Abstract
Background
High levels of estrogen are associated with increased risk of breast and endometrial cancer and have been suggested to also play a role in the development of ovarian cancer. Cancerogenic effects of estradiol, the most prominent form of estrogen, have been highlighted as a side effect of estrogen-only menopausal hormone therapy. However, whether high levels of endogenous estrogens, produced within the body, promote cancer development, has not been fully established.
Objective
We aimed to examine causal effects of estradiol on breast, endometrial, and ovarian cancer.
Methods
Here we performed a two-sample Mendelian randomization (MR) to estimate the effect of endogenous estradiol on the risk of developing breast, endometrial, and ovarian cancer, using the UK Biobank as well as 3 independent cancer cohorts.
Results
Using 3 independent instrumental variables, we showed that higher estradiol levels significantly increase the risk for ovarian cancer (OR = 3.18 [95% CI, 1.47-6.87], P = 0.003). We also identified a nominally significant effect for ER-positive breast cancer (OR = 2.16 [95% CI, 1.09-4.26], P = 0.027). However, we could not establish a clear link to the risk of endometrial cancer (OR = 1.93 [95% CI, 0.77-4.80], P = 0.160).
Conclusion
Our results suggest that high estradiol levels promote the development of ovarian and ER-positive breast cancer.
Keywords: estradiol, ovarian cancer, breast cancer, endometrial cancer, Mendelian randomization
Previous studies have shown that women with high blood levels of estradiol have an increased risk of breast cancer both before [1, 2] and after menopause [3, 4]. Ovarian cancer is the most fatal of the gynecological cancers worldwide, with no screening test and therefore typically late-stage diagnosis [5, 6]. Epidemiological studies have suggested a strong role of estrogen activity as well as the duration of exposure to estrogen in the initiation, pathogenesis, and progression of ovarian cancer [5]. Endometrial cancer, which is the most common gynecological cancer, is also known to be hormone dependent [7]. Endometrial cancer risk increases with use of menopausal hormonal treatment that includes estrogen only. However, this risk can be reduced if the treatment is combined with (opposed by) progesterone [5, 8], and a protective effect on both endometrial and ovarian cancer has been identified among oral contraceptive users [9, 10], most likely due to fewer ovulations [11].
Even though estrogen has been linked to all 3 cancer types, there is a lack of knowledge about whether the body’s own production of estrogen promotes the development of breast, endometrial, and ovarian cancer. One difficulty, when studying risk factors such as estrogen levels on cancer, is to distinguish correlation from causation. Mendelian randomization (MR) is an instrumental variable approach that can be used to disentangle the effect that estradiol exerts on the risk of developing cancer. In MR, germ-line genetic variants are used as instrumental variables. Thereby, the MR estimate is not affected by reversed causation, since genetic variants are not confounded by lifestyle or environmental factors. MR can therefore be used for estimating causal effects. In MR, the instrumental variables must fulfill 3 fundamental assumptions in order to be valid: 1) the variant must be associated with the exposure (estradiol); 2) the variant should not be associated with any potential confounder in the exposure-outcome relation; and 3) the variant should not be directly associated with the outcome (cancer) [12]. Two previous MR studies, both including only one genetic variant, close to the CYP19A1 gene, identified a causal effect of higher estradiol levels on endometrial cancer risk [13, 14]. An effect of estradiol levels on estrogen receptor (ER)-positive breast cancer, as well as a suggestive effect on ovarian cancer of the endometrioid subtype, was also identified in Larsson et al [14]. MR studies based on a single genetic variant may be greatly biased, as a result of undetectable pleiotropic effects [15]. To our knowledge, no previous study has identified a causal effect of estradiol on ovarian, breast, or endometrial cancer using multiple instruments for estradiol levels.
Here, we use MR, with 3 genetic variants, none of them included in previous MR studies for estradiol, aiming to establish and replicate a causal effect of estradiol on breast, endometrial, and ovarian cancer. We perform two-sample MR using 3 different independent cancer cohorts, with no overlapping samples with the estradiol genome-wide association study (GWAS) performed in the UK Biobank.
Methods
Study Samples
UK Biobank
The UK Biobank is a population-based cohort including 502 682 participants, of whom 273 404 are women, recruited from all across the United Kingdom. Participants were between 37 and 73 years old at the time of recruitment between 2006 and 2010. Health variables have been collected through questionnaires, interviews, and death and hospital records, as well as cancer registries. For all genetic analyses in the UK Biobank, the third release of the imputed genetic data was used.
Estradiol levels and instrumental variables for estradiol
Four instrumental variables to be used in the MR analyses were selected from our previously published GWAS for estradiol in UK Biobank [16]. Briefly, estradiol levels were measured from blood samples taken at the first assessment in association with the recruitment, using a two-step competitive analysis on a Beckman Coulter Unicel Dxl 800. Unfortunately, this measurement method was unable to detect estradiol levels below 175 pmol/L, which resulted in a substantial fraction of the participants without measured estradiol. In the discovery GWAS [16], estradiol was therefore analyzed as a binary variable (above or below detection limit). The GWAS included Caucasian UK Biobank participants, clustering with regards to their genetic principal components. Quality control and information on covariates have been described previously [16]. A total of 4 instrumental variables, the single nucleotide polymorphisms (SNPs) with the lowest P values (P < 1 × 10-7) in the previous GWAS in females (Table 1), all with an F-statistics > 10, were selected for the current MR study. All SNPs were nominally significant also when analyzed in post- and premenopausal women separately and the estimates were very similar between strata, except for 1 SNP, rs45446698 in CYP3A7, for which the effect was larger in postmenopausal women (see Supplementary Table S1 in Schmitz et al [16]). CYP3A7 is well known to be the key enzyme in metabolizing exogenous hormones (e.g., from hormone replacement therapy) [17]. Since hormone replacement therapy triggers the development of endometrial cancer [5, 8], rs45446698 might indeed influence cancer risk, through exogenous hormones rather than endogenous as we aim to investigate in this study. Consequently, rs45446698 could be regarded as pleiotropic and was excluded from the main MR analysis. Previous estradiol MR studies included the SNP rs727479 within the CYP19A1 gene as an instrument, selected from a previous GWAS performed in postmenopausal women [13]. The rs727479 SNP did not pass quality control in UK Biobank, but another CYP19A1 SNP (rs7175531), in perfect linkage disequilibrium (R2 = 1.0) with rs717479, was available in UK Biobank but not strongly associated with estradiol levels among females (P = 0.001). The CYP19A1 SNP has previously been highlighted as potentially pleiotropic in an MR for endometrial cancer [13] and for this reason, rs7175531 was excluded from our primary MR, too, and only included in sensitivity analyses. Excluding instruments used in previous studies gives us a unique set of estradiol instruments for replication. The variance explained by each genetic variant was estimated by calculating the difference between Nagelkerke’s pseudo-R2 for the full model, including both covariates and the SNP, and the reduced model, only including the covariates. The F-statistic for each SNP was estimated from the full model by computing the squared ratio of the SNP’s beta estimate and its standard error (Table 1).
Table 1.
Summary GWAS results for each instrument variable included in the MR analysis
| Breast cancer | Ovarian cancer | Endometrial cancer | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estradiol original GWASd | Estradiol tobite | BCAC | ER-positive, BCAC | ER-negative, BCAC | OCAC | ECAC | ||||||||||
| Chr:SNP Closest Gene |
Effect allele/ freqa | Delta R2c/F-statistics | Levels OR (95% CI) |
P value | Levels beta | P value | OR (95% CI) |
P value | OR (95% CI) |
P value | OR (95% CI) |
P value | OR (95% CI) |
P value | OR (95% CI) |
P value |
| Chr12: rs4764934 ASCL1 |
C/0.82 | 0.00015/26.42 | 1.09 (1.06-1.12) |
6.07 × 10-8 | 0.085 | 8.35 × 10-6 | 0.99 (0.96-1.01) |
0.26 | 0.99 (0.96-1.02) |
0.50 | 0.97 (0.92-1.02) |
0.18 |
1.04
(1.01-1.08) |
0.021 | 0.98 (0.95-1.02) |
0.42 |
| Chr19: rs10638101 TMEM150B |
A/0.51 | 0.00016/28.20 | 1.07 (1.04-1.09) |
6.28 × 10-8 | 0.014 | 0.00069 |
|
|
1.01 (0.99-1.04) |
0.34 | 1.00 (0.97-1.03) |
0.88 | ||||
| Chr19: rs897797b TMEM150B |
T/0.50 | 0.00016/28.52 | 1.07 (1.04-1.09) |
7.89 × 10-8 | 0.065 | 0.00073 |
1.02
(1.00-1.04) |
0.043 | 1.04 (1.01-1.06) | 0.0026 | 1.00 (0.94-1.04) | 0.96 | ||||
| Chr20: Rs16991615 MCM8 |
A/0.07 | 0.00016/11.4 | 28.94 (1.09-1.19) |
4.67 × 10-8 | 0.130 | 1.81 × 10-9 |
1.05
(1.01-1.09) |
0.024 | 1.04 (1.00-1.09) |
0.067 | 1.08 (1.00-1.16) |
0.048 | 1.05 (0.96-1.15) |
0.078 |
1.08
(1.02-1.15) |
0.013 |
|
Chr7:
rs45446698 CYP3A7 |
T/0.96 | 0.00026/45.16 |
1.23
(1.16-1.30) |
7.62 × 10 -12 | 0.206 | 7.96 × 10 -6 |
1.05
(0.99-1.10) |
0.09 |
1.05
(0.99-1.12) |
0.097 |
0.99
(0.90-1.10) |
0.89 |
0.97
(0.91-1.04) |
0.43 |
1.21
(1.10-1.32) |
3.09 × 10 -5 |
|
Chr15: rs7175531
CYP19A1 |
C/0.65 | 0.0002/9.49 | 1.04 (1.01-1.07) | 0.0018 | 0.014 | 0.0011 | 1.00 (0.93-1.09) | 0.90 | 1.02 (0.99-1.04) | 0.14 | 0.99 (0.96-1.03) | 0.78 | 1.01 (0.99-1.05) | 0.20 | 1.12 (1.09-1.16) | 8.07 × 10 -13 |
Nominally significant SNPs (P < 0.05) are highlighted in bold. Instruments only included in sensitivity analysis is highlighted in italics.
Abbreviations: BCAC, Breast Cancer Association Consortium; OCAC, Ovarian Cancer Association Consortium; ECAC, Endometrial Cancer Association Consortium.
aAllele frequency for the effect allele,
bProxy SNP, included since rs10638101 was not genotyped in BCAC,
cDelta R2 denotes the difference in Nagelkerke’s pseudo-R2 between the full model, including both covariates and SNP, and the reduced model, only including covariates,
dChange in odds for being above detection limit (>175 pmol/L) in estradiol levels, per effect allele,
eChange in rank-transformed estradiol levels (in SD units), per effect allele. Estradiol effect estimated quantitatively with the tobit method after excluding all current users of hormone replacement therapy and oral contraceptives as well as all participants with a prior cancer diagnosis (i.e., before assessment when blood was drawn). Observe that the sample size was reduced, which is reflected in the higher P value.
To perform analysis of estradiol levels as a quantitative exposure, including all participants below detection limit (175 pmol/L), we applied censored regression (Tobit-I) modeling [16, 18], and recalculated the effect estimates of the SNPs prior to the MR analyses. In this way, a potential problem of using dichotomized quantitative exposures in MR could be eliminated [19]. The tobit model incorporates a partially integrated error term, up to the detection limit, which enables censored individuals to be included. The VGAM package (version 1.1-2) in R was used to run the tobit regression models for each SNP, with estradiol levels being transformed using rank-based inverse normal transformation. For the tobit regression, women who reported a cancer diagnosis before assessment (Data field 2452), when blood samples were drawn (N = 14 635), were excluded. Also, current users of hormone replacement therapy (N = 9752) and oral contraceptives (N = 2681), and all women with unknown menopausal status, were removed, leaving 154 148 females (38 068 pre- and 116 080 postmenopausal). Of these, 30 044 had estradiol levels above the detection limit (25 111 pre- and 4933 postmenopausal). Finally, 136 487 women had both genotype and covariate information available and were included in the tobit analysis.
Two-Sample MR With Publicly Available GWAS Data
Two-sample MR was performed, since any weak instrument bias generally is directed toward null in the two-sample MR, in contrast toward the confounded association in a one-sample MR. Furthermore, the type I error is not inflated in a two-sample setting. For breast, endometrial, and ovarian cancer, respectively (Table 1), the effect sizes and standard errors for the SNPs were extracted from publicly available GWAS data, not including UK Biobank participants. For breast cancer, we used summary statistics from the Breast Cancer Association Consortium (BCAC) [20], including 122 977 breast cancer cases and 105 974 healthy controls of European ancestry (downloaded from http://bcac.ccge.medschl.cam.ac.uk/ on February 22, 2021). Using data from BCAC, we could also stratify for ER status. In the BCAC cohort, the SNP rs10638101 was not genotyped, and the proxy rs897797, in perfect linkage disequilibrium with rs10638101 (R2 = 1.0), was therefore included. For endometrial cancer, we used data from the Endometrial Cancer Association Consortium (ECAC), which includes 12 research cohorts based in Australia, Europe, and the USA. From the ECAC cohort, GWAS summary statistics from O’Mara et al (2018) [21], excluding participants from UK Biobank to avoid sample overlap, were used. This restricted ECAC dataset consisted of 12 270 cancer cases and 46 126 controls of European descent. Data from ECAC were available after request from the authors [21]. However, the effect of the instrument within CYP19A1 (used in the sensitivity analyses) was downloaded from the MR-base homepage (https://www.mrbase.org), on December 19, 2021, and did include UK Biobank participants. For ovarian cancer, we used GWAS summary statistics from the Ovarian Cancer Association Consortium (OCAC) [22]. For OCAC, genetic association analysis had been performed for 25 509 epithelial ovarian cancer cases and 40 941 healthy controls. Summary statistics were downloaded from http://ocac.ccge.medschl.cam.ac.uk/data-projects/results-lookup-by-region/, on February 26, 2021.
The main MR analyses were performed with the inverse-variance weighted (IVW) MR approach included in the “MendelianRandomization” package in R [23]. We further performed sensitivity analyses using weighted median and the MR-Egger methods, included in the same R package [23]. We also performed a sensitivity analysis including 2 potentially pleotropic instruments, rs45446698 and rs7175531, as well as running each of these instruments separately. Causal estimates were measured as the change in odds per 1 SD increase in rank-transformed estradiol levels.
Results
Out of the 3 instrumental variables selected for estradiol in the main analysis, none were strongly associated with any cancer phenotype. However, rs4764934 was weakly associated with ovarian cancer, rs897797 with breast cancer and ER-positive breast cancer, and rs16991615 with endometrial cancer (Table 1). rs45446698, located close to the CYP3A7 gene, and rs7175531 close to the CYP19A1, previously used as an instrument in a MR study [13], were both strongly associated with endometrial cancer (Table 1).
Mendelian Randomization
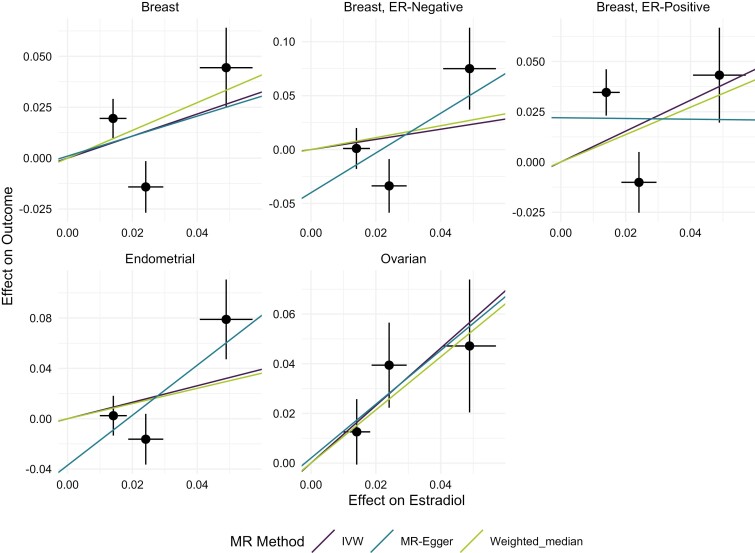
Using our primary MR-method, IVW, a significant effect of high estradiol levels on ovarian cancer was identified (OR = 3.18 per SD increase in rank-transformed estradiol levels [95% CI, 1.47-6.87], P = 0.003) (Fig. 1, Table 2). We also identified a nominally significant effect for ER-positive breast cancer (OR = 2.16 [95% CI, 1.09-4.26], P = 0.027) (Table 2, Fig. 1). However, ER-positive breast cancer did not hold for multiple testing (P = 0.05/4 = 0.0125). We could not establish a clear link to the risk of endometrial cancer (OR = 1.93 [95% CI, 0.77-4.80], P = 0.160), ER-negative breast cancer (OR = 1.50 [95% CI, 0.50-4.54], P = 0.471) or ER-negative and ER-positive breast cancer combined (OR = 1.72 [95% CI, 0.98-3.03], P = 0.061) (Table 2, Fig. 1).
Figure 1.
Results from the 3 different Mendelian randomization methods applied (IVW, MR-Egger, and weighted median) to estimate the causal effect of estradiol on breast, endometrial, and ovarian cancer. Breast cancer was also stratified for ER-positive and ER-negative breast cancer. The black dots represent the effect size of the SNPs in the GWAS for estradiol (x-axis) and cancer (y-axis) and the black lines are the standard errors. The lines represent the estimates from the different MR methods.
Table 2.
Mendelian Randomization results for each cancer and method
| Breast cancer | Ovarian cancer | Endometrial cancer | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BCAC | BCAC ER-positive breast cancer only | BCAC ER-negative breast cancer only | OCAC | ECAC | ||||||
| Method | ORa (95% CI) |
P value | ORa (95% CI) |
P value | ORa (95% CI) |
P value | ORa (95% CI) | P value | ORa (95% CI) | P value |
| IVW | 1.72 (0.98-3.03) | 0.061 | 2.16 (1.09-4.26) |
0.027 | 1.50 (0.50-4.54) | 0.471 | 3.18 (1.47-6.87) | 0.003 | 1.93 (0.77-4.80) |
0.160 |
| Weighted median | 1.98(0.92-4.26) | 0.081 | 1.96 (0.80-4.91) |
0.142 | 1.74 (0.42-7.15) |
0.45 | 2.91 (1.12-7.58) |
0.028 | 1.83 (0.57-5.92) |
0.313 |
| MR-Egger | 1.63(0.07-40.57) | 0.77 | 0.98 (0.02-40.89) |
0.99 | 6.30 (0.08-495) |
0.41 | 2.96 (0.56-15.61) |
0.202 | 7.31 (0.29-186.58) |
0.229 |
| MR-Egger intercept | 1.00(0.92-1.08) | 0.97 | 1.02 (0.93-1.12) |
0.64 | 0.96 (0.86-1.07) |
0.47 | 1.00 (0.96-1.04) |
0.92 | 0.96 (0.89-1.94) |
0.363 |
Abbreviations: BCAC, Breast Cancer Association Consortium; ECAC, Endometrial Cancer Association Consortium; IVW, inverse-variance weighted; OCAC, Ovarian Cancer Association Consortium.
aThe change in odds per 1 SD increase in rank-transformed estradiol levels.
Sensitivity Analyses
As sensitivity analyses, we applied the weighted median and the MR-Egger approach for each cancer. Comparing each method, we could see that all 3 cancers showed the same direction of effect (OR > 1), except for MR-Egger which showed a negative, but not significant OR for ER-positive breast cancer. The weighted median was nominally significant for ovarian cancer, which strengthened our main results (Fig. 1, Table 2).
Since rs45446698 and rs7175531 are likely to have pleiotropic effects in relation to cancer, especially endometrial cancer (see method section), those SNPs were not included in the primary MR analysis. Since we only include 3 to 5 instruments, we could not perform any formal test for heterogeneity among the MR instruments analysis [24]. However, we performed additional sensitivity analyses for each of the 2 possible pleiotropic SNPs in relation to all 3 cancers (Table 3). Including only rs45446698 as a single instrument, we identified a significant effect on endometrial cancer (OR = 54.43 [95% CI, 8.31-359.61], P < 0.0001), but no significant effect on other cancer phenotypes (Table 3), which agrees with the association results for this SNP (Table 1). Including only rs7175531 identified a strong effect on endometrial cancer (OR = 3561.72 [379.81-33400.29], P < 0.0001), which agrees with the association analysis (Table 1), as well as with 2 previous MR studies [13, 14]. We also performed sensitivity analyses by including both rs45446698 and rs7175531 as instruments in the MR analysis which resulted in similar MR estimates for ovarian cancer and ER-positive cancer. Here we also identified a nominally significant effect for breast cancer combined (Table 4). Finally, we included all 5 instruments for endometrial cancer. Even if the MR estimate was very high, in agreement with the strong effects by rs45446698 and rs7175531 individually (Table 1, Table 3), no significant effect was identified when including all 5 instruments (OR = 8.24 [95% CI, 0.59-115.03], P = 0.12), presumably due to heterogeneity among the instruments due to pleiotropy.
Table 3.
Possible pleiotropic instruments, rs45446698 and rs7175531, analyzed separately for breast, ovarian and endometrial cancer
| Method: IVW | Only rs45446698 | Only rs7175531 | ||
|---|---|---|---|---|
|
OR (95% CI) |
P value |
OR (95% CI) |
P value | |
| Breast cancer (BCAC) |
2.57 (0.86-7.63) | 0.090 | 3.21 (0.80-12.93) | 0.101 |
| ER-positive breast cancer (BCAC) | 3.01 (0.82-11.09) | 0.097 | 3.53 (0.67-18.76) | 0.138 |
| ER-negative breast cancer (BCAC) |
0.86 (0.10-7.18) | 0.887 | 0.68 (0.004-10.45) | 0.782 |
| Ovarian cancer (OCAC) |
0.56 (0.14-2.33) | 0.429 | 3.47 (0.51-23.48) | 0.203 |
| Endometrial cancer (ECAC) | 54.43 (8.31-356.61) | <0.0001 | 3561.72 (379.81-33400.29) | <0.0001 |
Table 4.
Sensitivity analysis for breast, ovarian, and endometrial cancer, running a two-sample analysis in BCAC, OCAC, and ECAC when including rs45446698 and rs7175531
| Breast cancer | Ovarian cancer | Endometrial cancerb | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BCAC | BCAC ER-positive breast cancer only | BCAC ER-negative breast cancer only | OCAC | ECAC | ||||||
| Method | ORa (95% CI) |
P value | ORa (95% CI) |
P value | ORa (95% CI) |
P value | ORa (95% CI) |
P value | ORa (95% CI) |
P value |
| IVW | 1.99 (1.02-3.87) |
0.042 | 2.43 (1.07-5.51) |
0.034 | 1.23 (0.41-3.69) |
0.707 | 2.26 (1.09-4.71) |
0.029 | 8.24 (0.59-115.03) |
0.12 |
| Weighted median | 2.51 (1.32-4.77) |
0.005 | 2.64 (1.21-5.73) |
0.014 | 0.97 (0.27-3.46) |
0.961 | 2.62 (1.10-6.25) |
0.030 | 4.24 (1.18-15.20) |
0.027 |
| MR-Egger | 1.74 (0.36-8.36) |
0.490 | 1.40 (0.23-8.58) |
0.717 | 3.32 (0.35-31.75) |
0.298 | 1.31 (0.27-6.44) |
0.741 | 2.96 (0.006-1480.30) |
0.73 |
| MR-Egger intercept | 1.00 (0.97-1.04) |
0.846 | 1.02 (0.97-1.06) |
0.495 | 0.97 (0.92-1.03) |
0.325 | 1.02 (0.98-1.05) |
0.440 | 1.03 (0.89-1.19) |
0.72 |
aThe change in odds per one SD increase in rank-transformed estradiol levels.
bFor ECAC, the effect of rs7175531 on endometrial cancer was estimated in a cohort that is partly overlapping with the UK Biobank cohort.
Discussion
In this study, we performed two-sample MR to show that high levels of estradiol in the body promote the development of ovarian cancer, as well as identify a nominally significant effect of estradiol on ER-positive breast cancer. We used 3 different instrumental variables, that were selected from a recent estradiol GWAS [16] and did not overlap with previous MR studies for estradiol [13, 14].
Our main estradiol instruments (SNPs) were annotated to ASCL1, TMEM150B, and MCM8. The MCM8 SNP rs16991615 has previously been associated with age at menopause [25]. However, we have shown previously that there is no significant difference in the effect of this SNP on estradiol levels, with or without adjustment for age at menopause in postmenopausal women [16], which suggests that the effect of the SNP on estradiol levels is not mediated by age at menopause. ASCL1 has previously been shown to promote tumor progression in lung adenocarcinoma [26] and overall survival in ovarian cancer patients [27]. The last instrument maps to TMEM150B, which has been associated with age at menopause [28] and with age at menarche [29].
To increase sample size and the power to identify strong instruments for the MR study, our prior estradiol GWAS included both pre- and postmenopausal women [16]. However, estradiol metabolism changes after menopause and that the cycle phase could dramatically influence estradiol prior to menopause. The previous study by Thompson et al [13], from which one of the instruments included in our sensitivity analysis was identified, only included postmenopausal women to limit the cycle phase bias. The estradiol estimates in our previous GWAS only differed between pre- and postmenopausal women for 1 significant SNP (rs45446698) close to CYP3A7 [16], and we consider that the other 3 instruments selected for the main analysis are valid instruments for both post- and premenopausal estradiol levels. Cancer often takes a long time to develop, and endometrial and ovarian cancer especially might be triggered during the reproductive period of a woman’s life. Since few postmenopausal women had detectable estradiol levels, our results might mainly reflect estradiol effects prior to menopause, even if most cancers are detected among postmenopausal women.
The previously used instrument rs7175531, within the CYP19A1 gene, was not strongly associated with estradiol in UK Biobank females (P = 0.001), and not included in our main analysis. One reason for this weak association in the UK Biobank might be that we combined pre- and postmenopausal women, while the previous association was identified in postmenopausal women only [13]. CYP19A1 encodes aromatase that synthesizes endogenous estrogens from testosterone in adipose tissue [30]. The lack of a strong association in the UK Biobank GWAS might also be due to the low-sensitivity estradiol measurement, making estradiol levels hard to measure in postmenopausal women. We further excluded an instrument within the CYP3A7 gene. The CYP3A7 gene encodes cytochrome P450 CYP3A7, which metabolizes dehydroepiandrosterone (DHEA), the main precursor of circulating estrogens in women [31, 32]. CYP3A7 is mainly expressed in the liver, which is one of the primary sites of estrogen metabolism [33]. However, CYP3A7 also metabolizes exogenous hormones [17], which agrees with a much less significant effect on estradiol levels when women using hormone replacement therapy were removed from the analyses (Table 1). When including the CYP19A1 SNP (rs7175531) as well as the CYP3A7 SNP (rs45446698) as instruments in our sensitivity analysis, the MR results for breast and ovarian cancer were similar to the primary approach. Also, the ER-positive and ER-negative cancers analyzed together were found to be nominally significant (P = 0.042), most probably driven by ER-positive cases. When analyzing the CYP19A1 instrument separately, as was done in previous MR studies, we confirm a significant effect on endometrial cancer (Table 3).
Even though we used a larger number of instrumental variables in our MR analyses than previous estradiol MR studies on cancer risk, one of the limitations of our study is still the low number of instruments, which reduces our ability to investigate and adjust for pleiotropy. However, we did use 3 instruments that were independently associated with estradiol levels, compared with the 1 used previously, which strengthens a true causal effect of estradiol levels on the risk of ovarian and breast cancer. Since estradiol is mainly produced by the ovaries during the reproductive years, and mainly by subcutaneous adipose tissues after menopause [34], another limitation of the present study is that the estradiol GWAS and MR includes both pre- and postmenopausal women and it is not possible to evaluate the timing of the harmful effects. Another limitation of this study is the hormonal fluctuations during the menstrual cycle and that estradiol is commonly measured at different time points during the menstrual cycle in different women. Unfortunately, we did not have information on cycle phase for all women and could not adjust for this. More detailed measurements of estradiol during different time points of the menstrual cycle would be beneficial to address the causal effects of estradiol pre menopause. Further, 4 of our instruments were selected from a previous GWAS (P < 1 × 10-7), each with an F-statistic > 10. However, when reanalyzing the GWAS, i.e., removing cancer cases and oral contraceptives and hormone replacement therapy users, the P values were less significant, which could lead to weak instrument bias.
It is important to consider that our analysis differs from previous MR studies in that we use different instruments, but it still supports previous studies to confirm that estradiol most probably have a causal effect on ovarian cancer as well as ER-positive breast cancer. However, our approach did not confirm the effect of estradiol on the risk of endometrial cancer as presented previously. Nonetheless, previous studies were confirmed when the CYP19A1 instrument was analyzed separately for endometrial cancer (Table 3). To further support the effect of estradiol on ovarian and ER-positive breast cancer risk, we also identified a significant effect in several of our sensitivity analyses, for example, when rs45446698 (CYP3A7) and rs7175531 (CYP19A1) were included as instrumental variables, and in the median weighted MR. This, together with previous studies, supports the possibility of a true causal effect of estradiol levels on ovarian and ER-positive breast cancer.
By identifying a causal link between estradiol and ovarian cancer, as well as replicating an effect on ER-positive breast cancer, our results further support carcinogenic effects of estrogen in these tissues. A deeper understanding of causal relations between estradiol levels and cancer risk could be of importance for novel interventions to prevent cancer in women.
Acknowledgments
We acknowledge participants and staff at UK Biobank. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Centre for Advanced Computational Science (UPPMAX) under project sens2017538. The breast cancer genome-wide association analyses were supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the “Ministère de l”Économie, de la Science et de l’Innovation du Québec’ through Genome Québec and grant PSR-SIIRI-701, The National Institutes of Health (U19 CA148065, X01HG007492), Cancer Research UK (C1287/A10118, C1287/A16563, C1287/A10710) and The European Union (HEALTH-F2-2009-223175 and H2020 633784 and 634935). All studies and funders are listed in Michailidou et al [20]. The endometrial cancer genome-wide association analyses were supported by the National Health and Medical Research Council of Australia (APP552402, APP1031333, APP1109286, APP1111246 and APP1061779, APP1173170), the U.S. National Institutes of Health (R01-CA134958), European Research Council (EU FP7 Grant), Wellcome Trust Centre for Human Genetics (090532/Z/09Z) and Cancer Research UK. OncoArray genotyping of ECAC cases was performed with the generous assistance of the Ovarian Cancer Association Consortium (OCAC), which was funded through grants from the U.S. National Institutes of Health (CA1X01HG007491-01 (C.I. Amos), U19-CA148112 (T.A. Sellers), R01-CA149429 (C.M. Phelan) and R01-CA058598 (M.T. Goodman); Canadian Institutes of Health Research (MOP-86727 (L.E. Kelemen)) and the Ovarian Cancer Research Fund (A. Berchuck). We particularly thank the efforts of Cathy Phelan. OncoArray genotyping of the BCAC controls was funded by Genome Canada Grant GPH-129344, NIH Grant U19 CA148065, and Cancer UK Grant C1287/A16563. All studies and funders included in ECAC are listed in O’Mara et al (2018). We also like to acknowledge Tracy O’Mara at QIMR Berghofer for sharing the ECAC data, after removing the UK Biobank.
Glossary
Abbreviations
- BCAC
Breast Cancer Association Consortium
- ECAC
Endometrial Cancer Association Consortium
- ER
estrogen receptor
- GWAS
genome-wide association study
- IVW
inverse-variance weighted
- MR
Mendelian randomization
- OCAC
Ovarian Cancer Association Consortium
- SNP
single nucleotide polymorphism
Contributor Information
Åsa Johansson, Department of Immunology, Genetics and Pathology, Science for Life Laboratory, Uppsala University, 75108 Uppsala, Sweden.
Daniel Schmitz, Department of Immunology, Genetics and Pathology, Science for Life Laboratory, Uppsala University, 75108 Uppsala, Sweden.
Julia Höglund, Department of Immunology, Genetics and Pathology, Science for Life Laboratory, Uppsala University, 75108 Uppsala, Sweden.
Fatemeh Hadizadeh, Department of Immunology, Genetics and Pathology, Science for Life Laboratory, Uppsala University, 75108 Uppsala, Sweden.
Torgny Karlsson, Department of Immunology, Genetics and Pathology, Science for Life Laboratory, Uppsala University, 75108 Uppsala, Sweden.
Weronica E Ek, Email: Weronica.ek@igp.uu.se, Department of Immunology, Genetics and Pathology, Science for Life Laboratory, Uppsala University, 75108 Uppsala, Sweden.
Funding
This work was mainly funded by The Swedish Cancer Society 21 0447 FE (W.E.E.) and 19 0383 Pj (Å.J.). It was also funded by the Swedish Research Council 2019-01497 (Å.J.), the Marcus Borgström Foundation (W.E.E.), K and O F Hedströms Foundation (W.E.E.), the Åke Wiberg Foundation M19-0349/M20-0057 (W.E.E.), A and M Rudbergs Foundation (W.E.E.), and Vleugels Foundation (WEE). The funding sources had no influence and took no part in the design or conduct of this research.
Author Contributions
W.E.E. and Å.J. designed the study; W.E.E. performed the MR analysis, D.S. generated the figure and performed the estradiol GWAS; W.E.E. and T.K. performed the statistical analysis; W.E.E., T.K., J.H., D.S., Å.J., and F.H. wrote the manuscript; D.S., J.H., T.K., W.E.E., F.H., and Å.J. interpreted the data and contributed to and reviewed the manuscript. All authors declare no conflicts of interest.
Disclosures
The authors declare no conflict of interest
Ethics Approval and Consent to Participate
The UK Biobank study was approved by the National Research Ethics Committee (REC reference 11/NW/0382). Informed consent to the study was given by all participants. An application for using data from UK Biobank has been approved (application nr: 41143) and analysis performed was approved by the Swedish Ethical Review Authority (dnr: 2020-04415). This study was performed in accordance with the Declaration of Helsinki.
Data Availability
The data used for this study is available for bona fide researchers from the UK Biobank Resource (http://www.ukbiobank.ac.uk/about-biobank-uk/) and can be accessed by an application to the UK Biobank. The estradiol data was taken from the supplementary material previously published at https://doi.org/10.5281/zenodo.4926701[16]. The BCAC data can be downloaded from http://bcac.ccge.medschl.cam.ac.uk/ and the OCAC can be downloaded from http://ocac.ccge.medschl.cam.ac.uk/data-projects/results-lookup-by-region/. The ECAC data excluding UK Biobank was approved after request to the authors, and summary statistics including UK Biobank can be downloaded from https://www.mrbase.org/. Summary statistics for the published MR study is found in Table 1.
References
- 1. Key T, Appleby P, et al. ; Endogenous Hormones and Breast Cancer Collaborative Group. Sex hormones and risk of breast cancer in premenopausal women: a collaborative reanalysis of individual participant data from seven prospective studies. Lancet Oncol. 2013;14(10):1009-1019. doi: 10.1016/S1470-2045(13)70301-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaaks R, Berrino F, Key T, et al. Serum sex steroids in premenopausal women and breast cancer risk within the European Prospective Investigation into Cancer and Nutrition (EPIC). J Natl Cancer Inst. 2005;97(10):755-765. doi: 10.1093/jnci/dji132 [DOI] [PubMed] [Google Scholar]
- 3. Zhang X, Tworoger SS, Eliassen AH, Hankinson SE. Postmenopausal plasma sex hormone levels and breast cancer risk over 20 years of follow-up. Breast Cancer Res Treat. 2013;137(3):883-892. doi: 10.1007/s10549-012-2391-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaaks R, Rinaldi S, Key TJ, et al. Postmenopausal serum androgens, oestrogens and breast cancer risk: the European prospective investigation into cancer and nutrition. Endocr Relat Cancer. 2005;12(4):1071-1082. doi: 10.1677/erc.1.01038 [DOI] [PubMed] [Google Scholar]
- 5. Mungenast F, Thalhammer T. Estrogen biosynthesis and action in ovarian cancer. Front Endocrinol (Lausanne). 2014;5:192. doi: 10.3389/fendo.2014.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stewart C, Raylea C, Lockwood S. Ovarian cancer: an integrated review. Semin Oncol Nurs . 2019;35(2):151-156. doi:10.1016/j.soncn.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 7. Rodriguez AC, Blanchard Z, Maurer KA, Gertz J. Estrogen signaling in endometrial cancer: a key oncogenic pathway with several open questions. Horm Cancer. 2019;10(2-3):51-63. doi: 10.1007/s12672-019-0358-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brinton LA, Felix AS. Menopausal hormone therapy and risk of endometrial cancer. J Steroid Biochem Mol Biol. 2014;142:83-89. doi: 10.1016/j.jsbmb.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karlsson T, Johansson T, Hoglund J, Ek WE, Johansson Å. Time-dependent effects of oral contraceptive use on breast, ovarian, and endometrial cancers. Cancer Res. 2021;81(4):1153-1162. doi: 10.1158/0008-5472.CAN-20-2476 [DOI] [PubMed] [Google Scholar]
- 10. Iversen L, Sivasubramaniam S, Lee AJ, Fielding S, Hannaford PC. Lifetime cancer risk and combined oral contraceptives: the Royal College of General Practitioners’ Oral Contraception Study. Am J Obstet Gynecol. 2017;216(6):580.e1-580.e9. doi: 10.1016/j.ajog.2017.02.002 [DOI] [PubMed] [Google Scholar]
- 11. Berchuck A, Scildkraut J. Oral contraceptive pills. Prevention of ovarian cancer and other benefits. N C Med J. 1997;58(6):405-407. [PubMed] [Google Scholar]
- 12. Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007;16(4):309-330. doi: 10.1177/0962280206077743 [DOI] [PubMed] [Google Scholar]
- 13. Thompson DJ, O’Mara TA, Glubb DM, et al. CYP19A1 fine-mapping and Mendelian randomization: estradiol is causal for endometrial cancer. Endocr Relat Cancer. 2016;23(2):77-91. doi: 10.1530/ERC-15-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsson SC, Siddharta K, Perry JRB, et al. Serum estradiol and 20 site-specific cancers in women: Mendelian randomization study. J Clin Endocrinol Metab. 2022;107(2):e467-e474. doi: 10.1210/clinem/dgab713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481-487. doi: 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- 16. Schmitz D, Ek WE, Berggren E, Höglund J, Karlsson T, Johansson Å. Genome-wide association study of estradiol levels, and the causal effect of estradiol on bone mineral density [published correction appears in J Clin Endocrinol Metab. 2021 Oct 26]. J Clin Endocrinol Metab. 2021;106(11):e4471-e4486. doi: 10.1210/clinem/dgab507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk. Genetic polymorphisms in phase I and phase II enzymes and breast cancer risk associated with menopausal hormone therapy in postmenopausal women. Breast Cancer Res. 2010;119(2):463-474. [DOI] [PubMed] [Google Scholar]
- 18. Tobin J. Estimation of relationships for limited dependent variables. Econometrica. 1958;26(1):24-36. doi: 10.2307/1907382 [DOI] [Google Scholar]
- 19. Burgess S, Labrecque JA. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur J Epidemiol. 2018;33:947-952. doi: 10.1007/s10654-018-0424-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Michailidou K, Lindström S, Dennis J, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92-94. doi: 10.1038/nature24284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O’Mara TA, Glubb DM, Amant F, et al. Identification of nine new susceptibility loci for endometrial cancer. Nat Commun. 2018;9(1):3166. doi: 10.1038/s41467-018-05427-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Phelan CM, Kuchenbaecker KB, Tyrer JP, et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat Genet. 2017;49(5):680-691. doi: 10.1038/ng.3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olena YO, Burgess S. Mendelian Randomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu Z, Zheng Z, Zhang F, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen CTL, Liu CT, Chen GK, et al. Meta-analysis of loci associated with age at natural menopause in African-American women. Hum Mol Genet. 2014;23(12):3327-3342. doi: 10.1093/hmg/ddu041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miyashita N, Horie M, Mikami Y, et al. ASCL1 promotes tumor progression through cell-autonomous signaling and immune modulation in a subset of lung adenocarcinoma. Cancer Lett. 2020;489:121-132. doi: 10.1016/j.canlet.2020.06.002 [DOI] [PubMed] [Google Scholar]
- 27. Moore KN, Tritchler D, Kaufman KM, et al. Genome-wide association study evaluating single-nucleotide polymorphisms and outcomes in patients with advanced stage serous ovarian or primary peritoneal cancer: An NRG Oncology/Gynecologic Oncology Group study. Gynecol Oncol. 2017;147(2):396-401. doi: 10.1016/j.ygyno.2017.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stolk L, Perry JRB, Chasman DI, et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet. 2012;44(3):260-268. doi: 10.1038/ng.1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48(7):709-717. doi: 10.1038/ng.3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Flote VG, Furberg AS, McTiernan A, et al. Gene variations in oestrogen pathways, CYP19A1, daily 17β-estradiol and mammographic density phenotypes in premenopausal women. Breast Cancer Res. 2014;16(1):499. doi: 10.1186/s13058-014-0499-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee AJ, Conney AH, Zhu BT. Human Cytochrome P450 3A7 Has a Distinct High Catalytic Activity for the 16α-Hydroxylation of Estrone but not 17β-Estradiol. Cancer Res. 2003;63(19):6532 - 66536. [PubMed] [Google Scholar]
- 32. Ohmori S, Nakasa H, Asanome K, et al. Differential catalytic properties in metabolism of endogenous and exogenous substrates among CYP3A enzymes expressed in COS-7 cells. Biochim Biophys Acta Gen Subj. 1998;1380(3):297-304. doi: 10.1016/S0304-4165(97)00156-6 [DOI] [PubMed] [Google Scholar]
- 33. Tsuchiya Y, Nakajima M, Yokoi T. Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett. 2004;227(2):115-124. [DOI] [PubMed] [Google Scholar]
- 34. Zhao H, Zhou L, Shangguan A, Bulun S. Aromatase expression and regulation in breast and endometrial cancer. J Mol Endocrinol. 2016;57(1):19-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used for this study is available for bona fide researchers from the UK Biobank Resource (http://www.ukbiobank.ac.uk/about-biobank-uk/) and can be accessed by an application to the UK Biobank. The estradiol data was taken from the supplementary material previously published at https://doi.org/10.5281/zenodo.4926701[16]. The BCAC data can be downloaded from http://bcac.ccge.medschl.cam.ac.uk/ and the OCAC can be downloaded from http://ocac.ccge.medschl.cam.ac.uk/data-projects/results-lookup-by-region/. The ECAC data excluding UK Biobank was approved after request to the authors, and summary statistics including UK Biobank can be downloaded from https://www.mrbase.org/. Summary statistics for the published MR study is found in Table 1.