytometry without washing. The concentration of 9EG7 Fab to saturably stabilize the two extended states and of Fn3 9–10 to saturably stabilize the EO state under each cation condition was determined in Supplemental Figure S2. Background-subtracted mean fluorescence intensity (MFI) as a function of Alexa647–12G10 Fab concentration was fitted to dose response curve for the Kd of 12G10 Fab for the different ensembles. (D–F) Enhancement of 10 nM Alexa488-labeled 9EG7 Fab binding to K562 cells by increasing Alexa647–12G10 Fab concentration under 1 mM Mg2+/1 mM Ca2+, D, 50 mM Mg2+/1 mM EGTA, E, and 2 mM Mn2+, F by flow cytometry without washing. MFI of 10 nM Alexa488-labeled 9EG7 Fab at different Alexa647–12G10 Fab concentrations were fitted to dose response curve for Kd values of 12G10 Fab for the basal ensemble. (G) The equations used to calculate the population of each conformational state from the affinities of Alexa647-12G10 Fab measured for the defined ensembles. (H) Tabulations of Alexa647–12G10 affinities determined for the defined ensembles, as well as the population of each conformational state calculated with Equations 4 and 5 shown in panel G, and the free energies of the BC and EC states relative to the EO state calculated from these populations based on the Boltzmann distribution.
Although we measured affinity for 12G10 Fab in these assays and not affinity for fibronectin or RGD, several findings validate this method for measuring the population of conformational states and their free energies (Figure 5G). First, the different assays in Figure 5, A–C compared with Figure 5, D–F gave almost identical results for the basal ensemble affinities in 50 mM Mg2+ and 2 mM Mn2+; their mean and difference from the mean are shown in Figure 5H. Second, the affinity of Alexa647–12G10 Fab for the EO state of α5β1 on K562 cells was almost identical (0.9 to 1.2 nM) and within the error of the fit values in all three metal ions (Figure 5H). Third, the populations of the α5β1 BC, EC, and EO states on K562 cells in 1 mM Mg2+/1 mM Ca2+ of 98.8%, 1%, and 0.13%, respectively, was similar to that previously determined with binding to the fibronectin Fn3 domain 9–10 fragment of 99.8%, 0.05%, and 0.11% (Li et al., 2017).
Overall, the results show that the population of the EO state in 1 mM Mg2+/1 mM Ca2+ of 0.13% increases to 1.8% in 50 mM Mg2+/1 mM EGTA and to 4.9% in 2 mM Mn2+ (Figure 5H). The EC state also was more populated in 50 mM Mg2+ and in 2 mM Mn2+. Similar to what was found with the ectodomain (Table 1), the EC state was more populated in 50 mM Mg2+ than in 2 mM Mn2+ (Figure 5H). On K562 cells, the energy of the BC state was increased relative to that of the EO state by ∼2 kcal/mol in both 50 mM Mg2+/1 mM EGTA and 2 mM Mn2+ (Figure 5H), which was very similar to the effect observed with the same cations on the soluble ectodomain (Table 1). The cations also increased the energy of the EC state by ∼1 kcal/mol (Figure 5H), which again was similar to the effect observed with the same cations on the soluble ectodomain (Table 1).
The ADMIDAS is important in activation of α5β1 by Mg 2+ and Mn 2+
To test the role of the ADMIDAS metal ion in regulating ligand-binding affinity and the integrin energy landscape, we mutated the only two residues with sidechains that coordinate the ADMIDAS metal ion in both the closed and open states, Asp137 and Asp138 (D8 and D9 in Figure 1B), to Ala (α5β1D137A, D138A).
The ADMIDAS was essential for the inhibitory effect of Ca2+. In the presence of a saturating concentration of 200 mM Mg2+ for WT α5β1, Ca2+ inhibited with an IC50 of 3.6 mM (Figure 6A). Calcium also reversed the activating effect of 2 mM Mn2+ and 50 mM Mg2+, with IC50 values of 2.2 and 0.2 mM, respectively (Supplemental Figure S3A). In contrast, Ca2+ had no inhibitory effect on the α5β1D137A, D138A ectodomain, consistent with the ADMIDAS being the binding site responsible for the inhibitory effect of Ca2+ (Figure 6B). In agreement, the inhibitory effect of Ca2+ on α5β1WT ectodomain was largely abolished by stabilizing its EO state with HUTS4 Fab, showing that Ca2+ inhibits by stabilizing closed states (Figure 6A). We further titrated Ca2+ in the presence of a physiological concentration of 1 mM Mg2+. Again, Ca2+ was inhibitory for α5β1WT but not α5β1D137A, D138A (Supplemental Figure S4).
FIGURE 6:
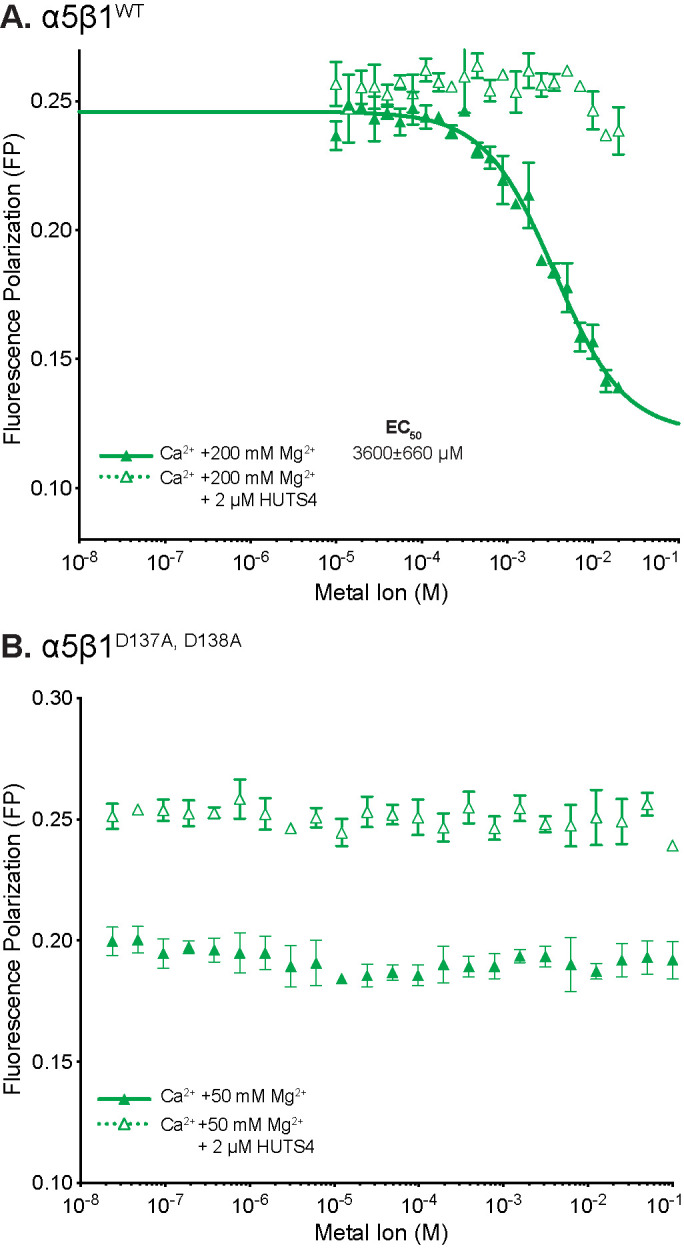
Metal-ion EC50 values for α5β1WT and α5β1D137A, D138A and inhibition by Ca2+ at the ADMIDAS. In FP assays containing 5 nM FITC-cyclic RGD peptide in the absence or presence of open-stabilizing HUTS4 Fab, Mg2+ alone was titrated or Ca2+ was titrated in the presence of saturating concentrations of Mg2+ with (A) 20 nM α5β1WT or (B) 200 nM α5β1D137A, D138A. Buffer was 20 mM Tris-HCL pH 7.4, 150 mM NaCl. Each point was in triplicate. Fit lines are shown only when fit values were greater than or similar to the fit error.
Influence of cations on intrinsic-ligand binding affinity and conformational equilibrium of soluble α5β1 D137A, D138A ectodomain
Before attempting measurements on the conformational equilibrium of α5β1D137A, D138A ectodomain, we measured EC50 values for Fabs to determine concentrations required to saturably populate the open, closed, and extended conformations (Figure 3, D–F). Results with mAb13 and 12G10, which stabilize the closed and open conformations of the βI domain by binding to the βI domain nearby the ligand binding site, were as expected. However, SG/19 inhibits by binding distally from the ligand binding site to an interface between the βI and hybrid domains that is present in the closed but not open conformations (Nagae et al., 2012). Most interestingly, in 1 mM Mg2+/Ca2+ and 50 mM Mg2+, SG/19 decreased FP with α5β1D137A, D138A ectodomain to a plateau level that was considerably higher than the plateau reached with mAb13 (Figure 3, D and E), whereas mAb13 and SG/19 reduced FP to identical levels with α5β1 WT (Figure 3, A and B). These results suggest that SG/19 stabilizes RGD-bound α5β1D137A, D138A ectodomain in two states, one with closed conformations of both the βI and hybrid domains, and the other with an open conformation of the βI domain at the ligand-binding site and with other portions of the βI and hybrid domains in the closed conformation. The disconnect between these open and closed portions of the βI domain may occur near the mutated ADMIDAS. Curiously, in 2 mM Mn2+, the SG/19 and mAb13 Fabs each decreased FP to similar levels (Figure 3F). However, while the effective concentrations of SG/19 and mAb13 Fab were similar under all other conditions (Figure 3, AºE), in 2 mM Mn2+ the effective concentrations of SG/19 and mAb13 Fab were much higher and lower, respectively (Figure 3F).
The results with SG/19 Fab show that an intact ADMIDAS binding site is important for coupling between the βI domain and the hybrid domain. Similar results were seen with HUTS4 Fab, which also binds to a βI/hybrid domain interface but stabilizes the open conformation. HUTS4 was also less effective in activating the α5β1D137A, D138A ectodomain than the WT α5β1 ectodomain (Figure 4, A–C compared with D–F). Two Fabs, 8E3 and 9EG7, that bind to different knee-proximal leg domains to stabilize the extended conformations also had less clear effects with the α5β1D137A, D138A than with the WT α5β1 ectodomain (Figure 3, D–F).
Therefore, we measured affinities of α5β1D137A, D138A for RGD and cRGD in the three metal-ion conditions only in the basal ensemble, the EO state, and the BC+EC states (Figure 4 and Supplemental Figure S5). EO affinities measured with cRGD with both mutant and WT α5β1 were nominally in the (0.1–2)-nM range, where free integrin was significantly depleted, which impeded accurate affinity measurement. Therefore, only RGD affinities for the EO state are reported in Table 1.
Compared with that of the α5β1WT ectodomain, the basal ensemble affinity of the α5β1D137A, D138A ectodomain was lower for both RGD and cRGD (Table 1), consistent with the lower FP maxima reached by α5β1D137A, D138A than α5β1 WT in Mn2+ and Mg2+ ion titrations (Supplemental Figure S3B). Interestingly, mutation of the ADMIDAS site changed metal-ion dependence for Mg2+ on affinity. Whereas 50 mM Mg2+ increased wild-type α5β1 affinity 30- to 37-fold for the two ligands compared with that in 1 mM Mg2+/1 mM Ca2+, basal ensemble α5β1D137A, D138A affinity was only increased 1.7-fold for RGD and showed indistinguishable affinities for cRGD (Table 1). Mn2+ (2 mM) was still activating for α5β1D137A, D138A with both ligands, but to a lesser extent, with a 41- and 26-fold increase in affinity for the RGD and cRGD ligands, respectively, compared with 1 mM Mg2+/1 mM Ca2+.
Although we were unable to fully characterize the conformational ensemble of α5β1D137A, D138A ectodomain, we were able to make measurements on the high-affinity EO state with RGD and the low-affinity BC+EC states with cRGD (Table 1). Mn2+ increased the affinity of the α5β1D137A, D138A EO state for RGD 12-fold and the affinity of the BC+EC states for cRGD 55-fold compared with 1 mM Mg2+/1 mM Ca2+. Contrasting results were obtained with 50 mM Mg2+. Compared with affinities in 1 mM Mg2+/1 mM Ca2+ of α5β1D137A, D138A, 50 mM Mg2+ decreased affinity of the EO state for RGD 6-fold and the affinity of the BC+EC states for cRGD 1.6-fold (Table 1). Most interestingly, the affinity for cRGD of α5β1D137A, D138A was higher than that of α5β1WT in the BC+EC states, by 9-fold in 1 mM Mg2+/1 mM Ca2+, by 1.2-fold in 50 mM Mg2+, and by 3-fold in 2 mM Mn2+.
DISCUSSION
For decades, manganese has been used extensively in studies on integrins for its ability to activate them, without proper understanding of its mechanism of action or how these Mn2+-activated integrins compare with physiologically activated integrins (Gailit and Ruoslahti, 1988; Ye et al., 2012). In this article, employing conformation-specific Fabs against the integrin β1 subunit to stabilize integrins into defined ensembles, we determined how cations influence the intrinsic ligand-binding affinity of each integrin conformational state and the equilibrium linkage between them. The conformational specificities of these Fabs were determined with negative stain EM (nsEM) with the integrin α5β1 ectodomain (Su et al., 2016) and further verified by ligand-binding affinity measurements in presence of saturating concentrations of Fab (Li et al., 2017). In the latter study, we used between two and four independent antibodies or antibody combinations, often binding to distinct domains, to stabilize α5β1 ectodomain ensembles containing the EO, EC, EO+EC, and EC+BC states. In all cases, independent antibodies that stabilized the same conformational state(s) as determined by EM yielded similar ensemble Kd values (Figure 3 in (Li and Springer, 2018; Li et al., 2017), validating stabilization of the same state(s), and supporting the assumption that these states resembled those in the absence of Fab. Quantitatively, the antibodies must be highly state-specific to give large shifts in affinities, to give consistent intrinsic affinities on constructs with large differences in basal affinities, and to give similar intrinsic affinities using Fabs to distinct epitopes. The conformational specificity of these antibodies and their use at sufficient concentration to saturate these states are important for our previous thermodynamic studies (Li et al., 2017) and the current study.
We show that integrin α5β1 activation by Mn2+ or Mg2+ is caused both by an intrinsic increase in affinity of the active site and by a conformational shift of the integrin’s ensemble toward the high-affinity EO state. By mutating ADMIDAS-binding residues, we also show how the ADMIDAS influences the intrinsic affinities of conformational states and the integrin conformational equilibria and links conformational change at the ligand-binding site to swingout of the hybrid domain at its interface with the βI domain. Our results provide important insights into the functions of all three metal ion–binding sites in the βI domain, which form the heart of the ligand-binding site and govern its remarkable ability to change the affinity of α5β1 for ligands by 4000-fold between the closed and open conformational states (Li et al., 2017; Li and Springer, 2018).
Our results change the paradigm for how metal ions activate integrins. We found that under activating conditions with 2 mM Mn2+ or 50 mM Mg2+, the conformational energy landscape was “flattened” compared with that at physiological concentrations of Ca2+ and Mg2+, which are each near 1 mM, causing a shift in the population from the low-affinity (BC and EC) states to the high-affinity (EO) state. Contrary to a notion common in the literature, Mn2+ does not fully open integrins. Shifting the conformational equilibrium is one of the mechanisms, but it is not complete. Integrin α5β1 ectodomain preparations were 3.1% in the high-affinity state (EO) in 1 mM Ca2+/1 mM Mg2+ (Li et al., 2017; Li and Springer, 2018) and were shifted in both 2 mM Mn2+ and 50 mM Mg2+ to 41% in the EO state. Intact α5β1 on cell surfaces is only 0.13% in the EO state and was shifted by 2 mM Mn2+ and 50 mM Mg2+ to 4.9% and 1.8% in the EO state, respectively. Intact α5β1 appears to be more stable than the ectodomain in the BC state, because integrins in this state on cell surfaces associate extensively through their α- and β-subunit TM and juxtamembrane domains (Lau et al., 2009; Zhu et al., 2009). Mn2+ and high Mg2+ increased the free energies of the BC and EC states of both intact and ectodomain forms of α5β1 by ∼2 kCal/mol. The ectodomain was shifted to the EO state more than the intact receptor on the cell surface because the free energy difference between the closed and EO states is less for the ectodomain. The similar effects on the BC and EC states suggest that 2 mM Mn2+ and 50 mM Mg2+ each exert their activating effect by favoring headpiece opening rather than integrin extension.
It is conceptually important to remember that the populations reported here are those in ensembles of unliganded integrins in the absence of ligand binding. Although our measurements use ligand, they measure the populations preexisting before addition of ligand. Also, “population” is just a measure of the percentage of molecules in a particular state, and does not imply any difference between the molecules other than what conformation they are in. Indeed, it is essential to the validity of our thermodynamic formalism that all of the molecules are equally able to equilibrate from one state to another. Our results on populations of states in Table 1 are consistent with negative stain electron microscopy (EM) results that in the absence of bound ligand, integrin αVβ3 ectodomain particles were predominantly in the BC state in 1 mM Mg2+/1 mM Ca2+, in both the EC and EO states in Mn2+, and predominantly in the EO state in Mn2+ and cyclic RGD (Takagi et al., 2002). The common use of Mn2+ to enhance ligand binding by integrins is explained by its ability to increase affinity of the α5β1 ectodomain for RGD and cRGD by ∼400-fold. Recent cryo-EM structures of intact α5β1 in nanodiscs revealed the BC state in Ca2+/Mg2+ in the absence of ligand and the EO state in Mn2+ when bound to a fibronectin fragment (Schumacher et al., 2021). While the authors were surprised to find that intact α5β1 was predominantly in the BC state in Mn2+ in the absence of ligand, their finding is consistent with the observation here that intact α5β1 on the cell surface in Mn2+ is 93% in the BC state.
The increase in integrin intrinsic affinities for ligand in Mn2+ and Mg2+ was not previously anticipated and emphasizes the fundamental importance of the metal–oxygen bond between the MIDAS metal ion and the Glu or Asp in all integrin ligands in determining integrin affinity for ligand. Mn2+ and Mg2+ show octahedral coordination (six coordination positions) and distances of 2.2 Å from oxygens that promote partially covalent interactions. Mn2+ and Mg2+ both activate by binding to the ADMIDAS, and assuming that their effects are equivalent at that site, the difference in intrinsic affinities is attributable to the difference in energy of the MIDAS metal-ion bond to the oxygen atom in the Asp sidechain of RGD. The energies of the Mn–O and Mg–O bonds will be influenced by their environments in solution and will be different yet at the MIDAS, where they are surrounded by a large network of hydrogen bonds. Nonetheless, a good approximation commonly used in chemistry for the difference in energies in solution is the difference in energy in the gas phase, which is 1.9 kcal/mol greater for Mn–O than for Mg–O (Speight, 2016). The relationship between free energy (ΔG) and the equilibrium constant (Keq) shows an expected increase in affinity from replacing Mg2+ with Mn2+ at the MIDAS of 25-fold. The measured increases in intrinsic affinity in 2 mM Mn2+ compared with 50 mM Mg2+ of 16-fold for the EO state binding to RGD peptide and 33-fold for the EC+BC states binding to cRGD peptide compare well with the calculated value of 25-fold, supporting the hypothesis that the difference in energy between the Mn2+ and Mg2+ ligand bonds with oxygen is largely responsible for the higher intrinsic integrin affinity for ligand in Mn2+.The increase in intrinsic affinity and shift in conformational equilibrium seen here in Mn2+ with α5β1 are expected to be similar in all other integrins, except for integrin αVβ8, which lacks an ADMIDAS.
Our results with α5β1D137A, D138A showed that alteration of the conformational equilibrium by Mn2+ and Mg2+ occurred primarily as a result of their ability to displace Ca2+ at the ADMIDAS. Our finding that the α5β1D137A, D138A mutant lost its ability to be inhibited by Ca2+ agreed with previous results on integrins including α4β7, α2β1, α5β1, and αLβ2 (Chen et al., 2003, 2006; Mould et al., 2003a; Valdramidou et al., 2008). Previously α5β1D137A and α5β1 D138A mutations were studied individually (Mould et al., 2003a); we mutated both of these residues in α5β1D137A, D138A to more completely remove metal ion binding.
We propose that differing coordination preferences of Mn2+ and Mg2+ compared with Ca2+ (Harding, 2001; Dokmanic et al., 2008), which are matched to ADMIDAS coordination in open and closed integrin conformations, explain activation by Mn2+ and Mg2+. Surveys of metal-ion binding to proteins (Harding, 2001; Dokmanic et al., 2008) show that as described above, Mn2+ and Mg2+ bind partially covalently to oxygen atom ligands at a typical distance of 2.2 Å, aligned octahedrally through electron orbitals (coordination number 6). In contrast, Ca2+ binds to oxygen atoms electrostatically at a typical distance of 2.4 Å, with a preference for pentagonal bipyramidal (coordination number 7) over octahedral (coordination number 6). High-resolution structures of integrin αIIbβ3 show that Ca2+ binds to the ADMIDAS with coordination number 7 in the closed conformation, whereas in the open conformation, both Ca2+ and Mn2+ bind with coordination number 6 (Springer et al., 2008; Zhu et al., 2012, 2013; Figure 1C). ADMIDAS metal-ion coordination to the backbone carbonyl group in the β6–α7 loop in the closed conformation is replaced by coordination to the sidechain carboxyl group of Asp-251 in the open conformation (Figure 1C). This change also favors the open conformation in Mn2+ and Mg2+, because both metal ions strongly favor carboxyl over carbonyl oxygens, whereas Ca2+ coordinates well with both types of oxygens. These coordination preferences provide a mechanistic explanation for the ability of Mn2+ and Mg2+ to stabilize integrins in the open conformation.
Integrin αvβ8 lacks an ADMIDAS metal ion and contains Asn residues in place of the ADMIDAS-coordinating Asp residues present in integrin β1 and the six other integrin β-subunits in mammals (Wang et al., 2019; Campbell et al., 2020). The βI domain β1–α1 loop and α1 helix contain the β-MIDAS motif, DXSXSXXDD (D1-S3-S5-D8-D9; Wang et al., 2019). Residues S5, D8, and D9 all coordinate the ADMIDAS metal ion, and movement of this loop and the ADMIDAS metal ion toward the open state is hindered by ADMIDAS metal-ion coordination to a backbone carbonyl oxygen in the βI domain β6–α7 loop (Figure 1B). When integrins are crystallized in the absence of ligand and then ligand is soaked in, crystal lattice contacts inhibit hybrid domain swingout and frustrate βI domain opening, so that no movement, or partial opening to intermediate states, predominates (Zhu et al., 2013). In crystals containing ADMIDAS-lacking integrin αvβ8, α1 helix movement is relatively large when ligand is soaked in. Furthermore, the affinity of the closed state of αvβ8 for ligand is higher than that of its sister integrin αvβ6, which has been attributed in part to lack of restraint by the ADMIDAS of α1 helix movement (Wang et al., 2019). The finding here that the closed state of α5β1D137A, D138A has an affinity ninefold higher than the closed state of α5β1WT suggests that in the closed state of α5β1D137A, D138A, the lack of ADMIDAS restraint allows its α1 helix to move further toward the MIDAS and thus reach higher affinity for the ligand, despite the overall closed state of the βI domain, which was enforced by direct binding of the mAb13 Fab to the βI domain.
The quantitative studies here, together with recent affinity measurements and structure–function mutational exploration of integrin αVβ8, which uniquely among mammalian integrins lacks an ADMIDAS, provide insights beyond the well-known role of the ADMIDAS in transmitting conformational change and regulating ligand binding (Mould et al., 2002, 2003a, b; Chen et al., 2003, 2006). Our work suggests that the ADMIDAS is key for binary behavior of integrins in being either on or off, that is, being in either high- or low-affinity states. Thus, we found that the low-affinity BC+EC state of the ADMIDAS mutant is 9-fold higher in affinity than that of WT, while its high-affinity EO state has 2.4-fold lower affinity than that of WT. Thus, while the high-affinity state of WT α5β1 is 4000-fold higher than its low-affinity state, the high-affinity state of the ADMIDAS mutant is only ∼200-fold higher in affinity than its low-affinity state. These results suggest that an important biological function of the ADMIDAS may be to provide a wide separation in affinity of integrin open and closed states. Our results with SG/19 and HUTS4 Fabs, which bind to interfaces between the βI and hybrid domains, also demonstrate that an intact ADMIDAS is essential for maintaining the fidelity of conformational communication between the ligand binding site in the βI domain and its interface with the hybrid domain. Thus, in the context of binding of integrins on cells to substrates and providing traction for supporting cell adhesion and cell migration, the ADMIDAS has an important role in ensuring fidelity in conformational communication within the integrin β-subunit and endows integrins with functional states that are akin to on and off states.
MATERIALS AND METHODS
Ligands
The ligands cyclic RGD peptide (ACRGDGWCG with N-terminal 6-aminohexanoic acid and cyclized by sidechain disulfide) and linear RGD peptide (GRGDSPK) were synthesized and labeled with FITC (fluorescein isothiocyanate) by GenScript. Stock solutions (10 μM) in DMSO were diluted with the appropriate buffer before use.
Integrin
Integrins used in these studies were expressed as the high-mannose glycan form using a stable HEK293S GnTI-/- (N-acetylglucosaminyltransferase I–deficient) cell line (Li et al., 2017). Both constructs (α5β1WT and α5β1D137A, D138A) contain α5 residues F1-Y954 and β1 residues Q1-D708. The ADMIDAS mutant was produced by replacement of a synthesized G-block (Integrated DNA Technologies) by 2-fragment Gibson assembly (HiFi, New England Biolabs) into the pD2529_CAG vector (ATUM). As previously described (Li et al., 2017), α5 and β1 subunits were coexpressed with a C-terminal His-tagged leucine zipper to aid expression and dimerization. Secreted proteins were purified from the clarified supernatant with a Ni-NTA column and eluted with high imidazole (300 mM). TEV cleavage removed the His-tag and leucine zipper and after a second run through a Ni-NTA column, the flowthrough was concentrated and buffer exchanged with pH 7.4, 20 mM Tris, 150 mM NaCl (TBS) containing 1 mM CaCl2 and 1 mM MgCl2. Further purification was accomplished by size-exclusion chromatography (Superdex 200, GE Healthcare) in the same buffer, resulting in a single peak, which was concentrated to 20 to 100 µM, flash frozen, and stored at –80°C for future use. Typical final yield after purification for the mutant was 8 mg from 1 L culture supernatant. Integrins were not stored in the absence of metal ions, because gel filtration using running buffer containing EDTA or EGTA to remove divalent cations showed that a significant amount of the protein eluted in the void volume.
Fabs
Hybridomas were HUTS4 (Luque et al., 1996), 12G10 (Mould et al., 1995b), 8E3 (Mould et al., 2005), 9EG7 (Lenter et al., 1993), SG/19 (Miyake et al., 1992), and mAb13 (Akiyama et al., 1989). IgG produced from hybridoma was purified by protein G and digested with papain (500:1 IgG:papain) in pH 7, 20 mM sodium phosphate, 150 mM NaCl with 10 mM EDTA and 10 mM L-Cys at 37°C for 18 h. After buffer exchange with 50 mM Tris (pH 9), the Fab was purified by anion exchange chromatography (HiTrap Q HP, GE Healthcare), after which the fractions containing Fab were concentrated, flash frozen, and stored at –80°C for future use.
FP assay
Fluorescence polarization measurements were made using a Synergy Neo plate reader (BioTek), with serial dilutions made in flat-bottomed black 384-well plates (Corning), with 10-µl samples. Samples were prepared in pH 7.4 buffer containing 20 mM Tris, 150 mM NaCl, and the given concentration of Ca2+, Mg2+, Mn2+, or EGTA (CaCl2, MgCl2, MnCl2, or ethylene glycol-bis[β-aminoethyl ether]-N,N,N’,N’-tetraacetic acid) with 5 nM ligand and 20 nM integrin, unless otherwise noted. Because the stock solutions of integrin were prepared in 1 mM Ca2+ and 1 mM Mg2+, for assays with different metal ions, samples were diluted more than 1000-fold into TBS with or without desired metal ions; alternatively, when used at less than 1000-fold dilution, integrins were exchanged with the desired metal ion in TBS by diluting >10-fold with the desired buffer and concentrating 10-fold a total of three times. For metal ion titrations, the integrin was diluted in TBS and metal ion titrations were then added. To ensure that the binding was completely equilibrated, most readings were taken at 2, 4, and 6 h, with negligible change found for the last two. When necessary, if excessively high concentrations of α5β1 were needed to saturate binding (e.g., α5β1WT closed ensemble and α5β1D137A,D138A basal ensemble), maximum values were shared with higher-affinity conditions in the same experiment. A detailed explanation of the fitting of FP data and calculation of Kd or ΔG values can be found in Li et al. (2017).
Cell surface binding assay
K562 cells (106 cells/mL in RPMI-1640 medium, 10% FBS) were washed twice with assay buffer (HBSS with 20 mM HEPES, pH 7.2, 1% BSA, and indicated cations) and suspended in assay medium. Each sample (50 µl) contained cells (2 × 106 cells/ml), Alexa647-12G10 Fab, and unlabeled 9EG7 Fab, or Fn39–10, or Alex488-9EG7 Fab in assay medium. The mixture was allowed to equilibrate for 1 h before flow cytometry (BD FACSCanto II) without washing. Direct measurement of Alexa647-12G10 Fab binding was recorded as mean fluorescence intensity ) of Alexa647; background MFI was subtracted and was measured under the same conditions, except with 500-fold higher concentration of unlabeled 12G10 Fab. Background-subtracted MFI at different concentrations of AF647-12G10 under each metal ion condition was fitted to a dosage response curve to yield the Kd value of intact α5β1 for AF647-12G10. For the basal condition, affinity of Alexa647–12G10 Fab was also determined from enhancement of 10 nM Alexa488–Fn39-10 binding and fitted to a dose–response curve.
Supplementary Material
Acknowledgments
Funding: National Institutes of Health 1R01HL131729: “Activation Trajectories of Integrin α5β1” (T.A.S. and J.L.).
Abbreviations used:
- ADMIDAS
adjacent to MIDAS
- BC
bent-closed
- EC
extended-closed
- ECM
extracellular matrix
- EO
extended-open
- FITC
fluorescein isothiocyanate
- FP
fluorescence polarization
- MIDAS
metal ion-dependent adhesion site
- RGD
Arg-Gly-Asp
- SyMBS
synergistic metal-ion binding site
- TBS
Tris-buffered saline.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-11-0536) on February 2, 2022.
REFERENCES
- Akiyama SK, Yamada SS, Chen WT, Yamada KM (1989). Analysis of fibronectin receptor function with monoclonal antibodies: Roles in cell adhesion, migration, matrix assembly, and cytoskeletal organization. J Cell Biol 109, 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimori T, Miyazaki N, Mihara E, Takizawa M, Taniguchi Y, Cabanas C, Sekiguchi K, Takagi J (2021). Structural mechanism of laminin recognition by integrin. Nat Commun 12, 4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MG, Cormier A, Ito S, Seed RI, Bondesson AJ, Lou J, Marks JD, Baron JL, Cheng Y, Nishimura SL (2020). Cryo-EM reveals integrin-mediated TGF-β activation without release from latent TGF-β. Cell 180, 490–501.e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Salas A, Springer TA (2003). Bistable regulation of integrin adhesiveness by a bipolar metal ion cluster. Nat Struct Biol 10, 995–1001. [DOI] [PubMed] [Google Scholar]
- Chen JF, Yang W, Kim M, Carman CV, Springer TA (2006). Regulation of outside–in signaling by the β2 I domain of integrin αLβ2. Proc Natl Acad Sci USA 103, 13,062–13,067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokmanic I, Sikic M, Tomic S (2008). Metals in proteins: correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination. Acta Crystallogr D Biol Crystallogr 64, 257–263. [DOI] [PubMed] [Google Scholar]
- Dong X, Zhao B, Iacob RE, Zhu J, Koksal AC, Lu C, Engen JR, Springer TA (2017). Force interacts with macromolecular structure in activation of TGF-β. Nature 542, 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dransfield I, Cabañas C, Craig A, Hogg N (1992). Divalent cation regulation of the function of the leukocyte integrin LFA-1. J Cell Biol 116, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailit J, Ruoslahti E (1988). Regulation of the fibronectin receptor affinity by divalent cations. J Biol Chem 263, 12,927–12,932. [PubMed] [Google Scholar]
- Harding MM (2001). Geometry of metal–ligand interactions in proteins. Acta Crystallogr D Biol Crystallogr 57, 401–411. [DOI] [PubMed] [Google Scholar]
- Hu DD, Barbas CF III, Smith JW (1996). An allosteric Ca2+ binding site on the β3-integrins that regulates the dissociation rate for RGD ligands. J Biol Chem 271, 21,745–21,751. [DOI] [PubMed] [Google Scholar]
- Kechagia JZ, Ivaska J, Roca-Cusachs P (2019). Integrins as biomechanical sensors of the microenvironment. Nat Rev Mol Cell Biol 20, 457–473. [DOI] [PubMed] [Google Scholar]
- Koivunen E, Wang B, Ruoslahti E (1995). Phage libraries displaying cyclic peptides with different ring sizes: ligand specificities of the RGD-directed integrins. Biotechnology (NY) 13, 265–270. [DOI] [PubMed] [Google Scholar]
- Labadia ME, Jeanfavre DD, Caviness GO, Morelock MM (1998). Molecular regulation of the interaction between leukocyte function-associated antigen-1 and soluble ICAM-1 by divalent metal cations. J Immunol 161, 836–842. [PubMed] [Google Scholar]
- Lau TL, Kim C, Ginsberg MH, Ulmer TS (2009). The structure of the integrin αIIbβ3 transmembrane complex explains integrin transmembrane signalling. EMBO J 9, 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenter M, Uhlig H, Hamann A, Jenö P, Imhof B, Vestweber D (1993). A monoclonal antibody against an activation epitope on mouse integrin chain β1 blocks adhesion of lymphocytes to the endothelial integrin α6β1. Proc Natl Acad Sci USA 90, 9051–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Springer TA (2017). Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc Natl Acad Sci USA 114, 4685–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Springer TA (2018). Energy landscape differences among integrins establish the framework for understanding activation. J Cell Biol 217, 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Su Y, Xia W, Qin Y, Humphries MJ, Vestweber D, Cabanas C, Lu C, Springer TA (2017). Conformational equilibria and intrinsic affinities define integrin activation. EMBO J 36, 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FY, Zhu J, Eng E, Hudson NE, Springer TA (2016). β-Subunit binding is sufficient for ligands to open the integrin αIIbβ3 headpiece. J Biol Chem 291, 4537–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Springer TA (2007). Structural basis of integrin regulation and signaling. Annu Rev Immunol 25, 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque A, Gomez M, Puzon W, Takada Y, Sanchez-Madrid F, Cabanas C (1996). Activated conformations of very late activation integrins detected by a group of antibodies (HUTS) specific for a novel regulatory region (355–425) of the common β1 chain. J Biol Chem 271, 11,067–11,075. [DOI] [PubMed] [Google Scholar]
- Marlin SD, Springer TA (1987). Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1). Cell 51, 813–819. [DOI] [PubMed] [Google Scholar]
- Miyake K, Hasunuma Y, Yagita H, Kimoto M (1992). Requirement for VLA-4 and VLA-5 integrins in lymphoma cells binding to and migration beneath stromal cells in culture. J Cell Biol 119, 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould AP, Akiyama SK, Humphries MJ (1995a). Regulation of integrin α5β1-fibronectin interactions by divalent cations. J Biol Chem 270, 26,270–26,277. [DOI] [PubMed] [Google Scholar]
- Mould AP, Askari JA, Barton S, Kline AD, McEwan PA, Craig SE, Humphries MJ (2002). Integrin activation involves a conformational change in the α1 helix of the β subunit A-domain. J Biol. Chem 277, 19,800–19,805. [DOI] [PubMed] [Google Scholar]
- Mould AP, Barton SJ, Askari JA, Craig SE, Humphries MJ (2003a). Role of ADMIDAS cation-binding site in ligand recognition by integrin α5β1. J Biol Chem 278, 51,622–51,629. [DOI] [PubMed] [Google Scholar]
- Mould AP, Barton SJ, Askari JA, McEwan PA, Buckley PA, Craig SE, Humphries MJ (2003b). Conformational changes in the integrin βA domain provide a mechanism for signal transduction via hybrid domain movement. J Biol Chem 278, 17,028–17,035. [DOI] [PubMed] [Google Scholar]
- Mould AP, Garratt AN, Askari JA, Akiyama SK, Humphries MJ (1995b). Identification of a novel anti-integrin monoclonal antibody that recognises a ligand-induced binding site epitope on the β1 subunit. FEBS Lett 363, 118–122. [DOI] [PubMed] [Google Scholar]
- Mould AP, Travis MA, Barton SJ, Hamilton JA, Askari JA, Craig SE, Macdonald PR, Kammerer RA, Buckley PA, Humphries MJ (2005). Evidence that monoclonal antibodies directed against the integrin β subunit plexin/semaphorin/integrin domain stimulate function by inducing receptor extension. J Biol Chem 280, 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M, Re S, Mihara E, Nogi T, Sugita Y, Takagi J (2012). Crystal structure of α5β1 integrin ectodomain: atomic details of the fibronectin receptor. J Cell Biol 197, 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher S, Dedden D, Nunez RV, Matoba K, Takagi J, Biertumpfel C, Mizuno N (2021). Structural insights into integrin α5β1 opening by fibronectin ligand. Sci Adv 7, eabe9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speight J (2016). Lange’s Handbook of Chemistry. Laramie, WY: McGraw Hill. [Google Scholar]
- Springer TA, Dustin ML. (2012). Integrin inside-out signaling and the immunological synapse. Curr Opin Cell Biol 24, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer TA, Zhu J, Xiao T (2008). Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin αIIbβ3. J Cell Biol 182, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staatz WD, Rajpara SM, Wayner EA, Carter WG, Santoro SA (1989). The membrane glycoprotein Ia-IIa (VLA-2) complex mediates the Mg2+-dependent adhesion of platelets to collagen. J Cell Biol 108, 1917–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Xia W, Li J, Walz T, Humphries MJ, Vestweber D, Cabañas C, Lu C, Springer TA (2016). Relating conformation to function in integrin α5β1. Proc Natl Acad Sci USA 113, E3872–E3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Costell M, Fassler R (2019). Integrin activation by talin, kindlin and mechanical forces. Nat Cell Biol 21, 25–31. [DOI] [PubMed] [Google Scholar]
- Takagi J, Petre BM, Walz T, Springer TA (2002). Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110, 599–611. [DOI] [PubMed] [Google Scholar]
- Valdramidou D, Humphries MJ, Mould AP (2008). Distinct roles of β1 metal ion-dependent adhesion site (MIDAS), adjacent to MIDAS (ADMIDAS), and ligand-associated metal-binding site (LIMBS) cation-binding sites in ligand recognition by integrin α2β1. J Biol Chem 283, 32,704–32,714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Su Y, Iacob RE, Engen JR, Springer TA (2019). General structural features that regulate integrin affinity revealed by atypical αVβ8. Nat Commun 10, 5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Takagi J, Wang J-H, Coller BS, Springer TA (2004). Structural basis for allostery in integrins and binding of fibrinogen-mimetic therapeutics. Nature 432, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J-P, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA (2001). Crystal structure of the extracellular segment of integrin αVβ3. Science 294, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA (2002). Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science 296, 151–155. [DOI] [PubMed] [Google Scholar]
- Ye F, Kim C, Ginsberg MH (2012). Reconstruction of integrin activation. Blood 119, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Choi W-S, McCoy JG, Negri A, Zhu J, Naini S, Li J, Shen M, Huang W, Bougie D, et al. (2012). Structure-guided design of a high affinity platelet integrin αIIbβ3 receptor antagonist that disrupts Mg2+ binding to the MIDAS. Sci Transl Med 4, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Luo BH, Barth P, Schonbrun J, Baker D, Springer TA (2009). The structure of a receptor with two associating transmembrane domains on the cell surface: integrin αIIbβ3. Mol Cell 34, 234–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhu J, Springer TA (2013). Complete integrin headpiece opening in eight steps. J Cell Biol 201, 1053–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.