Abstract
Microtubules establish the directionality of intracellular transport by kinesins and dynein through polarized assembly, but it remains unclear how directed transport occurs along microtubules organized with mixed polarity. We investigated the ability of the plus end–directed kinesin-4 motor KIF21B to navigate mixed polarity microtubules in mammalian dendrites. Reconstitution assays with recombinant KIF21B and engineered microtubule bundles or extracted neuronal cytoskeletons indicate that nucleotide-independent microtubule-binding regions of KIF21B modulate microtubule dynamics and promote directional switching on antiparallel microtubules. Optogenetic recruitment of KIF21B to organelles in live neurons induces unidirectional transport in axons but bidirectional transport with a net retrograde bias in dendrites. Removal of the secondary microtubule-binding regions of KIF21B or dampening of microtubule dynamics with low concentrations of nocodazole eliminates retrograde bias in live dendrites. Further exploration of the contribution of microtubule dynamics in dendrites to directionality revealed plus end–out microtubules to be more dynamic than plus end–in microtubules, with nocodazole preferentially stabilizing the plus end–out population. We propose a model in which both nucleotide-sensitive and -insensitive microtubule-binding sites of KIF21B motors contribute to the search and selection of stable plus end–in microtubules within the mixed polarity microtubule arrays characteristic of mammalian dendrites to achieve net retrograde movement of KIF21B-bound cargoes.
INTRODUCTION
Kinesins comprise a large superfamily of cytoskeletal motors that power transport in a polarized manner along microtubules (MTs) (Vale et al., 1985; Schnapp and Reese, 1989). Kinesin directionality is controlled by the organization of structurally polarized MT tracks (Erickson, 1974; Ludueña et al., 1977). Most kinesins move toward the more dynamic plus ends of MTs, resulting in net outward, or anterograde, transport of cargoes in cells with radially arrayed MTs. The mechanism of directional transport is more complicated in cell types in which the cytoskeleton is organized with mixed polarity (Muroyama and Lechler, 2017; Sanchez and Feldman, 2017). In mammalian neurons, MTs are organized with their plus ends out within the soma and axon but are oriented with mixed polarity in dendrites (Heidemann and McIntosh, 1980; Baas et al., 1988). Intriguingly, the plus end–directed kinesin-4 KIF21B motor is associated with cargoes moving predominantly in the retrograde direction in dendrites, despite the mixed polarity of microtubules (Ghiretti et al., 2016). However, the mechanism by which KIF21B achieves this retrograde directionality is not known.
KIF21B is expressed in a variety of tissues (Marszalek et al., 1999) including the brain, where mutations are associated with developmental disorders (Asselin et al., 2020). In neurons, KIF21B localizes mainly to dendrites (Marszalek et al., 1999), although accumulating data suggest that KIF21B is important for cargo transport and the regulation of MT organization in both axons and dendrites (Muhia et al., 2016; Morikawa et al., 2018; Asselin et al., 2020). Mutations in KIF21B impede axon growth and branching, resulting in abnormalities in brain development and connectivity (Asselin et al., 2020), while the dendrites of neurons lacking KIF21B are less complex and exhibit tighter packing of MTs (Muhia et al., 2016; Morikawa et al., 2018). At the cellular level, KIF21B is involved in the endocytic recycling of NMDA receptors (Gromova et al., 2018), the delivery of GABAA γ2-subunits (Labonté et al., 2014) to the cell periphery, and the retrograde transport of TrkB/BDNF cargoes (Ghiretti et al., 2016). Consistent with these roles in both cargo transport and MT organization, KIF21B-knockout mice display deficits in learning and memory (Muhia et al., 2016).
KIF21B interacts with MTs via a canonical N-terminal motor domain and secondary MT-binding regions (MTRs) within the coiled-coil stalk and WD40 tail domains (Ghiretti et al., 2016; van Riel et al., 2017). Dimeric motor domain constructs lacking the C-terminal MTRs move processively toward the MT plus end, indicating that the MTRs are not required for motility. Instead, the MTRs bind to MTs independently of the motor domain (Ghiretti et al., 2016; van Riel et al., 2017) and dampen or pause MT dynamics (Ghiretti et al., 2016; van Riel et al., 2017; Hooikaas et al., 2020), with the magnitude of these effects differing depending on experimental conditions.
In this study, we investigate how KIF21B motors establish long-range transport on mixed polarity MT networks. We started by examining KIF21B in simple in vitro systems, followed by in vitro reconstitution assays with added complexity, and finally live cell experiments using optogenetic motor recruitment to understand how KIF21B activity produces a net retrograde bias in neuronal dendrites. Consistent with a role in remodeling the MT cytoskeleton, recombinant KIF21B motors with intact MTRs stabilized MTs and promoted MT assembly at low nanomolar concentrations. The MTRs of KIF21B also promoted track switching and processive transport on engineered antiparallel MT bundles in vitro; both directional switching and run lengths were further enhanced by motor oligomerization. Directional switching was also observed for KIF21B motors moving on extracted and stabilized dendritic MT arrays, but this in vitro assay did not model the directional bias seen by KIF21B in live cells, suggesting that this bias requires active MT remodeling. Using optogenetics to acutely recruit KIF21B to peroxisomes in live neurons, we found that KIF21B-bound cargoes moved unidirectionally in axons but switched between anterograde and retrograde movement within dendrites. Strikingly, KIF21B recruitment induced net retrograde movement on the mixed polarity MT array in dendrites despite the higher fraction of plus end–out MTs. Using EB3 comet tails to assay MT dynamics, we noted a pronounced asymmetry in the organization of dynamic MTs in dendrites, with MTs oriented with plus ends toward the dendrite tip (plus end–out) more dynamic than those oriented with their plus ends toward the cell soma (plus end–in). Dampening these dynamics with low-dose nocodazole correlated directly with a loss in the retrograde bias of KIF21B motility, which was also lost upon truncation of the KIF21B MTRs. Together these results suggest a model in which KIF21B motors coordinate nucleotide-sensitive and -insensitive MT-binding sites to preferentially interact with stable plus end–in MTs to bias the net retrograde movement of KIF21B-bound cargoes.
RESULTS
Recombinant KIF21B motors stabilize MTs and promote MT assembly
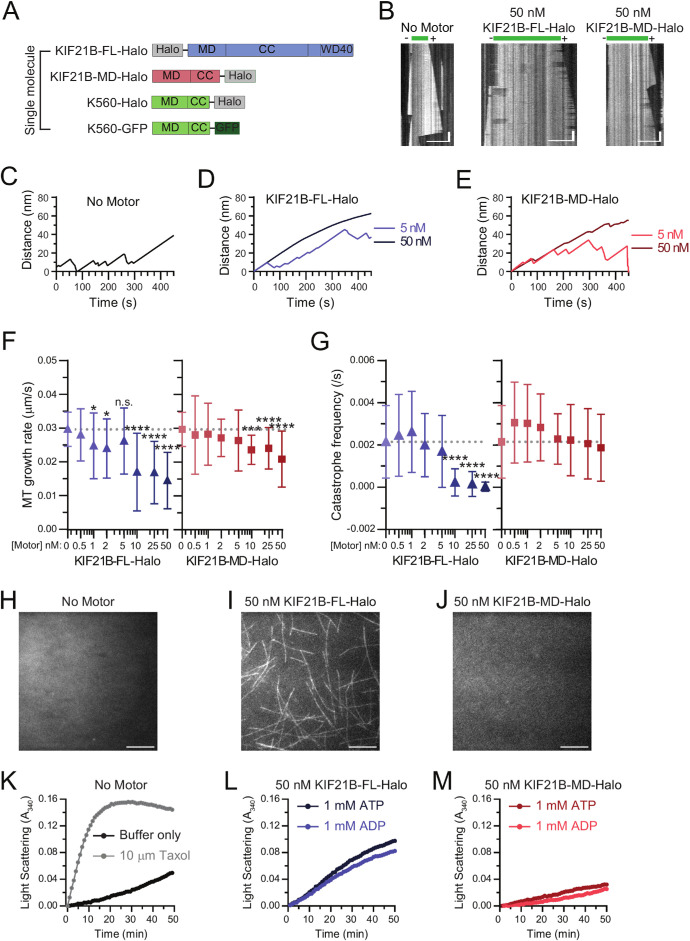
Live cell studies in neurons and T-cells indicate that KIF21B is involved in stabilizing MTs (Ghiretti et al., 2016; Hooikaas et al., 2020). However, in vitro experiments testing the effect of KIF21B on MT dynamics have produced different results, depending on assay conditions (Ghiretti et al., 2016; van Riel et al., 2017). At low ionic strength and high motor concentrations, KIF21B increased MT dynamics by promoting faster growth speeds and catastrophe frequencies (Ghiretti et al., 2016). In contrast, experiments performed in higher-ionic-strength buffer in the presence of the end-binding protein EB3 resulted in the accumulation of KIF21B at MT plus ends and pronounced pausing of growth and shortening (van Riel et al., 2017).
To better understand how KIF21B influences MT dynamics, we used total internal reflection fluorescence (TIRF) microscopy and an engineered system of single dynamic MTs to examine effects over a physiological range of motor concentrations (5–25 nM; Martens et al., 2006; Kim et al., 2014) in assays performed at physiological ionic strength. We compared the growth behavior of dynamic MT plus ends in the absence or presence of Halo-tagged full-length KIF21B (KIF21B-FL-Halo) or motor domain–truncated KIF21B (KIF21B-MD-Halo) motors (Figure 1, A–E, and Supplemental Figure S1). The average MT growth rate decreased with increasing concentrations of either KIF21B-FL-Halo or KIF21B-MD-Halo (Figure 1F), although the magnitude of the effect was more pronounced in experiments with KIF21B-FL-Halo. Increasing concentrations of KIF21B-FL-Halo also induced a significant decrease in the catastrophe frequency, which was reduced to almost zero at KIF21B-FL-Halo concentrations above 5 nM (Figure 1G). In contrast, the addition of KIF21B-MD-Halo had no significant effect on the catastrophe frequency over all concentrations tested. Of note, the motor domain of kinesin-1 (K560-Halo), known to stabilize MTs (Marceiller et al., 2005; Katsuki et al., 2014; Reuther et al., 2016; Peet et al., 2018) also suppressed the catastrophe frequency, but only at higher motor concentrations (≥25 nM; Supplemental Figure S2). These results indicate that the KIF21B motor domain is sufficient to modulate MT dynamics but inclusion of the C-terminal MTRs further enhances MT stabilization.
FIGURE 1:
KIF21B motors stabilize MT dynamics and promote MT assembly. (A) Motor constructs used for single molecule experiments. (B) Kymograph plots showing dynamic MTs polymerizing from stabilized MT seeds in the absence or presence of KIF21B-FL-Halo or KIF21B-MD-Halo motors. Scale bars: 10 µm (horizontal) and 30 s (vertical). (C–E) Average MT plus-end growth distances. Ten kymographs were averaged for each condition. N = 3 independent experiments. (F) MT plus-end growth rates in the presence of increasing concentrations of KIF21B-FL-Halo or KIF21B-MD-Halo motors. Plotted are means and SDs. Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (n.s. p > 0.05; *p < 0.05; ***p < 0.001; ****p < 0.0001). Data from 54–94 MTs and N = 3–6 independent experiments. G) MT plus-end catastrophe frequency in the presence of increasing concentrations of KIF21B-FL-Halo or KIF21B-MD-Halo motors. Plotted are means and SDs. Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (****p < 0.0001). Data from 54–94 MTs and N = 3–6 independent experiments. (H–J) TIRF microscopy images showing MT formation from free tubulin dimers incubated in the absence or presence of 50 nM KIF21B-FL-Halo or KIF21B-MD-Halo. Images were taken 10 min after solutions were introduced into flow chambers. Scale bar: 10 µm. (K–M) Averaged light scattering traces for solutions containing buffer, Taxol, or 50 nM KIF21B-FL-Halo or KIF21B-MD-Halo motors with 1 mM ATP or 1 mM ADP. Means were compared with Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (Buffer–Taxol ****p < 0.0001; Buffer–KIF21B-FL-Halo ****p < 0.0001; KIF21B-FL-Halo ATP–KIF21B-FL-Halo ADP n.s. p > 0.05; Buffer–KIF21B-MD-Halo n.s. p > 0.05; KIF21B-MD-Halo ATP–KIF21B-MD-Halo ADP n.s. p > 0.05). Data from 7–22 traces from N = 5 independent experiments. Graphs show mean values for each time point. See Supplemental Figure S2 for graphs with SDs.
Differences with previous work (Ghiretti et al., 2016) likely result from improved motor preparations, lower motor concentrations, and the use of higher-ionic-strength buffers that reduce the formation of KIF21B multimers and more closely model physiological conditions. We did not observe the prolonged attachment of KIF21B motors to dynamic MT plus ends or a motor preference for the stable GMPCPP seeds over the dynamic growing MT lattice, as reported by van Riel et al. (2017), who performed experiments in the presence of mCherry-EB3. This suggests that both the reduction in MT growth rate and the decreased catastrophe frequency induced by full-length KIF21B may be mediated by a stabilization of the MT lattice, rather than a specific stabilization or capping of dynamic MT plus ends.
In our TIRF experiments, in addition to the dampened MT dynamics induced by KIF21B, we observed that new MTs emerged over time at 50 nM KIF21B-FL-Halo, which may be due to nucleation of new filaments. To determine whether KIF21B motors promote the assembly of new MTs, we examined the effects of KIF21B on MT polymerization in the absence of preformed MT seeds. The addition of either buffer alone or KIF21B-MD-Halo was not sufficient to nucleate MTs, as assessed by TIRF microscopy and light scattering (Figure 1, H–M, and Supplemental Figure S3). However, the addition of KIF21B-FL-Halo resulted in the robust formation of MTs over 10 min (Figure 1I), similar to the assembly induced by the addition of K560-Halo (Supplemental Figure S2F). The rate and extent of MT growth in the presence of KIF21B-FL-Halo, as assessed by turbidity, were similar in the presence of MgATP or MgADP (Figure 1L and Supplemental Figure S3G). Because the addition of MgADP weakens the interaction between kinesin motor domains and MTs (Crevel et al., 1996) but does not inhibit nucleation in our assays, these results suggest that the MTRs promote MT assembly. Collectively, the effects on MT growth rate, catastrophe frequency, and induction of assembly induced by full-length KIF21B lead to a net stabilization of MT growth at physiologically relevant concentrations of KIF21B, estimated to be 5–25 nM by mass spectrometry (Martens et al., 2006; Kim et al., 2014).
Reconstitution of motor switching on engineered parallel and antiparallel MT bundles
Because KIF21B motors can effectively navigate the antiparallel MT arrays characteristic of dendrites in mammalian neurons (Ghiretti et al., 2016), we wanted to test whether the relative orientation of MTs within MT bundles affects the motile properties of KIF21B. As KIF21B-FL motors modulate MT dynamics (Figure 1), we reconstituted parallel and antiparallel arrays of dynamic MTs (Figure 2A). Most of the bundles were composed of two aligned MTs, oriented either parallel or antiparallel to one another, as evaluated by kymograph analysis of MT plus-end dynamics and fluorescence intensity measurements over time (Figure 2A and Supplemental Videos 1–3).
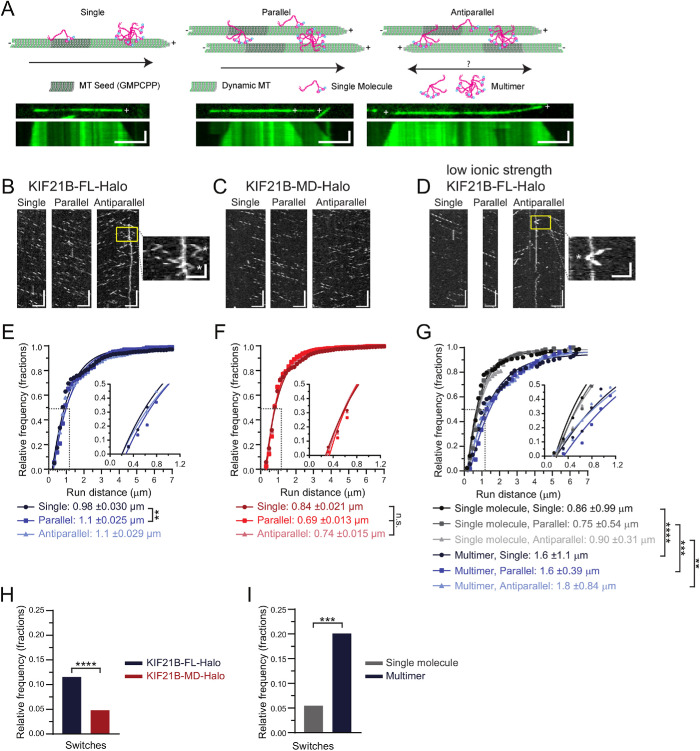
FIGURE 2:
Motor multimerization and KIF21B MTRs improve motor processivity and track switching. (A) Schematic of in vitro single molecule assays. Dynamic MTs were grown from surface-immobilized GMPCPP MT seeds. Some MTs remain isolated in space as they grow, while others overlap in parallel or antiparallel orientations with neighboring MTs. Kymographs show growing dynamic MT ends over time for each type of MT arrangement. Dynamic MT plus ends are indicated (+). KIF21B dimers or multimeric complexes contact MT lattices with their N-terminal motor domains, as well as MTRs within their C-terminal tails. Motor domains were labeled with TMR Halo ligands (cyan dot). Scale bars: 5 µm (horizontal) and 1 min (vertical). (B–D) Kymograph plots of KIF21B-FL-Halo, KIF21B-MD-Halo, and KIF21B-FL-Halo motility in low-ionic-strength buffering system along single MTs or parallel and antiparallel MT bundles. Zoom-in image highlights track switches observed on antiparallel MTs (indicated with white * in expanded images). Track switches were defined as a reversal in run direction exceeding three pixels. Scale bars: 5 µm (horizontal) and 20 s (vertical). Zoom-in scale bars: 2.5 µm (horizontal) and 5 s (vertical). (E–G) Cumulative distribution of total distance motor traveled along different MT arrays. Data points were fitted by a single exponential decay function. Differences in run distances can be more clearly seen in insets. Average distance traveled, as calculated by taking the inverse of the rate constant, and SDs are shown in the legends. Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (n.s. p > 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). (H, I) Frequency of track switches for single KIF21B-FL-Halo and -MD-Halo motor runs or single and multimeric KIF21B-FL-Halo motor run along antiparallel MTs. Two-tailed Mann–Whitney (***p < 0.001; ****p < 0.0001). Data for single molecule KIF21B-FL-Halo and KIF21B-MD-Halo from 400–1069 motor runs from n = 20–60 MTs and N = 6–7 independent experiments. Data for single and multimeric KIF21B-FL-Halo from 54–174 motor runs from n = 11–12 MTs and N = 4 independent experiments.
Movie S1.
Purified KIF21B-FL motor motility along single dynamic MTs. KIF21B-FL-Halo motors move towards the growing plus-end of the MT. Movie formatted at 30 FPS. Scale bar: 5 μm.
Movie S2.
Purified KIF21B-FL motor motility along dynamic MTs forming a parallel bundle. KIF21B-FL-Halo motors move towards the growing plus-ends of each MT. Movie formatted at 30 FPS. Scale bar: 5 μm.
Movie S3.
Purified KIF21B-FL motor motility along dynamic MTs forming an antiparallel bundle. KIF21B-FL-Halo motors move towards the growing plus-ends of each MT. Some motors switch from movement along one MT to the other to change direction. Movie formatted at 30 FPS. Scale bar: 5 μm.
We compared the motility of purified recombinant KIF21B-FL-Halo or -MD-Halo motors (Figure 1A) on single MTs and MT bundles organized in either parallel or antiparallel orientations, as assessed by monitoring the differing assembly and disassembly dynamics characteristic of MT plus and minus ends (Figure 2, A–D, and Supplemental Videos 1–3). We focused on motor particles whose intensity was consistent with that of single molecules, of which ∼50% were fully labeled. Both the full-length and motor domain constructs moved uniformly in the plus-end direction on single MTs (Figure 2B). We observed robust unidirectional movement on parallel MT bundles (Figure 2, B and C) and robust bidirectional movement on antiparallel MT bundles (Figure 2, B and C). The average speeds of KIF21B-FL-Halo and -MD-Halo motors along the three MT arrangements were very similar (0.43–0.5 µm/s; Supplemental Figure S4, A and B). However, KIF21B-MD-Halo displayed shorter run distances than KIF21B-FL-Halo, regardless of the MT arrangement (Figure 2, E and F). The run distance of KIF21B-FL-Halo was slightly, but significantly, longer on parallel MT bundles compared with single MTs, while the run distance of KIF21B-MD-Halo was not affected by MT track type. Strikingly, both KIF21B-FL-Halo and -MD-Halo motors exhibited track switching within antiparallel bundles (Figure 2B, zoom-in). Switching was more frequent for the full-length construct, measured as either total events observed (Figure 2H) or number of switches per distance traveled (Supplemental Figure S4C).
In the cell, cargoes are likely to be transported by more than one kinesin motor (Gross et al., 2007; Holzbaur and Goldman, 2010). Multimerization of KIF21B may also have been observed by van Riel et al. (2017), where KIF21B motor complexes formed at the plus ends of growing MTs. To test the effects of motor multimerization on motility and track switching, we introduced purified KIF21B-FL-Halo motors into low-ionic-strength buffer (12 mM PIPES, pH 6.8), which induced multimer formation. We used single molecule photobleaching to calibrate the relationship between fluorescence intensity and the number of molecules in a cluster (Supplemental Figure S5A) and used this calibration to distinguish between single molecules and multimers (estimated to be two to three motors) (Supplemental Figure S5B). KIF21B-FL-Halo multimers and single molecules displayed similar velocities on all MT arrays (Supplemental Figure S5D), but multimers moved longer distances than single molecules regardless of the type of MT arrangement (Figure 2G). Run distances positively correlated with intensity, while velocity did not correlate with intensity, suggesting that formation of motor teams leads to improved run distances with no effect on velocity (Supplemental Figure S6, A and B).
KIF21B-FL-Halo multimers switched tracks significantly more frequently per run than single molecules of KIF21B-FL-Halo (Figure 2I); multimeric motors also switched tracks more frequently per unit distance than single molecules (Supplemental Figure S5E). We observed a significant positive correlation between the number of track switches per distance traveled and the intensity of the oligomer (Supplemental Figure S6C), suggesting that the likelihood of track switching scales with multimerization. Collectively these results indicate that the MTRs in the C-termini of KIF21B enhance both processivity and track switching, while motor multimerization further contributes to both parameters.
Quantitative mapping of the dendritic MT network
We observed that bundles of only two MTs are sufficient to influence KIF21B motility. How does this scale to bundles of MTs organized into larger arrays, such as found in dendrites? Live cell imaging studies suggest that KIF21B drives endogenous cargoes with a net retrograde bias in dendrites (Ghiretti et al., 2016), a puzzling finding for a plus end–directed motor moving along an array in which the majority (∼60%; Ayloo et al., 2017) of MTs are oriented with their plus ends outward. A recent model proposed by Kapitein and colleagues (Tas et al., 2017) suggests that the dendritic MT network is composed of distinct unipolar MT bundles and that these bundles are marked by specific posttranslational modifications (PTMs) of the tubulin cytoskeleton. Tas et al. (2017) proposed that MT motors recognize this code, preferentially binding to and moving along selected MT populations to drive long-range transport in a specific direction. Does the MT code present in dendrites bias KIF21B motors to move in the retrograde direction?
To determine whether the native MT arrays in dendrites can bias motors toward movement in the retrograde direction, we used motor-PAINT with rat hippocampal neurons (Brawley and Rock, 2009; Tas et al., 2017), which allows superresolution tracking of single motors interacting with the cytoskeleton of fixed permeabilized cells. We used immunocytochemistry to compare the ratios of dendritic tubulin PTM and Microtubule-Associated protein (MAP) intensities in axons and dendrites in extracted arrays compared with those in permeabilized neurons and observed the general preservation of tubulin PTMs but a discernible reduction in some dendritic MAPs such as MAP2 after extraction (Supplemental Figure S7).
To extend the previous work of Tas et al. (2017) and generate a more quantitative map of the MT network within hippocampal dendrites, we used TIRF microscopy to track individual processive runs of K560-GFP (Figure 1A) along native MT arrays extracted from axons and dendrites (Figure 3, A and B, and Supplemental Videos 4 and 5), with the assumption that this motor provides a nonbiased readout of MT orientation. The resulting videos demonstrate the expected pattern of primarily unidirectional motility along axons and more bidirectional motility along dendrites, but the density of the native MT arrays makes unambiguous run detection more challenging. To more rigorously map the runs, we used an automated single particle segmentation and tracking program called Cega to track moving particles within a noisy environment (Masucci, Relich, et al., 2021; see Materials and Methods). Owing to the crossing of individual motor runs in both axons and dendrites, complete tracks could not be reliably determined. Instead, individual track segments from single motors connected frame-by-frame from motor movement captured every 0.2 s for 2 min were identified and used to construct a quantitative map of MT organization and polarity.
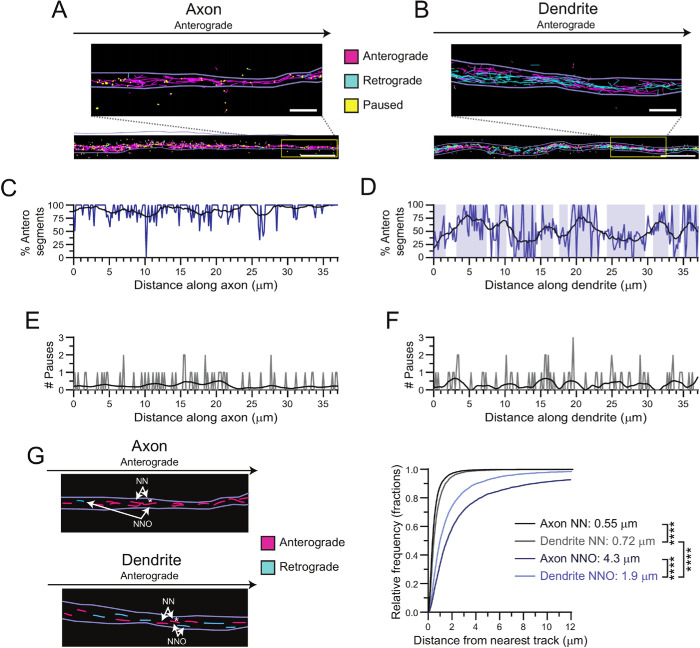
FIGURE 3:
Dendritic MTs are distributed heterogeneously. (A, B) Single particle motor track segments from a single TIRF movie. Light purple lines indicate bounds of axonal or dendritic process. Segments moving to the right (anterograde) are plotted in magenta, those moving to the left (retrograde) are cyan, and paused spots are shown in yellow. Zoom-in images show the separation between tracks. Scale bar: 5 µm. Zoom-in scale bar: 1 µm. (C, D) Percentage of anterograde track segments across the long axis of the axon and dendrite shown in A and B. Shaded blue areas indicate regions where the percentage of anterograde segments is above or below 50% in the dendritic process. Black lines show smoothed trendline over 2 µm window. (E, F) Number of paused track segments located across the long axis of the axon and dendrite shown in A and B. Black lines show smoothed trendline over 2 µm window. (G) Schematic of nearest neighbor (NN) track and nearest neighbor opposing (NNO) track and cumulative distribution of the distance between each anterograde moving track segment and its NN moving in any direction or NNO moving in the opposite direction in both axons and dendrites. A single exponential decay function was fitted to the data. Listed are the average distances traveled, as calculated by taking the inverse of the rate constant. Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (****p < 0.0001). Unless otherwise indicated, data from 59–80 processes from n = 49–50 neurons and N = 4 independent experiments.
Movie S4.
Purified K560-GFP motor motility along extracted axonal MT arrays. K560-GFP motors move mainly in the anterograde direction along axonal MTs. Movie formatted at 10 FPS. Scale bar: 5 μm.
Movie S5.
Purified K560-GFP motor motility along extracted dendritic MT arrays. K560-GFP motors move in both anterograde and retrograde directions along dendritic MTs. Some areas contain motor movement in mainly one direction. Movie formatted at 10 FPS. Scale bar: 5 μm.
Cega analysis demonstrates that K560-GFP motors moved along axonal MT arrays with >90% of the tracks in the anterograde direction (Figure 3, A and C, and Supplemental Video 4). On dendritic MT arrays, K560-GFP motors moved bidirectionally (Figure 3, B and D, and Supplemental Video 5), but strikingly, some regions showed preferred directionality as indicated by blue shading in Figure 3D; this local heterogeneity in MT organization is also apparent from the smoothed trend line that shows the average track direction within 2 µm (compare the trend lines from axons and dendrites in Figure 3, C and D). In contrast to the localized differences in MT orientation, pauses were randomly distributed throughout both axons and dendrites (Figure 3, E and F, and Supplemental Figure S8).
To investigate the spatial relationship of adjacent MT tracks, we measured the distance between anterograde moving track segments and their nearest neighboring (NN) track segment or nearest neighboring opposing (NNO) retrograde track segment in both axons and dendrites (Figure 3G). The distance between NN track segments in axons was significantly shorter than that of dendrites, consistent with tighter MT spacing in axons (Chen et al., 1992). If the dendritic MTs were homogenously mixed in orientation, then the distances between the anterograde track segments and their NN and NNO track segment should be similar. However, in both axons and dendrites, the distance between NN track segments was much shorter than that of NNO track segments. In axons, this is likely due to the very small number of MTs oriented with plus ends toward the soma (Baas et al., 1988). In dendrites, the increased NNO as compared with NN distance provides quantitative support for the model proposed by Tas et al. (2017), which suggests that MTs are locally organized in bundles with similar polarities, creating regions where similarly oriented MTs are located close together and those of opposite orientation are spatially removed. Further, our observations indicate that MT orientation in mammalian dendrites is distributed heterogeneously along the lengths of each process.
Recombinant KIF21B motors exhibit directional switches on extracted dendritic MT arrays
To determine whether the native MT PTM code together with the native organization of MTs in dendrites leads to a retrograde bias for KIF21B motility, we used the motor-PAINT assay to observe the movement of KIF21B molecules in axons and dendrites (Figure 4 and Supplemental Figure S9). We were able to directly compare the movement of either Alexa Fluor 660–labeled KIF21B-FL-Halo or KIF21B-MD-Halo to K560-GFP (Figure 1A) by mixing the differentially labeled motors and simultaneously tracking their movement on the same MT arrays (Figure 4, A and B). Individual runs along extracted MT arrays were tracked using Cega (Masucci, Relich, et al., 2021). Kymographs generated for the KIF21B constructs show that the full-length and truncated motors have similar directional patterns along axonal and dendritic MT arrays, and both closely resembled the patterns seen with K560-GFP (Figure 4, A and B). Again, we saw evidence for regional heterogeneity in the organization of MTs in dendrites, with some kymographs demonstrating motors moving in mainly one direction (Figure 4A, “Bi-directional motility” vs. “Anterograde motility”), supporting the idea that the MTs are locally organized into bundles with similarly oriented MTs (Tas et al., 2017).
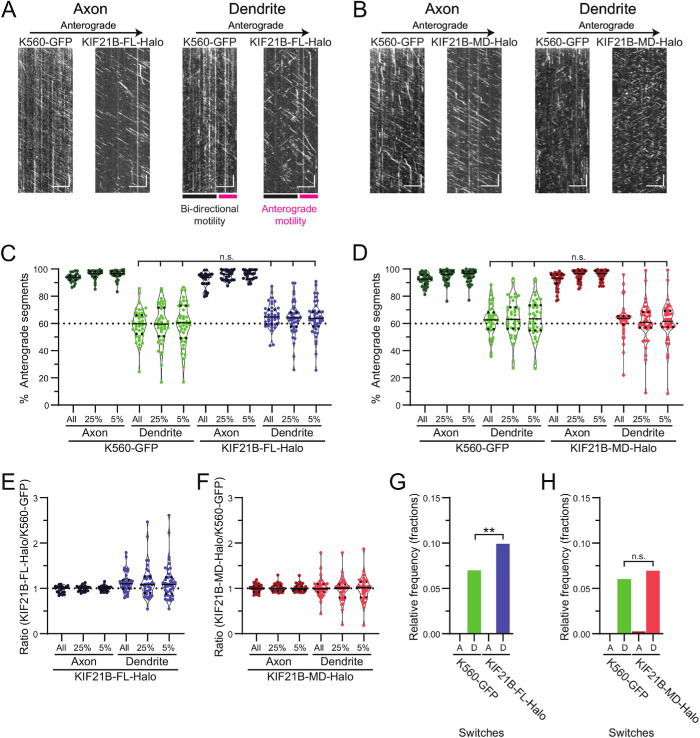
FIGURE 4:
KIF21B motors do not move with a directional bias on extracted dendritic MT arrays. (A, B) Kymographs of motor motility along axonal or dendritic MTs for K560-GFP and KIF21B-FL-Halo or KIF21B-MD-Halo motors moving on the same MTs. Colored bars in A indicate regions of the dendrite where motors move bidirectionally and regions where motors move primarily in one direction (anterograde motility). Scale bars: 10 µm (horizontal) and 10 s (vertical). (C, D) Percentage of track segments moving in the anterograde direction in axons and dendrites. Plotted are individual values in color, means in solid black lines, and 25th and 75th quartiles in dotted black lines. Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (n.s. p > 0.05). (E, F) Ratio of the percentage of KIF21B-FL-Halo or KIF21B-MD-Halo anterograde track segments to the percentage of K560-GFP anterograde tracks along the same axonal and dendritic arrays. Plotted are individual values in color, means in solid black lines, and 25th and 75th quartiles in dotted black lines. (G, H) Frequency of motors switching direction of movement along axonal (A) and dendritic (D) MT arrays. Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (n.s. p > 0.05; **p < 0.01). Data from 24–42 processes from n = 21–28 neurons and N = 4 independent experiments.
Along axonal MT arrays, >90% of all motors tested moved in the anterograde direction, while ∼60% moved in the anterograde direction on dendritic MTs (Figure 4, C and D). The standard deviations of directional segments in dendrites for all three motors were much larger than observed in axons, again consistent with local variability in MT organization along the length of the dendrite. We found no difference in direction of movement between K560-GFP and KIF21B-FL-Halo or KIF21B-MD-Halo in axons and dendrites (Figure 4, C–F). We further compared the percent anterograde directions of KIF21B-FL-Halo and KIF21B-MD-Halo motors in dendrites to 60%, because this is the direction of motility expected from the 60% plus end–out MT polarity measured by K560-GFP motors, and measured no significant difference. This comparison suggests that native, stabilized dendritic MT arrays do not bias KIF21B motors to move in the retrograde direction.
Our preparations of purified K560-GFP, KIF21B-FL-Halo, and KIF21B-MD-Halo motors produced mainly single, two-headed motor proteins, as assessed by controlled photobleaching of processing motors in motility assays (Supplemental Figure S5, A and B). However, ∼40% of the population of KIF21B-FL-Halo motors were multimers, as determined by fluorescence intensity measurements. We took advantage of this multimerization to test the idea that multiple motors are needed to confer retrograde bias to KIF21B. We compared the percentage of anterograde segments for each process for all motors to that of the brightest 25% or 5% of moving motors and found no significant difference between these groups and no significant difference against the 60% expected motility direction (Figure 4, C and D). We further quantified the ratio of the percentage of anterograde track segments of KIF21B motors compared with K560-GFP motors for each process and found that the ratio of KIF21B to K560-GFP segments was about 1 (Figure 4, E and F), regardless of the motor brightness, suggesting that motor multimerization does not contribute to directional bias in motility along extracted neuronal MT cytoskeletons.
In dendrites but not axons, we observed pronounced directional switching by KIF21B-FL-Halo. We measured the frequency of motor runs that switched direction of movement along dendritic and axonal MT arrays and observed that KIF21B-FL-Halo switched tracks more frequently than K560-GFP along the same dendritic MT tracks (Figure 4G). In contrast, KIF21B-MD-Halo and K560-GFP switched tracks at similar rates (Figure 4H). These results suggest that the MTRs in the C-terminal tail of KIF21B enhance track switching along dendritic MT bundles with native organization and spacing.
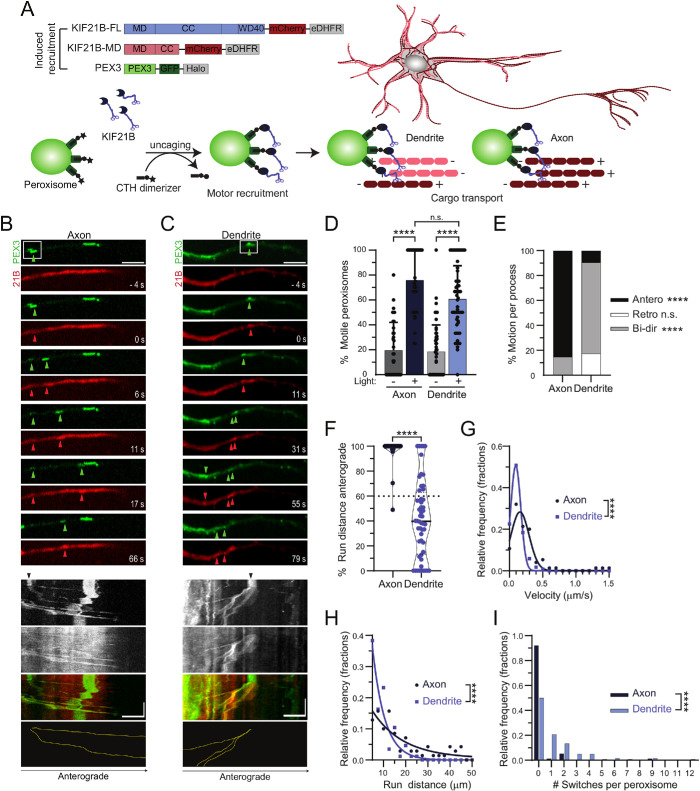
Optogenetic recruitment of KIF21B induces retrograde trafficking of dendritic cargoes in live neurons
As the native MT PTM code and cytoskeletal organization of extracted dendrites was insufficient to model the retrograde bias for KIF21B motors observed in live cell assays (Ghiretti et al., 2016), we wondered whether the key factor missing from our assay might be MT dynamics. As KIF21B has the ability to modulate MT dynamics both in vitro and in vivo, we hypothesize that this modulation may contribute to the establishment of net retrograde bias for KIF21B-driven cargoes seen in mammalian dendrites. To test this possibility, we used a previously validated optogenetic assay (Ayloo et al., 2017) to determine how KIF21B motors perform when recruited to a nonnative cargo in axons and dendrites of live neurons, under cellular conditions with robust MT dynamics (Figure 5A). Full-length KIF21B (KIF21B-FL) was recruited to peroxisomes, a largely immobile cargo in hippocampal neurons, via the heterodimerization of DHFR-fused motor and HaloTag (Halo)-fused peroxisome targeting sequence PEX3 in response to the photoinduced uncaging of a small organic compound (CTH) (Ballister et al., 2014, 2015; Ayloo et al., 2017; Zhang et al., 2017). The CTH was uncaged in defined regions of the dendrites and axons of neurons expressing both peroxisome and KIF21B constructs. Photoactivation induced the recruitment of mCherry-labeled KIF21B to peroxisomes labeled with GFP (Figure 5, B and C, and Supplemental Videos 6 and 7) and induced the processive motility of more than 60% of peroxisomes in axons and dendrites within ∼28 s (Figure 5D and Supplemental Figure S10).
FIGURE 5:
Induced recruitment of KIF21B motors to peroxisome cargo in live neurons promotes retrograde trafficking in dendrites. (A) Diagram of motor constructs and experimental schematic for induced recruitment experiments in live neurons. Dark red and pink lines and plus and minus signs indicate MT orientation within arrays; antiparallel MT arrays in dendrites and parallel arrays in axons. (B, C) Time series and corresponding kymograph showing movement of photoactivated (white box) peroxisomes in axons and dendrites. Green and red arrowheads indicate the locations of photoactivated peroxisomes with successful KIF21B recruitment in both the PEX3 and KIF21B fluorescence channel, respectively. Black arrows on kymographs indicate regions of photoactivation. Yellow traces indicate movement of the KIF21B–recruited peroxisomes identified in the time series. Scale bars: 5 µm (horizontal) and 1 min (vertical). (D) Percentage of peroxisomes that are motile in axons and dendrites with and without photoactivation. Plotted are means and SDs. Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (n.s. p > 0.05; ****p < 0.0001). (E) Percentage of peroxisome runs moving anterograde, retrograde, or bidirectionally for each axonal and dendritic process. Bidirectional (Bi-dir) movement is characterized by peroxisomes that move more than 0.4 µm in both anterograde (Antero) and retrograde (Retro) directions. Two-way ANOVA and Tukey multiple comparison (n.s. p > 0.05; ****p < 0.0001). (F) Percentage of total peroxisome run distance per process moving in the anterograde direction. Plotted are individual values in color, means in solid black line. and 25th and 75th quartiles in dashed black lines. Two-tailed Mann–Whitney test (****p < 0.0001). (G) Histogram of velocities of motile peroxisomes after photoactivation in axons and dendrites. Data points were fitted by a Gaussian function. Two-tailed Mann–Whitney test (****p < 0.0001). (H) Histogram of run distances of motile peroxisomes after photoactivation. Data points were fitted by a single exponential decay function. Two-tailed Mann–Whitney test (****p < 0.0001). (I) Histogram of number of track switches per peroxisome run in axons and dendrites. Track switches are characterized by a reversal in direction that exceeds 0.4 µm in both anterograde and retrograde directions. Two-tailed Mann–Whitney test (****p < 0.0001). Data from 76–120 peroxisomes from n = 34–52 neurons and N = 6 independent experiments.
Movie S6.
Induced recruitment of KIF21B motors to peroxisome cargo in axons of live neurons. KIF21B-FL recruited peroxisomes move in the anterograde direction immediately after photoactivation. White box indicates area of photoactivation. Green and red arrowheads indicate the locations of photoactivated peroxisomes with successful KIF21B-Fl recruitment in both the PEX3 and KIF21B fluorescence channel, respectively. Movie formatted at 10 FPS. Scale bar: 5 μm.
Movie S7.
Induced recruitment of KIF21B motors to peroxisome cargo in dendrites of live neurons. About 6 s after photoactivation, KIF21B-FL recruited peroxisomes in dendrites move in the retrograde direction. White box indicates area of photoactivation. Green and red arrowheads indicate the locations of photoactivated peroxisomes with successful KIF21B-FL recruitment in both the PEX3 and KIF21B fluorescence channel, respectively. Movie formatted at 10 FPS. Scale bar: 5 μm.
In axons, >85% of KIF21B-bound peroxisomes moved in the anterograde direction, consistent with the highly polarized nature of the axonal MT cytoskeleton (Figure 5E). KIF21B-bound peroxisomes in dendrites moved bidirectionally (Figure 5E), with 42% of the movement in the anterograde direction (Figure 5F), in contrast to the ∼60% plus end–out MT polarity measured in dendrites (Ayloo et al., 2017), consistent with a retrograde bias. The average velocities of KIF21B-induced motion were significantly slower in dendrites than in axons (Figure 5G), and the run distances were significantly shorter (Figure 5H). Moving peroxisomes also switched directions more frequently in dendrites than they did in axons (Figure 5I).
The retrograde bias in the motility of peroxisomes in dendrites following acute recruitment of KIF21B is remarkably similar to the movement of both KIF21B puncta and KIF21B-associated TrkB/BDNF cargo in the dendrites of live hippocampal neurons observed previously (Ghiretti et al., 2016). Of note, this retrograde bias is remarkably similar to that in parallel experiments acutely recruiting the minus end–directed dynein–dynactin motor complex, which induced retrograde trafficking of 60% of peroxisomes in dendrites (Ayloo et al., 2017).
MT dynamics drive the retrograde bias of KIF21B in dendrites
The retrograde bias in KIF21B motility observed in dendrites in cellular assays was not seen in our experiments on extracted neuronal cytoskeletal arrays, suggesting to us that MT dynamics may be necessary to promote this bias. This hypothesis is also consistent with our in vitro studies highlighting the importance of the MTRs of KIF21B in regulating motility and cytoskeletal remodeling.
To investigate this hypothesis, we first asked whether there is an asymmetry to the organization of dynamic MTs in dendrites. We mapped the dynamics of MTs present in dendrites of live neurons by tracking the movement of mCherry-EB3 comets. We compared the frequencies of anterograde-directed and retrograde-directed EB3 comets and found that plus end–out MTs were significantly more dynamic than plus end–in MTs (Figure 6A and Supplemental Figure S10A). Low-dose nocodazole treatment dampens MT dynamics without inducing net depolymerization or disrupting the integrity of the MT network (Vasquez et al., 1997; Guedes-Dias et al., 2019). Treatment of hippocampal neurons with 100 nM nocodazole reduced the number, run length, and velocity of MT plus-end tracking EB3 comets (Figure 6A and Supplemental Figure S10). Importantly in control conditions the speed and number of anterograde EB3 comets are significantly greater than those of the retrograde moving comets, while nocodazole treatment significantly reduced the dynamics of anterograde moving EB3 comets without significantly affecting the dynamics of retrograde comets, thus creating an environment in which the dynamics of plus end–out and plus end–in MTs are more similar.
FIGURE 6:
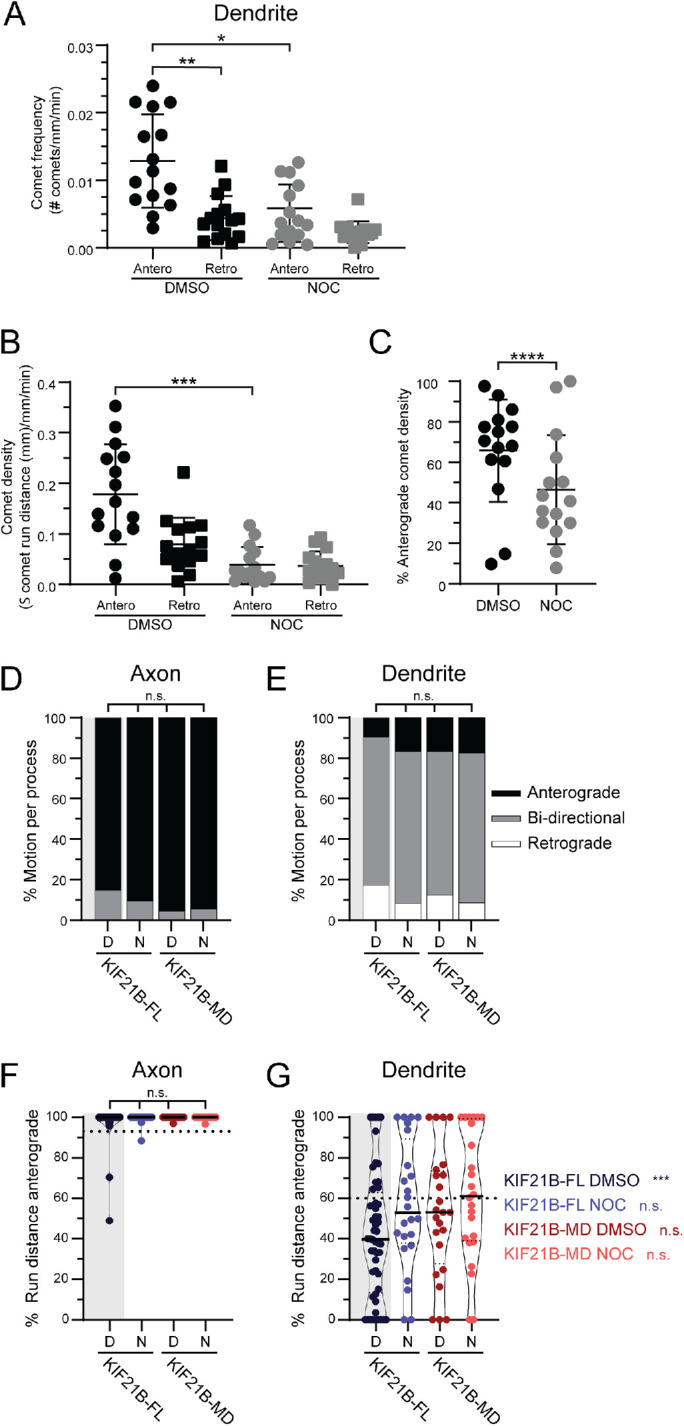
KIF21B-induced retrograde trafficking in dendrites requires C-terminal tail domains and MT dynamics. (A) Frequency of EB3 comets in live neurons treated with DMSO or nocodazole (Noc), separated by comet direction; anterograde (Antero) and retrograde (Retro). Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (*p < 0.05; **p < 0.01). (B) EB3 comet density in live neurons treated with DMSO or nocodazole (Noc) calculated by taking the relative sum of all EB3 comet run distances in both the anterograde (Antero) and retrograde (Retro) directions. Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (***p < 0.001). (C) Percent of EB3 comet density in the anterograde direction in DMSO control or nocodazole (Noc) conditions. Data from 63–476 comets from n = 6 neurons and N = 3 independent experiments. Two-tailed Mann–Whitney test (****p < 0.0001). (D, E) Percentage of peroxisomes moving anterograde, retrograde, or bidirectionally for individual axonal and dendritic processes. DMSO (D) or nocodazole (N) conditions are indicated at the bottom of the graph. Bidirectional movement is characterized by peroxisomes that move more than 0.4 µm in both anterograde and retrograde directions. Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (n.s. p > 0.05). (F, G) Percentage of total peroxisome run distance per process moving in the anterograde direction in both axons and dendrites. DMSO (D) or nocodazole (N) conditions are indicated at the bottom of the graph. Plotted are individual values in color, means in solid black line, and 25th and 75th quartiles in dashed black lines. For comparison of distributions, we used Kruskal–Wallis one-way ANOVA and Dunn’s multiple comparison (n.s. p > 0.05). In dendrites we used a one-sample Wilcoxon test to compare distributions against theoretical mean (60%). Significance is listed in color to the right side of the graph (n.s. p > 0.05; ***p < 0.001). Data from 37–120 peroxisomes from n = 18–52 neurons and N = 3–6 independent experiments, unless otherwise indicated.
To investigate the role of MT dynamics, we compared peroxisome motility following acute recruitment of KIF21B in neurons treated with 100 nM nocodazole or with dimethyl sulfoxide (DMSO). In this experiment, recruitment of KIF21B motors to peroxisomes induced the processive motility of 53%–76% of peroxisomes in axons and dendrites within 22–30 s, even in the presence of nocodazole (Supplemental Figure S11, A–F). In axons of both control and nocodazole-treated neurons, KIF21B-FL motors induced anterograde movement of >97% of peroxisomes (Figure 6, B and D). However, in dendrites, the addition of nocodazole led to a loss of retrograde bias, with the observed anterograde movement of 57% of peroxisomes (Figure 6, C and E), similar to the movement induced upon recruitment of K560 motors (Ayloo et al., 2017). We did note substantial scatter in the data, likely due to heterogeneity in MT polarity along the length of dendrites, as shown in Figure 4.
Finally, we asked whether the MTRs of KIF21B are important for retrograde trafficking within dendrites of live neurons, using our optogenetic recruitment assay to assess the direction and movement of motor domain–truncated (KIF21B-MD) motors when recruited to immobile peroxisomes in both axons and dendrites (Figures 5A and 6, B and C). In this system, recruitment of KIF21B motors to peroxisomes induced the processive motility of 60%–64% of peroxisomes in axons and dendrites within 26–40 s, even in the presence of nocodazole (Supplemental Figure S11, A–F). We found that 53% of KIF21B-MD–recruited peroxisomes moved in the anterograde direction (Figure 6E), indicating a loss of retrograde bias.
We compared the direction of motor-recruited peroxisomes in all conditions to the 60% plus end–out MT polarity measured in dendrites, which would be expected to induce 60% anterograde motility (Figure 6E). When MT dynamics were dampened with nocodazole, 57% of KIF21B-FL and 62% of KIF21B-MD–recruited peroxisomes moved in the anterograde direction, suggesting that both MT dynamics and the MTRs of KIF21B are important for biasing movement in the retrograde direction.
There was no significant difference in the switching behaviors of -FL or -MD–recruited peroxisomes in either axons or dendrites (Supplemental Figure S12, A and B), even when MT dynamics were dampened. The frequency of directional switches seen by peroxisomes in vivo was greater than that seen in vitro on engineered MT bundles, suggesting that the recruitment of multiple KIF21B motor domains on cargo allows for robust track switching. In addition, recruitment of both KIF21B-FL and KIF21B-MD motors to peroxisomes induced similar velocities and run lengths in the presence and absence of nocodazole (Supplemental Figure S12, C–F). However, both KIF21B-FL and KIF21B-MD–recruited cargoes moved slower and less processively in dendrites than in axons. Together, these results suggest that both MT dynamics and the secondary MT-binding regions of KIF21B are important for imparting retrograde biased movement in dendrites.
DISCUSSION
Most of our current understanding of MT motor function in cells comes from studies monitoring cargo movement within simple radial and axonal MT arrays (Hirokawa et al., 2010; Burute and Kapitein, 2019). However, there are many examples of cell types containing more complicated MT geometries on which molecular motors navigate (Muroyama and Lechler, 2017; Sanchez and Feldman, 2017). For example, kinesin and dynein motors function within the bipolar MT arrays of the mitotic spindle of dividing cells, where they work to pull and slide apart MTs, regulate MT dynamics, and transport materials and chromosomes to kinetochores and spindle poles (Wordeman, 2010; McIntosh, 2016; Dwivedi and Sharma, 2018).
The mixed MT polarity characteristic of mammalian dendrites represents a similar challenge to understand how unidirectional motors can provide net long-range movement of cargoes in a polarized manner. Kinesin motors have evolved specific properties to generate, maintain, and navigate these complex arrays (Sweeney and Holzbaur, 2018). Some kinesin motors contain MT-binding domains, in addition to their canonical motor domains, that have been implicated in MT sliding or MT transport (Navone et al., 1992; Andrews et al., 1993; Furuta and Toyoshima, 2008; Fink et al., 2009; Jolly et al., 2010; Seeger and Rice, 2010; Reinemann et al., 2017). In addition, many kinesin motors are known to influence the MT network by regulating MT dynamics (Acharya et al., 2013; Gudimchuk et al., 2013; Arellano-Santoyo et al., 2017; Trofimova et al., 2018; Chen et al., 2019). Neuronal motors likely have evolved specific properties to locally regulate MT dynamics and/or control their directionality in dendrites.
Recent studies have advanced our understanding of the MT organization differences within dendrites and axons (Sánchez-Huertas et al., 2016; Sanchez and Feldman, 2017; Wu and Akhmanova, 2017; Cunha-Ferreira et al., 2018; Cao et al., 2020; Liang et al., 2020). Initial measurements using the “hook” method showed that axons contain uniform plus end–out oriented MTs, while only ∼60% of the MTs within dendrites are oriented with their plus ends extending away from the cell body (Baas et al., 1988; Burton, 1988). Studies focused on dynamic MTs show that ∼65% of dendritic MTs are oriented plus end–out, as assessed by EB3 dynamics in live Purkinje neurons (Stepanova et al., 2003; Kleele et al., 2014). Measurements of EB3 comets in hippocampal neurons, at the same developmental stage as those used in this paper, indicated that 64% of MTs in dendrites are oriented plus end–out (Ayloo et al., 2017). The ∼60% anterograde motility observed for K560 motors moving on stabilized extracted dendritic MTs (Figure 4) is strikingly similar to the orientation of dynamic MTs within dendrites measured through EB3 comets (Ayloo et al., 2017). A model proposed by Kapitein and colleagues (Tas et al., 2017) suggests that the dendritic MT network is composed of distinct unipolar MT bundles and that these bundles are marked by specific PTMs of the tubulin cytoskeleton. Using the same technique of motor-PAINT to track the movement of K560 motors, we acquired a detailed map of MT orientation within the dendrites of rat hippocampal neurons and found that MTs are organized heterogeneously into regions characterized by bundles with uniform polarity (Figure 3). The MT organization within axons and dendrites provides a clear way to differentiate between the two compartments (Bentley and Banker, 2016; Nirschl et al., 2017; Gumy and Hoogenraad, 2018). However, the mixed MT organization present within dendrites sets up problems for motors to move productively/efficiently within MT arrays.
KIF21B has been shown to move with net retrograde bias within the dendrites of live neurons while affecting MT dynamics (Ghiretti et al., 2016; Muhia et al., 2016). The in vitro assays reported here confirm that KIF21B strongly stabilizes MTs. We found that at physiologically relevant motor concentrations and ionic strength, KIF21B slows growth and potently suppresses catastrophe, leading to a net stabilization of the assembled MT (Figure 1). Using in vitro engineered MT bundles, we observed that both motility and track switching by KIF21B were enhanced by MT bundling. Motor multimerization also had a robust effect on KIF21B run distance and track switching. KIF21B motors moving within native extracted dendritic MT arrays showed the directional switching seen along engineered MT bundles.
Surprisingly, however, despite the pronounced track switching observed on extracted cytoskeletal arrays, KIF21B exhibited no directional bias in extracted dendritic MT arrays. In contrast, in live cells, acute recruitment of full-length KIF21B motors to nonmotile peroxisomes using optogenetics was sufficient to induce both directional switching and active transport with a pronounced retrograde bias. These observations suggest that track switching may be necessary but not sufficient to establish a retrograde bias.
What controls KIF21B’s directional bias in dendrites? An important difference between the live cell and extracted network assays is the presence of dynamic MTs in live cells. When MT dynamics were dampened in dendrites of live neurons, the retrograde bias induced by KIF21B recruitment was eliminated. These results favor a model where modulation of MT dynamics by KIF21B is necessary for retrograde bias of cargo transport. In live neurons, KIF21B affects MT dynamics (Ghiretti et al., 2016; Muhia et al., 2016). In agreement, we observed that KIF21B robustly inhibits MT dynamics and assembly in vitro, which opens the possibility that this motor controls the remodeling of its own tracks. This remodeling requires the C-terminal domains of KIF21B containing the MTRs. Indeed, removal of the MTRs abolished retrograde biased cargo movement in live dendrites, suggesting that the MT dynamic regulation effect of the C-terminal MTRs is important for biasing retrograde biased movement. There is potential for this dynamic regulation to be different for more stable acetylated MTs oriented plus end–in over more dynamic tyrosinated MTs oriented plus end–out due to differences in lattice stability (Tas et al., 2017; Park and Roll-Mecak, 2018; Kelliher et al., 2019; Janke and Magiera, 2020). In this case, KIF21B might differentiate between MT orientations by selectively walking on more stable plus end–in MTs, resulting in net retrograde bias. Indeed, KIF21B lost the ability to select for plus end–in MTs for retrograde movement in dendrites with the addition of nocodazole, which dampened the dynamics of plus end–out MTs and rendered similar dynamics between plus end–in and plus end–out MTs. The synergistic combination of KIF21B-driven motility and modulation of MT dynamics may be necessary to direct KIF21B-bound cargoes in a net retrograde direction.
An alternate possibility is that the net retrograde bias observed for KIF21B is mediated by dendritic MAPs that differentially associate with dynamic versus stable MTs. Extracting native neuronal MT arrays affected MAP decoration and could explain why we did not observe a significant directional bias of motors in this environment. However, KIF21B directional bias was lost in dendrites of live cells treated with nocodazole where dendritic MAPs are generally retained to the same level as in control cells, suggesting that MAPs may have less of an influence on KIF21B directionality. However, we note that the immunofluorescence staining used to compare relative MAP levels between extracted MTs and live cells in control and nocodazole conditions may not be sufficiently sensitive to detect small changes in MAP decoration. We did note that both KIF21B and K560 motor run distances were shorter on native and extracted dendritic MT arrays compared with axonal arrays. This suggests that the native MAP code and the underlying PTM code, which differ in axons and dendrites, may affect long-distance motor motility without affecting motor directionality.
In summary, our work provides evidence for a model where KIF21B motor teams utilize MT track switching and regulation of MT dynamics to traffic cargoes with directional retrograde bias in the dendrites of neurons. The ability of this motor to both recognize and remodel the MT cytoskeleton sheds light on the morphological defects observed in both axon and dendrites where KIF21B is disrupted (Muhia et al., 2016; Morikawa et al., 2018; Asselin et al., 2020). Few kinesins have been directly implicated in dendritic transport, suggesting that specialized mechanisms are required to successfully navigate the complex MT organization within this cellular compartment. It will be of interest to determine whether other dendritic kinesin motors navigate mixed MT arrays with a mechanism similar to that described here for KIF21B, or whether other motors have adopted alternative strategies.
MATERIALS AND METHODS
Reagents
Constructs.
For in vitro single molecule experiments, HaloTag-KIF21B donor plasmids were formed by inserting KIF21B sequences (full-length 1–1624 and motor domain 1–657) from prior constructs (Ghiretti et al., 2016) into pFastBac vectors. Baculovirus containing bacmid DNA for these constructs was produced at the Protein Expression Facility at The Wistar Institute.
The K560-GFP plasmid (Addgene #15219) was acquired from the Vale Lab (HHMI Janelia Farm; Woehlke et al., 1997). The K560 sequence was inserted into the pHTC HaloTag CMV neo vector (Promega, Madison, WI; G7711) to build a K560-Halo plasmid. The rigor-kinesin (K560 E236A mutant; Addgene #60909) plasmid was acquired from the Vale lab (Tanenbaum et al., 2014).
For tracking of dynamic MT plus ends in neurons, we used the EB3-mCherry construct described in Boecker et al. (2021).
For peroxisome recruitment, the KIF21B-mCherry-eDHFR construct was derived by inserting the KIF21B sequence from the mCherry-KIF21B construct (gift of Matthias Kneussel, University of Hamburg) into the mCherry-eDHFR plasmid described in Ayloo et al. (2017). The GFP-Halo-Pex3 construct was used as described in Ayloo et al. (2017).
Protein expression and purification.
To express KIF21B-FL-Halo and -MD-Halo proteins, Sf9 insect cells (Expression Systems, Davis, CA; 94-001F) were grown to a density of 4 × 106 cells/ml in ESP 921 media (Expression Systems; 96-001-01) at 27°C, at which point they were infected with high-titer baculovirus for 48 h. Cells were pelleted, flash frozen in liquid nitrogen, and stored at −80°C. On the day of purification, cells were lysed in lysis buffer (10 mM Tris, 200 mM NaCl, 2 mM ATP, 4 mM MgCl2, 5 mM dithiothreitol [DTT], 1 mM ethylene glycol-bis(2-aminoethylether)-tetraacetic acid [EGTA], 0.5% Igepal, 1 mM 2-mercaptoethanol [β-ME], and 0.01 mg/ml aprotinin [GoldBio, St. Louis, MO; A-655-100] and leupeptin [Peptides International, Louisville, KY; ILP-4041-100 mg], pH 7.5) and clarified through centrifugation at 42,000 rpm for 1 h. Lysate was flowed over an Anti-FLAG resin (GenScript, Piscataway, NJ; L00432) column at 1 ml/min. After bound protein was washed with 10 column volumes (CVs) wash buffer (10 mM Tris, 200 mM NaCl, 2 mM ATP, 4 mM MgCl2, 5 mM DTT, 1 mM EGTA, 1 mM β-ME, and 0.01 mg/ml aprotinin and leupeptin, pH 7.5), protein was eluted by incubating one CV of elution buffer (10 mM Tris, 200 mM NaCl, 1 mM DTT, 1 mM EGTA, 0.2 mg/ml FLAG peptide [Sigma-Aldrich, St. Louis, MO; SAB1306078-400UL], pH 8.0) with resin beads for 1 h, followed by a second one CV incubation for 30 min and three more quick one CV incubations for a total of five CVs elution volume. Eluted protein was labeled with either 1.75 µM Alexa Fluor 660 or 2.5 µM TMR HaloTag Ligands (Promega; G8471 and G8251) for 2 h on ice. Labeled protein was dialyzed in PEM buffer (100 mM NaPIPES, 50 mM NaCl, 1 mM MgSO4, 1 mM EGTA, pH 6.8) for 2–18 h. Protein was then flowed over a Q sephaorse (GE Healthcare, Chicago, IL; 17051001) column and eluted in high-salt buffer (100 mM NaPIPES, 600 mM NaCl, 1 mM MgSO4, 1 mM EGTA, pH 6.8). MT affinity/dead-head spin was performed by binding motor protein to newly polymerized MTs (unlabeled, 2 µM paclitaxel [Taxol, Cytoskeleton, Denver, CO; TXD01]-stabilized) in binding buffer (12 mM NaPIPES, 200 mM KCl, 5 mM MgCl2, 2 mM EGTA, 1 mM AMPPNP, and 20 µM Taxol, pH 6.8) for 30 min. Motors and MTs were pelleted through centrifugation at 18,000 rpm for 20 min. Unbound motors were rinsed using wash buffer (12 mM NaPIPES, 200 mM KCl, 5 mM MgCl2, 2 mM EGTA, and 20 µM Taxol, pH 6.8). Bound motors were released from MTs through a 5 min incubation in release buffer (12 mM NaPIPES, 200 mM KCl, 5 mM MgCl2, 2 mM EGTA, 5 mM ATP, and 20 µM Taxol, pH 6.8), followed by centrifugation at 18,000 rpm for 10 min to remove any MTs and nonactive kinesin motors from the supernatant. MT-released protein was aliquoted, flash frozen, and stored in liquid nitrogen.
To produce K560-Halo and K560-GFP protein, plasmids were transformed into BL21(DE3)pLysE bacteria (Sigma-Aldrich; CMC0015-20 × 40UL) and grown in Terrific Broth (TB) supplemented with 50 µg/ml kanamycin and 33 µg/ml chloramphenicol at 37°C until an OD600 of 0.4 was reached. Cultures were cooled to 18°C, and protein expression was induced with 0.15 mM Isopropyl beta-d-1-thiogalactopyranoside (IPTG) for 18 h. Cells were pelleted and flash frozen in liquid nitrogen and stored at −80°C. On the day of purification, cells were lysed by microfluidizer (Microfluidics, Westwood, MA) after resuspension in a lysis buffer (50 mM NaPO4, 250 mM NaCl, 20 mM imidazole, 1 mM MgCl2, 0.5 mM ATP, 1 mM β-ME, 0.01 mg/ml aprotinin and leupeptin, pH 6.0) and clarified through centrifugation at 42,000 rpm for 30 min. Lysate was run over a Co2+ agarose bead (GoldBio; H-310-25) column at 1 ml/min. After bound protein was washed with 10 CVs wash buffer (50 mM NaPO4, 300 mM NaCl, 10 mM imidazole, 1 mM MgCl2, 0.1 mM ATP, 0.01 mg/ml aprotinin and leupeptin, pH 7.4), protein was eluted with five × one CV of elution buffer (50 mM NaPO4, 300 mM NaCl, 150 mM imidazole, 1 mM MgCl2, 0.1 mM ATP, pH 7.4). Fractions were pooled and concentrated. Buffer was exchanged to BRB80 (80 mM NaPIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.8) by loading protein over NAP-10 (GE Healthcare; 17-0854-01) and PD-10 (GE Healthcare; 17-0851-01) desalting columns. MT affinity/dead-head spin was performed as described above for KIF21B motors. MT-released protein was aliquoted, flash frozen, and stored in liquid nitrogen.
To produce rigor-kinesin protein, the construct plasmid was transformed into BL21(DE3)pLysE bacteria (Sigma-Aldrich; CMC0015-20 × 40UL) and grown in TB supplemented with 100 µg/ml ampicillin and 33 µg/ml chloramphenicol at 37°C until an OD600 of 0.4 was reached. Cultures were cooled to 18°C, and protein expression was induced with 0.1 mM IPTG for 18 h. Cells were pelleted and flash frozen in liquid nitrogen and stored at −80°C. On the day of purification, cells were lysed by microfluidizer after resuspension in a lysis buffer (25 mM KPO4, 300 mM NaCl, 40 mM imidazole, 1 mM MgCl2, 10% glycerol, 100 µM ATP, 670 mM phenylmethylsulfonyl fluoride (PMSF), pH 8.0) and clarified through centrifugation at 42,000 rpm for 30 min. Lysate was run over a Co2+ agarose bead column at 1 ml/min. After bound protein was washed with 10 CVs of wash buffer at pH 8.0 (300 mM NaCl, 40 mM imidazole, 1 mM MgCl2, 10% glycerol, 100 µM ATP, 5 mM β-ME, pH 8.0) and 10 CVs of wash buffer at pH 7.0, protein was eluted with five CVs of elution buffer (300 mM NaCl, 250 mM imidazole, 1 mM MgCl2, 10% glycerol, 100 µM ATP, pH 7.0). Elution fractions were pooled and concentrated. Protein was aliquoted, flash frozen, and stored at –80°C.
Neuronal cell culture.
The day before plating neurons, 35-mm glass-bottom dishes (MatTek, Ashland, MA; P35G-1.5-14-C) or 25-mm round coverslips were coated with 0.5 mg/ml poly-l-lysine (Sigma-Aldrich; P1274). E18 Sprague–Dawley rat hippocampal neurons were received from the Neuron Culture Service Center at the University of Pennsylvania and plated in attachment media (MEM [Life Technologies, ThermoFisher, Waltham, MA; 1109-072] supplemented with 10% horse serum [Life Technologies; 26050-070], 33 mM glucose [Corning, Corning, NY; 25-037-CIR], and 1 mM sodium pyruvate [Life Technologies; 11360-070]) at a density of 100,000 cells per coverslip or 500,000 cells per dish. After 4–6 h of attachment, neurons were cultured at 37°C with 5% CO2 and maintained in either maintenance media (Neurobasal [Life Technologies; 21103-049] supplemented with 33 mM glucose [Corning; 25-037-CIR], 2 mM GlutaMAX [Life Technologies; 35050-061], 100 U/ml penicillin and 100 µg/ml streptomycin [Life Technologies; 15140-122], and 2% B27 [Life Technologies; 17504-044]) or BrainPhys Neuronal Medium with NeuroCult SM1 Neuronal Supplement (BrainPhys Primary Neuron Kit [StemCell Technologies, Vancouver, Canada; 05794]). AraC (Sigma-Aldrich; C6645) was added at 10 µM 24 h after initial plating to prevent glial cell division.
Antibodies.
Antibodies and dilutions used in immunofluorescence assays were chicken anti–β3 tubulin (Chemicon; AB9354; 1:500), rat anti–tyrosinated α-tubulin, clone YL1/2 (EMD Millipore Corporation, Billerica, MA; MAB1864; 1:500), mouse anti–acetylated tubulin, clone 6-11B-1 (Sigma-Aldrich; T7451; 1:1000), mouse anti-MAP2 (Chemicon; MAB3418; 1:200), rabbit polyclonal anti-MAP7 (Proteintech, Rosemont, IL; 13446-1-AP; 1:100), mouse anti-MAP1, clone HM-1 (Chemicon; MAB362; 1:100), rabbit polyclonal anti-MAP9 (Proteintech 2678-1-AP; 1:250), mouse anti-DCX, clone 3E1 (Abcam, Cambridge, MA; ab254133; 1:1000), rabbit polyclonal anti-DCLK1 (Abcam; ab31704; 1:100), goat anti-chicken immunoglobulin Y (IgY) (H+L) Alexa Fluor 488 (ThermoFisher; A-11039; 1:1000), goat anti-mouse IgG (H+L) Alexa Fluor 555 (ThermoFisher; A-21424; 1:1000), goat anti-rat IgG (H+L) Alexa 633 (ThermoFisher; A-21094; 1:1000), and goat anti-rabbit IgG (H+L) Alexa 633 (ThermoFisher; A-21070; 1:1000).
Experimental procedures
In vitro single molecule assay.
Flow cells were assembled from attaching silane (PlusOne Repel Silane; GE Healthcare; 17-1332-01)-coated coverslips and cleaned glass slides together with double-side stick tape to form ∼10 μl flow chambers. Each flow cell was treated as follows: 1) 10 μl of 1 µM rigor-kinesin incubated for 5 min; 2) 30 μl of 5% pluronic F-127 (Sigma-Aldrich; P2443-250G) incubated for 5 min; 3) 30 μl of casein wash buffer (30 mM DTT, 1 mg/ml filtered casein [Sigma-Aldrich; C5890-500G] in BRB80, pH 6.8); 4) 50 μl of 1 μM MT seeds incubated for 2 min. Seeds were formed by shearing larger MTs (GMPCPP-stabilized, labeled 1:40 with HiLyte 488 tubulin (Cytoskeleton; TL488M-B) with a Hamilton syringe (Avanti Polar Lipids, Alabaster, AL; 610015). Seeds were aligned in the direction of flow by flowing in the seed solution with the chamber glass at a low-grade angle to keep the flow slow and continuous; 5) 30 μl of casein wash buffer; 6) 20 μl of final flow mixture incubated while imaging. Final flow: 0–50 nM kinesin motor, 10 µM tubulin (labeled 1:20 with HiLyte 488 tubulin), 1 mM ATP, 1 mM GTP, 0.32 mg/ml casein, 0.32 mg/ml bovine serum albumin [BSA] [Fisher Scientific, ThermoFisher Scientific; 50-253-90], 9.7 mM DTT, 15.5 glucose, 119 U/ml glucose oxidase [Sigma-Aldrich; G2133-250KU], 278 U/mg catalase [Sigma-Aldrich; C100-500MG], 0.244 mg/ml creatine phosphokinase [Sigma-Aldrich; C3755-35KU], 11.5 mM phosphocreatine [Sigma-Aldrich; P7936-1G]. and 0.2% methylcellulose, diluted in BRB80, pH 6.8. For KIF21B-FL multimerization experiments, the final flow was diluted in P12T (12 mM NaPIPES, 2 mM MgCl2, 1 mM EGTA, pH 6.8).
TIRF nucleation assay.
Flow cells were assembled by attaching silane-coated coverslips and cleaned glass slides together with double-side stick tape to form ∼10 μl flow chambers. Each flow cell was blocked with 5% pluronic F-127 incubated for 5 min and washed with casein wash buffer (30 mM DTT, 1 mg/ml filtered casein in BRB80, pH 6.8) before the addition of the final flow solution. A final flow (0–50 nM kinesin motor, 10 µM tubulin [labeled 1:20 with HiLyte 488 tubulin], 1 mM ATP, 1 mM GTP, 0.32 mg/ml casein, 0.32 mg/ml BSA, 9.7 mM DTT, 15.5 glucose, 119 U/ml glucose oxidase, 278 U/mg catalase, 0.244 mg/ml creatine phosphokinase, 11.5 mM phosphocreatine, and 0.2% methylcellulose, diluted in BRB80, pH 6.8) was added to chambers and heated to 35°C with an objective collar. Chambers were imaged 10 min after incubation with the final flow solution.
Light scattering assay.
Wells of a 96-well half-area UV transparent plate (Corning; 3679) were used to mix solutions containing 50 nM kinesin motor, 10 µM Taxol or BRB80 in assay buffer (10 µM tubulin, 10% glycerol [vol/vol], 4 mM DTT, 1 mM GTP, 1 mM ATP, and 0.133 mg/ml casein diluted in BRB80, pH 6.8). Solutions were pipetted in triplicate. The absorbance values of each well were measured at 340 nm every minute at 37°C using a prewarmed SynergyMx platereader (BioTek).
Motor-PAINT assay.
MT arrays in neurons were fixed and stabilized as described (Tas et al., 2017) with slight modifications. Briefly, membranes were removed from neurons cultured in MatTek dishes at 8–10 days in vitro (DIV) by incubating cells with extraction buffer (1 M sucrose, 0.15% Triton X-100 [Roche, Basel, Switzerland; MilliporeSigma, 10789704001] in BRB80, pH 6.8, at 37°C) for 1 min. An equal amount of fixation buffer (1% paraformaldehyde [PFA] [Affymetrix, ThermoFisher Scientific; 199431LT] in BRB80, pH 6.8, at 37°C) was added for 1 min with gentle swirling. Dishes were rinsed three times with wash buffer (2 µM Taxol in BRB80, pH 6.8, at 37°C). Dishes were washed once more right before imaging. To image, wash buffer was replaced with imaging buffer (1 mM ATP, 2 µM Taxol, 0.133 mg/ml casein, 0.133 mg/ml BSA, 4 mM DTT, 6 mg/ml glucose, 49 U/ml glucose oxidase, 115 U/mg catalase, 0.21 mg/ml creatine phosphokinase, and 4.76 mM phosphocreatine in BRB80, pH 6.8) containing 5–10 nM KIF21B-Halo-660 motor and 5–10 nM K560-GFP motor.
Immunofluorescence.
At 8–10 DIV, hippocampal neuron–plated coverslips were treated by either extraction followed by fixation as described for the motor-PAINT assay or treatment with 10 nM nocodazole for 30 min or left alone. All neurons were then fixed with 37°C warmed 4% PFA in phosphate-buffered saline (PBS) (50 mM NaPO4, 150 mM NaCl, pH 7.4) for 10 min, followed by three PBS washes and permeabilization in 0.1% Triton X-100 in PBS for 10 min. Coverslips were blocked with cell block (0.2% Triton X-100 and 3% BSA in PBS) for 2 h at room temperature. Primary antibodies were diluted as described above in cell block and incubated on coverslips for 2 h at room temperature. Coverslips were then washed three times, for 10 min each, with PBS. Alexa-conjugated secondary antibodies were diluted in cell block and incubated on coverslips for 1 h at room temperature. Coverslips were washed again three times with PBS, each for 10 min. Coverslips were rinsed in dH2O and mounted on glass slides with ProLong Gold antifade reagent (Invitrogen; P36930).
Induced recruitment assay.
Induced recruitment experiments with KIF21B motors were performed in DIV 8–10 hippocampal neurons as previously described (Ayloo et al., 2017), with the exception of transfecting hippocampal neurons with DNA plasmids for PEX3-GFP-Halo and either full-length KIF21B(aa1-1624)-mCherry-eDHFR or motor domain–truncated KIF21B(aa1-657)-mCherry-eDHFR. Cells were incubated with 100 nM nocodazole or DMSO as well as 10 µM cTMP-Htag (Ballister et al., 2015) or CTH (Zhang et al., 2017) dimerizer 30 min before imaging.
EB3 monitoring of MT dynamics.
Experiments tracking MT plus-end growth in neurons via EB3 dynamic tracking were performed in DIV 8–10 hippocampal neurons as previously described (Ghiretti et al., 2016), with the exception of transfecting hippocampal neurons with EB3-mCherry. Cells were incubated with DMSO (ACROS Organics, Geel, Belgium; 326881000) before 10 min of imaging and were subsequently treated with 100 nM nocodazole (Sigma-Aldrich; M1404-10MG) for 30 min before another 10 min of imaging.
Microscopy.
Single molecule and motor-PAINT assays with dynamic MTs were performed at 37°C with a PerkinElmer Nikon Eclipse Ti TIRF system, using a Nikon Apo TIRF 100 × 1.49 NA oil-immersion objective and a Hamamatsu ImagEM C9100-13 EMCCD camera operated by Volocity software (PerkinElmer, Waltham, MA; version 6.4.0). TIRF MT nucleation experiments were performed at 35°C using a dual-view Leica TIRF microscope, with an Olympus UplanApo 60 × 1.45 NA oil-immersion objective and an Andor iXon Ultra EMCCD camera operated with Metamorph software (Molecular Devices, San Jose, CA; version 7.10.3.279). Immunofluorescence images were acquired with a Leica DMI6000 inverted microscope, using an HCX PL APO 40 × 1.25 NA oil-immersion objective and a Hamamatsu ORCA-R2 CCD camera operated with Leica Application Suite X (LAS X) software (Leica Microsystems, Buffalo Grove, IL; version 3.0.3.279). EB3 dynamics and induced recruitment assays were performed at 37°C using a PerkinElmer Nikon Eclipse spinning-disk confocal Ultraview VoX system equipped with a 405 nm Ultraview Photokinesis accessory, using a Hamamatsu ImagEM C9100-50 EMCD camera operated by Volocity software.
Movies in each experiment were taken with different frame rates. In vitro assay movies of KIF21B multimers and single molecules were obtained by acquiring motor images at 5 frames/s (FPS) while acquiring MT track images at 1 FPS for 2 min. Every 6th frame of these movies was skipped to image the growing MTs. In vitro assays of dynamic MTs were obtained by continuously acquiring MT images at 2 FPS for 10 min. Motor-PAINT movies were obtained by continuously acquiring motor images at 5 FPS for 2 min. EB3 comet movies were taken by imaging EB3 at 2 s/frame (SPF) for 10 min. Peroxisome-motor–induced recruitment movies were taken by imaging both peroxisome and motor images at 2 FPS for 20 s before photoactivation, followed by 2 FPS for 2–5 min after photoactivation. In vitro assay bleaching movies of KIF21B multimers were acquired by continuously acquiring motor images at 5 FPS for 5–15 min.
Quantification and statistical analysis
Protein composition.
Recombinant protein concentration was assessed using a Pierce BCA Protein Assay Kit (ThermoFisher; 23225). Protein purity was assessed through Western blotting with the Odyssey CLx Imaging System (LI-COR Biosciences, Lincoln, NE) using Image Studio Lite version 5.2.5.
Motor multimerization calibration.
We measured the number of TMR Halo tag ligands bound to each KIF21B peptide through stepwise bleaching as well as the corresponding intensity values for each step. We then fitted a linear equation to the data to describe the number of TMR dyes and the measured intensity and calculated the intensity for two TMR dyes, corresponding to one motor dimer to within 1 SD. We applied the ratio of this value to the mean intensity of the population of all spots measured to that of the moving spot data to determine the intensity threshold limit of a motor dimer.
Single particle tracking.
Single kinesin particles moving along extracted axonal and dendritic MT arrays were segmented out with Cega (Masucci, Relich, et al., 2021) and tracked with a modified version of the software used in Schwartz et al. (2017) and described in Relich (2016), which is a tracking algorithm inspired by u-track (Jaqaman et al., 2008). Cega modified the initial candidate motor-finding step to greatly enhance the quality of the downstream tracking processes when analyzing data that have heterogeneous background fluorescence and nuisance particles. To eliminate particles moving along arrays belonging to another nearby process, a dilated binary mask was applied. Once the candidates were identified, the original movie images were analyzed by the localization algorithm with tracking performed by an algorithm similar to that of Schwartz et al. (2017). Analysis of track segment directionality was performed for segments that moved within 50–400 nm and belonged to a track with more than three connected segments. Intensity information for track segments was calculated from the original unmodified movie data.
Image/kymograph analysis.
For single molecule experiments using dynamic MTs, growth and catastrophe events along growing MTs were characterized by a change in the MT plus-end position that exceeded 0.5 µm. Dynamic MT tracing was accomplished using MATLAB R2019a software (MathWorks, Natick, MA; version 9.6.0.1174912). Purified KIF21B motility and dynamic MT behavior were analyzed with kymographs generated along the MT long axis using the Multi Kymograph plug-in for Fiji version 1.8.0-66 (ImageJ 1.53c; National Institutes of Health, Bethesda, MD). Unless specifically stated, we focused on analyzing only motor particles whose intensity was consistent with single molecules, of which ∼50% were fully labeled. Run distances and velocities were quantified for motor runs that extend 0.5 µm (three pixels) or more. Run distances were measured as the sum of run lengths in each direction for an individual motor. Track switches are characterized by a reversal in run direction that exceeds 0.5 µm. Along extracted MT arrays the direction switches for each kinesin were quantified by analyzing kymographs of motor movement along the midline of each process. The frequency of switching was calculated by counting the motors that switched direction or did not switch direction along the center line of each kymograph. Switches were quantified for motor runs that extended 0.5 µm or more.
For neuron experiments, axons and dendrites were identified based on morphologic criteria as previously outlined (Kaech and Banker, 2006). At 8–10 DIV, dendrite lengths in our cultures were ∼50–100 µm. For EB3 comet tracking experiments, neurons expressing EB3 at a low level were imaged. EB3 comet motility was analyzed using kymographs generated along the length of each dendritic process. Run distances and velocities were quantified for EB3 comets moving more than 3 µm. For motor-induced recruitment experiments, only neurons expressing both of the cotransfected GFP and mCherry markers were imaged. Peroxisome motility was analyzed using kymographs generated along the length of each process. Peroxisome diameter was measured along the long axis of each corresponding process. Run distances and velocities were quantified for peroxisomes that moved more than 3 µm. Run distances were measured as the total sum of distances run in each direction for an individual organelle. Peroxisome movement was considered bidirectional if the organelle moved more than 0.4 µm in both the anterograde and retrograde directions. Peroxisome switches were quantified for organelles that moved more than 0.4 µm in the opposing direction.
Statistical analysis.
Data from each experiment were analyzed from a minimum of three independent replicates, unless otherwise noted. Our statistical analysis was performed in GraphPad Prism (version 9.0.1 (151); Prism 9, San Diego, CA). We used a two-tailed Mann–Whitney test to compare two variables and Kruskal–Wallis analysis of variance (ANOVA) with Dunn’s multiple comparisons or ordinary one-way ANOVA to compare multiple variables. A Wilcoxon signed-rank test was used to determine whether values were significantly different from 60%, and an F test was used to determine whether values were significantly different from zero. Statistical values are mentioned in the figure captions.
Data availability
Excel files and example images and movies for all figures are available at doi:10.5281/zenodo.4587529. Metadata will be provided upon direct contact with the authors.
Supplementary Material
Acknowledgments
We acknowledge Mariko Tokito for help designing and constructing DNA constructs; Daniel Safer for help producing recombinant proteins; and Amy Ghiretti for help optimizing and performing induced recruitment experiments. This work was supported by the Center for Engineering MechanoBiology NSF Science and Technology Center, CMMI:15-48571, and National Institutes of Health grants RM1 GM136511 (to E.M.O., E.L.F.H., and M.L.) and R35 GM126950 (to E.L.F.H.).
Abbreviations used:
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- BDNF
brain-derived neurotrophic factor
- BSA
bovine serum albumin
- CTH
caged trimethoprim(TMP)-HaloTag ligand
- CVs
column volumes
- DHFR
dihydrofolate reductase
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- EB3
end binding protein 3
- EGTA
ethylene glycol-bis(2-aminoethylether)-tetraacetic acid
- FPS
frames per second
- GABA
gamma-aminobutyric acid
- GMPCPP
guanosine-5’-[(α,β)-methyleno]triphosphate
- Halo
HaloTag
- IPTG
Isopropyl beta- d-1-thiogalactopyranoside
- MAPs
microtubule associated protein
- β-ME
2-mercaptoethanol
- MTRs
MT-binding regions
- MTs
microtubules
- NMDA
N-methyl- d-aspartate
- NN
nearest neighboring
- NNO
nearest neighboring opposing
- Noc
nocodazole
- PBS
phosphate-buffered saline
- PEX3
peroxisomal biogenesis factor 3
- PFA
paraformaldehyde
- PSF
phenylmethylsulfonyl fluoride
- PTMs
post-translational modifications
- SDs
standard deviations
- SPF
seconds per frame
- TB
Terrific Broth
- TIRF
total internal reflection fluorescence
- trkB
tropomyosin receptor kinase B.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-10-0480) on October 27, 2021.
REFERENCES
Boldface names denote co–first authors.
- Acharya BR, Espenel C, Kreitzer G (2013). Direct regulation of microtubule dynamics by KIF17 motor and tail domains. J Biol Chem 288, 32302–32313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SB, Gallant PE, Leapman RD, Schnapp BJ, Reese TS (1993). Single kinesin molecules crossbridge microtubules in vitro. Proc Natl Acad Sci USA 90, 6503–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano-Santoyo H, Geyer EA, Stokasimov E, Chen G-Y, Su X, Hancock W, Rice LM, Pellman D (2017). A tubulin binding switch underlies Kip3/kinesin-8 depolymerase activity. Dev Cell 42, 37–51.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin L, Rivera Alvarez J, Heide S, Bonnet CS, Tilly P, Vitet H, Weber C, Bacino CA, Baranaño K, Chassevent A, et al. (2020). Mutations in the KIF21B kinesin gene cause neurodevelopmental disorders through imbalanced canonical motor activity. Nat Commun 11, 2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayloo S, Guedes-Dias P, Ghiretti AE, Holzbaur ELF (2017). Dynein efficiently navigates the dendritic cytoskeleton to drive the retrograde trafficking of BDNF/TrkB signaling endosomes. Mol Biol Cell 28, 2543–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW, Deitch JS, Black MM, Banker GA (1988). Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci USA 85, 8335–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister ER, Aonbangkhen C, Mayo AM, Lampson MA, Chenoweth DM (2014). Localized light-induced protein dimerization in living cells using a photocaged dimerizer. Nat Commun 5, 5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister ER, Ayloo S, Chenoweth DM, Lampson MA, Holzbaur ELF (2015). Optogenetic control of organelle transport using a photocaged chemical inducer of dimerization. Curr Biol 25, R407–R408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley M, Banker G (2016). The cellular mechanisms that maintain neuronal polarity. Nat Rev Neurosci 17, 611–622. [DOI] [PubMed] [Google Scholar]
- Boecker CA, Goldsmith J, Dou D, Cajka GG, Holzbaur ELF (2021). Increased LRRK2 kinase activity alters neuronal autophagy by disrupting the axonal transport of autophagosomes. Curr Biol 31, 2140–2154.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley CM, Rock RS (2009). Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. Proc Natl Acad Sci USA 106, 9685–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PR (1988). Dendrites of mitral cell neurons contain microtubules of opposite polarity. Brain Res 473, 107–115. [DOI] [PubMed] [Google Scholar]
- Burute M, Kapitein LC (2019). Cellular logistics: unraveling the interplay between microtubule organization and intracellular transport. Annu Rev Cell Dev Biol 35, 29–54. [DOI] [PubMed] [Google Scholar]
- Cao Y, Lipka J, Stucchi R, Burute M, Pan X, Portegies S, Tas R, Willems J, Will L, MacGillavry H, et al. (2020). Microtubule minus-end binding protein CAMSAP2 and kinesin-14 motor KIFC3 control dendritic microtubule organization. Curr Biol 30, 899–908.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G-Y, Cleary JM, Asenjo AB, Chen Y, Mascaro JA, Arginteanu DFJ, Sosa H, Hancock WO (2019). Kinesin-5 promotes microtubule nucleation and assembly by stabilizing a lattice-competent conformation of tubulin. Curr Biol 29, 2259–2269.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kanai Y, Cowan NJ, Hirokawa N (1992). Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature 360, 674–677. [DOI] [PubMed] [Google Scholar]
- Crevel IM-TC, Lockhart A, Cross RA (1996). Weak and strong states of kinesin and Ncd. J Mol Biol 257, 66–76. [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Chazeau A, Buijs RR, Stucchi R, Will L, Pan X, Adolfs Y, van der Meer C, Wolthuis JC, Kahn OI, et al. (2018). The HAUS complex is a key regulator of non-centrosomal microtubule organization during neuronal development. Cell Rep 24, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi D, Sharma M (2018). Multiple roles, multiple adaptors: dynein during cell cycle. In: Biochemical and Biophysical Roles of Cell Surface Molecules, ed. Chattopadhyay K, Basu SC, Singapore: Springer, 13–30. [DOI] [PubMed] [Google Scholar]
- Erickson HP (1974). Microtubule surface lattice and subunit structure and observations on reassembly. J Cell Biol 60, 153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S (2009). The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol 11, 717–723. [DOI] [PubMed] [Google Scholar]
- Furuta K, Toyoshima YY (2008). Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr Biol 18, 152–157. [DOI] [PubMed] [Google Scholar]
- Ghiretti AE, Thies E, Tokito MK, Lin T, Ostap EM, Kneussel M, Holzbaur ELF (2016). Activity-dependent regulation of distinct transport and cytoskeletal remodeling functions of the dendritic kinesin KIF21B. Neuron 92, 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromova KV, Muhia M, Rothammer N, Gee CE, Thies E, Schaefer I, Kress S, Kilimann MW, Shevchuk O, Oertner TG, Kneussel M (2018). Neurobeachin and the kinesin KIF21B are critical for endocytic recycling of NMDA receptors and regulate social behavior. Cell Rep 23, 2705–2717. [DOI] [PubMed] [Google Scholar]
- Gross SP, Vershinin M, Shubeita GT (2007). Cargo transport: two motors are sometimes better than one. Curr Biol 17, R478–R486. [DOI] [PubMed] [Google Scholar]
- Gudimchuk N, Vitre B, Kim Y, Kiyatkin A, Cleveland DW, Ataullakhanov FI, Grishchuk EL (2013). Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat Cell Biol 15, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes-Dias P, Nirschl JJ, Abreu N, Tokito MK, Janke C, Magiera MM, Holzbaur ELF (2019). Kinesin-3 responds to local microtubule dynamics to target synaptic cargo delivery to the presynapse. Curr Biol 29, 268–282.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Hoogenraad CC (2018). Local mechanisms regulating selective cargo entry and long-range trafficking in axons. Curr Opin Neurobiol 51, 23–28. [DOI] [PubMed] [Google Scholar]
- Heidemann SR, McIntosh JR (1980). Visualization of the structural polarity of microtubules. Nature 286, 517–519. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y (2010). Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638. [DOI] [PubMed] [Google Scholar]
- Holzbaur E, Goldman Y (2010). Coordination of molecular motors: from in vitro assays to intracellular dynamics. Curr Opin Cell Biol 22, 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooikaas PJ, Damstra HG, Gros OJ, van Riel WE, Martin M, Smits YT, van Loosdregt J, Kapitein LC, Berger F, Akhmanova A (2020). Kinesin-4 KIF21B limits microtubule growth to allow rapid centrosome polarization in T cells. eLife 9, e62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM (2020). The tubulin code and its role in controlling microtubule properties and functions. Nat Rev Mol Cell Biol 21, 307–326. [DOI] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G (2008). Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods 5, 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly AL, Kim H, Srinivasan D, Lakonishok M, Larson AG, Gelfand VI (2010). Kinesin-1 heavy chain mediates microtubule sliding to drive changes in cell shape. Proc Natl Acad Sci USA 107, 12151–12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Banker G (2006). Culturing hippocampal neurons. Nat Protoc 1, 2406–2415. [DOI] [PubMed] [Google Scholar]
- Katsuki M, Drummond DR, Cross RA (2014). Ectopic A-lattice seams destabilize microtubules. Nat Commun 5, 3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher MT, Saunders HA, Wildonger J (2019). Microtubule control of functional architecture in neurons. Curr Opin Neurobiol 57, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-S, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. (2014). A draft map of the human proteome. Nature 509, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleele T, Marinković P, Williams PR, Stern S, Weigand EE, Engerer P, Naumann R, Hartmann J, Karl RM, Bradke F, et al. (2014). An assay to image neuronal microtubule dynamics in mice. Nat Commun 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté D, Thies E, Kneussel M (2014). The kinesin KIF21B participates in the cell surface delivery of γ2 subunit-containing GABAA receptors. Eur J Cell Biol 93, 338–346. [DOI] [PubMed] [Google Scholar]
- Liang X, Kokes M, Fetter RD, Sallee MD, Moore AW, Feldman JL, Shen K (2020). Growth cone-localized microtubule organizing center establishes microtubule orientation in dendrites. eLife 9, e56547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludueña R, Shooter E, Wilson L (1977). Structure of the tubulin dimer. J Biol Chem 252, 7006–7014. [PubMed] [Google Scholar]
- Marceiller J, Drechou A, Durand G, Perez F, Poüs C (2005). Kinesin is involved in protecting nascent microtubules from disassembly after recovery from nocodazole treatment. Exp Cell Res 304, 483–492. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Weiner JA, Farlow SJ, Chun J, Goldstein LSB (1999). Novel dendritic kinesin sorting identified by different process targeting of two related kinesins: KIF21A and KIF21B. J Cell Biol 145, 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens L, Müller M, Stephan C, Hamacher M, Reidegeld KA, Meyer HE, Blüggel M, Vandekerckhove J, Gevaert K, Apweiler R (2006). A comparison of the HUPO Brain Proteome Project pilot with other proteomics studies. Proteomics 6, 5076–5086. [DOI] [PubMed] [Google Scholar]
- Masucci EM, Relich PK, Ostap EM, Holzbaur ELF, Lakadamyali M (2021). Cega: a single particle segmentation algorithm to identify moving particles in a noisy system. Mol Biol Cell 32, 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR (2016). Mitosis. Cold Spring Harb Perspect Biol 8, a023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Tanaka Y, Cho H-S, Yoshihara M, Hirokawa N (2018). The molecular motor KIF21B mediates synaptic plasticity and fear extinction by terminating Rac1 activation. Cell Rep 23, 3864–3877. [DOI] [PubMed] [Google Scholar]
- Muhia M, Thies E, Labonté D, Ghiretti AE, Gromova KV, Xompero F, Lappe-Siefke C, Hermans-Borgmeyer I, Kuhl D, Schweizer M, et al. (2016). The kinesin KIF21B regulates microtubule dynamics and is essential for neuronal morphology, synapse function, and learning and memory. Cell Rep 15, 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroyama A, Lechler T (2017). Microtubule organization, dynamics and functions in differentiated cells. Development 144, 3012–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navone F, Niclas J, Hom-Booher N, Sparks L, Bernstein HD, McCaffrey G, Vale RD (1992). Cloning and expression of a human kinesin heavy chain gene: interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J Cell Biol 117, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirschl JJ, Ghiretti AE, Holzbaur ELF (2017). The impact of cytoskeletal organization on the local regulation of neuronal transport. Nat Rev Neurosci 18, 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Roll-Mecak A (2018). The tubulin code in neuronal polarity. Curr Opin Neurobiol 51, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet DR, Burroughs NJ, Cross RA (2018). Kinesin expands and stabilizes the GDP-microtubule lattice. Nat Nanotechnol 13, 386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinemann DN, Sturgill EG, Das DK, Degen MS, Vörös Z, Hwang W, Ohi R, Lang MJ (2017). Collective force regulation in anti-parallel microtubule gliding by dimeric Kif15 kinesin motors. Curr Biol 27, 2810–2820.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relich P (2016). Single Particle Tracking: Analysis Techniques for Live Cell Nanoscopy, https://digitalrepository.unm.edu/phyc_etds/142. [Google Scholar]
- Reuther C, Diego AL, Diez S (2016). Kinesin-1 motors can increase the lifetime of taxol-stabilized microtubules. Nat Nanotechnol 11, 914–915. [DOI] [PubMed] [Google Scholar]
- Sanchez AD, Feldman JL (2017). Microtubule-organizing centers: from the centrosome to non-centrosomal sites. Curr Opin Cell Biol 44, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Huertas C, Freixo F, Viais R, Lacasa C, Soriano E, Lüders J (2016). Non-centrosomal nucleation mediated by augmin organizes microtubules in post-mitotic neurons and controls axonal microtubule polarity. Nat Commun 7, 12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp BJ, Reese TS (1989). Dynein is the motor for retrograde axonal transport of organelles. Proc Natl Acad Sci USA 86, 1548–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SL, Cleyrat C, Olah MJ, Relich PK, Phillips GK, Hlavacek WS, Lidke KA, Wilson BS, Lidke DS (2017). Differential mast cell outcomes are sensitive to FcεRI-Syk binding kinetics. Mol Biol Cell 28, 3397–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger MA, Rice SE (2010). Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules. J Biol Chem 285, 8155–8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, Zeeuw CID, Grosveld F, van Cappellen G, Akhmanova A, Galjart N (2003). Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J Neurosci 23, 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney HL, Holzbaur ELF (2018). Motor proteins. Cold Spring Harb Perspect Biol 10, a021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD (2014). A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas RP, Chazeau A, Cloin BMC, Lambers MLA, Hoogenraad CC, Kapitein LC (2017). Differentiation between oppositely oriented microtubules controls polarized neuronal transport. Neuron 96, 1264–1271.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofimova D, Paydar M, Zara A, Talje L, Kwok BH, Allingham JS (2018). Ternary complex of Kif2A-bound tandem tubulin heterodimers represents a kinesin-13-mediated microtubule depolymerization reaction intermediate. Nat Commun 9, 2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Schnapp BJ, Mitchison T, Steuer E, Reese TS, Sheetz MP (1985). Different axoplasmic proteins generate movement in opposite directions along microtubules in vitro. Cell 43(Part 2), 623–632. [DOI] [PubMed] [Google Scholar]
- van Riel WE, Rai A, Bianchi S, Katrukha EA, Liu Q, Heck AJ, Hoogenraad CC, Steinmetz MO, Kapitein LC, Akhmanova A (2017). Kinesin-4 KIF21B is a potent microtubule pausing factor. eLife 6, e24746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez RJ, Howell B, Yvon AM, Wadsworth P, Cassimeris L (1997). Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol Biol Cell 8, 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD (1997). Microtubule interaction site of the kinesin motor. Cell 90, 207–216. [DOI] [PubMed] [Google Scholar]
- Wordeman L (2010). How kinesin motor proteins drive mitotic spindle function: lessons from molecular assays. Semin Cell Dev Biol 21, 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Akhmanova A (2017). Microtubule-organizing centers. Annu Rev Cell Dev Biol 33, 51–75. [DOI] [PubMed] [Google Scholar]
- Zhang H, Aonbangkhen C, Tarasovetc EV, Ballister ER, Chenoweth DM, Lampson MA (2017). Optogenetic control of kinetochore function. Nat Chem Biol 13, 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Excel files and example images and movies for all figures are available at doi:10.5281/zenodo.4587529. Metadata will be provided upon direct contact with the authors.