Abstract
Clathrin-mediated endocytosis (CME) robustness under elevated membrane tension is maintained by actin assembly–mediated force generation. However, whether more actin assembles at endocytic sites in response to increased load has not previously been investigated. Here actin network ultrastructure at CME sites was examined under low and high membrane tension. Actin and N-WASP spatial organization indicate that actin polymerization initiates at the base of clathrin-coated pits and that the network then grows away from the plasma membrane. Actin network height at individual CME sites was not coupled to coat shape, raising the possibility that local differences in mechanical load feed back on assembly. By manipulating membrane tension and Arp2/3 complex activity, we tested the hypothesis that actin assembly at CME sites increases in response to elevated load. Indeed, in response to elevated membrane tension, actin grew higher, resulting in greater coverage of the clathrin coat, and CME slowed. When membrane tension was elevated and the Arp2/3 complex was inhibited, shallow clathrin-coated pits accumulated, indicating that this adaptive mechanism is especially crucial for coat curvature generation. We propose that actin assembly increases in response to increased load to ensure CME robustness over a range of plasma membrane tensions.
INTRODUCTION
Actin networks produce force for a wide variety of cellular processes through a Brownian ratchet mechanism (Mogilner and Oster, 1996, 2003; Pollard, 2016). Live-cell studies of lamellipodia (Mueller et al., 2017), biochemical reconstitutions (Bieling et al., 2016; Funk et al., 2021; Li et al., 2021), and modeling studies (Akamatsu et al., 2020) have shown that actin networks nucleated by the Arp2/3 complex respond to increased load by becoming more dense, which enhances force production. This phenomenon has been demonstrated in the context of actin networks producing force on the lamellipodum, whose membrane is essentially flat on the length scale of an individual actin filament branch. However, actin networks can also produce force on membranes that change shape over time, for example during vesicle formation. Whether actin networks in the latter context show load adaptation has not been investigated.
From yeast to humans, transient actin assembly is associated with the formation of clathrin-coated endocytic vesicles. In yeast cells, actin assembly is required to generate forces to invaginate the plasma membrane against a high intrinsic turgor pressure for clathrin-mediated endocytosis (CME) (Kaksonen et al., 2005; Aghamohammadzadeh and Ayscough, 2009; Idrissi et al., 2012; Kukulski et al., 2012). When actin assembly is perturbed in mammalian cells, CME typically slows in a manner that depends on cell type (Fujimoto et al., 2000; Merrifield et al., 2005; Yarar et al., 2005, 2007; Grassart et al., 2014; Dambournet et al., 2018; Schöneberg et al., 2018). A potential cause of this reported variation between cell types might be differences in plasma membrane tension (Pontes et al., 2017; Djakbarova et al., 2021). Indeed, in mammalian cells, actin assembly becomes increasingly critical as plasma membrane tension increases (Yarar et al., 2005; Liu et al., 2009; Batchelder and Yarar, 2010; Boulant et al., 2011). Actin perturbation results in the accumulation of “U-shaped” membrane invaginations, reflecting difficulty in progressing to the subsequent “Ω-shaped” membrane stage (Fujimoto et al., 2000; Yarar et al., 2005; Boulant et al., 2011; Almeida-Souza et al., 2018). In total, these findings suggest that actin assembly improves the efficiency of CME in mammalian cells, potentially compensating for changes in plasma membrane tension.
Despite the fact that actin assembly appears to be associated with CME in all eukaryotes, and over a large range of membrane tension values, whether it adapts to changes in membrane tension is not known. Knowing whether the actin cytoskeleton at CME sites adapts to changes in membrane tension will facilitate elucidation of the fundamental mechanisms by which cytoskeletal complexes produce force.
Platinum replica electron microscopy of cultured cells led to the proposal that actin networks assemble in a collar-like arrangement around the vesicle neck (Collins et al., 2011). This actin organization would imply that a constricting force is generated toward the neck of the pit, supporting fission. However, actin filaments interact not only with the vesicle neck but also with the bud surface, given that the clathrin coat is impregnated with actin-binding linker proteins like Hip1R and Epsin (Engqvist-Goldstein et al., 2001; Messa et al., 2014; Sochacki et al., 2017; Clarke and Royle, 2018). Such an arrangement and evidence from photobleaching studies imply that actin filaments also apply a force that pulls the clathrin-coated pit (CCP) into the cell interior (Kaksonen et al., 2003; Akamatsu et al., 2020).
In yeast, such a pulling mechanism is likely. Actin filaments are nucleated in a ring surrounding the pit and grow from the plasma membrane toward the cell interior. The resulting actin filaments are coupled to the clathrin coat surface, generating an inward force orthogonal to the plane of the plasma membrane (Kaksonen et al., 2003; Carroll et al., 2012; Skruzny et al., 2012, 2015; Picco et al., 2015; Mund et al., 2018). Because the endocytic machinery is highly conserved from yeast to mammals, a similar mechanism for actin force generation during CME seems likely (Akamatsu et al., 2020). However, ultrastructural evidence for actin organization through different stages of mammalian CME, and for how this organization might respond to changing membrane tension, is lacking.
Several competing models for actin organization at CME sites in mammalian cells have been proposed, so it is important to distinguish between these models (Engqvist-Goldstein et al., 2001; Boulant et al., 2011; Messa et al., 2014; Sochacki et al., 2017; Clarke and Royle, 2018). Recent advances in superresolution microscopy (SRM) permit examination of cellular ultrastructure with large sample sizes, low invasiveness, and high molecular specificity to reveal the ultrastructure of membrane cytoskeletal systems in mammalian cells (Xu et al., 2012, 2013; Hauser et al., 2018).
Here, we combined two-color, three-dimensional stochastic optical reconstruction microscopy (2c-3D STORM) with live-cell fluorescence imaging to determine how filamentous actin is organized at CME sites with different coat geometries under various values of plasma membrane tension. These measurements led to the conclusion that the actin network grows from the base of the pit inward, supporting a pulling mechanism for mammalian endocytic actin filaments. The size of the actin network is not tightly coupled to the geometry of the endocytic coat, implying that assembly is modulated by factors uncoupled from coat shape. Importantly, under elevated membrane tension, the actin network grows higher relative to the pit at all endocytic coat geometries. These observations support a mechanism in which the actin network adapts to load by increasing in size to enhance force production and ensure robust CME across a wide range of membrane tension values.
RESULTS
Actin organization at CME sites suggests that force generation can be either parallel or orthogonal to the axis of CCP formation, or both, at different sites in the same cell
We used 2c-3D STORM (Rust et al., 2006; Huang et al., 2008) to determine the ultrastructural organization of the actin cytoskeleton at sites of CME. Henceforth, we refer to clathrin-coated structures (CCSs) as relatively flat clathrin structures and CCPs as curved, invaginating clathrin structures. Our 2c-3D STORM method preserves cellular ultrastructure by chemical fixation of intact cells, provides high molecular specificity due to immunofluorescence labeling, and allows large numbers of sites to be imaged (Xu et al., 2012). We conducted our experiments on a skin-melanoma cell line (SK-MEL-2) wherein ∼87% of dynamin2-eGFPEN (DNM2-eGFPEN) spots coaccumulate with actin (Grassart et al., 2014). We used this SK-MEL-2 cell line endogenously expressing DNM2-eGFPEN and clathrin light chain A-tagRFP-T (CLTA-TagRFP-TEN) for both live-cell and superresolution experiments (Doyon et al., 2011).
In these cells, we resolved CF680-labeled CCSs in the X-Y plane as discrete, round or elliptical shapes on the ventral surface (Figure 1A). Most of the CCSs appeared connected to filamentous actin visualized by Alexa Fluor 647–tagged phalloidin. These superresolution reconstructions resolve the association between clathrin coats and actin networks for hundreds of pits with high resolution in all three dimensions. The SDs of positions of single fluorophores were 10 nm in plane for XY and 19 nm in depth for the Z dimension (Supplemental Figure S1; Materials and Methods).
FIGURE 1:
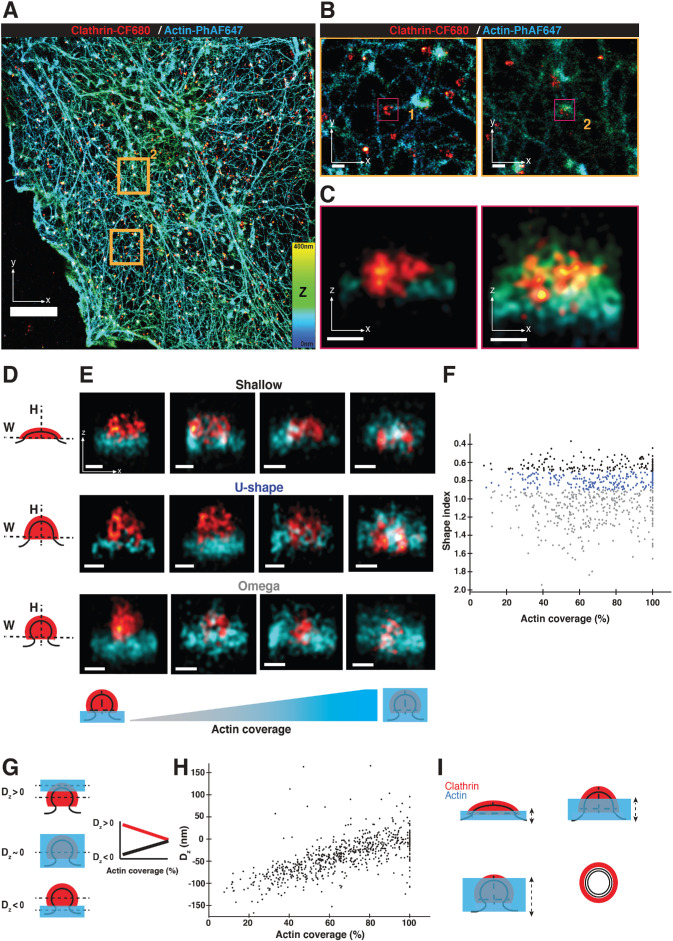
2c-3D STORM resolves clathrin structures highly connected to actin networks at different stages of endocytosis. (A) STORM image of the ventral surface of an SK-MEL-2 cell immunolabeled with the CF-680 antibody (clathrin coats in red) and phalloidin-AF647 (actin in cyan). Orange squares are areas shown in panel B. Color bar shows the z position of actin. Scale bar: 5 µm. (B) Magnification of highlighted areas 1 and 2 in panel A. Magenta squares are shown in panel C. Scale bars: 250 nm. (C) X-Z projections of the regions highlighted in panel B. Scale bars: 100 nm. (D) Illustration of binning clathrin coats (red) into three geometric stages based on their aspect ratio (shape index SI). Shallow: SI < 0.7; U-shape: 0.7 < SI > 0.9 and Ω: SI > 0.9. (E) X-Z projections of representative STORM images showing clathrin coats (red) with different actin (cyan) coverages around clathrin. Calculated SI of shallow CCSs from left to right image: 0.56, 0.53, 0.51, 0.55; for U-shaped CCPs from left image to right image: 0.87, 0.89, 0.86, 0.82; for Ω-shaped CCPs from left image to right image: 1.31, 1.06, 1.31, 1.52. Scale bars: 100 nm. (F) Graph of endocytic coat SI as a function of actin coverage for shallow (black dots), U-shaped (blue dots), and Ω-shaped (gray dots) pits. Categories of shape indices are chosen similar to E. Pits with actin coverage >5% are shown. R = –0.04, n = 719. Events accumulated from six cells. (G) Cartoon depicting the clathrin coat with actin either at the tip of the coat (top), covering the clathrin coat completely (middle), or at the base of the clathrin coat (bottom). Dashed black lines indicate the average Z position of actin and clathrin. Dz is the difference between average actin and clathrin Z positions. Dz < 0 is defined as the average actin Z position nearer the base of the pit. Schematic is a hypothetical plot of Dz vs. actin coverage for scenarios in which actin grows from the tip of the coat (red line) or the base of the pit (black line). (H) Dz as a function of actin coverage (for actin coverage >5%, R = 0.66, n = 719, N = 6 cells). (I) Cartoon of actin (blue) growing from the base of the pit (black lines) to cover clathrin coat (red) from a shallow membrane invagination to a fully formed membrane vesicle. X-Z projection (side profile) is shown. Dashed arrows indicate that growth of the actin network is not tightly coupled to the endocytic coat geometry and is variable in extent.
Knowing how actin networks are organized spatially in three dimensions at CME sites provides insights into its force generation mechanisms. It was important to show that we could distinguish actin specifically associated with CCSs from actin in the cell cortex. In Supplemental Figure S2, we show STORM images to compare actin at CCSs with actin at randomly selected regions of the cell cortex. We found examples of actin that specifically accumulates at the CCP (Supplemental Figure S2, D and I). Here, actin builds up higher into the cell interior compared with the actin extending horizontally away from the CCP and at randomly selected sites on the cell cortex (Supplemental Figure S2, E and J). This observation is consistent with recent cryoelectron tomograms in this cell type distinguishing branched actin networks around CCPs from the largely unbranched cortical actin network (Serwas et al., 2021).
To then investigate actin organization at multiple CCSs, we rendered the CCSs in three dimensions by cropping a tight area around each clathrin and actin structure in x-y STORM image projections and generated an x-z STORM image projections from the selected regions of interest (Figure 1, B and C). To our surprise, we observed strikingly different actin filament spatial organizations in the clathrin coats that we examined, even when they were near each other in the same cell and even when they were at the same morphological stage of CME (see below) (Figure 1C). In the first example shown, a thin layer of actin filaments resided along the base of the clathrin coat (Figure 1C, inset 1), reminiscent of structures observed in electron micrograph by Collins et al. (2011). In contrast, in the second example, actin filaments covered the CCP completely (Figure 1C, inset 2, and Supplemental Figure S2D). This organization resembles actin interacting with the entire clathrin coat as in yeast (Mulholland, 1994; Idrissi et al., 2008; Ferguson et al., 2009; Kukulski et al., 2012; Buser and Drubin, 2013; Mund et al., 2018) and in recent cryoelectron tomograms (Akamatsu et al., 2020). These micrographs indicate that distinct CME-associated actin structures can coexist in the same cell, consistent with models for force generation both parallel and orthogonal to the invagination axis.
CME site–associated actin networks grow from the CCP base to the tip of the coat, and their organization is not coupled to clathrin coat geometry
We used quantitative analysis of these STORM reconstructions to determine how actin is organized at CME sites and how actin organization relates to coat geometry. We first selected 989 high-resolution clathrin coats by visual inspection based on quality control criteria explained in Materials and Methods and determined which ones had associated actin filaments. When cropping CCSs for analysis, we could clearly distinguish single sites from double CCSs. In Supplemental Figure S3 we present views of 20 sites identified as singles and 20 sites identified as doubles in this manner. We excluded 86% of the visually inspected images (in total 6858) that accounted for double CCSs and CCSs of noncircular shape, as well as very large and very small clathrin antibody–positive immunostained structures in the XY plane (Supplemental Figure S4A). We detected actin associated with 74% of the clathrin coats selected for further analysis, which is comparable to previous measurements for the same cell type that we made from endocytic traces for events in live cells in which dynamin2-GFP associated with actin-RFP, considering that our superresolved images are snapshots of time-lapse events (Grassart et al., 2014).
We classified the coats by stage based on their shape in the XZ dimension, similar to earlier analyses of electron micrographs (Avinoam et al., 2015). For presentation purposes, clathrin coat shapes were binned into three geometrical categories based on their aspect ratio (which we call the shape index [SI]): shallow (flatter coat), U-shaped (intermediate aspect ratio), or Ω-shaped (coat with height similar to its width) (Figure 1D; Supplemental Figure S4, B and C). This measurement of SI was consistent whether we carried out the measurement on XZ or YZ projections of the coat (r2 = 0.92) (Supplemental Figure S5). We found a surprisingly wide variety of actin organizations in each of the three geometries (Figure 1E and Supplemental Figure S4D). To quantify the relationship between actin organization and endocytic coat geometry, the SI of the coat was plotted as a function of the extent of actin covering clathrin. When the actin network is larger in size than the clathrin coat, we define this state as 100% coverage (see Materials and Methods; Supplemental Figure S4B). There was no significant correlation between actin/coat coverage and coat shape for any of the three geometries (Figure 1F). We also observed a lateral mean displacement between the peak signals of clathrin and actin of 74 ± 42 nm, indicating an asymmetry in actin localization around the pits (Yarar et al., 2005; Collins et al., 2011; Jin et al., 2021; Serwas et al., 2021). This asymmetry value did not significantly change between CME geometries (Supplemental Figure S4, E and F). We conclude that irrespective of coat geometry, some pits have a thin actin network at the base of the pit, others have an intermediate level of coverage around the clathrin coat, and others have actin completely covering the pit. Thus, actin assembly at CCSs is not coupled to a particular stage of coat curvature development but might be recruited to promote curvature development.
In our reconstructions, we observed that whenever actin only partially covers the clathrin coat, the network is always located at the base of the pit (Figure 1E). This observation suggests that actin polymerization is nucleated at the base of the pit and that the network then grows inward around and over the tip of the clathrin coat. To test the generality of this observation, we calculated the difference in average axial (Z) position between the clathrin and actin signals for each site. We define this difference, Dz, such that negative Dz corresponds to actin residing nearer the base of the pit (Figure 1G). To determine whether actin grows from the base or tip of the pit, we plotted Dz as a function of the extent of coverage between actin and the clathrin coat. Increasing values of Dz would indicate that the network grows from the base of the pit toward the cell interior (Figure 1G). Indeed, as a function of actin/coat coverage, Dz increased from negative values to near zero (Figure 1H). We conclude that actin polymerization is initiated at the base of clathrin coats.
Given our finding that actin growth originates from the base of the pit, we next investigated the spatial distribution of the actin nucleation factor N-WASP at clathrin coats by 2c-3D STORM. Consistent with our conclusions about where actin assembly occurs at CCPs, N-WASP localized to the base of both shallow and highly curved clathrin coats (Supplemental Figure S6A). More unexpectedly, at some CME sites, N-WASP covered the entire clathrin coat irrespective of coat geometry (Supplemental Figure S6B).
In summary, we conclude that actin polymerization is nucleated at the base of CCPs and grows toward the coat’s tip (Figure 1I). Unexpectedly, actin nucleation is not coupled to coat geometry. A possible explanation for the variety of actin organizations that we observed associated with clathrin coats is that actin network organization responds to differences in load at CME sites.
Dynamics of CME slow down under elevated membrane tension
We next combined osmotic shock with live-cell fluorescence microscopy and 2c-3D STORM to determine how actin-mediated force generation contributes to CCP formation under elevated membrane tension (Figure 2A). Previous EM studies identified a requirement for actin filaments at the “U” to “Ω” transition (Boulant et al., 2011). However, for a mechanistic understanding, the quantitative relationship between membrane tension and endocytic dynamics must be elucidated (Akamatsu et al., 2020). Our quantitative light microscopy–based analysis of a large number of sites at different CME stages provided the necessary sensitivity to detect effects throughout the process. We first needed to establish conditions under which CME dynamics are affected by elevated membrane tension in these cells.
FIGURE 2:
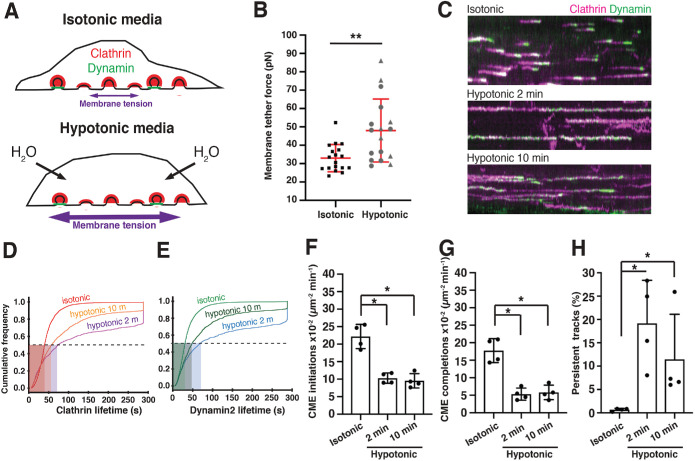
Quantitative analysis of CME mechanosensitivity under elevated membrane tension. (A) Schematic of cells in isotonic media (top) or hypotonic media (bottom), which causes water influx and stretches the cell membrane. In this figure, hypotonic treatment is 75 mOsm. (B) Mean membrane tether force values measured by AFM of cells in isotonic media (n = 18) or in hypotonic media (n = 17). Mean values were obtained by pulling at least three tethers (three independent experiments). In hypotonic treatment, circles are mean tether values from 2 to 10 min after hypotonic media exchange and triangles are mean tether values obtained between 10 and 16 min after hypotonic media exchange. Bars are mean ± SD. p = 0.002 by two-tailed Mann–Whitney test. (C) Kymographs of TIRF micrographs of live SK-MEL-2 cells endogenously expressing CLTA-TagRFP-TEN (magenta) and DNM2-eGFPEN (green). Time is on the X axis. Kymographs are 4.8 min long. Cells were imaged in isotonic media (top) or hypotonic media for 2 min (middle) or 10 min (bottom). (D) Cumulative distribution plot of clathrin lifetimes marked by CLTA-TagRFP-TEN in isotonic media (red), hypotonic media for 2 min (violet), and hypotonic media for 10 min (orange). These tracks were associated with DNM2-eGFPEN. (E) Cumulative distribution plot of dynamin2 lifetime marked by DNM2-eGFPEN in isotonic media (light green), hypotonic media for 2 min (blue), and hypotonic media for 10 min (dark green). These tracks were associated with CLTA-TagRFP-TEN. n = 5831 tracks in 17 cells across four experiments for D–H. (D, E) Detailed statistics in Supplemental Table S1. (F) Plot of endocytic initiation rate for the three conditions. p < 0.05 by two-tailed Mann–Whitney test for both comparisons. (G) Endocytic completion rate in the three conditions. p < 0.05 by two-tailed Mann–Whitney for both comparisons. (H) Percentage of persistent tracks (defined as tracks lasting the entirety of the image acquisition) for the three conditions. p < 0.05 by two-tailed Mann–Whitney for both comparisons. (F–H) Barplots show mean ± SD. Statistics to plots in Supplemental Figure S7H.
To determine how membrane tension is affected by changes in media osmolarity, we performed membrane tether pulling experiments by atomic force microscopy (AFM) on SK-MEL-2 cells cultured under isotonic and hypotonic conditions (75 mOsm) (Figure 2B). In isotonic media, the force required to maintain a pulled membrane tether was 33.0 ± 7.4 pN. Under hypotonic conditions the tether force increased to 48.0 ± 17.1 pN (Figure 2B). These measurements allowed us to quantitatively relate a given hypotonic environment in these cells to changes in membrane tension.
To decipher the relationship between CME dynamics and membrane tension, we used total internal reflection fluorescence (TIRF) microscopy to image SK-MEL-2 cells under isotonic and hypotonic conditions (Figure 2C). CLTA-TagRFP-TEN and DNM2-eGFPEN fluorescence lifetimes were determined by single-particle tracking (Hong et al., 2015). In isotonic media most clathrin tracks were relatively short (47 ± 32 s) with a burst of DNM2-eGFPEN signal peaking at the end (DNM2 lifetime 39 ± 32 s) (Figure 2C, Supplemental Movie 1, and Supplemental Table S1). Neither isotonic media nor exchange to slightly hypotonic 225 mOsm media noticeably affected DNM2-eGFPEN and CLTA-TagRFP-TEN lifetimes or CME initiation and completion rates (Supplemental Figure S7, A–D). Only 1–2% of these CLTA-TagRFP-TEN and DNM2-eGFPEN fluorescence tracks persisted over the entire 4.8 min movie under these conditions (Supplemental Figure S7, B and D).
Movie S1.
Movie of CLTA-TagRFP-TEN (magenta) and DNM2-eGFPEN (green) in SK-MEL-2 cells in isotonic media.
Upon treatment with moderately hypotonic (150 mOsm) media for 10 min, CLTA-TagRFP-TEN lifetime in cells increased by 20% (62 ± 51 vs. 49 ± 31 s) (Supplemental Figure S7E). This moderate treatment also had mild effects on the CME initiation rate (15 ± 5 × 10–2 μm–2 min–1 vs. 25 ± 6 × 10–2 μm–2 min–1), completion rate (11 ± 4 × 10–2 μm–2 min–1 vs. 19 ± 6 × 10–2 μm–2 min–1), and stalling rate (events that persist the entire length of our 4.8 min movies) (3 ± 1% vs. 0.8 ± 0.9% (Supplemental Figure S7F).
Treatment of cells with more strongly hypotonic (75 mOsm) media dramatically perturbed CME dynamics. After 2 min in 75 mOsm hypotonic media, tracks elongated and very often lasted the entire duration of the 4.8 min movie (Figure 2C, Supplemental Movie 2, and Supplemental Table S1). The mean lifetimes of CLTA-TagRFP-TEN and DNM2-eGFPEN tracks were 128 ± 112 s (Figure 2D) and 125 ± 112 s (Figure 2E), respectfully. We also observed a substantial decrease in the CME initiation rate (10.3 ± 1.5 × 10–2 μm–2 min–1 vs. 22.2 ± 3.5 × 10–2 μm–2 min–1) and completion rate (5.3 ± 1.7 × 10–2 μm–2 min–1 vs. 17.8 ± 3.4 × 10–2 μm–2 min–1) and an increase in track stalling (19 ± 9% vs. 0.7 ± 0.3%) (Figure 2, F–H; Supplemental Figure S7H). After 10 min of culturing, the CLTA-TagRFP-TEN and DNM2-eGFPEN lifetimes began to recover, most likely reflecting cellular adaptation to the hypotonic treatment (Figure 2, C–H, and Supplemental Movie 3, 4, and 5). We did not detect effects of hypotonic media treatment on lifetimes of tracks containing only CLTA-TagRFP-TEN, characteristic of clathrin structures not associated with CME (Supplemental Figure S7, A, C, E, and G) (Hong et al., 2015). DNM2-eGFPEN-only events, presumably representing non–clathrin mediated endocytosis events, showed a moderate response to elevated membrane tension with 75 mOsm hypotonic media (Supplemental Figure S7G). We conclude that elevating plasma membrane tension with hypotonic shock perturbs CME dynamics in a dose-dependent manner.
Movie S2.
Movie of CLTA-TagRFP-TEN (magenta) and DNM2-eGFPEN (green) in SK-MEL-2 cells after 2 minutes of media exchange to 75 mOsm media.
Movie S3.
Movie of CLTA-TagRFP-TEN (magenta) and DNM2-eGFPEN (green) in SK-MEL-2 cells after 10 minutes of media exchange to 75 mOsm media.
Movie S4.
Movie of CLTA-TagRFP-TEN (magenta) and DNM2-eGFPEN (green) in SK-MEL-2 cells in isotonic media before treatment with CK666 and 150 mOsm hypotonic media.
Movie S5.
Movie of CLTA-TagRFP-TEN (magenta) and DNM2-eGFPEN (green) in SK-MEL-2 cells in 150 mOsm hypotonic media with 100 μ M CK666. Movie acquisition started 2 minutes after media exchange.
Actin force generation assists clathrin coat remodeling under elevated membrane tension
Next, we determined the role of branched actin filament assembly on endocytic outcome under elevated membrane tension. Branched actin networks are generated by activated Arp2/3 complex (Mullins et al., 1998). We inhibited Arp2/3 complex–mediated actin polymerization using the small molecule CK666 (Nolen et al., 2009; Hetrick et al., 2013). We reasoned that inhibition of the Arp2/3 complex might have less effect on membrane tension than global actin inhibitors such as latrunculin and jasplakinolide often used in previous CME studies. Nevertheless, because Arp2/3 complex inhibition can affect membrane tension (Diz-Muñoz et al., 2016), we first carefully established experimental conditions in which effects of high membrane tension or CK666 would not mask one another. Treatment of cells with 100 µM CK666 did not affect the membrane tether force in these cells (p > 0.5) during the time the experiment was performed (Supplemental Figure S8A). Under these optimized conditions, we performed live-cell fluorescence microscopy and STORM to learn more about the clathrin coat remodeling steps at which branched actin network assembly is required.
We first titrated CK666 and monitored the effect on CME dynamics after 2 min of treatment. We aimed to identify a minimal CK666 concentration that induced a rapid effect on CME dynamics. CLTA-TagRFP-TEN lifetimes to 79 ± 66 s after 2 min of 100 μM CK666 treatment compared with 56 ± 40 s for the dimethyl sulfoxide (DMSO) control (Supplemental Figure S8B and Supplemental Table S2). Similarly, DNM2-EGFPEN lifetimes increased to 65 ± 64 s upon CK666 treatment, compared with 49 ± 38 s for the DMSO control (Supplemental Figure S8B; Supplemental Table S2). CK666 at 100 μM did not affect CME completion frequency or the percentage of persistent CLTA-TagRFP-TEN tracks (Supplemental Figure S8, C and D), though we observed a small decrease in the CME initiation rate (Supplemental Figure S8E).
The elongation of DNM2-eGFPEN and CLTA-TagRFP-TEN lifetimes upon 100 μM CK666 treatment was exacerbated upon simultaneous incubation in moderately hypotonic media (150 mOsm). Compared with controls, the combination of 100 μM CK666 and 150 mOsm hyopotonic media markedly lengthened the clathrin lifetimes to 96 ± 86 s (compared with DMSO control, 59 ± 52 s) and dynamin2 to 84 ± 85 s (compared with DMSO control, 47 ± 49 s) (Figure 3, A–F, and Supplemental Table S3). We conclude that in these cells, Arp2/3 complex–mediated actin assembly is required to maintain normal CME dynamics under elevated membrane tension.
FIGURE 3:
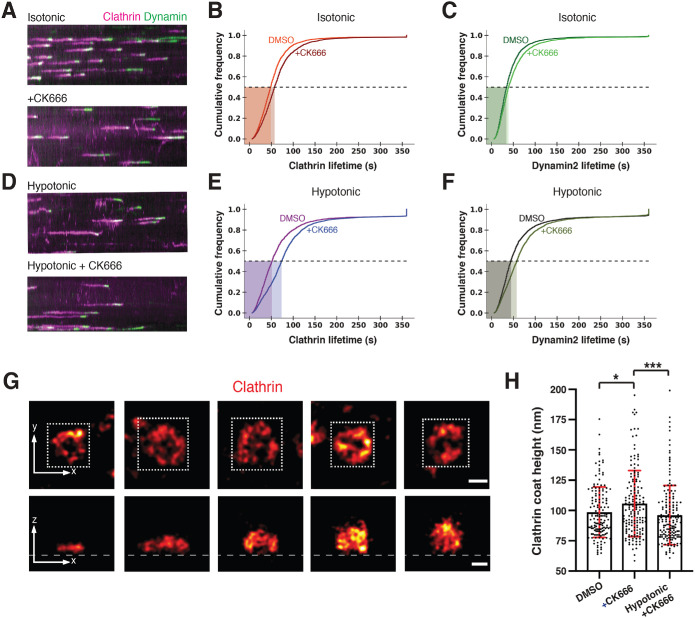
Importance of Arp2/3 complex–mediated actin polymerization during CME increases under elevated membrane tension. In this figure, CK666 (Arp2/3 complex inhibitor) treatment is 100 µM and hypotonic shock is 150 mOsm. (A) Kymographs of cells expressing CLTA-TagRFP-TEN (magenta) and DNM2-eGFPEN (green) imaged by TIRF. Cells were imaged in isotonic media. The media was then exchanged to include CK666 (bottom panel) and imaged after 4 min. (B) Cumulative distribution plots of clathrin lifetimes in control, DMSO-treated conditions (orange, n = 4219), and CK666-treated (red, n = 3124) conditions. (C) Cumulative distribution plots for control, DMSO-treated (dark green, n = 4219), and CK666-treated (light green, n = 3124) dynamin2 lifetimes associated with clathrin lifetimes in B. (BC) Control, N = 10 cells, and CK666 treatment, N = 10 cells, measured in three independent experiments. Complete statistics in Supplemental Table S3. (D) Kymographs of cells in hypotonic media. In the top panel, cells were placed in hypotonic media and imaged after 4 min. In the bottom panel, cells were treated with CK666 in hypotonic media and imaged after 4 min. (E) Cumulative distribution plots of clathrin lifetimes for control, DMSO-treated conditions (magenta, n = 1405), and CK666-treated conditions (blue, n = 2783) in hypotonic media. (F) Cumulative distribution plots of DMSO-treated (black, n = 1405) and CK666-treated (olive, n = 2783) dynamin2 lifetimes in hypotonic media associated with clathrin lifetimes in E. (E, F) Control, N = 9 cells, and CK666 treatment, N = 10 cells, measured in three independent experiments. Complete statistics in Supplemental Table S3. (G) Representative STORM images of immunolabeled clathrin-coated structures in control cells arranged by coat height. Top panel shows the x-y projections and bottom panel the corresponding x-z projections. The white square in the x-y projections shows the area that was cropped to generate the x-z projections. The heights of clathrin coats in the x-y projection from left to right image are 61, 96, 124, 158, and 177 nm. Scale bars: 100 nm. (H) Clathrin coat heights when cells were treated with DMSO (n = 154) or CK666 (n = 158) in isotonic media or CK666 in hypotonic media (n = 159). Clathrin coat images for quantitative analysis were collected from at least three different cells for each condition from a single experiment. Statistics are given in Supplemental Figure S8F. p < 0.05 in both comparisons by Mann–Whitney test.
Next, we used STORM to determine which endocytic coat geometries are enriched upon a combination of Arp2/3 complex inhibition and osmolarity treatment. Cells were cultured under the CK666 and osmolarity conditions described above, chemically fixed, and then immunolabeled for clathrin. Clathrin coat height served as a metric for the membrane invagination stage. As in the above 2c-3D STORM experiments, the full progression from a flat clathrin coat to a rounded vesicle could be clearly resolved in 3D (Figure 3G, bottom image panel). Control cells treated with DMSO showed an average clathrin coat height of 98 ± 21 nm (Figure 3H and Supplemental Figure S8F). The average height increased to 106 ± 27 nm when cells were treated with 100 μM CK666 (Figure 3H and Supplemental Figure S8F). Thus, when Arp2/3 complex activity was inhibited in these cells, clathrin pits accumulated at a greater height. This observation suggests that more clathrin pits may stall at a later stage of progression, consistent with observations of accumulated coat geometries from actin inhibitor studies (Yarar et al., 2005; Boulant et al., 2011; Yoshida et al., 2018).
Interestingly, when Arp2/3-mediated actin polymerization was inhibited in cells with elevated membrane tension, the average clathrin coat height decreased to 96 ± 24 nm (Figure 3, G and H, and Supplemental Figure S8F). This height decrease was also reflected in an accumulation of smaller shape indices and a very mild effect on clathrin coat width (Supplemental Figure S8H). This result, along with our observation of slowed CME dynamics, suggests that upon membrane tension elevation in response to hypotonic conditions in SK-MEL-2 cells, inhibition of Arp2/3 complex activity may cause an enrichment of shallow endocytic coat geometries.
Actin organization adapts to elevated membrane tension by increasing clathrin coat coverage
Finally, we used STORM to determine the relationship between elevated membrane tension and actin cytoskeleton organization at CME sites (Figure 4). We treated cells with strong (75 mOsm) hypotonic shock for 5 min and then chemically fixed them for 2c-3D STORM. When we superresolved clathrin and actin by 2c-3D STORM in these cells, the actin cytoskeleton remained intact (Pan et al., 2019) and associated with CCSs (Supplemental Figure S9, A–D).
FIGURE 4:
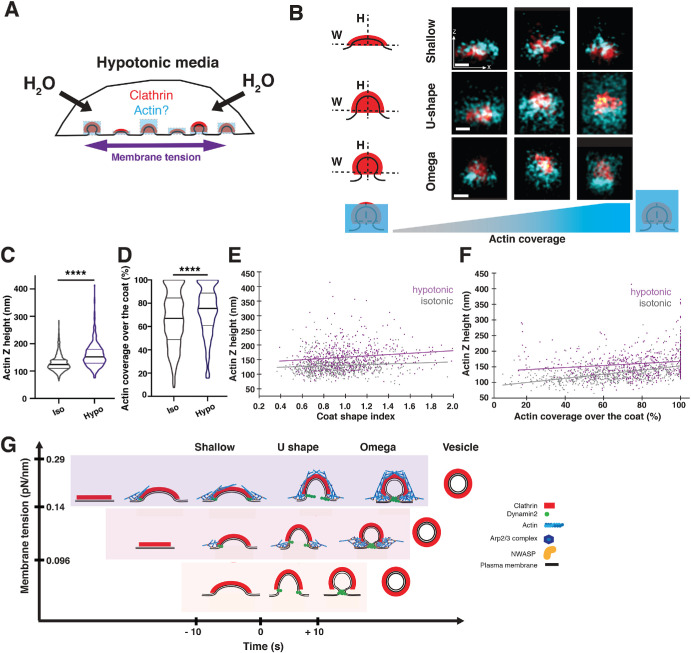
The actin network at CME sites increases in size in response to elevated membrane tension. In this figure, hypotonic refers to 75 mOsm media. (A) Schematic of cells in hypotonic media, which increases plasma membrane tension. The response of the actin network (blue) to elevated plasma membrane tension (purple) was previously unknown. (B) Representative STORM images of clathrin (red) and actin (cyan) in x-z projections for cells fixed after treatment in the hypotonic media for 5 min (bottom). Coated pits are classified as shallow, U-shaped, or Ω-shaped based on the aspect ratio of the coat. Scale bars: 100 nm. (C) Plots of actin Z height at CCPs from cells in the isotonic (n = 736) and hypotonic (n = 527) media measured from STORM x-z projections. Lines are median ± interquartile range. p < 0.0001 determined by Mann–Whitney test. (D) Plots of actin coverage over the clathrin coat in pits found in STORM x-z projection images in isotonic (n = 719) and hypotonic (n = 509) conditions. Pits with actin coverage >1% are plotted. Lines are median ± interquartile range. p < 0.0001 determined by Mann–Whitney test. (E) Actin Z height as a function of coat shape in isotonic (gray, n = 736) and hypotonic (purple, n = 527) conditions. (F) Actin Z height as a function of actin coverage over the clathrin coat in isotonic (gray, n = 719) and hypotonic (purple, n = 509) conditions. The data for isotonic conditions were also used to generate the plots in Figure 1. Three independent STORM experiments with N_ = 6 cells in isotonic and N_ = 7 cells in hypotonic media. (G) Cartoon depicting an adaptive actin force–generating mechanism that counteracts elevated membrane tension to ensure robust CME progression. This schematic describes three scenarios in which membrane tension is low, intermediate, or high and how CME is accomplished efficiently by adaptive actin network organization. Under low tension (bottom), the clathrin coat provides sufficient force to advance CME. At intermediate tension (middle), actin polymerization is required for the transition from U to Ω shape. At high tension (top), endocytic progression slows. More pits stall at the shallow conformation. In response to increased resistance, the actin network grows to envelop the coat and provide additional force against high membrane tension.
Strikingly, in response to hypotonic media treatment, the average actin height increased for all endocytic geometries (Figure 4, B, C, and E; Supplemental Figure S9, E–G). When we quantified actin cell cortex height in randomly selected regions of the cell cortex, we did not observe any increase in cortical actin height, reinforcing the conclusion that any observed effects are specific for CME sites (Supplemental Figure S10, A–J and P–R). Overall, the average actin height at CME sites increased from 130 ± 30 nm under isotonic media conditions to 160 ± 40 nm (Figure 4C). This increase corresponded to an increase of actin growth over the clathrin coat from covering 66 ± 23% of the coat to covering 73 ± 21% under 75 mOsm hypotonic media (Figure 4D). The increase in actin height was observed for all coat shapes (e.g., shallow or curved pits) (Figure 4E). Similarly, for different extents of clathrin coverage by actin, the average actin height was greater in hypotonic conditions across different levels of coat coverage (Figure 4F). However, the extent of asymmetry between actin and clathrin signals did not significantly change compared with that in the isotonic condition (Supplemental Figure S9H). These observations of greater average actin height and coverage over the clathrin coat suggest that the force contribution of actin at CME sites is increased when membrane tension is elevated such that a greater load is carried by the actin network.
Overall, these observations led us to conclude that under elevated plasma membrane tension, actin grows higher in the z-dimension around clathrin coats (Figure 4G). Such an adaptive mechanism for actin organization presumably generates the required forces to ensure the efficient progression of mammalian CME under various levels of membrane tension.
DISCUSSION
By combining 2c-3D STORM imaging and quantitative live-cell TIRF microscopy for thousands of CME sites, with AFM tether pulling membrane tension measurements, and manipulations of membrane tension and Arp2/3-mediated actin assembly, we showed that actin assembly adapts to changes in load at individual CME sites. This mechanism likely ensures that sufficient forces are generated for robust endocytic progression over a range of membrane tension regimes. While STORM of individual CCSs cannot attain the resolution of EM, our approach had several advantages that allowed us to gain new mechanistic insights: 1) it allowed us to sample much larger numbers of CME sites with a preserved actin network than is possible by EM, thus permitting rigorous quantitative analysis, 2) we imaged CME sites and associated actin in intact cells that had not been subjected to unroofing or extraction protocols, and 3) we were able to use antibodies and fluorescent phalloidin to unambiguously identify specific proteins at CME sites.
Actin assembly and organization adapt to elevated membrane tension
Elevating membrane tension can have a dramatic impact on CME progression in mammalian cells (Raucher and Sheetz, 1999; Boulant et al., 2011; Ferguson et al., 2016, 2017; Willy et al., 2017; Bucher et al., 2018). However, how cells adapt to compensate for increased load at CME sites has not been elucidated. Our results provide critical mechanistic insights into how the CME machinery adapts to elevated membrane tension to maintain robust CME progression. We showed quantitatively that actin assembly and organization adapt to changes in membrane tension, which we measured by AFM membrane tether pulling (Figure 2). Changes in membrane tension can in principle occur globally (entire cells) or locally (in different regions of one cell, or even within different regions of a single endocytic site) (Houk et al., 2012; Rangamani et al., 2014; Shi et al., 2018). Because we detect different actin organizations at individual CME sites with similar clathrin coat geometries within a single cell, such differences might reflect subcellular, local load variation resulting from variance in such factors as membrane tension and cell adhesion. The observation in other studies that actin assembles late in CME progression (Merrifield et al., 2002; Grassart et al., 2014; Akamatsu et al., 2020; Jin et al., 2021), coupled with the observation here that actin can associate with clathrin coats of different geometries, is consistent with the possibility that clathrin lattices of low curvature can develop high curvature under the “constant area” model (Avinoam et al., 2015; Sochacki et al., 2021). Moreover, our data provide evidence that the low to high coat curvature transition becomes more dependent on actin assembly forces as load increases.
Measurement of membrane tension changes using AFM.
Using AFM, we measured the membrane tether force for SK-MEL-2 cells in isotonic and hypotonic conditions (Figure 2). We measured tension on the dorsal cell surface while imaging endocytic events on the ventral surface. Local differences in membrane tension have been observed in some cell types (Shi et al., 2018) but not others (Houk et al., 2012). Nevertheless, AFM remains the gold standard for measuring apparent membrane tension in cells (Sitarska and Diz-Muñoz, 2020). In isotonic conditions, the tether force was 33 ± 7 pN (Figure 2B). This tether force is within an intermediate range measured for other cell types such as NIH3T3 cells, HeLa cells, and macrophages (Sitarska and Diz-Muñoz, 2020; Roffay et al., 2021). Assuming a 100 pN*nm bending rigidity of the plasma membrane, this value corresponds to a membrane tension of 0.14 ± 0.04 pN/nm (Diz-Muñoz et al., 2013). In hypotonic conditions, the membrane tether force increased to 48 ± 17 pN, which corresponds to a doubling of membrane tension to 0.29 ± 0.04 pN/nm. Higher membrane tether values have been reported for other cell types (Sitarska and Diz-Muñoz, 2020). Below, we describe implications of earlier results and our new results reported here on actin’s role in CME under three different plasma membrane tension regimes (Figure 4G).
Low-membrane-tension regime.
When membrane tension is low, clathrin coat assembly provides sufficient energy to bend the underlying plasma membrane into a full spherical shape, similar to the constant-curvature model (Figure 4G) (Saleem et al., 2015; Willy et al., 2021). We indeed found in our STORM data that 26% of clathrin coats lack actin in shallow, U-shape and Ω-shape CME geometries. This observation is consistent with mathematical modeling, which indicates that the coat can provide sufficient energy to bend the plasma membrane when membrane tension is low (0.002 pN/nm) (Hassinger et al., 2017). Here, we consider low membrane tension to be of a value lower than those we measured for SK-MEL-2 cells in standard media (Figure 2). Given that actin polymerization appears to be dispensable for CME in some cell types, even though it still might make CME more efficient, we hypothesize that the basal membrane tension may be lower in those cell types. However, we urge caution with the interpretation of experiments with harsher actin drug treatments, as prolonged actin inhibitor treatment of cells can perturb the actin cortex and reduce the tether force measurements (membrane tension) by a factor of ∼50% (Pontes et al., 2017). In future studies, strategies should be developed to relate local membrane tension to specific endocytic site geometry and actin organization.
Intermediate tension regime.
In the intermediate tension regime, defined as the resting tension of SK-MEL-2 cells in isotonic conditions (Figure 2), clathrin coat assembly and membrane curvature–inducing proteins still appear to provide sufficient energy to drive clathrin coats to adopt the U shape (Figure 4G, intermediate membrane tension). When we inhibited Arp2/3-mediated actin polymerization using CK666, clathrin coat progression stalled at an aspect ratio most likely reflecting the U-shaped stage, consistent with the effects of actin assembly inhibition reported for other cell types (Yarar et al., 2005; Boulant et al., 2011; Almeida-Souza et al., 2018). Thus, at intermediate membrane tension it appears that actin force generation is primarily required for the U- to Ω-shaped clathrin coat transition. The actin network observed near the base of the pit may reflect a role driving plasma membrane neck constriction and scission by generating forces orthogonal to the direction of membrane invagination as proposed previously (Bucher et al., 2018; Scott et al., 2018; Mund et al., 2021).
High-membrane-tension regime.
Our live-cell and STORM observations indicate that as membrane tension is elevated further, coat deformation and membrane invagination become increasingly dependent on actin force generation (Figure 4). When membrane tension was elevated to an intermediate level (e.g., 150 mOsm media), CME lifetimes slowed modestly (Supplemental Figure S7). When we inhibited Arp2/3-mediated actin polymerization using CK666 in cells treated with 150 mOsm media, the average clathrin coat height was lower than in CK666-treated cells cultured under isotonic conditions, likely reflecting an enrichment of shallow pits (Figure 3H). Under elevated membrane tension, we observed no change in actin cortex thickness, while others reported that the actually actin cortex tends to become thinner and less dense (Supplemental Figure S10) (Houk et al., 2012; Chugh et al., 2017; Roffay et al., 2021). However, our study shows that the actin network associated with CCPs instead increases in height under elevated membrane tension (Figure 4). Actin assembly from the base of the CCP continues until the network covers the clathrin coat completely, allowing it to interact with proteins linking the actin network to the clathrin coat (Engqvist-Goldstein et al., 2001; Messa et al., 2014; Sochacki et al., 2017). Actin-binding linker proteins such as Hip1R and Epsin1 cover the clathrin coat completely and are thus positioned to provide high internalization efficiency by harnessing actin assembly forces perpendicular to the plasma membrane (Akamatsu et al., 2020; Serwas et al., 2021). It may be that the “constant area” model (Avinoam et al., 2015; Sochacki et al., 2021), in which a flat or shallow coat grows to full size and then bends, applies under the high load regime (Bucher et al., 2018; Scott et al., 2018; Mund et al., 2021), and it is here that actin assembly forces are most important for remodeling the coat into a curved vesicle.
An important question for future studies concerns the nature of the adaptive mechanism that increases actin assembly in response to elevated membrane tension. Possible mechanisms include the following: 1) stalling works passively to allow actin to assemble longer; or 2) high load works actively through mechanisms documented for other contexts, for example by increasing contact of filaments associated with the coated pit with membrane-associated N-WASP-Arp2/3 complex, leading to more assembly, alongside load-dependent decreases in the rate of filament capping (Bieling et al., 2016; Akamatsu et al., 2020; Funk et al., 2021; Li et al., 2021). As endocytosis and other actin-mediated trafficking events operate within a membrane geometry and local protein and lipid environment different from that in lamellipodia, a high priority is now to determine whether similar adaptive mechanisms operate during such local membrane bending processes.
When the actin network fully covers the clathrin coat, it resembles the radial organization described by mathematical modeling for mammalian CME and the actin network organization described for budding yeast (Ferguson et al., 2009; Hassinger et al., 2017; Mund et al., 2018; Akamatsu et al., 2020). In yeast this actin organization drives endocytic membrane invagination against the high resistance that results from turgor pressure. Mathematical modeling showed that this organization produces high forces perpendicular to the plasma membrane (Hassinger et al., 2017). Actin-generated forces parallel and orthogonal to the membrane invagination at high tension may coexist to drive membrane invagination followed by neck constriction and scission.
CME dynamics dramatically slow down when cells are in this high-membrane-tension regime, resulting in only ∼40% of endocytic lifetimes shorter than 50 s compared with 80% at low membrane tension (Figure 2D). We also detected ∼19% of lifetimes being longer than the 4.8 min movies that we captured, a 20-fold increase in stalled (persistent) tracks (Figure 2H). We suggest that this tension regime pushes this adaptive mechanism to the limit.
N-WASP spatial organization suggests an actin force generation control mechanism
N-WASP spatial organization at CCSs and CCPs provides valuable mechanistic insight into how actin network assembly contributes to force generation during CME. We found that N-WASP localizes at the base of early and late CCPs, where it likely interacts with SH3 domain–containing proteins present at the endocytic membrane neck (Schöneberg et al., 2017; Sochacki et al., 2017; Almeida-Souza et al., 2018). This organization is similar to that of the homologous nucleation-promoting factor Las17 in budding yeast (Mund et al., 2018). Filaments nucleated at the base of CCPs would be able to interact with coat proteins such as Hip1R and Epsin1/2/3 to generate forces to invaginate the plasma membrane (Hassinger et al., 2017; Mund et al., 2018; Akamatsu et al., 2020; Joseph et al., 2020).
Intriguingly, we also sometimes observed a strikingly different N-WASP spatial organization in which it was distributed over the full clathrin coat. The type II nucleation factors Abp1 and cortactin bind to actin filaments and to the Arp2/3 complex and could serve as binding partners for N-WASP around the CCP (Le Clainche et al., 2007; Pinyol et al., 2007; Helgeson and Nolen, 2013; Guo et al., 2018). WASP located on the clathrin coat might reflect a distinct path for filament nucleation, originating from the coat rather than the base, that is potentially important to generate higher forces when actin already surrounds the CCP.
A late burst of actin assembly was shown previously to often accompany CME (Merrifield et al., 2002) and to facilitate CME progression, especially when membrane tension is high (Fujimoto et al., 2000; Yarar et al., 2005; Batchelder and Yarar, 2010; Boulant et al., 2011; Grassart et al., 2014; Li et al., 2015; Ferguson et al., 2017; Yoshida et al., 2018; Akamatsu et al., 2020; Jin et al., 2021). In this study we observed that load-adapted, CME-associated actin assembly buffers changes in plasma membrane tension, thereby ensuring clathrin-coated vesicle (CCV) formation over a range of membrane tension regimes. We expect that the ability of actin assembly to respond to load is a common feature critical to many membrane remodeling processes.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture
SK-MEL-2 cells from clone Ti13 (hCLTAEN–1/hDNM2EN–1) were cultured in DMEM/F12 with GlutaMax supplement (10565-018; Thermo Fisher Scientific) containing 10% fetal bovine serum (FBS) and a 1000 U/ml penicillin–streptomycin mix (15140122; Thermo Fisher Scientific) and kept in a 37°C humidified incubator with 5% CO2 (cell source information in Doyon et al., 2011). After each cell vial was thawed, cells were checked after two passages for mycoplasma contamination. Cell line authentication was performed by short tandem repeat validation.
Antibodies and reagents
The primary antibodies used were mouse anti-clathrin light chain (AB CON.1, MA5-11860; Thermo Fisher Scientific), mouse anti-clathrin heavy chain (AB X-22, MA1-065; Thermo Fisher Scientific), and rabbit anti–N-WASP (ab126626; Abcam). The secondary antibodies used were Alexa Fluorophore 647 chicken anti-rabbit (A21443; Thermo Fischer Scientific), goat anti-mouse (115-005-205; Jackson ImmunoResearch) conjugated to CF680-NHS ester (Biotium; 92139). Reagents and small molecule inhibitors used were DMSO (D2650; Sigma Aldrich), CK666 (SML0006, batch #0000012761; Sigma Aldrich), and phalloidin-AF647 (A22287; Fisher Scientific).
Preparation of CF680-labeled secondary goat anti-mouse antibody
CF680 NHS ester was dissolved at a concentration of 3 mM in anhydrous DMSO. One microliter of dye solution, 80 μl of a 1.25 mg/ml suspension of unlabeled goat anti-mouse immunoglobulin G1 secondary antibody (115-005-205; Jackson ImmunoResearch Laboratories), and 10 μl of 1 M sodium bicarbonate solution were mixed and allowed to react for 15 min at room temperature. The reaction mixture was added to an equilibrated NAP-5 column (Sigma; GE17-0853-01) and flushed with phosphate-buffered saline (PBS). The dye-conjugated antibody was collected from the first colored eluent fraction, and a concentration of 0.12 mg/ml was determined with a NanoDrop spectrophotometer.
Sample preparation for two-color clathrin and actin imaging
Round coverslips (18 mm) were cleaned 20 min in 70% ethanol (Electron Microscopy Science; Cat #72222-01). Cells were detached with 500 µl 0.05% trypsin (25300-054; Life Technologies), washed once in DMEM/F12, and collected by centrifugation. Cells were counted using a hemocytometer,and 20,000 cells/ml were seeded onto 18 mm round coverslips in 12-well plates. Cells were incubated for 16–24 h in culture media before preparation for imaging.
Cells were fixed first for 1–2 min in 0.3% (vol/vol) glutaraldehyde (GA) solution containing 0.25% (vol/vol) Triton in cytoskeleton buffer (CB: 10 mM MES, 150 mM NaCl, 5 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), 5 mM glucose, 5 mM MgCl2, 0.005% NaN3, pH 6.1) and then immediately fixed for 10 min in 2% (vol/vol) GA solution in CB. Both solutions were prepared fresh from a 70% GA stock (Electron Microscopy Science; cat #16365) (protocol follows Xu et al., 2012). After fixation, samples were washed once in CB and then incubated for 7 min in freshly prepared CB containing 0.1% (wt/vol) NaBH4. Subsequently, samples were washed three times for 10 min in CB with gentle agitation on a shaker. Samples were then blocked for 30 min in 5% (wt/vol) bovine serum albumin (BSA) in CB (Sigma Aldrich; A3733). For dense clathrin labeling, light (diluted 1:200) and heavy (diluted 1:200) chain antibodies were used together in a 1% (wt/vol) BSA CB solution. Primary antibody immunostaining was performed overnight at 4°C. The next day, samples were washed twice in 1% (wt/vol) BSA CB for 5 min. The mouse secondary antibody CF680 was used at a final concentration of 0.40–0.60 μg/ml in a 1% BSA–1× CB solution. Samples were stained for 30 min at room temperature in the dark and washed twice for 5 min in 1% (wt/vol) BSA CB solution and then for 10 min in CB solution. Samples were then placed into a solution of CB containing 0.5 μM phalloidin-AF647 and kept at room temperature in the dark for a minimum of 2 h. Samples were washed once with PBS before STORM imaging.
Sample preparation for single-color clathrin and dual-color N-WASP imaging
Cells were prepared as for the two-color sample preparation on coverslips and then fixed for 20 min in 3% (vol/vol) paraformaldehyde (PFA; 15710; Electron Microscopy Sciences) in CB (protocol follows Li et al., 2018). Samples were washed quickly in CB and subsequently were incubated for 7 min in freshly prepared 0.1% (wt/vol) NaBH4 in CB solution. Subsequently, samples were washed three times for 10 min in CB with gentle agitation on a shaker and permeabilized afterward in a 0.1% Triton-PBS solution for 1–2 min. For single-antibody clathrin staining, subsequent washing, blocking, and antibody incubation steps were similar to those of the two-color clathrin and actin sample preparation protocol.
Dual-color immunolabeling was performed with primary antibody against N-WASP (diluted 1:200) and clathrin heavy and clathrin light chain (diluted 1:600–1:1000) in 1% (wt/vol) BSA in PBS overnight at 4°C. Samples were washed the next day twice for 5 min in 1% (wt/vol) BSA in PBS. Secondary antibody staining was first performed with Alexa Fluorophore 647 anti-rabbit antibody (diluted 1:200) in 1% BSA (wt/vol) in PBS for 30 min at room temperature and kept in the dark. After two 10-min-long washes in PBS containing 1% (wt/vol) BSA, secondary antibody staining was performed with CF680 anti-mouse antibody (diluted 1:600). The samples were given three final washes in PBS for 10 min each.
SRM imaging
Dye-labeled cell samples were mounted on glass slides in a standard STORM imaging buffer consisting of 5% (wt/vol) glucose, 100 mM cysteamine, 0.8 mg/ml glucose oxidase, and 40 µg/ml catalase in 1 M Tris-HCl (pH 7.5) (Rust et al., 2006; Huang et al., 2008). Coverslips were sealed using Cytoseal 60. STORM imaging was performed on a homebuilt setup (Wojcik et al., 2015) based on a modified Nikon Eclipse Ti-U inverted fluorescence microscope using a Nikon CFI Plan Apo λ 100× oil immersion objective (NA 1.45). Dye molecules were photoswitched to the dark state and imaged using a 647-nm laser (MPB Communications); this laser was passed through an acousto-optic tunable filter and introduced through an optical fiber into the back focal plane of the microscope and onto the sample at an intensity of ∼2 kW cm2. A translation stage was used to shift the laser beam toward the edge of the objective so the light reached the sample at incident angles slightly smaller than the critical angle of the glass–water interface. A 405-nm laser was used concurrently with the 647-nm laser to reactivate fluorophores into the emitting state. The power of the 405-nm laser (typical range 0–1 W cm–2) was adjusted during image acquisition so that at any given instant, only a small, optically resolvable fraction of the fluorophores in the sample was in the emitting state. For 3D STORM imaging, a cylindrical lens was inserted into the imaging path so that images of single molecules were elongated in opposite directions for molecules on the proximal and distal sides of the focal plane (Huang et al., 2008). The raw STORM data were analyzed according to previously described methods (Rust et al., 2006; Huang et al., 2008). Data were collected at a frame rate of 110 Hz for a total of ∼80,000 frames per image. Single- and two-color imaging was performed on cells labeled with Alexa Fluor 647 only or Alexa Fluor 647 and CF680 with 647-nm excitation based on a ratiometric detection scheme (Bossi et al., 2008; Testa et al., 2010; Gorur et al., 2017). In the two-color imaging scheme, light emitted from the AF647 and CF680 fluorophores was collected concurrently and split into two light paths using a long-pass dichroic mirror (T685lpxr; Chroma). Each light path was projected onto half of an Andor iXon Ultra 897 EM-CCD camera. Dye assignment was performed by localizing and recording the intensity of each single molecule in each channel. Conventional imaging of 560- and 488-nm dyes was performed immediately before STORM imaging using the appropriate laser and filter set. Emission data were collected through the short-wavelength reflected path of the aforementioned optical setup and overlaid directly onto the final STORM image. Details of selection and analysis of SRM images are found in the Supplemental Information.
Our 3D-STORM setup was based on the same design as in Huang et al. (2008) and so we achieved comparable resolutions. The experimental STORM resolution was measured by repeatedly measuring the position of a single fluorophore and determining the SD of the localization distribution (Huang et al., 2008; Xu et al., 2015). We accordingly examined our STORM data in this work and overlaid the localization distributions of 24 representative single molecules from three different samples, as shown (Supplemental Figure S1). Gaussian fits (red curves) gave SDs of 10 nm in plane for the XY directions and 19 nm in depth for the Z direction. These results are similar to those reported in Figure 1C of Huang et al. (2008), where SDs are 9 nm in X, 11 nm in Y, and 22 nm in Z.
TIRF microscopy
TIRF imaging was carried out on a Nikon Eclipse Ti2 inverted microscope with a CFI60 60× Apo TIRF objective and a Hamamatsu Orca-Flash 4.0 V2 sCMOS camera. eGFP and Tag.RFP-T fluorescence was excited using 488 and 561 nm lasers and detected using a Chroma HC TIRF Quad Dichroic (C-FL TIRF Ultra Hi S/N 405/488/561/638) and Chroma HC Quad emission filters BP 525/50 and BP600/50, respectively (Bellows Falls, VT). Unless mentioned specifically, channels were acquired sequentially at a 1.2 s interval and 400 ms exposure time over 4.8 to 6 min. Real-time acquisition was achieved by a National Instruments (PXI 1033; Austin, TX) controller. The system was controlled with NIS-Elements software and maintained at 37°C by an OkoLab environmental chamber (Burlingame, CA).
Hypo-osmotic media treatment
SK-MEL-2 cells were plated on glass coverslips 1 d before osmotic treatment and imaging: 20,000 cells/m were seeded 16–24 h before the experiment on 25 mm round #1.5 glass coverslips that had been cleaned with 70% ethanol (Warner Instruments; 64-0715). Isotonic imaging media contained DMEM and Ham’s F-12 medium (DMEM/F12) without phenol red (11039; Thermo Fisher Scientific) with 5% vol/vol FBS. The media was diluted with an inorganic salt solution containing 10 mM CaCl2 0.3 mM MgCl2 and 0.1 mM MgSO4 (CMM) to maintain concentrations of critical ions, while obtaining hypo-osmotic conditions by diluting the media containing components such as d-glucose. The 225 mOsm hypotonic imaging media contained 1:4 vol/vol CMM solution in DMEM/F12, the 150 mOsm hypotonic imaging media contained 1:1 vol/vol CMM solution in DMEM/F12, and the 75 mOsm hypotonic imaging media contained 4:1 vol/vol CMM solution in DMEM/F12. Five percent vol/vol FBS was present in all hypotonic solutions.
For live-cell fluorescence microscopy
CLTA-TagRFP-TEN and DNM2-eGFPEN fluorescence in SK-MEL-2 cells was acquired first in isotonic media over a course of 4.8 min. Subsequently, media was exchanged on the stage to hypotonic media (either 225, 150, or 75 mOsm) and movies were acquired for 4.8 min, starting 2 and 10 min after media exchange. Media exchange on the stage did not affect CME initiation rates or fluorescence lifetimes beyond the existing experimental intrinsic variability (Supplemental Figure S7, A and B).
For STORM imaging, 75 mOsm hypotonic buffer treatment was performed in the cell culture dish for 5 min. Cells were immediately chemically fixed after the 5 min treatment and further treated with the STORM sample preparation protocol as described above. Methods to analyze TIRF data can be found in the Image analysis section.
CK666 concentration titration
SK-MEL-2 cells (20,000/ml) were seeded in eight-well chambers 16 h—24 h before the experiment (80826; ibidi, Fitchburg, WC). A CK666 (SML0006, batch #0000012761; Sigma Aldrich) stock solution was prepared at 50 mM in DMSO and kept at –20°C. CK666 (25, 50, and 100 μM) and equivalent 0.5% vol/vol DMSO, 1% vol/vol DMSO, and 2% DMSO vol/vol solutions for controls were prepared fresh in DMEM/F12 containing 5% FBS and kept at 37°C until used. Cells were first imaged in DMEM/F12 containing 5% FBS solution as a baseline control for 4.8 min. Subsequently, imaging solution was exchanged on the microscopy stage to CK666- or DMSO-containing imaging solution and another 4.8-min movie was acquired after 2 min of treatment. Each treatment was repeated twice, and an area of 1024 pixels × 1024 pixels was used to record 3–6 cells per experiment.
CK666 in combination with hypo-osmotic media
Cells were prepared as for the CK666 concentration titration experiment described above. Solutions of 2% vol/vol DMSO in DMEM/F12, 100 μM CK666 in DMEM/F12, 2% vol/vol DMSO in 1:1 vol/vol CMM solution in DMEM/F12 (150 mOsm hypotonic media), and 100 μM CK666 1:1 vol/vol CMM solution in DMEM/F12 (150 mOsm hypotonic media) were prepared fresh and kept at 37°C until used. All solutions contained 5% FBS. Cells were first imaged in DMEM/F12-5% FBS solution as a baseline control for 6 min. Subsequently, the imaging solution was exchanged on the microscopy stage to the desired experimental solutions and a 6-min movie was recorded after 4 min of incubation.
Tether pulling experiments using AFM
Custom-cut 35-mm glass-bottom dishes (Greiner Bio-One;#627860) were coated with fibronectin (50 µg/ml; Corning #356008) for 30 min and washed with Dulbecco’s phosphate buffered saline (DPBS) shortly before use. SK-MEL-2 cells were seeded at a density of 0.15–0.20 × 105 cells/ml in DMEM/F12 GlutaMax supplement with 1% FBS and a penicillin–streptomycin mix (Gibco; #15140-122) in a 37°C humid incubator with 5% CO2 for 2–4 h and used directly for membrane tether pulling experiments. OBL-10 cantilevers (Bruker) were mounted on a CellHesion 200 AFM (Bruker) integrated into an Eclipse Ti inverted light microscope (Nikon), calibrated using the thermal noise method and coated with 2.5 mg/ml concanavalin A (C5275; Sigma) for 1 h at 30°C. After the cantilever was rinsed with DPBS, it was positioned at any location over the cell for tether pulling using bright-field imaging. The approach velocity was set to 1 μm/s, contact force to 100–300 pN, contact time to 300 ms–10 s, and retraction speed to 10 μm/s. After a 10 μm tether was pulled, the cantilever position was held constant until the moment of tether breakage and at least 2 s afterward. The sampling rate was set to 2000 Hz. After measurements of tether forces in control conditions, an inorganic salt solution containing 10 mM CaCl2, 0.3 mM MgCl2 , and 0.1 mM MgSO4 was added to the medium (4:1 vol/vol) to achieve 75 mOsm hypotonic treatment. Tether forces were measured after media dilution for 2–16 min. Tether forces per cell are the average of at least three tethers. Cells were not used longer than 1 h for data acquisition. Force-time curve analysis was performed using JPKSPM Data Processing Software.
Data analysis, statistical analysis, and data plotting
For statistical analysis, data plotting, and image analysis, Prism version 7.0e and python module matplotlib, numpy, pandas modules, and scikit library in Jupyter notebook (Python 3.8.3) were used. For some parts of the image analysis we used FIJI/ImageJ (version 2.1.0/1.53c).
Image analysis
Selection of clathrin-coated superresolved structures for image analysis.
Clathrin-containing structures were extracted from processed STORM images using a custom MATLAB routine. Briefly, a kernel convolution with a disk of ∼80 nm radius was performed on the superresolved clathrin image channel to accentuate possible CCPs. The resulting image was median filtered, and peaks were detected by sequentially identifying and cropping out regions corresponding to the local image maxima. Regions 310 nm × 310 nm wide square centered at these peaks were cropped from all color channels and aligned. Subsequently, we selected superresolved clathrin coats by visual inspection for further quantitative image analysis based on the following criteria: We excluded clathrin-labeled structures for image analysis that appeared deformed, that covered nearly the entire 310 nm × 310 nm wide square, or that contained small punctuated structures that were not distinguishable from background noise or small clathrin seeds (Supplemental Figure S4A). Clathrin-coated structures selected for analysis were first inspected to determine whether they appeared round-shaped, elliptical-shaped, or triangle-shaped in the x-y projected superresolved image. These images were 3D rendered to determine whether the x-z projection resulted in the typical wide-crescent shape, U-shape, or round-shape appearance for the clathrin coat. If we could identify the stereotypical clathrin-coat shapes in both projections, we included the clathrin coat in the pool for further image analysis. We define stereotypical shapes of individual clathrin coats based on platinum replica EM data of CCPs in the same cell type (Supplemental Figure S3) (courtesy of Justin Taraska). We then assigned the clathrin coats as being actin negative when the number of localizations in the actin channel in the 310 nm × 310 nm region of interest (ROI) was below 50, which is close to the background level. Clathrin-coated structures were classified as actin positive when the number of localizations in the actin channel in the 310 nm × 310 nm ROI was above 50 and actin signal overlapped with the clathrin signal (Supplemental Figure S9, E–G).
STORM image data display in figures.
Reconstructed superresolution images were visualized using the “insight” software package and saved in a .png file format (Huang et al., 2008). These images were loaded into ImageJ, converted from an RGB image into an 8 bit image, pseudocolored for clathrin = red hot lookup table, actin = cyan lookup table, and then converted back into an RGB image.
Image analysis of reconstructed clathrin and actin side views.
Selected reconstructed superresolved images of clathrin and actin were treated as digital images for further analysis. A custom-written Jupyter notebook script was used to project reconstructed side views of clathrin and actin onto their x- and z-axes to obtain histograms of the clathrin and actin pixel intensity signals normalized to their respective maximum intensity value (Supplemental Figure S4, B and C). From these normalized z-axis and x-axis pixel intensity histograms, we read out the height and width of clathrin and actin at the 30th percentile. This process resulted in more robust results than the readout at the full width at half maximum (Supplemental Figure S4, B and C).
Actin and clathrin coat height: The z-axis intensity histograms were used to report the actin and clathrin coat heights in the x-z projections. Before extracting the values, a mean filter set to 50 was run over the histograms to reduce signal fluctuations that interfere with the size measurement (Supplemental Figure S4, B and C).
Actin-clathrin overlap calculation: We calculated the total overlap between clathrin and actin using the indices obtained at the upper position of clathrin and lower position of actin at the 30th percentile of the respective z axis–projected intensity histograms (Supplemental Figure S4B). We then reported the overlap relative to the clathrin height in units of percentage.
Clathrin-coat width: We used the 30th percentile of x-axis intensity histogram to report the clathrin coat width in the x-z projection. Before extracting the values, a median filter set to100 was run over the histogram to smooth out signal fluctuations that interfere with the correct size determination (Supplemental Figure S4, B and C).
Shape index calculation: Shape indices of individual clathrin structures are defined as the ratio between clathrin coat height and clathrin coat width. This value allowed us to sort flat (low shape index) and rounded (high shape index) clathrin coats from each other.
Asymmetry of actin signal around clathrin coat: To evaluate the asymmetry of the spatial actin organization around the clathrin coat, we determined the difference in the positions of the peak actin and clathrin signals on both x-z and y-z projections of our images (Supplemental Figure S4E). We obtained the center of the clathrin coat in nanometers by rendering the clathrin superresolved image into a diffraction-limited image, as explained in the section Selection of clathrin-coated superresolved structures for image analysis. We first identified the position of the actin maximum intensity in the x-z projection profile and y-z projection profile in nanometers. Then we measured the distance of the obtained position to the middle position of the clathrin intensity profile. This distance measurement is proportional to the asymmetry of the actin position with respect to clathrin; namely, a low distance corresponds to high symmetry and a high distance corresponds to high asymmetry in the position of the actin signal.
Image analysis of TIRF live-cell microscopy data.
Fluorescent diffraction-limited spots of DNM2-eGFPEN and CLTA-TagRFP-TEN in SK-MEL-2 cells were tracked in each channel over the course of a movie using the detection and tracking features of the cmeAnalysis software package in Matlab (Aguet et al., 2013). Subsequently, we cleaned up the background signal and, on the basis of the x and y positions of associated CLTA-TagRFP-TEN and DNM2-eGFPEN fluorescent tracks, separated them from CLTA-TagRFP-TEN and DNM2-eGFPEN that were not associated with each other, using a custom-written Matlab script (10, 36, 42). We measured fluorescence lifetimes for DNM2-eGFPEN and CLTA-TagRFP-TEN tracks that were associated and not associated with each other and that appeared and disappeared within the duration of a movie. We classified the tracks as “persistent” when they were already present in the first movie frame and lasted longer than the movie.
CME initiation rate and completion rate measurement.
We defined a track as initiated if it appeared within the course of the acquisition, excluding the first frame. We classified a track as complete when it appeared and disappeared within the course of the acquisition. To calculate rates of initiation and completion, we used these values along with measurement of the area of the tracked region, which was obtained from the binary cell mask image generated during the detection step of the cmeAnalysis software that highlights the cell area in which particle tracking was performed. ImageJ was then used to calculate the area of the cell from the binary mask image. The final metric is defined as the number of initiations (or completions) per area per time..
Actin cortex height quantification.
To measure the height of the actin cortex in our 2c-3D STORM images, we cropped an approximately 6 μm × 6 μm ROI of the superresolved clathrin image in a cell in which both actin and clathrin were imaged. Using this mask we removed all coordinates from the actin X, Y, and Z localization fluorophore list that overlapped with clathrin in that ROI. Then, we reported the SD of those Z coordinates not associated with clathrin and their median as a metric for actin cortex height.
The clathrin mask was generated as follows: An approximately 4 μm–6 μm × 4 μm–6 μm ROI of the superresolved clathrin image of a cell was convolved with Gaussian distribution (sigma = 5) (Supplemental Figure S2K). We ran a local maxima finder on the Gaussian-rendered image (Scikit Image, Python). We generated a 2D histogram array of the local maxima and saved these as an image file. To this image we applied an outline finding algorithm (FIJI), followed by a maximum filter (px = 3, FIJI), and finally with a hole-filling algorithm (FIJI). The hole-filled image was inverted such that all clathrin areas had pixel value zero and all nonclathrin areas had pixel value 1 (Supplemental Figure S2L). This mask was subsequently applied to a 2D histogram–generated array of actin X, Y, and Z superresolved coordinates from the same ROI (Supplemental Figure S2, M and O). Both the clathrin mask and the 2D histogram actin array were adjusted to the same pixel size and rescaled by a factor of four. We then calculated the SDs of binned Z coordinates. Bins, respectively pixels, with fewer than four localizations were rejected for the analysis. The SDs of binned Z coordinates were converted from pixels into nanometers for data visualization and plotted in histograms in Supplemental Figure S2, P and Q. In Supplemental Figure S2R we report the median SD of the actin Z coordinates as a metric for actin cortex height.
Supplementary Material
Acknowledgments
C.K. was funded by the German Research Foundation (DFG KA4305/1-1). D.G.D. was funded by National Institutes of Health (NIH) grant R35GM118149. K.X. was funded by the National Science Foundation under CHE-1554717 and the Pew Biomedical Scholars Award. A.D.M. was funded by the European Molecular Biology Laboratory (EMBL), Human Frontiers Science Program (HFSP) grant number RGY0073/2018, and Deutsche Forschungsgemeinschaft (DFG) grant numbers DI 2205/2-1 and DI 2205/3-1. M.A. was funded by NIH grant 1 K99 GM132551-01. E.S. was funded by the EMBL and the Joachim Herz Stiftung Add-on Fellowship for Interdisciplinary Science. We thank Justin Taraska and Kem Sochacki (National Heart, Lung, and Blood Institute, NIH) for use of the platinum replica EM images for comparison in Supplemental Figure S3. We thank Yidi Sun, Padmini Rangamani, and Ross T.A. Pedersen for critical reading and discussions on the manuscript. We thank Sungmin Son and Daniel A. Fletcher for valuable input and discussion on the manuscript.
Abbreviations used:
- CCP
clathrin-coated pit
- CCS
clathrin-coated structure
- CME
clathrin-mediated endocytosis
- SI
shape index.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-11-0589) on April 7, 2022.
REFERENCES
- Aghamohammadzadeh S, Ayscough KR (2009). Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol 11, 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu M, Vasan R, Serwas D, Ferrin MA, Rangamani P, Drubin DG (2020). Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis. eLife 9, e49840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida-Souza L, Frank RAW, Gracía-Nafría J, Colussi A, Gunawardana N, Johnson CM, Yu M, Howard G, Andrews B, Vallis Y, et al. (2018). A flat BAR protein promotes actin polymerization at the base of clathrin-coated pits. Cell 174, 325–337.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avinoam O, Schorb M, Beese CJ, Briggs JAG, Kaksonen M (2015). ENDOCYTOSIS. Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science 348, 1369–1372. [DOI] [PubMed] [Google Scholar]
- Batchelder EM, Yarar D (2010). Differential requirements for clathrin-dependent endocytosis at sites of cell-substrate adhesion. Mol Biol Cell 21, 3070–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Li T-D, Weichsel J, McGorty R, Jreij P, Huang B, Fletcher DA, Mullins RD (2016). Force feedback controls motor activity and mechanical properties of self-assembling branched actin networks. Cell 164, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi M, Fölling J, Belov VN, Boyarskiy VP, Medda R, Egner A, Eggeling C, Schönle A, Hell SW (2008). Multicolor far-field fluorescence nanoscopy through isolated detection of distinct molecular species. Nano Lett 8, 2463–2468. [DOI] [PubMed] [Google Scholar]
- Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T (2011). Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol 13, 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D, Frey F, Sochaki KA, Kummer S, Bergeest JP, Godinez WJ, Kräusslich HG, Rohr K, Taraska JW, Schwarz US, et al. (2018). Clathrin-adaptor ratio and membrane tension regulate the flat-to-curved transition of the clathrin coat during endocytosis. Nat Commun 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser C, Drubin DG (2013). Ultrastructural imaging of endocytic sites in Saccharomyces cerevisiae by transmission electron microscopy and immunolabeling. Microsc Microanal 19, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SY, Stimpson HEM, Weinberg J, Toret CP, Sun Y, Drubin DG (2012). Analysis of yeast endocytic site formation and maturation through a regulatory transition point. Mol Biol Cell 23, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P, Clark AG, Smith MB, Cassani DAD, Dierkes K, Ragab A, Roux PP, Charras G, Salbreux G, Paluch EK (2017). Actin cortex architecture regulates cell surface tension. Nat Cell Biol 19, 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke NI, Royle SJ (2018). FerriTag is a new genetically-encoded inducible tag for correlative light-electron microscopy. Nat Commun 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Warrington A, Taylor KA, Svitkina T (2011). Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr Biol 21, 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambournet D, Sochacki KA, Cheng AT, Akamatsu M, Taraska JW, Hockemeyer D, Drubin DG (2018). Genome-edited human stem cells expressing fluorescently labeled endocytic markers allow quantitative analysis of clathrin-mediated endocytosis during differentiation. J Cell Biol 2, jcb.201710084-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diz-Muñoz A, Fletcher DA, Weiner OD (2013). Use the force: membrane tension as an organizer of cell shape and motility. Trends Cell Biol 23, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diz-Muñoz A, Thurley K, Chintamen S, Altschuler SJ, Wu LF, Fletcher DA, Weiner OD (2016). Membrane tension acts through PLD2 and mTORC2 to limit actin network assembly during neutrophil migration. PLoS Biol 14, e1002474-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakbarova U, Madraki Y, Chan E, Kural C (2021). Dynamic interplay between cell membrane tension and clathrin-mediated endocytosis. Biol Cell 113, 344–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon JB, Zeitler B, Cheng J, Cheng AT, Cherone JM, Santiago Y, Lee AH, Vo TD, Doyon Y, Miller JC, et al. (2011). Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat Cell Biol 13, 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Warren RA, Kessels MM, Keen JH, Heuser J, Drubin DG (2001). The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J Cell Biol 154, 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson JP, Huber SD, Willy NM, Aygün E, Goker S, Atabey T, Kural C (2017). Mechanoregulation of clathrin-mediated endocytosis. J Cell Sci 130, 3631–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson JP, Willy NM, Heidotting SP, Huber SD, Webber MJ, Kural C (2016). Deciphering dynamics of clathrin-mediated endocytosis in a living organism. J Cell Biol 282, jcb.201604128-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, Destaing O, Ko G, Takasaki J, Cremona O, et al. (2009). Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell 17, 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto LM, Roth R, Heuser JE, Schmid SL (2000). Actin assembly plays a variable, but not obligatory role in receptor-mediated endocytosis in mammalian cells. Traffic 1, 161–171. [DOI] [PubMed] [Google Scholar]
- Funk J, Merino F, Schaks M, Rottner K, Raunser S, Bieling P (2021). A barbed end interference mechanism reveals how capping protein promotes nucleation in branched actin networks. Nat Commun 12, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorur A, Yuan L, Kenny SJ, Baba S, Xu K, Schekman R (2017). COPII-coated membranes function as transport carriers of intracellular procollagen I. J Cell Biol 216, 1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassart A, Cheng AT, Hong SH, Zhang F, Zenzer N, Feng Y, Briner DM, Davis GD, Malkov D, Drubin DG (2014). Actin and dynamin2 dynamics and interplay during clathrin-mediated endocytosis. J Cell Biol 205, 721–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Sokolova OS, Chung J, Padrick S, Gelles J, Goode BL (2018). Abp1 promotes Arp2/3 complex-dependent actin nucleation and stabilizes branch junctions by antagonizing GMF. Nat Commun 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassinger JE, Oster G, Drubin DG, Rangamani P (2017). Design principles for robust vesiculation in clathrin-mediated endocytosis. Proc Natl Acad Sci USA 114, E1118–E1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M, Yan R, Li W, Repina NA, Schaffer DV, Xu K (2018). The spectrin-actin-based periodic cytoskeleton as a conserved nanoscale scaffold and ruler of the neural stem cell lineage. Cell Rep 24, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson LA, Nolen BJ (2013). Mechanism of synergistic activation of Arp2/3 complex by cortactin and N-WASP. eLife 2, e00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick B, Han MS, Helgeson LA, Nolen BJ (2013). Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem Biol 20, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Cortesio CL, Drubin DG (2015). Machine-learning-based analysis in genome-edited cells reveals the efficiency of clathrin-mediated endocytosis. Cell Rep 12, 2121–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, Weiner OD (2012). Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell 148, 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, Zhuang X (2008). Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi FZ, Blasco A, Espinal A, Geli MI (2012). Ultrastructural dynamics of proteins involved in endocytic budding. Proc Natl Acad Sci USA 109, E2587–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi F-Z, Grötsch H, Fernández-Golbano IM, Presciatto-Baschong C, Riezman H, Geli MI (2008). Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J Cell Biol 180, 1219–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Shirazinejad C, Wang B, Yan A, Schöneberg J, Upadhyayula S, Xu K, Drubin DG (2021). Rescue of stalled clathrin-mediated endocytosis by asymmetric Arp2/3-mediated actin assembly. bioRxiv, 10.1101/2021.07.16.452693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph JG, Osorio C, Yee V, Agrawal A, Liu AP (2020). Complimentary action of structured and unstructured domains of epsin supports clathrin-mediated endocytosis at high tension. Commun Biol 3, 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, 475–487. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG (2005). A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123, 305–320. [DOI] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Kaksonen M, Briggs JAG (2012). Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell 150, 508–520. [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Pauly BS, Zhang CX, Engqvist-Goldstein ÅEY, Cunningham K, Drubin DG (2007). A Hip1R-cortactin complex negatively regulates actin assembly associated with endocytosis. EMBO J 26, 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Shao L, Chen BC, Zhang X, Zhang M, Moses B, Milkie DE, Beach JR, Hammer JA 3rd, Pasham M, et al. (2015). ADVANCED IMAGING. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349, aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T-D, Bieling P, Weichsel J, Dyche Mullins R, Fletcher DA (2021). The molecular mechanism of load adaptation by branched actin networks. bioRxiv, 10.1101/2021.05.24.445507. [DOI] [PMC free article] [PubMed]
- Li Y, Mund M, Hoess P, Deschamps J, Matti U, Nijmeijer B, Sabinina VJ, Ellenberg J, Schoen I, Ries J (2018). Real-time 3D single-molecule localization using experimental point spread functions. Nat Methods 15, 367–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AP, Loerke D, Schmid SL, Danuser G (2009). Global and local regulation of clathrin-coated pit dynamics detected on patterned substrates. Biophys J 97, 1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield CJ, Feldman ME, Wan L, Almers W (2002). Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol 4, 691–698. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Perrais D, Zenisek D (2005). Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell 121, 593–606. [DOI] [PubMed] [Google Scholar]
- Messa M, Fernández-Busandiego R, Sun EW, Chen H, Czapla H, Wrasman K, Wu Y, Ko G, Ross T, Wendland B, et al. (2014). Epsin deficiency impairs endocytosis by stalling the actin-dependent invagination of endocytic clathrin-coated pits. eLife 3, e03311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Oster G (1996). Cell motility driven by actin polymerization. Biophys J 71, 3030–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Oster G (2003). Force generation by actin polymerization II: the elastic ratchet and tethered filaments. Biophys J 84, 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J, Szep G, Nemethova M, Vries ID, Lieber AD, Winkler C, Kruse K, Small JV, Schmeiser C, Keren K, et al. (2017). Load adaptation of lamellipodial actin networks. Cell 171, 188–200.e16. [DOI] [PubMed] [Google Scholar]
- Mulholland J (1994). Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol 125, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD (1998). The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. PNAS 95, 6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund M, Tschanz A, Wu Y-L, Frey F, Mehl JL, Kaksonen M, Avinoam O, Schwarz US, Ries J (2021). Superresolution microscopy reveals partial preassembly and subsequent bending of the clathrin coat during endocytosis. bioRxiv, 10.1101/2021.10.12.463947. [DOI]
- Mund M, van der Beek JA, Deschamps J, Dmitrieff S, Hoess P, Monster JL, Picco A, Nédélec F, Kaksonen M, Ries J (2018). Systematic nanoscale analysis of endocytosis links efficient vesicle formation to patterned actin nucleation. Cell 174, 884–896.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD (2009). Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Zhang P, Hu F, Yan R, He M, Li W, Xu J, Xu K (2019). Hypotonic stress induces fast, reversible degradation of the vimentin cytoskeleton via intracellular calcium release. Adv Sci (Weinh) 6, 1900865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picco A, Mund M, Ries J, Nédélec F, Kaksonen M (2015). Visualizing the functional architecture of the endocytic machinery. eLife 4, e04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyol R, Haeckel A, Ritter A, Qualmann B, Kessels MM (2007). Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS One 2, e400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD (2016). Actin and actin-binding proteins. Cold Spring Harb Perspect Biol 8, a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes B, Monzo P, Gauthier NC (2017). Membrane tension: a challenging but universal physical parameter in cell biology. Semin Cell Dev Biol 71, 30–41. [DOI] [PubMed] [Google Scholar]
- Rangamani P, Mandadap KK, Oster G (2014). Protein-induced membrane curvature alters local membrane tension. Biophys J 107, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raucher D, Sheetz MP (1999). Membrane expansion increases endocytosis rate during mitosis. J Cell Biol 144, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffay C, Molinard G, Kim K, Roux A (2021). Passive coupling of membrane tension and cell volume during active response of cells to osmosis. Proc Natl Acad Sci USA 118, e2103228118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X (2006). Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3, 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M, Morlot S, Hohendahl A, Manzi J, Lenz M, Roux A (2015). A balance between membrane elasticity and polymerization energy sets the shape of spherical clathrin coats. Nat Commun 6, 6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J, Dambournet D, Liu T-L, Forster R, Hockemeyer D, Betzig E, Drubin DG (2018). 4D cell biology: big data image analytics and lattice light-sheet imaging reveal dynamics of clathrin-mediated endocytosis in stem cell-derived intestinal organoids. Mol Biol Cell 29, 2959–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J, Lehmann M, Ullrich A, Posor Y, Lo W-T, Lichtner G, Schmoranzer J, Haucke V, Noé F (2017). Lipid-mediated PX-BAR domain recruitment couples local membrane constriction to endocytic vesicle fission. Nat Commun 8, 15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott BL, Sochacki KA, Low-Nam ST, Bailey EM, Luu QA, Hor A, Dickey AM, Smith S, Krekvliet JG, Taraska JW, et al. (2018). Membrane bending occurs at all stages of clathrin-coat assembly and defines endocytic dynamics. Nat Commun 9, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwas D, Akamatsu M, Moayed A, Vegesna K, Vasan R, Hill JM, Schöneberg J, Davies KM, Rangamani P, Drubin DG (2021). Actin force generation in vesicle formation: mechanistic insights from cryo-electron tomography, bioRxiv, 10.1101/2021.06.28.450262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Graber ZT, Baumgart T, Stone HA, Cohen AE (2018). Cell membranes resist flow. Cell 175, 1769–1779.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitarska E, Diz-Muñoz A (2020). Pay attention to membrane tension: mechanobiology of the cell surface. Curr Opin Cell Biol 66, 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skruzny M, Desfosses A, Prinz S, Dodonova SO, Gieras A, Uetrecht C, Jakobi AJ, Abella M, Hagen WJH, Schulz J, et al. (2015). An organized co-assembly of clathrin adaptors is essential for endocytosis. Dev Cell 33, 150–162. [DOI] [PubMed] [Google Scholar]
- Skruzny M, Brach T, Ciuffa R, Rybina S, Wachsmuth M, Kaksonen M (2012). Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc Natl Acad Sci USA 109, E2533–E2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochacki KA, Dickey AM, Strub M-P, Taraska JW (2017). Endocytic proteins are partitioned at the edge of the clathrin lattice in mammalian cells. Nat Cell Biol 12, 517–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochacki KA, Heine BL, Haber GJ, Jimah JR, Prasai B, Alfonzo-Méndez MA, Roberts AD, Somasundaram A, Hinshaw JE, Taraska JW (2021). The structure and spontaneous curvature of clathrin lattices at the plasma membrane. Dev Cell 56, 1131–1146.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa I, Wurm CA, Medda R, Rothermel E, von Middendorf C, Fölling J, Jakobs S, Schönle A, Hell SW, Eggeling C (2010). Multicolor fluorescence nanoscopy in fixed and living cells by exciting conventional fluorophores with a single wavelength. Biophys J 99, 2686–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy NM, Ferguson JP, Akatay A, Huber S, Djakbarova U, Silahli S, Cakez C, Hasan F, Chang HC, Travesset A, et al. (2021). De novo endocytic clathrin coats develop curvature at early stages of their formation. Dev Cell 56, 3146–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy NM, Ferguson JP, Huber SD, Heidotting SP, Aygün E, Wurm SA, Johnston-Halperin E, Poirier MG, Kural C (2017). Membrane mechanics govern spatiotemporal heterogeneity of endocytic clathrin coat dynamics. Mol Biol Cell 28, 3480–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik M, Hauser M, Li W, Moon S, Xu K (2015). Graphene-enabled electron microscopy and correlated super-resolution microscopy of wet cells. Nat Commun 6, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Babcock HP, Zhuang X (2012). Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat Methods 9, 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Shim S-H, Zhuang X (2015). Super-resolution imaging through stochastic switching and localization of single molecules: an overview. In: Far-Field Optical Nanoscopy, ed. Tinnefeld P, Eggeling C, Hell SW, Berlin: Springer, 27–64. [Google Scholar]
- Xu K, Zhong G, Zhuang X (2013). Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL (2005). A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol Biol Cell 16, 964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL (2007). SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev Cell 13, 43–56. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Sakai N, Uekusa Y, Imaoka Y, Itagaki Y, Suzuki Y, Yoshimura SH (2018). Morphological changes of plasma membrane and protein assembly during clathrin-mediated endocytosis. PLoS Biol 16, e2004786-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.