Abstract
While fluorescence microscopy has proven to be an exceedingly useful tool in bioscience, it is difficult to offer simultaneous high resolution, fast speed, large volume, and good biocompatibility in a single imaging technique. Thus, when determining the image data required to quantitatively test a complex biological hypothesis, it often becomes evident that multiple imaging techniques are necessary. Recent years have seen an explosion in development of novel fluorescence microscopy techniques, each of which features a unique suite of capabilities. In this Technical Perspective, we highlight recent studies to illustrate the benefits, and often the necessity, of combining multiple fluorescence microscopy modalities. We provide guidance in choosing optimal technique combinations to effectively address a biological question. Ultimately, we aim to promote a more well-rounded approach in designing fluorescence microscopy experiments, leading to more robust quantitative insight.
INTRODUCTION
Cell and developmental biologists have historically relied on light microscopy to visualize the complex, minute structures and their behavior that govern living organisms (Amos, 2000). As advances in optical engineering (Power and Huisken, 2017; Girkin and Carvalho, 2018; Sigal et al., 2018; Schermelleh et al., 2019; Wan et al., 2019), fluorescent probe development (Dempsey et al., 2011; Lavis, 2017, 2021; Specht et al., 2017), and computation (Arganda-Carreras et al., 2017; Rueden and Eliceiri, 2017; McQuin, Goodman, Chernyshev, Kamentsky L, Cimini, Karhohs, et al., 2018; Berg et al., 2019; Haase, Royer, et al., 2020) have progressed in the ensuing years, so too has the power and utility of light microscopy to answer increasingly difficult biological questions. Indeed, fluorescence microscopy has progressively broadened its reach from an observational, often supplementary, tool into an essential analytical platform for hypothesis-driven discovery (Wait et al., 2020).
The term “fluorescence microscopy” encompasses a broad variety of technologies and techniques, from comparatively simple wide-field (WF) microscopes to complex superresolution (SR) (Sigal et al., 2018; Schermelleh et al., 2019) and light-sheet imaging systems (Huisken et al., 2004; Power and Huisken, 2017; Girkin and Carvalho, 2018). Indisputably, the enormous technological advancements in optical microscopy have revolutionized many aspects of bioscience research. However, no single technique offers the ability to image a specimen at combined high resolution, fast speed, long duration, large field of view (FOV) and depth, all while preserving signal strength and sample health. Maximizing any one of these parameters requires a sacrifice in one or more of the others (Jonkman et al., 2020). As a result, a technique that excels in one aspect—for example, by achieving extremely high spatial resolution—will be lacking in another regard, such as being limited by poor imaging speed (Marx, 2013). For many imaging-savvy researchers, combining multiple fluorescence microscopy techniques makes intuitive sense to address the limitations of any single method. This reinforces the notion that a single “best” microscopy technique does not necessarily exist for addressing a particular biological question. This Technical Perspective aims to guide readers toward a more holistic appreciation of how to leverage the increasingly diverse set of optical microscopy technologies for biological inquiry. We will not provide an in-depth discussion of the principles behind individual fluorescence microscopy technologies themselves, as there are numerous reviews and guides to which readers may refer (Lichtman and Conchello, 2005; North, 2006; Waters, 2009; Combs, 2010; Lambert and Waters, 2016; Demmerle et al., 2017; Lemon and McDole, 2020). Rather, we will illustrate, by highlighting recent exemplary studies, how multiple fluorescence microscopy techniques can be effectively combined to yield insight that no single technique may offer. These approaches are distinct from other “multi-modal” approaches such as correlative light and electron microscopy (CLEM) (Razi and Tooze, 2009; Lucas et al., 2012; Timmermans and Otto, 2015; Hoffman et al., 2020) or combined optical and atomic force microscopy (AFM) (Kassies et al., 2005; Harke et al., 2012; Odermatt et al., 2015; Gómez-Varela et al., 2020; Hirvonen et al., 2020; Nelsen et al., 2020; Miranda, Gómez-Varela, et al., 2021). These approaches merge imaging modalities with wholly different underlying principles, and while powerful, they have been discussed elsewhere. Rather, we seek to fill an apparent gap in the literature by discussing how multiple forms of fluorescence microscopy can complement one another, particularly when facing the task of understanding complex biological phenomena.
To guide readers, we divide the following discussion into three experimental classes and showcase studies in each category from which readers can extrapolate to formulate their own multitechnique studies. The first of these experimental scenarios involves interrogating related, but not necessarily the same, specimens using multiple microscopy techniques (Figure 1, left). The complementary results obtained from the data collected with each method can be combined and synthesized to provide a more thorough working model of a biological process. But while studies that utilize multiple microscopy techniques are commonly encountered in the literature, a judicious choice of which instruments to utilize, and how best to merge and interpret the results, can remain elusive for many researchers. It requires appreciating the weaknesses inherent to a technique and selecting the appropriate complementary method to compensate. In this scenario, however, images from each microscopy system do not correspond to the same FOV, or even the same specimen. Thus, each image type cannot be directly compared or combined but only the results derived from these data.
FIGURE 1:
Three classes of multitechnique microscopy experiments. (Left) Scenario 1: multi-instrument experiments image related, but not the same, biological specimens using multiple instruments, after which the results are combined. (Middle) Scenario 2: correlative light microscopy uses two or more techniques to image the same FOV in a specimen; the images from each instrument are subsequently registered, forming a correlated data set. (Right) Scenario 3: dynamic, multitechnique acquisition rapidly switches between microscopy methods on a single system, enabling correlated time-series imaging.
We then explore a second experimental scenario that can ameliorate the weaknesses found in the first case. In many instances, multiple microscopes can be used to interrogate the same FOV within a specimen, followed by spatial registration of the image data—termed correlative light microscopy (CLM) (Figure 1, middle). Capturing and registering multiple images of the same FOV can provide tremendous advantages; however, it also requires more complex experimental, data processing, and analysis procedures. For instance, samples may need to be labeled in an “orthogonal” manner, such that the fluorophores used in one modality do not adversely affect those used for another technique. Furthermore, spatially registering images is a nontrivial computational and experimental undertaking. More fundamentally, this scenario provides limited dynamic information, due to the usual requirement that specimens be immobilized before moving between imaging systems.
Finally, we will detail a third scenario wherein the same FOV is examined using a single instrument that is capable of dynamically switching between multiple imaging modalities (Figure 1, right)—ideally in a manner that is guided by the biology itself. By combining multiple techniques into a single imaging system, computational image registration is often much simpler or even unnecessary. Furthermore, it allows researchers to dynamically correlate image data in a way that is impossible with the preceding two scenarios. While relatively underutilized compared with the other two cases, these capabilities are currently possible using commercially available systems. Furthermore, expanding these capacities is an active forefront of microscopy development. From each of these three experimental scenarios and corresponding examples, we will draw useful guidelines and themes that researchers may refer to when contemplating multitechnique microscopy experiments.
SCENARIO 1: MULTI-INSTRUMENT EXPERIMENTS
Utilizing multiple types of optical microscopy to dissect a biological problem can seem like an obvious–even trivial-way to build a more convincing body of data to support a hypothesis. A temptation exists, however, to simply utilize a combination of either the most readily accessible instruments or the most advanced systems available, with the assumption that the combined results will necessarily provide a “fuller picture.” In reality, an effective combination of microscopy techniques requires an appreciation of their relative strengths and weaknesses and how they can be leveraged to produce the desired results.
For example, an often-repeated desire among many life scientists is to see their specimens in greater detail. Indeed, the dawn of SR microscopy has borne out many hypotheses that would have been difficult or impossible to prove otherwise (Baddeley and Bewersdorf, 2018; Sigal et al., 2018). Yet, imaging at higher spatial resolution often comes at the cost of a smaller FOV (Mahecic et al., 2019). This limitation is significant in two ways. First, it can make imaging a statistically robust number of biological replicates considerably more time-consuming, or even impractical. Second, it can force researchers to focus on relatively small subregions within a specimen. However, a study by Cai et al. (2019) is a useful example that combines confocal imaging and photoactivation and localization microscopy (PALM) to manage the trade-off between resolution and FOV. In this report, confocal microscopy revealed that the transcriptional coactivator Yes-associated protein (YAP) exhibits strong phase condensation behavior in response to hyperosmotic conditions. The ubiquity and replicability of this molecular behavior was established across many dozens of biological replicates. Furthermore, confocal imaging permitted measurements of condensate number and colocalization with various effector molecules throughout the cellular volume. PALM imaging, in contrast, allowed the authors to precisely characterize how YAP condensation altered the nanoscale distribution of transposase-accessible chromatin regions in the nucleus. While SR imaging was restricted to only three biological replicates, combining these results with those from confocal microscopy supported the hypothesis that YAP phase separation was responsible for altering genome topology during hyperosmosis.
Similarly, Stubb, Guzmán, et al. (2019) conducted a study of focal adhesion (FA) morphology in human pluripotent stem cells (hPSCs). Lower-resolution, live-cell imaging allowed quantification of the micron-scale FA morphology throughout a hPSC colony and across multiple biological replicates. These data identified so-called “cornerstone” FAs, located at the hPSC colony periphery, that were larger, less mobile, and more resistant to turnover than canonical adhesions (Figure 2, A and B). Subsequently, ultrahigh resolution interferometric photoactivation and localization microscopy (iPALM) (Shtengel et al., 2009) was employed to better characterize cornerstone FA nanoscale architecture, albeit with a much smaller number of samples and FOV. These results (Figure 2, C–G) unexpectedly showed that vinculin contained in cornerstone hPSC FAs was oriented in a “head over tail” orientation as compared with non-hPSC cells (Kanchanawong, Shtengel, et al., 2010; Xia et al., 2019). By interpreting both their conventional and SR data collectively, the authors were able to draw a possible connection between the dynamic, micron-scale behavior of cornerstone FAs and their nanoscale architecture.
FIGURE 2:
Multi-instrument microscopy experiments examine FA dynamics and architecture in hPSCs. (A) Representative SD confocal image depicting paxillin-positive FAs located at the cell edge (green) and cell interior (magenta). Selected time points of 0 min (green), 50 min (cyan), and 100 min (magenta) are shown for both edge and center FAs. White regions indicate highly stable FAs. (B) Corresponding images of the inset square region shown in A. (C–F). Lateral (top) and axial (bottom) projections of iPALM images of (C) Eos-tagged integrin β5, (D) paxillin, (E) Eos-tagged actin, and (F) Eos-tagged α-actinin. Color scale in each bottom panel represents axial position as noted. Scale bar = 1 µm. (G) Average (and standard deviation) axial positions of hPSC cornerstone FA components as determined from iPALM images. Images are reproduced with permission from Stubb, Guzmán, et al. (2019).
From these examples and others (Fabrowski, Necakov, et al., 2013; Sato et al., 2019; Nava, Miroshnikova, et al., 2020; Costa, Rodia, et al., 2021; Moore et al., 2021), the trade-off between resolution and FOV is brought into relief. The studies by Cai et al. (2019) and Stubb, Guzmán, et al. (2019) show microscopy technique combinations that effectively bring together the exquisite structural detail afforded by SR microscopy with the larger FOV that is possible with conventional microscopy. This general strategy is useful in multiple contexts. First, natural questions of statistical significance, possible selection bias, and overall replicability often arise when analyzing SR data sets representing only a few biological replicates (Jost and Waters, 2019). However, conventional techniques can be used to support or complement lower-throughput SR microscopy data by establishing the presence (if not the fine structural details) of a biological phenomenon over a statistically relevant number of samples. Second, a small FOV can make finding biological structures of interest difficult, if not impossible. Thus, it is advantageous to use conventional microscopy to first identify objects of interest based on their micron-scale phenotype or dynamic behavior. These signatures can then be used to readily find similar structures when employing SR imaging. The FOV of a chosen microscopy technique should be carefully assessed before using it. If, as is often the case with SR microscopy, a technique is found to impose unacceptable limits on the amount or extent of data that can be practically acquired, other techniques should be paired with it to compensate. Such higher-throughput techniques may include confocal microscopy, total internal reflection fluorescence (TIRF) microscopy, or even simple WF microscopy.
These studies showcase how the superior image detail provided by SR microscopy combined with the larger FOV available with more conventional imaging techniques can successfully compensate for each other’s shortcomings. In a similar way, life scientists are often tasked with needing to probe fine structural detail, while at the same time characterizing the highly dynamic behavior of a biological process. Owing to the trade-off between spatial resolution and imaging speed, however, multiple microscopy methods are often required to accomplish these tasks. A study by Jaumouillé et al. (2019) serves as a noteworthy example. Here, the authors characterized both the dynamic behavior and fine structures related to macrophage complement receptor (CR)-mediated phagocytosis using a well-designed selection of microscopy techniques. Initially, spinning-disk (SD) confocal microscopy indicated a tantalizing mechanism whereby actin-containing macrophage protrusions “reached” around an opsonized particle to engulf and internalize it (Figure 3A). However, more information was needed to better characterize this process. Higher imaging speed was needed to uncover the nature of the actin flow. At the same time, better spatial resolution was needed to characterize the actin structure and its associated adhesion complexes. To accomplish the first aim, the authors employed fluorescence speckle microscopy (FSM) (Danuser and Waterman-Storer, 2006), revealing that actin flowed in only one direction toward the extending protrusion, without retrograde flow (Figure 3B). While fast, FSM gives almost no structural detail. Thus, a frustrated phagocytosis assay was performed using TIRF structured illumination microscopy (TIRF-SIM) (Figure 3C). Although lacking dynamic information, it permitted a twofold resolution increase beyond the diffraction limit, allowing the authors to delineate the denser peripheral actin network from the sparser interior network that notably lacked stress bundles. It also revealed that phosphorylated paxillin occurred in sub–100-nm puncta, reminiscent of focal complexes formed by macrophages on extracellular matrix (ECM) substrates (Pixley, 2012). Together, the results highlighted the role of the “molecular clutch,” most often seen in the context of cellular migration (Case and Waterman, 2015), in phagocytosis, albeit with important differences.
FIGURE 3:
Multi-instrument microscopy experiments inform complement receptor–mediated phagocytosis. (A) SD confocal microscopy images of a RAW 264.7 macrophage expressing F-tractin-eGFP (green) engulfing an iC3b-opsonized polystyrene bead (red). (B) Fluorescence speckle microscopy of RAW macrophages expressing actin-mEos3.2 during phagocytosis of opsonized target beads. Red circles and triangles depict detected speckles, and red lines show tracks of speckles through time. (C) Immunofluorescence TIRF-SIM images of a RAW macrophage engaging with an anti-αMβ2–coated coverslip. Images of actin labeled with fluorescent phalloidin (green) and immunostained for phosphorylated paxillin (red) were collected. Right images depict the square inset region shown in the left image. Images are reproduced with permission from Jaumouillé et al. (2019).
This example highlights a nearly ubiquitous challenge faced by microscopists–that is, the tension between achieving good image detail and high imaging speed. However, by subjecting specimens to multiple techniques that span the spectrum of resolution and speed, better biological insight is possible (Nixon-Abell, Obara, Weigel, et al., 2016; Barger et al., 2019; Pfisterer et al., 2020; dos Santos et al., 2021; Moore et al., 2021). As was shown in the preceding example, a technique with moderate spatial resolution and speed such as SD confocal microscopy can be useful to identify a biological process of interest. Inevitably, however, more nuance is nearly always needed. The questions of actin flow as well as actin network and adhesion arrangement were critical and required both higher temporal and spatial resolution, respectively. The key to answering these related questions was the realization that a single microscopy technique could not meet these demands; rather, the hypothesis was best served by a multipronged approach. Yet, in each of the previous studies noted so far, a critical limitation remains: the images collected from each microscopy technique do not spatially correspond to each other. So, while the results from multiple microscopy experiments were effectively combined, the data from one technique cannot be directly considered within the context of another.
SCENARIO 2: CORRELATIVE LIGHT MICROSCOPY
In many instances, microscopy data from a single technique may not be interpretable by itself—that is, a biological process must be contextualized within its surroundings. Such cases may then necessitate acquiring and overlaying images of identical areas within the same specimen using additional microscopy techniques, termed CLM (Figure 1, middle). In contrast to merely combining the results from different modalities, CLM involves spatially registering images from multiple techniques with respect to each other to create information that may be otherwise unavailable. In principle, any combination of fluorescence microscopy techniques may be used in tandem. As discussed in the preceding section, however, these choices should be made judiciously to best overcome the limitations of individual techniques. One practical consideration in CLM is that generally the specimen must be fixed and rendered static before being transferred between microscopes to allow accurate image registration. As a result, using CLM to study biological processes across multiple timescales is generally not feasible. It is, however, achievable to use CLM to juxtapose multiple length scales within a biological specimen. Therefore, CLM very often takes the form of a diffraction-limited (DL) microscopy, such as WF or confocal, combined with SR microscopy, such as STED (stimulated emission depletion), PALM, and STORM (stochastic optical reconstruction microscopy) (Hauser, Wojcik, Kim, et al., 2017).
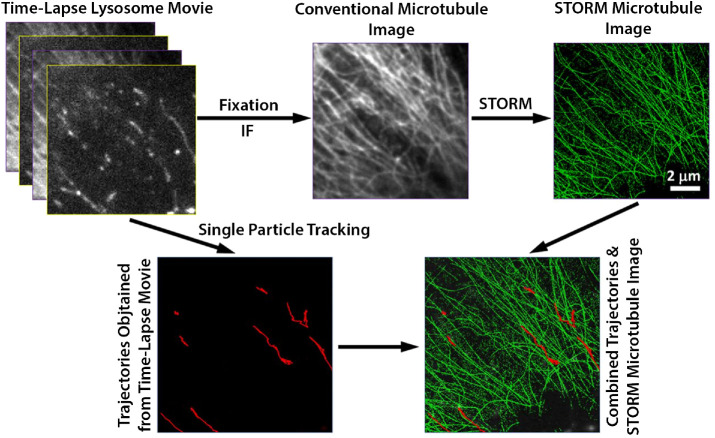
Biology is intrinsically dynamic, and many relevant questions require tracking structures of interest over time. Unfortunately, visualizing a single biological component within the context of its microenvironment is often needed to tell the full story. Furthermore, this surrounding spatial context may need to be resolved on length scales below the diffraction limit. These types of scenarios are well-suited to be addressed by correlative DL-SR microscopy. Such experiments require using conventional microscopy to capture a dynamic process before fixation, followed by SR imaging of surrounding structures (Bálint et al., 2013; Tam et al., 2014). The work of Bálint et al. (2013) expertly demonstrates coupling of time series DL microscopy with SR imaging (Figure 4). Lysosomes were first tracked using high-speed WF microscopy, after which the cells were fixed in situ and the nanoscale microtubule network was subsequently reconstructed using three-dimensional (3D) STORM. By directly correlating the two data sets, the investigators were able to map lysosome trajectories onto individual microtubules. Doing so showed how lysosomes pause at the intersection of two microtubules before either navigating across the junction or transitioning to the crossing microtubule (Bálint et al., 2013). Importantly, such analyses are possible only using CLM methods. The DL imaging was necessary to follow the lysosome trajectories at high speed, and the SR microscopy provided the necessary resolving power to trace the intricate cytoskeletal network, thereby providing spatial context to the lysosome trafficking data.
FIGURE 4:
Correlative fluorescence microscopy allows detailed measurements of lysosome mobility along microtubules. First, a WF time-lapse movie of lysosomes was captured and used for tracking. The sample was then fixed in situ and imaged via dSTORM to delineate the microtubule network at the nanoscale. Lysosome tracks were then overlaid with the SR microtubule image. Images are reproduced with permission from Bálint et al. (2013).
CLM can also serve to contextualize SR images of one structure with DL images of other relevant cellular components (Crossman et al., 2015; Dudok, Barnav, et al., 2015; Vangindertael et al., 2015; Barna et al., 2016; Soeller et al., 2017; Xiang, Julia Roberti, et al., 2018). As was discussed previously, single molecule localization methods (SMLM) such as PALM and STORM are generally limited by smaller FOVs (Lelek et al., 2021), while conventional WF and confocal microscopy allow a user to quickly image a relatively large FOV. Practically speaking, it is then grossly inefficient to use SR methods to both locate a region of interest as well as image it. In this sense, correlative “screening” with a DL imaging modality not only allows a user to select an optimal region of interest but also allows for assessing sample health in a more global sense. This is exemplified by Xu, Zhong, et al. (2013) wherein DL imaging was first used to locate a dendrite within a fixed hippocampal neuron. Subsequently, 3D STORM was used to create a “zoomed-in” image of actin within the selected dendrite and ultimately show that it is distributed as extended, cortical filaments along the long axis of the dendrite (Xu, Zhong, et al., 2013). Beyond its use in correlative screening, DL imaging also allows one to visualize multiple structures within the same specimen, which is currently challenging using SR methods due to the relative lack of dyes and labels compatible with such methods (Dempsey et al., 2011; Li and Vaughan, 2018). Crossman et al. (2015) showed how the additional color channels accessible through conventional microscopy facilitate analysis of correlative imaging (Figure 5). By visualizing the surface of cardiac myocytes with confocal microscopy, the investigators were able to segment the cardiac myocytes into “plasma membrane” and “cytoplasm” regions (Figure 5A). Combining this mask with dSTORM (direct stochastic optical reconstruction microscopy) imaging of junctophilin (JPH) and ryanodine receptor (RyR), they showed that these macromolecular complexes have a higher colocalization near the plasma membrane as opposed to the intracellular compartments (Figure 5, B–E) (Crossman et al., 2015). The spatial context provided by DL microscopy enabled a much more biologically relevant interpretation of the SR images; indeed, without CLM, conclusions such as this may not be possible. Importantly, however, correlative DL and SR microscopy is not the only useful form of CLM.
FIGURE 5:
Correlative confocal microscopy and dSTORM provides spatial context for macromolecular complexes. (A) Confocal microscopy image of a cardiac myocyte labeled with wheat germ agglutinin (WGA). (B) Segmented cell surface and t-tubules (gray) overlaid with confocal microscopy image of JPH (red) and RyR (green). (C) dSTORM image of JPH (red) and RyR (green) overlaid with the cell membrane (cyan) as segmented from the confocal microscopy data. (D) Colocalization analysis of the image shown in C indicating the distance between RyR and the nearest region of JPH. (E) Colocalization analysis shown in D, separated into plasma membrane- (black) and cytoplasm-associated (white) regions. Analysis shows that RyR and JPH have a stronger colocalization within the plasma membrane than the cytoplasm. Images are reproduced with permission from Crossman et al. (2015).
Several groups have correlated SR images with “enhanced” resolution microscopy techniques, such as through correlative STORM and 3D SIM (Hamel et al., 2014; Mönkemöller, Øie, et al., 2015; Burri et al., 2017; Pinnington et al., 2018; Reinhard et al., 2019). The benefit of such combinations is that many enhanced-resolution techniques are capable of live-cell imaging, after which cells can be fixed and imaged with molecular-scale precision using SMLM. In another example, STED was combined with fluorescence lifetime imaging microscopy– Förster resonance energy transfer (FLIM-FRET) to incorporate molecular interaction information into SR images (Günther et al., 2019). Furthermore, not all correlative microscopy involves SR microscopy. Light-sheet fluorescence microscopy (LSFM) images have been correlated with confocal microscopy to show how sample mounting conditions alter blood vessel morphology in a murine retinal model (Prahst, Ashrafzadeh, Mead, et al., 2020). Additionally, correlative LSFM and two-photon microscopy has been used to combine in vivo mouse brain imaging with the enhanced clarity of cleared tissue imaging (Silvestri et al., 2014a,b).
As can be seen, CLM offers a powerful means to directly merge image data from multiple microscopy methods—and thereby support biological conclusions that could not otherwise be supported. While this represents a clear advantage over the examples discussed in Scenario 1, there are significant technical hurdles that can complicate CLM. Chiefly, these revolve around how best to properly register images from each imaging method (Pitkeathly et al., 2012). From a sample preparation perspective, image registration can most readily be achieved by embedding fiducial markers throughout the specimen that act as common fixed points to align each image type (Tam et al., 2014; Burri et al., 2017). Introducing foreign fiducial markers such as polymer beads, however, can be both experimentally challenging and potentially harmful to the biological specimen. Therefore, other algorithms have been developed to use structures within the images themselves to enable image registration (Lowe, 1999; Reinhard et al., 2019), though this may require dual labeling one or more structure within the sample. In either case, image registration necessitates some form of digital transformation to map one image onto the spatial coordinate system of another. In the simplest case, this may only require translating and/or rotating one image to match another. Indeed, such transformations are virtually guaranteed to be necessary due to the transportation of the specimen from one microscope to another. Unfortunately, what may be considered an innocuous difference between microscopes during correlative imaging can noticeably complicate the registration processes afterward. For example, changes in the objective lens magnification—as is commonplace in correlative SR and DL imaging—require stretching or shrinking one image (in addition to translation and rotation) to match the other. To complicate matters further, CLM generally requires fixation of the specimen during the transition between modalities. As has been shown (Gusnard and Kirschner, 1977; Lee et al., 1989; Cross and Williams, 1991; Talman and Boughner, 1995; Zhu et al., 2021), fixation can locally “warp” the specimen. Therefore, it cannot always be assumed that relatively simple transformations such as shifting or stretching will achieve the needed alignment accuracy. Yet, computing optimal image warping parameters not only greatly increases the computational complexity but also requires more fiducial markers or internal structures to accurately register images.
While image registration may appear to be a daunting task, a variety of freely available tools have been built into Fiji (Schindelin et al., 2012) with varied levels of intricacy, which can be readily accessed through online documentation (https://imagej.net/imaging/registration). The available tools range in scope and usability depending on the application at hand. For instance, some tools are designed for fiducial-free image registration (Lowe, 1999; Cardona et al., 2012), and are capable of correcting beyond just translational motion. One particular tool of interest is Fijiyama (Fernandez and Moisy, 2021), which has been developed with multi-instrument microscopy data in mind. In the case of images that require significant warping for effective registration, BigWarp (Bogovic et al., 2016) is available within Fiji and has been used previously with great success (Gao et al., 2019; Wan et al., 2019; Hoffman et al., 2020). However, even with proper image correlation in hand, quantitative analysis of correlated data sets is still nontrivial. Several groups have developed software packages for specific correlative techniques to quantify molecular abundance, clustering, and distance, among other parameters (Barna et al., 2016). As can be seen, image registration represents a major challenge for a CLM approach. Many of these issues can be ameliorated, however, by using a single microscope to perform imaging with multiple modalities.
SCENARIO 3: DYNAMIC MULTITECHNIQUE IMAGING
The preceding scenarios highlighted the benefits of using multiple instruments to visualize either the same sample at multiple length scales or the same biological process in multiple specimens across modalities. Unfortunately, there are certain scenarios where neither of these approaches is sufficient for understanding the animate and hierarchical nature of biological phenomena. This necessitates a single system capable of both dynamic and correlative imaging: that is, multimodal light microscopy in its truest sense (Figure 1, right). Technological complexity has rendered such systems predominantly elusive until recent years, wherein both commercial and custom microscopes can now enable rapid transitions between modalities for correlative, time-series imaging. This is usually achieved by using “modules.” A single module houses all the necessary and unique aspects of a given modality, and it can be added to or removed from the light path by a simple switching mechanism. Such modules are becoming more commonplace among commercial systems and can be readily added to existing systems to extend their capabilities. While the notion of dynamically switching imaging modalities during a single experiment may seem unnecessary, there are several key scenarios wherein dynamic multitechnique imaging may be required to properly characterize a biological process.
The first such scenario occurs when a specimen needs to be optically manipulated during an imaging experiment. Photomanipulation modules enable a user to do just this by illuminating a small portion of their specimen to activate a specific biological process of interest and are commonly found in commercial systems. The work of Liu, Hobson, et al. (2019) is an informative example of using modular designs for photomanipulation on a custom-built instrument. This system utilized a unique combination of electrically tunable lenses and steering mirrors to dynamically switch between TIRF microscopy and localized photoactivation. In doing so, the local activation of Rac1 was shown to alter the dynamics of thin lamellipodia protrusions at the site of activation (Figure 6). This same multimodal system has more recently been used for controlling the small GTPase Rap1 (Elston, Pablo, Pimenta, et al., 2021). A version of this experiment could indeed be performed on a single-mode commercial system. For example, a laser-scanning confocal microscope could be used to both image and photoactivate; however, this means that the benefits of low phototoxicity and high speed associated with TIRF microscopy are lost. Similarly, a TIRF microscope could be used for both imaging and activation, but this comes at the cost of no 3D spatial specificity for the activation region. It is therefore clear that the dynamic switching between TIRF illumination and point activation captured the benefits of both techniques simultaneously to optimize the imaging experiment at hand. This benefit is not unique to combined photomanipulation modules and TIRF microscopy. Photomanipulation modules can readily be adopted and integrated with conventional microscopes such as the SD confocal (Sorkina et al., 2013; Ch’ng et al., 2015; Hayer et al., 2016) and even more complex systems such as light-sheet microscopes (Rieckher et al., 2015; Ducros et al., 2019; Pfisterer et al., 2020; Sapoznik et al., 2020) to leverage the benefits unique to each modality. However, photomanipulation modules are useful only in activating and hindering specific processes. Performing correlative, multimodal imaging wherein each module is used for image acquisition rather than stimulation or activation requires additional considerations.
FIGURE 6:
Dynamic, multitechnique approach for studying cell adhesions during activation of Rac1. Images of a mouse embryonic fibroblast (MEF) transiently transfected with mCherry-Paxillin (black) were collected with variable angle (va)TIRF microscopy. This cell line also stably expressed mVenus–photoactivatable Rac1. Dynamic switching to a laser point-scanning mode enabled localized photoactivation of Rac1 within the dashed circle (blue-filled circles represent images collected during localized photoactivation). Subsequent switching to TIRF mode showed that activation at the cell edge was immediately followed by a protrusion and the formation of adhesions, after which the cell retracted and the adhesions dissipated. Images were reproduced with permission from Liu, Hobson, et al. (2019).
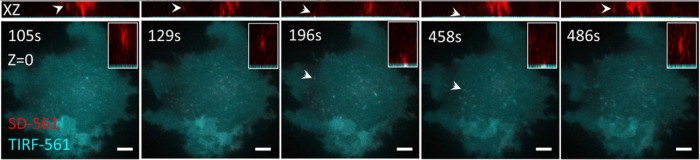
As discussed in depth previously, each imaging modality has its unique strengths and weaknesses in balancing speed, resolution, sample viability, and depth. Similarly, different biological components can exhibit heterogeneity in depth, size, motility, and light tolerance. Therefore, some imaging modalities are simply better suited than others for imaging certain biological structures. In experiments where two very different structures must be imaged within the same specimen, dynamic multitechnique imaging can dramatically improve the interpretable results garnered from an imaging experiment. For example, Zobiak and Failla (2018) sought to localize the interaction of intracellular vesicles with the basal cell membrane. SD confocal microscopy was an excellent modality choice for imaging vesicle movement as it provides suitable optical sectioning over a full 3D volume with acceptable speed. However, the basal cell membrane is more appropriately imaged by TIRF microscopy as it provides superior contrast and lower photobleaching directly at the sample–coverslip interface. Thus, the authors adopted a dynamic, multitechnique approach capturing the benefits of both modalities. This allowed the investigators to track vesicle movements throughout the cell body and precisely detect interaction events with the basal cell membrane (Figure 7). Similar approaches have also been developed to dynamically combine TIRF microscopy and WF microscopy (Smith et al., 2009; Walker et al., 2009; Ellefsen et al., 2015; Chen et al., 2018; Liu et al., 2019). These cases are illustrative because the biological structures and their behavior are matched to the most suitable microscopy modality, thus extracting the most information out of each imaging experiment.
FIGURE 7:
Dynamic TIRF and confocal microscopy facilitates studying vesicle and basal cell membrane interactions. Z-stacks of HEK293 cells were collected first via SD confocal microscopy, after which a single TIRF microscopy image was collected. A β1AR vesicle (red) was tracked in 3D (arrowhead and insert) and observed to interact with a Snx27 vesicle (blue) at the basal cell membrane. Scale bar = 5 µm. Images are reproduced with permission from Zobiak and Failla (2018).
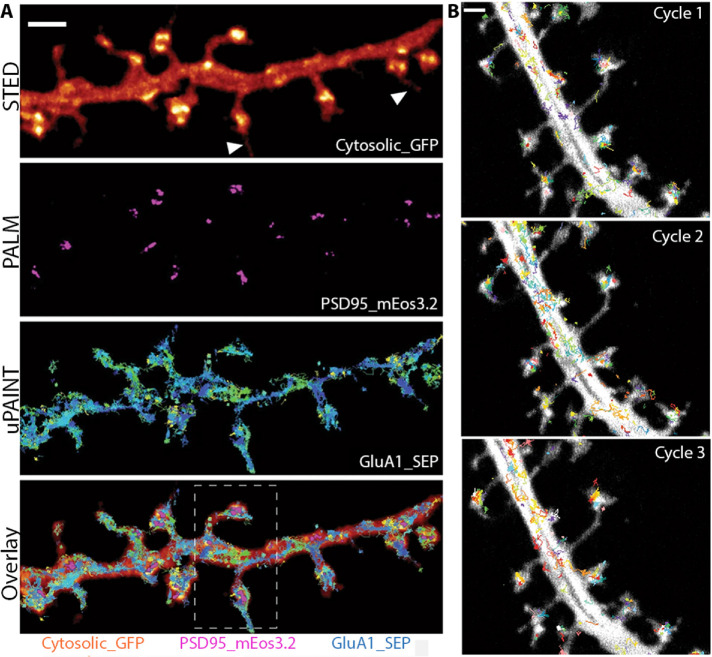
Our previous discussion of static correlative microscopy highlighted the benefits of using DL-SR microscopy to provide spatial context to an imaging experiment. In some instances, however, the spatial context is dynamic, requiring a different approach. Some SR techniques are live-cell compatible, and therefore allow for dynamic, correlative SR microscopy. Such a system was recently demonstrated by Inavalli et al. (2019) wherein they combined three separate SR techniques into a single system: 3D-STED, sptPALM (single particle tracking photoactivation and localization microscopy), and uPAINT (Universal Point Accumulation for Imaging in Nanoscale Topography). Though predominantly limited by the long acquisition times of SMLM methods and the high phototoxicity of STED microscopy, Inavalli et al. (2019) successfully demonstrated the feasibility of multimodal SR microscopy. In doing so, the investigators could image the mobility of the synaptic protein PSD95 within the dendrite with resolution beyond the diffraction limit (Figure 8). The additional temporal information of multimodal SR microscopy opens new opportunities for studying cell biology at the nanoscale; however, exchanging one SR module for a more conventional DL imaging modality may prove more beneficial in leveraging the strengths of multiple modalities within a single experiment. An enhanced resolution technique such as SIM (Gustafsson, 2000, 2008) or image scanning microscopy (ISM) (Sheppard, 1988; Müller and Enderlein, 2010; Sheppard et al., 2013) can easily be combined with TIRF and confocal imaging, respectively, to better leverage their live-cell imaging capabilities at the cost of some resolving power. In either case, this will mitigate some of the issues surrounding phototoxicity and acquisition time associated with multiple SR techniques.
FIGURE 8:
Dynamic, correlative SR microscopy of synaptic proteins within neurons. (A) Correlative STED, PALM, and uPAINT (trajectories) of a dendritic segment of a hippocampal neuron. Scale bar = 2 µm. (B) Time-lapse correlative STED and sptPALM microscopy of PSD95-mEos3.2 trajectories within the dendrite. Scale bar = 1 µm. Images are reproduced with permission from Inavalli et al. (2019).
Along with SR microscopy, LSFM has made tremendous strides over the past two decades. Light-sheet imaging is positioned to be the method of choice for fast, 3D microscopy with low phototoxicity across diverse length- and timescales. Traditional LSFM necessitates two orthogonal objective lenses positioned within the sample space: one for illuminating the sample with a thin sheet of light and a second for detecting the emitted fluorescence (Huisken et al., 2004; Girkin and Carvalho, 2018). This geometry can be a major hindrance in creating LSFM modules or integrating LSFM with other techniques. However, two LSFM designs may remedy this issue. The first is known as oblique plane microscopy (OPM) (Dunsby, 2008; Bouchard et al., 2015; Yang et al., 2019; Sapoznik et al., 2020), which is an LSFM design that can be incorporated into a traditional epifluorescence microscope. OPM utilizes a separate space and two additional objectives for remote refocusing (Botcherby et al., 2007, 2008), thus leaving the sample space with only a single objective. Such a configuration allows one to incorporate additional modules as previously described. The second solution is through external modules that generate light sheets that can be merged with commercial systems (Fadero et al., 2018). This “plug-and-play” scheme may be more immediately realizable for most users; however, they are more limited in their flexibility. While LSFM currently is underutilized in the sense of multimodal microscopy, its widespread use for single-mode imaging and recent designs enabling more flexibility promise future integration with other techniques. Multimodal microscopy experiments—whether they are multi-instrument, correlative, or dynamic multitechnique—demonstrate the benefits of combining methods to leverage the unique capacity of multiple tools to gain insight into biological phenomena across diverse length- and timescales.
DISCUSSION
Fluorescence microscopy has, and continues to be, a platform for open-ended discovery and exploration. Likewise, it plays an equally important role as a quantitative means of validating hypotheses. As articulated by Wait et al. (2020), it is in this second regard that careful microscopy experimental design is vital. At its heart, an imaging experiment is guided by the results that are needed to address the central question(s) at hand. These results, in turn, determine the critical parameters that a microscope must achieve. From these requirements, a suitable imaging technique can be selected. Yet, the preceding examples show that complex biological hypotheses are often composed of multiple interrelated questions that a single set of imaging parameters—and therefore a single technique—cannot adequately address. By using multiple microscopy methods in combination, more complete insight can often be achieved.
The value inherent to multipronged approaches for studying biological phenomena is not a new revelation; in fact, it is often accepted as good standard practice within bioscience to leverage multiple techniques with each other. For example, gel electrophoresis and mass spectrometry are routinely combined for proteomic analysis (Beranova-Giorgianni, 2003) due to their complementary capabilities. In much the same way, multiple forms of microscopy can be synergistically combined. Indeed, combinatorial imaging techniques such as CLEM have been an active area of development for at least two decades (Razi and Tooze, 2009). Nevertheless, it has been underappreciated that combining multiple fluorescence microscopy methods can yield valuable advantages when faced with complex biological phenomena. This in part can be attributed to the difficulty in purchasing, building, and accessing a wide range of imaging techniques. Microscopy core facilities and their specialized expertise represent valuable resources for researchers in this regard. Yet, the rapid pace of microscope innovation has revealed a critical gap between the development of new imaging techniques and their commercial availability. For that reason, unique facilities such as the Advanced Imaging Center at Janelia Research Campus (George, 2014) and the Advanced Bioimaging Center located at Berkeley are well-poised to offer access to unique precommercial microscopy technologies—including those that can dynamically leverage multiple image techniques in tandem.
In addition to technology access, effective experimental design is equally critical, as exemplified by the various studies cited here. Yet, in each of these cases, the experimental approach and parameters were decided a priori. Despite this, biological specimens develop, move, and interact with their environment, often in unpredictable ways. Relying on human perception and predetermined experimental designs therefore runs the risk of missing critical phenomena. Imaging systems that adapt their configuration and acquisition parameters to changes in the biological specimen itself represent a new frontier and are the topic of recent research (Royer et al., 2016; Alvelid et al., 2021; Mahecic et al., 2021). Such an automated and adaptive microscope could, in principle, change its imaging modality and/or parameters at the onset of a detected biological event. However, such adaption necessitates fast and robust biological event detection. Machine learning approaches have already been successful in this regard, such as being able to accurately detect cellular mitosis (Liu et al., 2017) and other behaviors (Zinchuk and Grossenbacher-Zinchuk, 2020). Combining these computational capabilities with advanced multifunctional imaging systems may well prove to be exceptionally powerful going forward for studying biological processes with intrinsically variant length- and timescales.
Acknowledgments
The Advanced Imaging Center at the Janelia Research Campus is generously supported by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation.
Abbreviations used:
- AFM
atomic force microscopy
- CLEM
correlative light and electron microscopy
- CLM
correlative light microscopy
- CR
complement receptor
- DL
diffraction limited
- dSTORM
direct stochastic optical reconstruction microscopy
- ECM
extracellular matrix
- FA
focal adhesion
- FLIM
fluorescence lifetime imaging microscopy
- FOV
field of view
- FRET
Förster resonance energy transfer
- FSM
fluorescence speckle microscopy
- hPSC
human pluripotent stem cell
- iPALM
interferometric photoactivation and localization microscopy
- ISM
image scanning microscopy
- JPH
junctophilin
- LSFM
light sheet fluorescence microscopy
- OPM
oblique plane microscopy
- PALM
photoactivation and localization microscopy
- RyR
ryanodine receptor
- SD
spinning disk
- SIM
structured illumination microscopy
- SMLM
single molecule localization microscopy
- sptPALM
single particle tracking photoactivation and localization microscopy
- SR
superresolution
- STED
stimulated emission depletion
- STORM
stochastic optical reconstruction microscopy
- TIRF
total internal reflection fluorescence
- uPAINT
Universal Point Accumulation for Imaging in Nanoscale Topography
- WF
wide field
- YAP
Yes-associated protein.
Footnotes
REFERENCES
Boldface names denote co–first authors.
- Alvelid J, Damenti M, Testa I (2021). Event-triggered STED imaging. BioRxiv, 2021.10.26.465907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos B (2000). Lessons from the history of light microscopy. Nat Cell Biol 2, E151–E152. [DOI] [PubMed] [Google Scholar]
- Arganda-Carreras I, Kaynig V, Rueden C, Eliceiri KW, Schindelin J, Cardona A, Sebastian Seung H (2017). Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426. [DOI] [PubMed] [Google Scholar]
- Baddeley D, Bewersdorf J (2018). Biological insight from super-resolution microscopy: What we can learn from localization-based images. Annu Rev Biochem 87, 965–989. [DOI] [PubMed] [Google Scholar]
- Bálint Š, Vilanova IV, Álvarez ÁS, Lakadamyali M (2013). Correlative live-cell and superresolution microscopy reveals cargo transport dynamics at microtubule intersections. Proc Natl Acad Sci USA 110, 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger SR, Reilly NS, Shutova MS, Li Q, Maiuri P, Heddleston JM, Mooseker MS, Flavell RA, Svitkina T, Oakes PW, et al. (2019). Membrane-cytoskeletal crosstalk mediated by myosin-I regulates adhesion turnover during phagocytosis. Nat Commun 10, 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna L, Dudok B, Miczán V, Horváth A, László ZI, Katona I (2016). Correlated confocal and super-resolution imaging by VividSTORM. Nat Protoc 11, 163–183. [DOI] [PubMed] [Google Scholar]
- Beranova-Giorgianni S (2003). Proteome analysis by two-dimensional gel electrophoresis and mass spectrometry: strengths and limitations. TrAC Trends Anal Chem 22, 273–281. [Google Scholar]
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, et al. (2019). ilastik: interactive machine learning for (bio)image analysis. Nat Methods 16, 1226–1232. [DOI] [PubMed] [Google Scholar]
- Bogovic JA, Hanslovsky P, Wong A, Saalfeld S (2016). Robust registration of calcium images by learned contrast synthesis. In: Proc—Int Symp Biomed Imaging, Piscataway, NJ: IEEE, 1123–1126. [Google Scholar]
- Botcherby EJ, Juskaitis R, Booth MJ, Wilson T (2007). Aberration-free optical refocusing in high numerical aperture microscopy. Opt Lett 32, 2007–2009. [DOI] [PubMed] [Google Scholar]
- Botcherby EJ, Juškaitis R, Booth MJ, Wilson T (2008). An optical technique for remote focusing in microscopy. Opt Commun 281, 880–887. [Google Scholar]
- Bouchard, MB, Voleti, V, Mendes, CS, Lacefield C, Grueber WB, Mann RS, Bruno RM, Hillman EMC (2015). Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nat Photonics 9, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri O, Laroche T, Guiet R, Seitz A (2017). Correlative SIM-STORM microscopy. Methods Mol Biol 1663, 95–103. [DOI] [PubMed] [Google Scholar]
- Cai D, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N, Sukenik S, Liu Z, Lippincott-Schwartz J (2019). Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat Cell Biol 21, 1578–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, Douglas RJ (2012). TrakEM2 software for neural circuit reconstruction. PLoS One 7, e38011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Waterman CM (2015). Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol 17, 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cao R, Liu W, Zhu D, Zhang Z, Kuang C, Liu X (2018). Widefield and total internal reflection fluorescent structured illumination microscopy with scanning galvo mirrors. J Biomed Opt 23, 1. [DOI] [PubMed] [Google Scholar]
- Ch’ng TH, DeSalvo M, Lin P, Vashisht A, Wohlschlegel JA, Martin KC (2015). Cell biological mechanisms of activity-dependent synapse to nucleus translocation of CRTC1 in neurons. Front Mol Neurosci 8, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs CA (2010). Fluorescence microscopy: A concise guide to current imaging methods. Curr Protocols Neurosci 50, 2.1.1–2.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R, Rodia MT, Zini N, Pegoraro V, Marozzo R, Capanni C, Angelini C, Lattanzi G, Santi S, Cenacchi G (2021). Morphological study of TNPO3 and SRSF1 interaction during myogenesis by combining confocal, structured illumination and electron microscopy analysis. Mol Cell Biochem 476, 1797–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross AR, Williams RC (1991). Kinky microtubules: bending and breaking induced by fixation in vitro with glutaraldehyde and formaldehyde. Cell Motil Cytoskeleton 20, 272–278. [DOI] [PubMed] [Google Scholar]
- Crossman DJ, Hou Y, Jayasinghe I, Baddeley D, Soeller C (2015). Combining confocal and single molecule localisation microscopy: a correlative approach to multi-scale tissue imaging. Methods 88, 98–108. [DOI] [PubMed] [Google Scholar]
- Danuser G, Waterman-Storer CM (2006). Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu Rev Biophys Biomol Struct 35, 361–387. [DOI] [PubMed] [Google Scholar]
- Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X (2011). Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods 8, 1027–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmerle J, Innocent C, North AJ, Ball G, Müller M, Miron E, Matsuda A, Dobbie IM, Markaki Y, Schermelleh L (2017). Strategic and practical guidelines for successful structured illumination microscopy. Natu Proto 12, 988–1010. [DOI] [PubMed] [Google Scholar]
- dos Santos A, Cook AW, Gough RE, Schilling M, Olszok NA, Brown I, Wang L, Aaron J, Martin-Fernandez ML, Rehfeldt F, et al. (2021). DNA damage alters nuclear mechanics through chromatin reorganization. Nucleic Acids Res 49, 340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducros M, Getz A, Arizono M, Pecoraro V, Fernandez Monreal M, Letellier M, Nägerl V, Choquet D (2019). Lattice light sheet microscopy and photo-stimulation in brain slices. In: Neural Imaging and Sensing 2019, ed. Q Luo, J Ding, and L Fu, SPIE, 8.
- Dudok B, Barnav L, Ledri M, Szabó SI, Szabadits E, Pintér B, Woodhams SG, Henstridge CM, Balla GY, Nyilas R, et al. (2015). Cell-specific STORM super-resolution imaging reveals nanoscale organization of cannabinoid signaling. Nat Neurosci 18, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsby C (2008). Optically sectioned imaging by oblique plane microscopy. Opt Express 16, 20306. [DOI] [PubMed] [Google Scholar]
- Ellefsen KL, Dynes JL, Parker I (2015). Spinning-spot shadowless TIRF microscopy. PLoS One 10, e0136055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston RN, Pablo M, Pimenta FM, Hahn KM, Watanabe T (2021). Optogenetic inhibition and activation of Rac and Rap1 using a modified iLID system. BioRxiv, 2020.12.11.421990. [Google Scholar]
- Fabrowski P, Necakov AS, Mumbauer S, Loeser E, Reversi A, Streichan S, Briggs JAG, De Renzis S (2013). Tubular endocytosis drives remodelling of the apical surface during epithelial morphogenesis in Drosophila. Nat Commun 4, 2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadero TC, Gerbich TM, Rana K, Suzuki A, DiSalvo M, Schaefer KN, Heppert JK, Boothby TC, Goldstein B, Peifer M, et al. (2018). LITE microscopy: Tilted light-sheet excitation of model organisms offers high resolution and low photobleaching. J Cell Biol 217, 1869–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez R, Moisy C (2021). Fijiyama: a registration tool for 3D multimodal time-lapse imaging. Bioinformatics 37, 1482–1484. [DOI] [PubMed] [Google Scholar]
- Gao R, Asano SM, Upadhyayula S, Pisarev I, Milkie D, Liu T-L, Singh V, Graves A, Huynh GH, Zhao Y, et al. (2019). Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363, eaau8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RA (2014). Opening the doors to pre-commercial technology. J Biomol Tech 25, S9. [Google Scholar]
- Girkin JM, Carvalho MT (2018). The light-sheet microscopy revolution. J Opt 20, 053002. [Google Scholar]
- Gómez-Varela AI, Stamov DR, Miranda A, Alves R, Barata-Antunes C, Dambournet D, Drubin DG, Paiva S, De Beule PAA (2020). Simultaneous co-localized super-resolution fluorescence microscopy and atomic force microscopy: combined SIM and AFM platform for the life sciences. Scientific Reports 10, 1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther E, Klauß A, Toro-Nahuelpan M, Schüler D, Hille C, Faivre D (2019). The in vivo mechanics of the magnetotactic backbone as revealed by correlative FLIM-FRET and STED microscopy. Sci Rep 9, 19615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusnard D, Kirschner RH (1977). Cell and organelle shrinkage during preparation for scanning electron microscopy: effects of fixation, dehydration and critical point drying. J Microsc 110, 51–57. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL (2000). Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc 198, 82–87. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL, Shao L, Carlton PM, Wang CJR, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW (2008). Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J 94, 4957–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase R, Royer LA, Steinbachv P, Schmidt D, Dibrov A, Schmidt U, Weigert M, Maghelli N, Tomancak P, Jug F, et al. (2020). CLIJ: GPU-accelerated image processing for everyone.. Nat Methods 17, 5–6. [DOI] [PubMed] [Google Scholar]
- Hamel V, Guichard P, Fournier M, Guiet R, Flückiger I, Seitz A, Gönczy P (2014). Correlative multicolor 3D SIM and STORM microscopy. Biomed Opt Express 5, 3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harke B, Chacko JV, Haschke H, Canale C, Diaspro A (2012). A novel nanoscopic tool by combining AFM with STED microscopy. Opt Nanoscopy, 1, 3. [Google Scholar]
- Hauser M, Wojcik M, Kim D, Mahmoudi M, Li W, Xu K (2017). Correlative super-resolution microscopy: new dimensions and new opportunities. Chem Rev 117, 7428–7456. [DOI] [PubMed] [Google Scholar]
- Hayer A, Shao L, Chung M, Joubert LM, Yang HW, Tsai FC, Bisaria A, Betzig E, Meyer T (2016). Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat Cell Biol 18, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvonen LM, Marsh RJ, Jones GE, Cox S (2020). Combined AFM and super-resolution localisation microscopy: Investigating the structure and dynamics of podosomes. Eur J Cell Biol 99, 151106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DP, Shtengel G, Xu CS, Campbell KR, Freeman M, Wang L, Milkie DE, Pasolli HA, Iyer N, Bogovic JA, et al. (2020). Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367, eaaz5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EHK (2004). Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009. [DOI] [PubMed] [Google Scholar]
- Inavalli VVGK, Lenz MO, Butler C, Angibaud J, Compans B, Levet F, Tønnesen J, Rossier O, Giannone G, Thoumine O, et al. (2019). A super-resolution platform for correlative live single-molecule imaging and STED microscopy. Nat Methods 16, 1263–1268. [DOI] [PubMed] [Google Scholar]
- Jaumouillé V, Cartagena-Rivera AX, Waterman CM (2019). Coupling of β2 integrins to actin by a mechanosensitive molecular clutch drives complement receptor-mediated phagocytosis. Nat Cell Biol 21, 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkman J, Brown CM, Wright GD, Anderson KI, North AJ (2020). Tutorial: guidance for quantitative confocal microscopy. Nat Protocols, 1–27. [DOI] [PubMed] [Google Scholar]
- Jost AP-T, Waters JC (2019). Designing a rigorous microscopy experiment: validating methods and avoiding bias. J Cell Biol 218, 1452–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM (2010). Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassies R, Van der Werf KO, Lenferink A, Hunter CN, Olsen JD, Subramaniam V, Otto C (2005). Combined AFM and confocal fluorescence microscope for applications in bio-nanotechnology. J Microsc 217, 109–116. [DOI] [PubMed] [Google Scholar]
- Lambert TJ, Waters JC (2016). Navigating challenges in the application of superresolution microscopy. J Cell Biol 216, 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavis LD (2017). Teaching old dyes new tricks: biological probes built from fluoresceins and rhodamines. Annu Rev Biochem 86, 825–843. [DOI] [PubMed] [Google Scholar]
- Lee JM, Haberer SA, Boughner DR (1989). The bovine pericardial xenograft: I. Effect of fixation in aldehydes without constraint on the tensile viscoelastic properties of bovine pericardium. J Biomed Mater Res 23, 457–475. [DOI] [PubMed] [Google Scholar]
- Lelek M, Gyparaki MT, Beliu G, Schueder F, Griffié J, Manley S, Jungmann R, Sauer M, Lakadamyali M, Zimmer C (2021). Single-molecule localization microscopy. Nat Rev Methods Prim 1, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon WC, McDole K (2020). Live-cell imaging in the era of too many microscopes. Curr Opin Cell Biol 66, 34–42. [DOI] [PubMed] [Google Scholar]
- Li H, Vaughan JC (2018). Switchable fluorophores for single-molecule localization microscopy. Chem Rev 118, 9412–9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Conchello J-A (2005). Fluorescence microscopy. Nat Methods 2, 910–919. [DOI] [PubMed] [Google Scholar]
- Liu A, Lu Y, Chen M, Su Y (2017). Mitosis detection in phase contrast microscopy image sequences of stem cell populations: a critical review. IEEE Trans Big Data 3, 443–457. [Google Scholar]
- Liu B, Hobson CM, Pimenta FM, Nelsen E, Hsiao J, O’Brien T, Falvo MR, Hahn KM, Superfine R (2019). VIEW-MOD: a versatile illumination engine with a modular optical design for fluorescence microscopy. Opt Express 27, 19950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe DG (1999). Object recognition from local scale-invariant features. In: Proceedings of the Seventh IEEE International Conference on Computer Vision, vol. 2, IEEE, 1150–1157.
- Lucas MS, Günthert M, Gasser P, Lucas F, Wepf R (2012). Chapter 17 - Bridging microscopes: 3d correlative light and scanning electron microscopy of complex biological structures. In: Correlative Light and Electron MIcroscopy, ed. Müller-Reichert T, Verkade P, Academic Press, 325–356. [DOI] [PubMed] [Google Scholar]
- Mahecic D, Stepp WL, Zhang C, Griffié J, Weigert M, Manley S (2021). Event-driven acquisition for content-enriched microscopy. BioRxiv, 2021.10.04.463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahecic D, Testa I, Griffié J, Manley S (2019). Strategies for increasing the throughput of super-resolution microscopies. Curr Opin Chem Biol 51, 84–91. [DOI] [PubMed] [Google Scholar]
- Marx V (2013). Is super-resolution microscopy right for you? Nat Methods 10, 1157–1163. [DOI] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al. (2018). CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol 16, e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda A, Gómez-Varela AI, Stylianou A, Hirvonen LM, Sánchez H, De Beule PAA (2021). How did correlative atomic force microscopy and super-resolution microscopy evolve in the quest for unravelling enigmas in biology? Nanoscale 13, 2082–2099. [DOI] [PubMed] [Google Scholar]
- Mönkemöller V, Øie C, Hübner W, Huser T, McCourt P (2015). Multimodal super-resolution optical microscopy visualizes the close connection between membrane and the cytoskeleton in liver sinusoidal endothelial cell fenestrations. Sci Rep 5, 16279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AS, Coscia SM, Simpson CL, Ortega FE, Wait EC, Heddleston JM, Nirschl JJ, Obara CJ, Guedes-Dias P, Boecker CA, et al. (2021). Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 591, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller CB, Enderlein J (2010). Image scanning microscopy. Phys Rev Lett 104, 198101. [DOI] [PubMed] [Google Scholar]
- Nava MM, Miroshnikova YA, Biggs LC, Whitefield DB, Metge F, Boucas J, Vihinen H, Jokitalo E, Li X, García Arcos JM, et al. (2020). Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell 181, 800-817.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen E, Hobson CM, Kern ME, Hsiao JP, O’Brien ET, Watanabe T, Condon BM, Boyce M, Grinstein S, Hahn KM, et al. (2020). Combined atomic force microscope and volumetric light sheet system for correlative force and fluorescence mechanobiology studies. Sci Rep 10, 8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon-Abell J, Obara CJ, Weigel AV, Li D, Legant WR, Xu CS, Pasolli HA, Harvey K, Hess HF, Betzig E, et al. (2016). Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 354, aaf3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North AJ (2006). Seeing is believing? A beginners’ guide to practical pitfalls in image acquisition. J Cell Biol 172, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odermatt PD, Shivanandan A, Deschout H, Jankele R, Nievergelt AP, Feletti L, Davidson MW, Radenovic A, Fantner GE (2015). High-resolution correlative microscopy: bridging the gap between single molecule localization microscopy and atomic force microscopy. Nano Letters 15, 4896–4904. [DOI] [PubMed] [Google Scholar]
- Pfisterer K, Levitt J, Lawson CD, Marsh RJ, Heddleston JM, Wait E, Ameer-Beg SM, Cox S, Parsons M (2020). FMNL2 regulates dynamics of fascin in filopodia. J Cell Biol 219, e201906111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnington SJL, Marshall JF, Wheeler AP (2018). Correlative 3D structured illumination microscopy and single-molecule localization microscopy for imaging cancer invasion. Methods Mol Biol 1764, 253–265. [DOI] [PubMed] [Google Scholar]
- Pitkeathly WTE, Poulter NS, Claridge E, Rappoport JZ (2012). Auto-align—multi-modality fluorescence microscopy image co-registration. Traffic 13, 204–217. [DOI] [PubMed] [Google Scholar]
- Pixley FJ (2012). Macrophage migration and its regulation by CSF-1. Int J Cell Biol 2012, 501962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power RM, Huisken J (2017). A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat Methods 14, 360–373. [DOI] [PubMed] [Google Scholar]
- Prahst C, Ashrafzadeh P, Mead T, Figueiredo A, Chang K, Richardson D, Venkaraman L, Richards M, Martins Russo A, Harrington K, et al. (2020). Mouse retinal cell behaviour in space and time using light sheet fluorescence microscopy. eLife 9, e49779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M, Tooze, SA (2009). Correlative light and electron microscopy. Methods Enzymol 452, 261–275. [DOI] [PubMed] [Google Scholar]
- Reinhard S, Aufmkolk S, Sauer M, Doose S (2019). Registration and visualization of correlative super-resolution microscopy data. Biophys J 116, 2073–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckher M, Kyparissidis-Kokkinidis I, Zacharopoulos A, Kourmoulakis G, Tavernarakis N, Ripoll J, Zacharakis G (2015). A customized light sheet microscope to measure spatio-temporal protein dynamics in small model organisms. PLoS One 10, e0127869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer LA, Lemon WC, Chhetri RK, Wan Y, Coleman M, Myers EW, Keller PJ (2016). Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat Biotechnol 34, 1267–1278. [DOI] [PubMed] [Google Scholar]
- Rueden CT, Eliceiri KW (2017). The imagej ecosystem: an open and extensible platform for biomedical image analysis. Microsc Microanal 23, 226–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapoznik E, Chang B-J, Huh J, Ju RJ, Azarova EV, Pohlkamp T, Welf ES, Broadbent D, Carisey AF, Stehbens SJ, et al. (2020). A versatile oblique plane microscope for large-scale and high-resolution imaging of subcellular dynamics. eLife 9, 1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Hilbert L, Oda H, Wan Y, Heddleston JM, Chew T-L, Zaburdaev V, Keller P, Lionnet T, Vastenhouw N, et al. (2019). Histone H3K27 acetylation precedes active transcription during zebrafish zygotic genome activation as revealed by live-cell analysis. Development 146, dev179127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Ferrand A, Huser T, Eggeling C, Sauer M, Biehlmaier O, Drummen GPC (2019). Super-resolution microscopy demystified. Nat Cell Biol 21, 72–84. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard CJR (1988). Super-resolution in confocal imaging. Opt 80, 53–54. [Google Scholar]
- Sheppard CJR, Mehta SB, Heintzmann R (2013). Superresolution by image scanning microscopy using pixel reassignment. Opt Lett 38, 2889. [DOI] [PubMed] [Google Scholar]
- Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, et al. (2009). Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci USA 106, 3125–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal Y, Zhou R, Zhuang X (2018). Visualizing and discovering cellular structures with super-resolution microscopy. Science 361, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri L, Allegra Mascaro AL, Costantini I, Sacconi L, Pavone FS (2014a). Correlative two-photon and light sheet microscopy. Methods 66, 268–272. [DOI] [PubMed] [Google Scholar]
- Silvestri L, Allegra Mascaro AL, Costantini I, Sacconi L, Pavone FS (2014b). Exploring the brain on multiple scales with correlative two-photon and light sheet microscopy. In: Multiphoton Microscopy in the Biomedical Sciences XIV, ed. A Periasamy, PTC So, and K Königed, 89480I. [DOI] [PubMed]
- Smith IF, Wiltgen SM, Parker I (2009). Localization of puff sites adjacent to the plasma membrane: functional and spatial characterization of Ca2+ signaling in SH-SY5Y cells utilizing membrane-permeant caged IP3. Cell Calcium 45, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeller C, Hou Y, Jayasinghe ID, Baddeley D, Crossman D (2017). Correlative single-molecule localization microscopy and confocal microscopy. Methods Mol Biol 1663, 205–217. [DOI] [PubMed] [Google Scholar]
- Sorkina T, Caltagarone J, Sorkin A (2013). Flotillins regulate membrane mobility of the dopamine transporter but are not required for its protein kinase C dependent endocytosis. Traffic 14, 709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht EA, Braselmann E, Palmer AE (2017). A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annu Rev Physiol 79, 93–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubb A, Guzmán C, Närvä E, Aaron J, Chew T-L, Saari M, Miihkinen M, Jacquemet G, Ivaska J (2019). Superresolution architecture of cornerstone focal adhesions in human pluripotent stem cells. Nat Commun 10, 4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talman EA, Boughner DR (1995). Glutaraldehyde fixation alters the internal shear properties of porcine aortic heart valve tissue. Ann Thorac Surg 60, S369–S373. [DOI] [PubMed] [Google Scholar]
- Tam J, Cordier GA, Bálint Š, Sandoval Álvarez Á, Borbely JS, Lakadamyali M (2014). A microfluidic platform for correlative live-cell and super-resolution microscopy. PLoS One 9, e115512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans FJ, Otto C (2015). Contributed Review: Review of integrated correlative light and electron microscopy. Rev Sci Instrum 86, 11501. [DOI] [PubMed] [Google Scholar]
- Vangindertael J, Beets I, Rocha S, Dedecker P, Schoofs L, Vanhoorelbeke K, Hofkens J, Mizuno H (2015). Super-resolution mapping of glutamate receptors in C. elegans by confocal correlated PALM. Sci Rep 5, 13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wait EC, Reiche MA, Chew T-L (2020). Hypothesis-driven quantitative fluorescence microscopy—the importance of reverse-thinking in experimental design. J Cell Sci 133, jcs250027. [DOI] [PubMed] [Google Scholar]
- Walker S, Cunniffe N, Bootman M, Roderick HL, Ahrent E (2009). Dynamic imaging of calcium and STIM1 in the same cell using wide-field and TIRF microscopy. Biotechniques 45, 43. [DOI] [PubMed] [Google Scholar]
- Wan Y, Wei Z, Looger LL, Koyama M, Druckmann S, Keller PJ (2019). Single-cell reconstruction of emerging population activity in an entire developing circuit. Cell 179, 355–372.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC (2009). Accuracy and precision in quantitative fluorescence microscopy. J Cell Biol 185, 1135–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Yim EKF, Kanchanawong P (2019). Molecular organization of integrin-based adhesion complexes in mouse embryonic stem cells. ACS Biomater Sci Eng 5, 3828–3842. [DOI] [PubMed] [Google Scholar]
- Xiang W, Julia Roberti M, Hériché JK, Huet S, Alexander S, Ellenberg J (2018). Correlative live and super-resolution imaging reveals the dynamic structure of replication domains. J Cell Biol 217, 1973–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhong G, Zhuang X (2013). Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Chen X, Wang Y, Feng S, Pessino V, Stuurman N, Cho NH, Cheng KW, Lord SJ, Xu L, et al. (2019). Epi-illumination SPIM for volumetric imaging with high spatial-temporal resolution. Nat Methods 16, 501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Rajendram M, Huang KC (2021). Effects of fixation on bacterial cellular dimensions and integrity. iScience 24, 102348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinchuk V, Grossenbacher-Zinchuk O (2020). Machine learning for analysis of microscopy images: a practical guide. Curr Protoc Cell Biol 86, e101. [DOI] [PubMed] [Google Scholar]
- Zobiak B, Failla AV (2018). Advanced spinning disk-TIRF microscopy for faster imaging of the cell interior and the plasma membrane. J Microsc 269, 282–290. [DOI] [PubMed] [Google Scholar]