Abstract
Phosphatase and tensin homolog (PTEN)-induced kinase 1 (Pink1) is regarded as a tumor suppressor and plays an important role in cancer cell biology, while relatively few studies have examined Pink1 in breast cancer, especially in vivo. The aims of this study were to investigate Pink1 expression in different subtypes of breast cancer tissues and cell lines and explore the effect of Pink1 protein on breast cancer. In these experiments, Pink1 expression was investigated using the tissue microarray immunohistochemistry (TMA-IHC) method in 150 samples of breast cancer tissues with different subtypes, and strong staining of Pink1 was significantly correlated with the histological grade of breast cancer (p = 0.015). In addition, Pink1 messenger RNA (mRNA) displayed much higher expression levels in breast cancer cell lines than in MCF-10A breast epithelial cells. Moreover, proteomic data obtained by isobaric tags for relative and absolute quantification (iTRAQ) showed that Pink1 deletion induced a distinct proteomic profile in MDA-MB-231 cells, and enrichment analysis showed that the differential proteins were concentrated mainly in energy metabolism-related pathways. Moreover, Seahorse XF analysis showed that Pink1 knockout reduced the glycolytic ability of MDA-MB-231 cells. Our findings indicated that Pink1 may be an indicator of malignancy in breast cancer and that it presents oncogenic properties in breast cancer, which raises another perspective for understanding the regulatory role of Pink1 in breast cancer.
Keywords: Pink1, breast cancer, immunohistochemistry, MDA-MB-231, glycolysis
Impact Statement
Pink1 was regarded as a tumor suppressor and plays an important role in cancer cell biology while relatively few researches was done on Pink1 in breast cancer especially in vivo. This study investigated the expression of Pink1 in 150 samples of breast cancer tissues and the results suggested that Pink1 was an indicator of malignancy in breast cancer, which was the first time to detect the expression of Pink1 protein in different subtypes of breast cancer up to now. In addition, proteomic data and Seahorse XF analysis revealed that Pink1 affected glycolysis in MDA-MB-231 cell lines, which was regarded as a representative cell line of basal-like subtype of breast cancer. The conclusion of this study indicated that Pink1 promotes the progression of breast cancer and presented oncogenic properties in breast cancer, which raised another perspective for understanding the regulation role of Pink1 in breast cancer.
Introduction
Phosphatase and tensin homolog (PTEN)-induced kinase 1 (Pink1), a serine/threonine protein kinase, was initially identified as a gene upregulated by overexpression of a major tumor suppressor-PTEN. 1 The Pink1 gene encodes 581 amino acids and the N-terminal and C-terminal of this protein includes a mitochondrial-targeting sequence and an autoregulatory sequence, respectively; besides, it also contains a transmembrane domain and a highly conserved kinase domain homologous to the Ca2+/calmodulin family.2,3 These structural compositions may be related to diverse functions of Pink1.
Starting shortly after Pink1 was identified, many studies on the function of Pink1 have been carried out in recent decades. Since loss-of-function mutations in Pink1 were discovered to cause autosomal recessive forms of Parkinson’s disease (PD), 4 subsequent studies have focused primarily on the role of Pink1 in PD and other neurodegenerative diseases, such as Alzheimer’s disease (AD), for a period of time.5,6 Pink1 was also found to play an important role in cancer cell biology.7,8 Some epidemiologic studies have indicated an inverse association between the prevalence of neurodegenerative diseases and the development of some cancers.9,10 Studies over the past decades have demonstrated that the Pink1 signaling system has been shown to control several processes key to cancer cell biology and presents pro- and anti-tumorigenic properties with context-dependent tumor as it displays different expression across cancer cell types.11–13 Mechanistically, Pink1 acts as a cross point connecting different pathways to regulate cell survival, death, and other cell activities. Recent data illustrated that Pink1 suppresses colon tumor growth via activation of the P53 signaling pathway 14 and regulates hepatocellular carcinoma cell growth through the hypoxia-inducible factor (HIF)/HEY1/Pink1 pathway. 15 Moreover, previous studies have demonstrated that Pink1 negatively regulates cell growth and the Warburg effect in glioblastoma, 16 and silencing Pink1 is lethal in DNA mismatch repair-deficient cancers. 17 Several studies have reported that Pink1 is involved in modulating inflammatory responses,18–20 which also indicates Pink1’s multifunctional features. Furthermore, evidence has shown the dual function of Pink1 in breast cancer, although the specific mechanism needs further elucidation.13,21,22
Breast cancer is one of the most common malignant tumors and is divided into four intrinsic subtypes based on the recognition of intrinsic biological subtypes within the breast cancer spectrum, 23 of which basal-like subtype was regarded as the most serious subtype with high metastasis and poor prognosis. As studies on Pink1 in breast cancer have been controversial, it is essential to further specify the role of Pink1 in breast cancer, especially in basal-like subtype. Therefore, this investigation intends to detect Pink1 expression in breast cancer subtypes and tries to explore the role of Pink1 on basal-like subtype of breast cancer cells.
Materials and methods
Formalin-fixed paraffin-embedded breast tissue specimen, tissue microarray, and immunohistochemistry for Pink1
A total of 150 samples of formalin-fixed paraffin-embedded (FFPE) breast cancer tissues and 18 samples of breast fibroma were obtained from the Department of Pathology of the Huzhou Central Hospital (ZheJiang, China), and subtypes of these breast cancer tissues were examined by immunohistochemistry (IHC) profiles as we described previously: luminal A subtype (estrogen receptor (ER)-positive) and/or progesterone receptor (PR)-positive, human epidermal growth factor receptor (HER)-positive with 2+, or ER-positive and/or PR-positive, HER2-negative and index of Ki67 < 14%), luminal B subtype (ER-positive and/or PR-positive, HER2-negative and index of Ki67 ≧ 14%), basal-like subtype (ER-negative, PR-negative, HER2-negative and cytokeratin 5/6-positive and/or estimated glomerular filtration rate (EGFR)-positive), and HER2-overexpression subtypes were defined as 0, 1+, 2+, and 3+ referred to the guidelines from American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP). All samples of breast tissues were stained with hematoxylin and eosin (HE) and evaluated by two experienced pathologists. Each breast tumor block was perforated one core at the tumor location identified by the pathologist and the tissue microarray (TMA) consisting of 168 FFPE breast tissues was generated. All of the specimens were taken from the primary lesions and none of these patients had received radiotherapy or chemotherapy before surgical operation; besides, clinicopathological information of these patients including age, tumor size, histopathology grade, lymph node metastasis, and TNM (tumor (T), nodes (N), and metastases (M)) stage were also analyzed in this experiment. The study was permitted by the Huzhou Central Hospital Ethics Committee and informed consent of patients.
The immunohistochemical process was as follows: rabbit polyclonal anti Pink1 antibody was purchased from Abcam Company at a dilution ratio of 1:100, and a DAB staining kit (ZSGQ-BIO) was used for staining. After antibody staining, the nuclei were labeled with hematoxylin. The staining results were determined according to the staining depth as follows: mild staining (1+), moderate staining (2+), and severe staining (3+); a total of 1000 cells were counted in each section at 400× magnification randomly selected from five fields (200 cells in each field) to calculate the percentage of positive cells as the low expression group (number of positive cells < 50%), medium expression group (number of positive cells between 50 and 75%) and high expression group (number of positive cells > 75%). Finally, the samples were divided into a high expression group and a weak/moderate expression group based on the evaluation of staining intensity and percentage of positive cells.
Cell lines and cell culture
The breast cancer cell line MDA-MB-231 was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in complete medium: Dulbecco’s Modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), and penicillin/streptomycin (100 U/mL and 100 μg/mL; Gibco, Life Technologies) in a humidified incubator with 5% CO2 at 37 °C.
Pink1 Double Nickase Plasmid (h) and Control CRISPR/Cas9 Plasmid were purchased from Santa Cruz Biotechnology (Heidelberg, Germany), and the target-specific guide RNA sequences were 5′-tatctgatagggcagtccat-3′ and 5′-gcaagcgtctcgtgtccaac-3′. MDA-MB-231 cells were grown in six-well plates until they reached 80% confluence and then transfected with Pink1 Double Nickase Plasmid (h) and Control CRISPR/Cas9 Plasmid with Lipofectamine 3000 according to the instructions. The cells were expanded 24–72 h post-transfection and selected with complete medium containing puromycin (10 μg/mL) until the nontransfected cells were all killed. Then, surviving cells were seeded in 96-well plates and grown in complete medium with the lowest puromycin (1 μg/mL) for isolation of the single-cell colonies.
For MTT assay, MDA-MB-231con cells and MDA-MB-231Pink1−/−cells (3×103 cells/well) were seeded in 96-well plates and cultured in complete medium. The cells were treated with WST-8 at 6, 24, 48, 72, 96 and 120 h to measure cell viability. Each group was treated biologically in triplicate, and at least five wells of cells were tested at each time point.
Protein extraction
MDA-MB-231con and MDA-MB-231Pink1−/− cells were collected with cell scrapers and homogenized in RIPA lysis buffer (Beyotime, containing 50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) on ice for 30 min and then centrifuged at 16,000 g at 4 °C for 10 min to harvest the protein samples. Protein content was measured with a BCA Protein Assay Kit (Beyotime) and stored at −80 °C until use.
Protein digestion and iTRAQ labeling
One hundred micrograms of protein solution of each sample was digested with 2.5 μg trypsin at 37 °C for 4 h, and then another 2.5 μg trypsin was added to the protein mixture to continue incubation for 8 h at 37 °C. The acquired peptide mixture was desalted on a strata X C18 column (Phenomenex) and vacuum-dried.
Some iTRAQ labeling reagents were recovered to ambient temperature, and then 50 μL isopropanol was added to each sample with vortex agitation. The vacuum-dried peptides were dissolved in 30 μL of 0.5 M triethylammonium bicarbonate (TEAB) buffer and then added to different iTRAQ labeling tags. Peptide labeling was performed by an iTRAQ Reagent 8-plex Kit according to the manufacturer’s protocol. The labeled peptides with different tags were combined and desalted with a Strata X C18 column (Phenomenex) and dried with a rotary vacuum concentrator.
HPLC fractionation and LC-MS/MS analysis
The labeled peptides were separated on a Shimadzu LC-20AB HPLC Pump system coupled with a high pH Gemini column. The peptides were redissolved with 2 ml buffer A (5% acetonitrile (ACN), 95% H2O, pH at 9.8) and passed through the column at a flow rate of 1 mL/min. Then, the peptides were eluted in order with a gradient of 5%, 5–35%, and 35–95% buffer B (5% H2O, 95% ACN, pH = 9.8) for 10, 40, and 1 min. Then, the system was maintained in 95% buffer B for 3 min, and then the gradient of elution was decreased to 5% within 1 min followed by equilibration for 10 min. Eluting peptides were monitored by measuring absorbance at 214 nm during the separation process, and fractions were collected every 1 min. Finally, 20 fractions were obtained by combining chromatographic elution peaks with samples and vacuum-dried as above.
The vacuum-dried fraction was resuspended in buffer A (2% (ACN), 0.1% FA) and separated by a Shimadzu LC-20AD NASL liquid chromatograph system after centrifugation at 20,000 g for 10 min. The supernatant was loaded on a trap column for sample enrichment and desalination and then separated at a flow rate of 300 nL/min on a C18 column under the following conditions. First, the gradient of 5% buffer B (98% ACN, 0.1% FA) was applied for 8 min, and then the gradient of buffer B was increased from 5% to 35% in 40 min followed by another 40 min for a gradient of buffer B from 35% to 60%. Next, the samples were eluted in 50 min for a gradient of buffer B from 60% to 80% followed by maintenance at 80% buffer B for 50 min. Finally, the fractions were equilibrated at 5% buffer B for 50 min followed by mass spectrometry.
The separated peptides from nanoHPLC were subjected to tandem mass spectrometry Q EXACTIVE HF X (Thermo Fisher Scientific, San Jose, CA, USA) for DDA (data-dependent acquisition) detection by nanoelectrospray ionization. The parameters of MS analysis were as follows: electrospray voltage at 1.6 kV; mass-to-charge ratio of precursor scan ranges from 350 to1600 m/z at a resolution of 70,000 in Orbitrap; MS/MS fragment scan range > 100 m/z at a resolution of 17,500 in HCD mode and normalized collision energy at 30% and dynamic exclusion time set for 15 s. Automatic gain control (AGC) for full MS target and MS2 target was 3E6 and 1E5, respectively.
Glycolysis assay and ATP measurement
Cells were counted and plated at 1.0×104 cells per well in a Seahorse XF24 Cell Culture Microplate for all experiments (Agilent). Cells were then prepared for glycolysis stress testing and real-time adenosine triphosphate (ATP) rate testing after cell adhesion according to the instructions. For the glycolysis stress test, the cells were washed twice with Agilent Seahorse XF DMEM (Agilent) without glucose and pyruvate, and then a final volume of 525 μL was placed in each well. The cells were then incubated in a chamber free of CO2 at 37 °C for 1 h before being placed into a Seahorse XFe24 Analyzer (Agilent). Then, the cells were treated by automatic injection into the wells with glucose (2 mg/mL), oligomycin (1 μM), and 2-deoxy-D-glucose (2-DG, 100 mM), successively. For the real-time ATP rate assay, the cells were incubated with test solution containing glucose and pyruvate and were treated with 1.5 μM oligomycin and 0.5 μM rotenone/antimycin A. The values of OCR and ECAR were measured three times after each compound was administered. All Seahorse experiments were repeated at least three times and OCR and ECAR data were normalized to cell number using CyQUANT (Thermo Fisher Scientific).
Quantitative real-time analysis
Total RNA was extracted with TRIzol reagent (Life Technologies, USA), and 1 μg of total RNA was used to synthesize cDNA with the PrimeScriptTM RT Reagent Kit with gDNA Eraser according to the kit directions. Quantitative real-time PCR (qRT-PCR) was performed on an ABI7500 system (Life Technologies, USA) using SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) reagents (Takara, Japan) and specific primer sets (Table 1). The amplification reactions were performed with the following parameters: 30 s at 95 °C, 40 cycles of 95 °C for 5 s, and 60 °C for 34 s. The relative expression levels of the targeted gene were normalized to the expression value of the internal control gene β-actin. Relative transcript levels of Pink1 in breast cell lines were analyzed with the ∆Ct value, and the relative expression of four important catalytic enzymes of glycolysis was evaluated by the 2−∆∆Ct method. All experiments were performed in triplicate biologically, and each contained three technical replicates.
Table 1.
Primer sets used in the experiment.
| Gene name | Primer sequences(5′–3′) | Product length (bp) |
|---|---|---|
| Pink1 | For: TGGACACGAGACGCTTGCAG Rev: TCCTGGTGCACTGGTACCTG |
179 |
| PDH | For: GAGTGACCCTGGAGTCAGTTAC Rev: CTGGGCAGCATCCTCAATCTC |
172 |
| PKM | For: CGATCAGTGGAGACGTTGAAG Rev: CAGCTCCACCTCTGCAGTG |
237 |
| HKII | For: CACCAAGCGTGGACTACTCTTC Rev: GGCCACCACAGTGCACACCTC |
199 |
| LDH | For: GCAGGTGGTTGAGAGTGCTTAT Rev: GGCCTCTTCCTCAGAAGTCAGA |
250 |
| β-actin | For: ACGCCAACACAGTGCTGTCTG Rev: GGCCGGACTCGTCATACTCC |
219 |
Pink1: Phosphatase and tensin homolog–induced kinase 1; for: forward; rev: reverse; PDH: pyruvate dehydrogenase; PKM: pyruvate kinase; HKII: hexokinase 2; LDH: lactate dehydrogenase.
Western blot analysis
MDA-MB-231con and MDA-MB-231Pink1−/− cells were collected with cell scrapers, homogenized in RIPA lysis buffer (Beyotime, China) on ice for 30 min, and then centrifuged at 16,000 g at 4 °C for 10 min to harvest the protein samples. Protein content was measured with a BCA Protein Assay Kit (Beyotime). Each protein sample mixed with an appropriate amount of loading buffer was incubated at 95 °C for 5 min, and 10 μg of protein from each sample was spotted for 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) for 2 h at 120 V constant pressure. The proteins were transferred onto polyvinylidene difluoride (PVDF) membranes for 30 min at 25 V with Bio-Rad Trans-Blot Turbo, and the PVDF membranes were blocked with 5% skimmed milk for 1 h at room temperature followed by incubation with primary antibodies overnight at 4 °C. The next day, the membranes were immersed in secondary antibody for 2 h at room temperature followed by the addition of reagent to scan the strips. Primary antibodies against HKII and PDHA were from Abcam (Cambridge, UK), β-actin was from Bioker (Hangzhou, China), and secondary antibodies against rabbits were from Cell Signaling Technology (MA, USA).
Bioinformatics and statistical analysis
In this study, the original mass spectrum data were converted into MGF files and searched using the Mascot engine against the Uni-Prot database and the decoy database. For protein identification, differentially expressed proteins (DEPs) were screened out by the cutoff value of fold change and p-value, and DAVID Database (version 6.7) was applied to perform all of the pathways and gene ontology (GO) enrichment analyses of these DEPs. Relative mRNA expression of target genes was analyzed by GraphPad Prism 5.0 software and t-tests were used to evaluate statistical significance with p < 0.05.
Results
Pink1 expression in different subtypes and clinicopathological and histopathological features of breast cancer using the TMA-IHC method
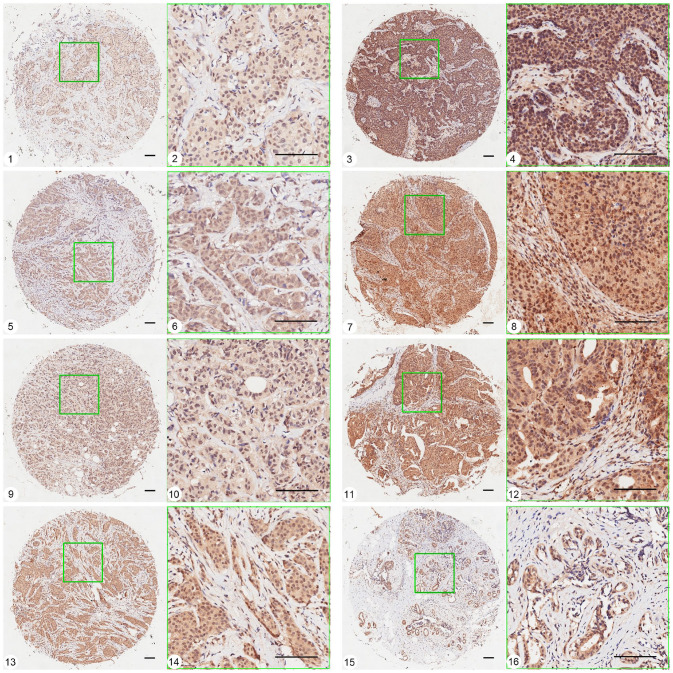
Pink1 was universally expressed in all breast cancer subtypes, and 147 of 150 cases of invasive breast ductal carcinoma exhibited positive staining (Figure 1). Yellow Pink1 particles were observed to be distributed diffusely in the nucleus and cytoplasm, and different subtypes and fibroma of breast tissue all displayed nonuniform intensity of staining even within the same group of tissue samples.
Figure 1.
Representative pictures of Pink1 expression in different subtypes of breast cancer tissues and breast adenosis using TMA-ISH method. 1, 2 and 3, 4: expression of Pink1 in basal-like subtype tissue, grade I (weak staining) and grade III (strong staining). 5, 6 and 7, 8: expression of Pink1 in HER2 over-expression subtype tissue, grade I (weak staining) and grade III (strong staining). 9, 10 and 11, 12: expression of Pink1 in Luminal B subtype tissue, grade I (weak staining) and grade III (strong staining). 13, 14 and 15, 16: expression of Pink1 in Luminal A subtype tissue, grade I–II and breast adenosis. (A color version of this figure is available in the online journal.)
Ruler scale = 100.8 μm.
The relationships between Pink1 expression in different subtypes of breast cancer and the clinicopathological parameters are analyzed and presented in Table 2. The expression of Pink1 in breast cancer is associated significantly with histopathology grade of breast cancer tissues, as 40 of 117 cases with well-differentiated cancer cells (grade I–II, 34.2%) and 19 of 33 cases with poorly differentiated cancer cells (grade III, 57.6.2%; χ2 = 5.90, p = 0.015) display strong staining of Pink1 protein. Besides, there was no significant difference in Pink1 expression among the four subtypes, and also no significant associations between Pink1 expression and other clinicopathological parameters, including age (t = −0.97, p = 0.33), tumor size (t = 0.92, p = 0.36), lymph node metastasis (χ2 = 0.129, p = 0.719), and TNM stage (χ2 = 2.831, p = 0.092).
Table 2.
Relationship between Pink1 expression and clinicopathologic features in patients with breast tumor.
| Pathological factors | Cases (n) | Pink1 expression (%) | p-value | |
|---|---|---|---|---|
| Weak/moderate | Strong | |||
| Malignant/benign | ||||
| Benign | 18 | 16 (88.9) | 2 (11.1) | 0.014* χ2 = 5.990 |
| Malignant | 150 | 89 (59.3) | 61 (40.7) | |
| Subtype of carcinoma | ||||
| Luminal A | 18 | 12 (66.7) | 6 (33.3) | 0.331 χ2 = 3.423 |
| Luminal B | 20 | 14 (70) | 6 (30) | |
| HER-2 | 24 | 17 (70.8) | 7 (29.2) | |
| Basal-like | 88 | 48 (54.5) | 40 (45.5) | |
| TNM staging | ||||
| 0–I | 42 | 30 (71.4) | 12 (28.6) | 0.092 χ2 = 2.831 |
| II–IV | 108 | 61 (56.5) | 47 (43.5) | |
| Lymph node metastasis | ||||
| − | 79 | 49 (62) | 30 (38) | 0.719 χ2 = 0.129 |
| + | 71 | 42 (59.2) | 29 (40.8) | |
| Grade | ||||
| I–II | 117 | 77 (65.8) | 40 (34.2) | 0.015* χ2 = 5.900 |
| III | 33 | 14 (42.4) | 19 (57.6) | |
| Age (years) | 150 | 52.96 ± 10.75 | 54.70 ± 11.16 | 0.330 t = −0.97 |
| Tumor size (cm) | 150 | 2.83 ± 1.23 | 2.63 ± 1.40 | 0.360 t = 0.920 |
HER-2: human epidermal growth factor receptor 2; TNM: tumor, nodes, and metastases.
*p < .05.
Pink1 mRNA expression in different subtypes of breast cell lines
At present, the qRT-PCR assay is applied to detect the mRNA levels of Pink1 in different subtypes of breast cell lines and the data are normalized to β-actin and presented as the ΔCt value (ΔCt = CT Pink1 − CTβ-actin). In this study, breast cancer cell lines were classified to different subtypes based on the IHC staining of ER, PR, HER-2, CK5/6, and EGFR, as presented in Table 3. MCF-10A and MDA-MB-231 cells were identified as representatives of the basal-like subtype. MCF-7 and SK-BR-3 cells belonged to the luminal A and HER2 overexpression subtypes, respectively, and MDA-MB-453 cells belonged to the unclassified subtype. 24 As the data indicated, relative expression of Pink1 mRNA was the lowest in MCF-10A cells (ER−, PR−, EGFR 2+), followed by MCF-7 cells (ER+, PR+, EGFR 1+) and SK-BR-3 cells (ER−, PR−, EGFR 2+, HER 3+), while MDA-MB-231 cells (ER−, PR−, EGFR 1+) and MDA-MB-453 cells (ER−, PR−, EGFR−) displayed relatively high expression levels of Pink1, as smaller ΔCt values were obtained in these two cell lines. It is interesting that the expression of Pink1 mRNA in breast cell lines was essentially consistent with the results obtained from breast tissue IHC in this study: Pink1 expression was positively correlated with the histological grade of breast cancer tissues.
Table 3.
Pink1 expression and histopathological features in different breast cell lines.
| Cell lines | Subtype | Pink1((Δct)) | ER | PR | HER2 | EGFR | CK5/6 | Ki-67 |
|---|---|---|---|---|---|---|---|---|
| MCF-10A | Basal-like | 17.10 ± 0.06 | − | − | 0–1+ | 2+ | + | 30% |
| MCF-7 | Lumina A | 15.16 ± 1.00 | + | + | 0–1+ | 1+ | − | 90% |
| SK-BR-3 | HER2 over-expression |
11.15 ± 0.51 | − | − | 3+ | 2+ | − | 20% |
| MDA-MB-453 | Unclassified | 10.70 ± 0.20 | − | − | 0 | 0 | − | 80% |
| MDA-MB-231 | Basal | 10.99 ± 0.18 | − | − | 0–1+ | 1+ | − | 100% |
ER: estrogen receptor; PR: progesterone receptor; HER-2: HER-2: human epidermal growth factor receptor 2; EGFR: estimated glomerular filtration rate.
Δct = CTPink1 − CTβ-actin, the smaller of Δct value means the higher expression of Pink1.
Effect of Pink1 deletion on the MDA-MB-231 cells
To investigate the role of Pink1 in breast cancer cells, the Pink1 gene was deleted with the CRISPR/Cas9 gene editing method (Figure 2(B)) in MDA-MB-231 cells. Compared to MDA-MB-231con cells, the MDA-MB-231Pink1−/− cells appeared to be round, lost cell processes, and connected with each other closely and almost no individual cell outline could be clearly observed (Figure 2(A)). In addition, the MTT assay revealed that Pink1 deletion also affected cell viability, and the proliferation rate of MDA-MB-231Pink1−/− cells was almost 30% lower than that of MDA-MB-231con cells (p < 0.05; Figure 2(C)). These results demonstrated that Pink1 may affect the growth of MDA-MB-231 cells.
Figure 2.
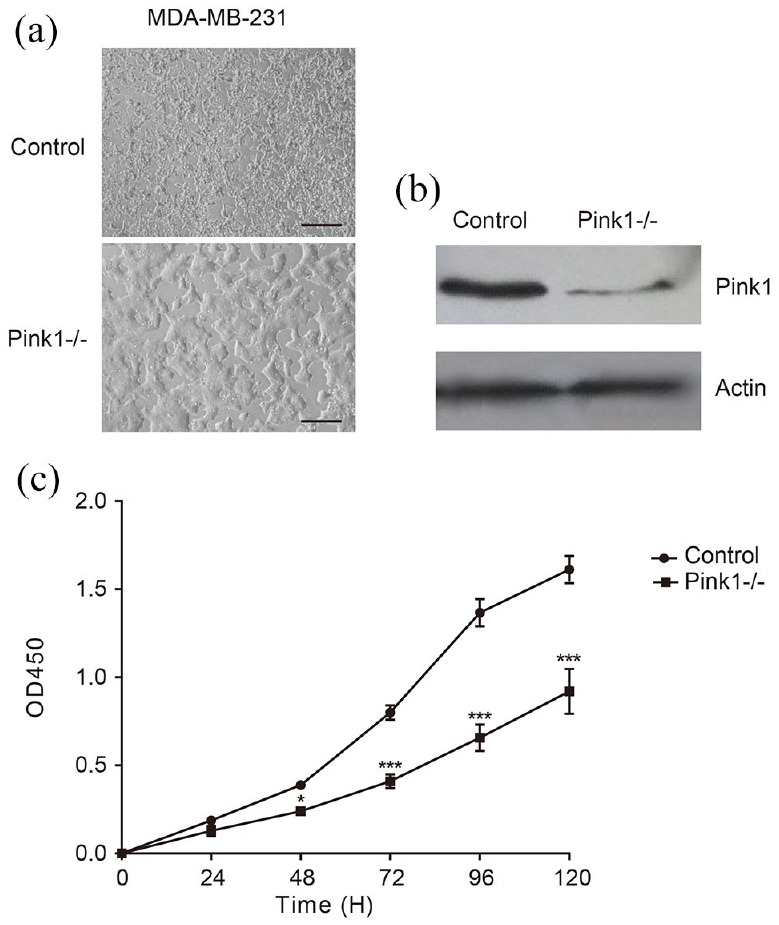
Deletion of Pink1 in MDA-MB-231 cells. (A) Morphology of MDA-MB-231 cells loss of Pink1. Scale bar = 100 μm. (B) Verification of deletion of Pink1 by Western blot. (C) Cell proliferation rate of MDA-MB-231Pink1−/− was decreased compared to control.
*p < 0.05, **p < 0.01, ***p < 0.001.
Protein profiling of MDA-MB-231 cells and protein responses to Pink1 deletion
To explore the role of Pink1 deletion in breast cells in vitro, the iTRAQ technique was applied to compare the comprehensive protein expression profiles between MDA-MB-231con and MDA-MB-231Pink1−/− cells. After processing the MS/MS spectra using Mascot software, we obtained a high-quality dataset, of which a total of 232,405 spectra were generated, and 11,522 peptides (11,179 unique peptides) and 3252 proteins were identified with 1% false discovery rate (FDR). According to the cutoff of a fold change ⩾ 1.2 or ⩽ 0.83 and p < 0.05, 426 proteins were identified to be significant in MDA-MB-231 cells in response to Pink1 ablation, including 257 upregulated proteins and 169 downregulated proteins (Supplemental Table S1 and Table S2). These results indicated that Pink1 deletion induced a distinct proteomic profile in MDA-MB-231 cells.
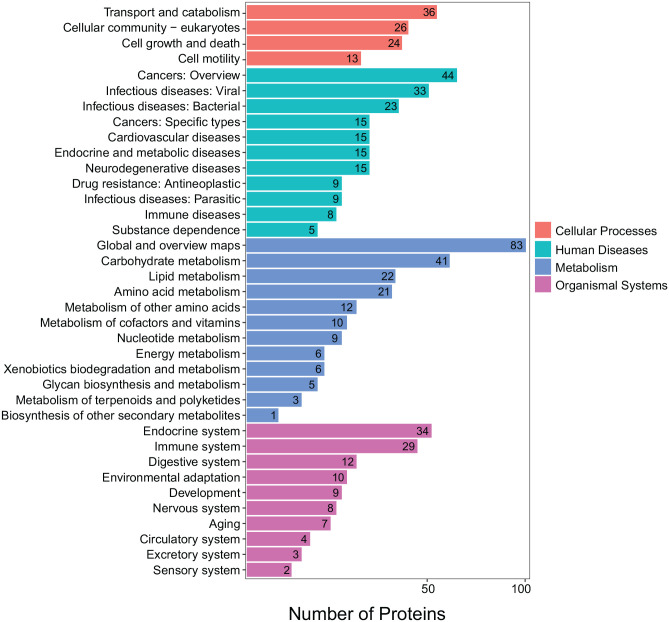
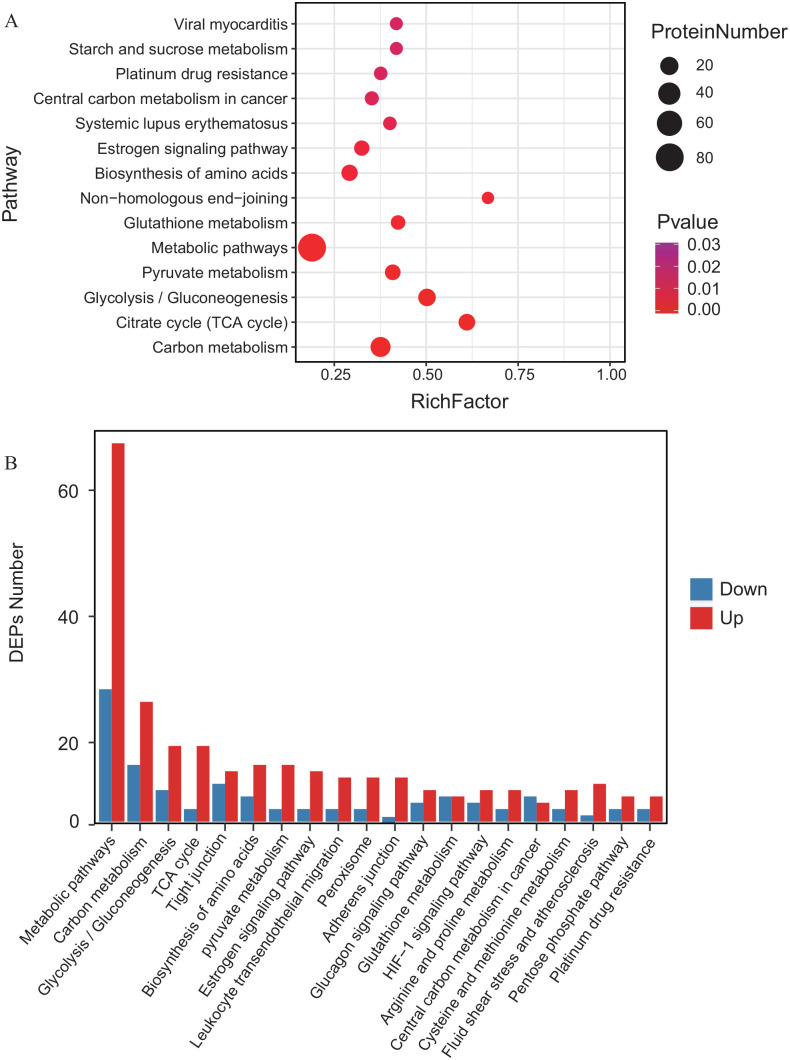
All DEPs were subjected to Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis and they were found to be involved in cellular processes, metabolism, organismal systems, and human diseases (Figure 3). Similarly, the significantly up- and downregulated DEPs were also annotated with pathway enrichment analysis. Obviously, most of the DEPs were assigned to pathways related to energy metabolism: carbon metabolism, glycolysis/gluconeogenesis, citrate cycle, pyruvate metabolism, and glutathione metabolism (Figure 4). Together, the DEP analysis results indicated that Pink1 is related to cell metabolism, especially glucose metabolism, in MDA-MB-231 cells.
Figure 3.
KEGG annotation of differently expressed proteins induced by deletion of Pink1 in MDA-MB-231 cells. (A color version of this figure is available in the online journal.)
Figure 4.
Enrichment analysis of differently expressed proteins induced by deletion of Pink1 in MDA-MB-231 cells. (A) Pathway enrichment of differently expressed proteins induced by deletion of Pink1 in MDA-MB-231 cells. (B) Classification of up- and down regulated proteins, respectively, enriched in MDA-MB-231 cells loss of Pink1. (A color version of this figure is available in the online journal.)
Validation of gene expression of metabolic reprogramming in MDA-MB-231Pink1−/− cells
To further explore the effect of Pink1 on glucose metabolism in MDA-MB-231 cells, we investigated four important catalytic enzymes of glycolysis that were found to be significantly up- or downregulated proteins by real-time PCR and Western blotting, including hexokinase 2 (HK II), pyruvate kinase (PKM), pyruvate dehydrogenase (PDH), and lactate dehydrogenase (LDH). As the results shown by qPCR analysis, the mRNA expression levels of HKII, PKM, and LDH were significantly decreased by silencing Pink1, while PDH was increased in MDA-MB-231Pink1−/− cells (Figure 5(A)). In addition, Western blot analysis further verified that the expression of HKII was reduced, and PDH was slightly increased in MDA-MB-231Pink1−/− cells compared to control cells (Figure 5(B) and (C)), which was also in agreement with the proteomic data. These results demonstrated that deletion of Pink1 weakened glycolytic activity in MDA-MB-231 cells.
Figure 5.
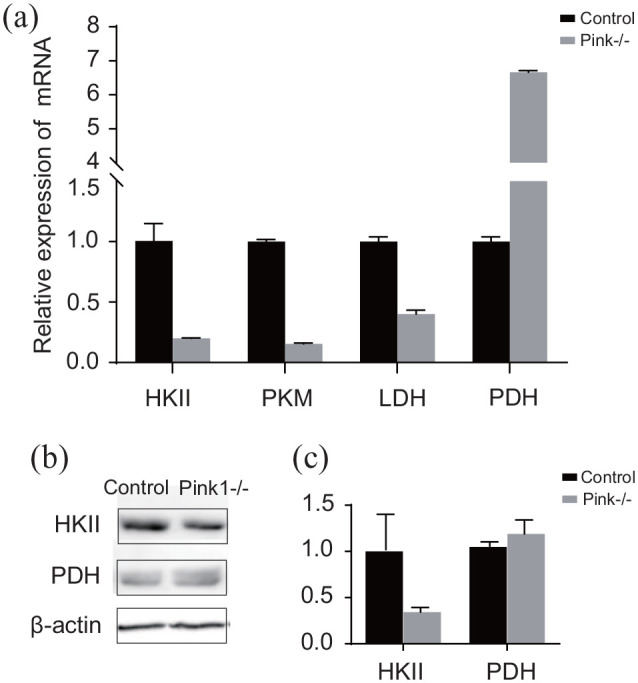
Q-PCR and Western blot analysis in MDA-MB-231con cells and MDA-MB-231Pink1−/− cells.
*p < 0.05, **p < 0.01, ***p < 0.001.
Pink1 regulates metabolic reprogramming in MDA-MB-231Pink1−/− cells
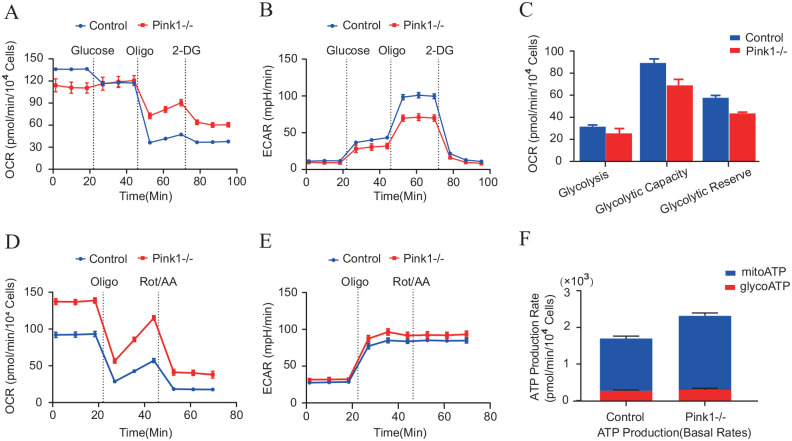
As validated by the proteomic data above, Pink1 may be involved in the regulation of glycolysis in MDA-MB-231 cells, and we employed Seahorse XF Extracellular Flux Analyzers to measure the extracellular acidification rate (ECAR) and oxygen consumption rates (OCRs), which could reflect the overall glycolytic flux. Interestingly, we found that Pink1 deletion in MDA-MB-231 cells led to decreased glycolysis, including glycolytic capacity and glycolytic reserve (Figure 6(A) to (C)). In addition, real-time ATP rate assays revealed that deletion of Pink1 in MDA-MB-231 cells led to increased ATP production, which was generated mainly by mitochondrial respiration (Figure 6(D) to (F)). These findings further indicated that Pink1 may influence metabolic patterns in MDA-MB-231 cells.
Figure 6.
Pink1 drives metabolic reprogramming in vitro. Seahorse XF (A to C) glycolysis stress test and (E and F) real-time ATP rate assay in MDA-MB-231con cells (n = 3) and MDA-MB-231Pink1−/− cells (n = 3). (A color version of this figure is available in the online journal.)
*p < 0.05.
Taken together, the proteomic data and DEP verification experiments suggested that Pink1 promoted cell growth and affected glycolysis in breast cancer cells.
Discussion
Pink1 is a serine/threonine kinase and is expressed ubiquitously in most human tissues. During the past decade, a wealth of research on Pink1 has demonstrated the role of Pink1 in cell survival, stress resistance, mitochondrial function, and bioenergetics. However, relatively fewer studies on Pink1 in breast cancer have been reported compared with other tumor types. This study detected Pink1 expression in breast cancer and explored the role of Pink1 in MDA-MB-231 cell growth. The IHC results demonstrated that Pink1 presented at relatively higher levels in all breast cancer subtypes compared to breast fibroma tissues, and was distributed in the nuclei and cytoplasm, which was inconsistent with a previous report, 13 inferring that the different subcellular localizations were probably associated with the heterogeneity of specimens in breast cancer. In addition, we found that staining intensity was statistically dependent on the differentiation grade, suggesting that Pink1 may be an indicator of malignancy and that Pink1 mRNA expression in breast cells of different subtypes confirmed the results obtained from histological examination. Then, the Pink1 gene was knocked down to explore the effect of Pink1 on MDA-MB-231 cells, which are regarded as a representative cell line of basal-like subtype, the most serious subtype with high metastasis and poor prognosis.
As deletion of Pink1 reduced the proliferation rate of MDA-MB-231 cells, proteomics experiments presented a relatively comprehensive profile of proteins induced by deletion of Pink1 in MDA-MB-231 cells, which provided large-scale information to investigate the role of Pink1 in cell line of basal-like subtype of breast cancer. Interestingly, signaling pathways enriched in upregulated and downregulated DEPs demonstrated that metabolic reprogramming, including glycolysis, citrate cycle, and pyruvate metabolism, may play an important role in the progression of basal-like subtype. These interesting findings were then verified by detection of key proteins related to glycolysis and Seahorse analysis.
Accelerated glucose metabolism under aerobic conditions is one of the features of cancer cells, 25 and cellular metabolism make the switch from mitochondrial oxidative phosphorylation (OXPHOS) to glycolysis, which lead to accumulation of lactic acid and facilitate tumor cell metastasis. 26 Molecularly, abnormal activities of some catalytic enzymes occur in glycolysis pathway, such as HK II and PDH. Increased expression of HKII has been found in several cell types, such as in nonsmall-cell lung cancer and breast cancer.27,28 It also has been reported that in breast cancer cells, elevated expression of HKII is closely associated with a high glycolytic rate, which lead to apoptosis inhibition and drug resistance. 29 PDH is a key catalytic enzyme that couples glycolysis and OXPHOS as it modulate pyruvate flux from the cytoplasm to the mitochondria, 30 and previous studies have suggested that attenuation of PDH activity may be associated with metabolic changes in a variety of tumor cells. 31 In this study, deletion of Pink1 in MDA-MB-231 cells led to the decreased expression of key catalytic in glycolysis pathways, while increased the expression of PDH, indicating that Pink1 affects the glycolysis and Pink1 deletion drive metabolic mode from glycolysis to OXPHOS, which was also proven by measurements of ECAR as well as observed ATP production levels. Together, these data indicate that Pink1 affects the metabolism of MDA-MB-231 cells, and further investigations are needed to elucidate the mechanism of specific mechanism of Pink1 in breast cancer.
Breast cancer is a heterogeneous disease and this study suggested that Pink1 promotes the cell growth and affect the glycolysis of breast cancer, which are inconsistent with the results reported by Yin et al. 14 in colon tumor. However, recent study reported by Rabas et al. 32 suggested that Pink1 promotes invasiveness in breast cancer was related to the package of mitochondrial (mtDNA)-containing extracellular vesicles, but Zhu et al. 33 obtained the conclusion by Pan-analysis that Pink1 presents anti-tumorigenic property in breast cancer but plays a detrimental role in colorectal cancer, and remains controversial in ovarian cancer, and they also pointed out these discrepancies of PINK1 expression cross cancers might be due to the heterogeneity of data collection and different cancer type presents specific biological properties. Therefore, Pink1 is worthy of further study in tumor biology as a multifunctional protein.
Conclusions
In summary, this study demonstrated that Pink1 expression was related to the histological grade of breast cancer tissues, which suggested that Pink1 may be an indicator of malignancy of breast cancer. Moreover, deletion of Pink1 suppressed cell proliferation in MDA-MB-231 cells, and proteomics data revealed the role of Pink1 in glycolysis regulation during the progression of basal-like subtype of breast cancer, which raised another perspective for understanding the regulatory role of Pink1 in breast cancer. However, further investigations are needed to elaborate the specific mechanism of Pink1 in breast cancer.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221082613 for Pink1 promotes cell proliferation and affects glycolysis in breast cancer by Jing Li, Xuting Xu, Huilian Huang, Liqin Li, Jing Chen, Yunfeng Ding and Jinliang Ping in Experimental Biology and Medicine
Acknowledgments
The microarray of human breast and breast cancer tissue specimens in this study were generously prepared by Biobank of Huzhou Central Hospital, Huzhou, 313000.
Footnotes
Authors’ contributions: All authors participated in this study, including experimental design and performance, data analysis and manuscript preparation. YD JP, JL, and JC conducted the experiments and JL wrote the manuscript. HH and XX performed data analysis and image process. LL designed and supervised the research.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the Project of Health Commission of Zhejiang Province [2019KY677] and the Project of Science and Technology of Huzhou City [2018GYB26].
Data availability: The proteomics data involved in this study have been successfully submitted to the ProteomeXchange Consortium via the PRIDEpartner repository (Perez-Rivero et al., 2019) and the identification number of this dataset is PXD027405.
ORCID iD: Jing Li  https://orcid.org/0000-0002-8045-6010
https://orcid.org/0000-0002-8045-6010
Supplemental material: Supplemental material for this article is available online.
References
- 1. Unoki M, Nakamura Y. Growth-suppressive effects of BPOZ and EGR2, two genes involved in the PTEN signaling pathway. Oncogene 2001;20:4457–65 [DOI] [PubMed] [Google Scholar]
- 2. Silvestri L, Caputo V, Bellacchio E, Atorino L, Dallapiccola B, Valente EM, Casari G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet 2005;14:3477–92 [DOI] [PubMed] [Google Scholar]
- 3. Cardona F, Sánchez-Mut JV, Dopazo H, Pérez-Tur J. Phylogenetic and in silico structural analysis of the Parkinson disease-related kinase PINK1. Hum Mutat 2011;32:369–78 [DOI] [PubMed] [Google Scholar]
- 4. Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, González-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004;304:1158–60 [DOI] [PubMed] [Google Scholar]
- 5. Michiorri S, Gelmetti V, Giarda E, Lombardi F, Romano F, Marongiu R, Nerini-Molteni S, Sale P, Vago R, Arena G, Torosantucci L, Cassina L, Russo MA, Dallapiccola B, Valente EM, Casari G. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ 2010;17:962–74 [DOI] [PubMed] [Google Scholar]
- 6. Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H, Hattori N. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett 2010;584:1073–9 [DOI] [PubMed] [Google Scholar]
- 7. Lee KS, Lu B. Targeting PINK1 and MQC in brain tumors. Oncotarget 2014;5:2864–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee KS, Wu Z, Song Y, Mitra SS, Feroze AH, Cheshier SH, Lu B. Roles of PINK1, mTORC2, and mitochondria in preserving brain tumor-forming stem cells in a noncanonical Notch signaling pathway. Genes Dev 2013;27:2642–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Catalá-López F, Suárez-Pinilla M, Suárez-Pinilla P, Valderas JM, Gómez-Beneyto M, Martinez S, Balanzá-Martínez V, Climent J, Valencia A, McGrath J, Crespo-Facorro B, Sanchez-Moreno J, Vieta E, Tabarés-Seisdedos R. Inverse and direct cancer comorbidity in people with central nervous system disorders: a meta-analysis of cancer incidence in 577,013 participants of 50 observational studies. Psychother Psychosom 2014;83:89–105 [DOI] [PubMed] [Google Scholar]
- 10. Ong EL, Goldacre R, Goldacre M. Differential risks of cancer types in people with Parkinson’s disease: a national record-linkage study. Eur J Cancer 2014;50:2456–62 [DOI] [PubMed] [Google Scholar]
- 11. Fujiwara M, Marusawa H, Wang HQ, Iwai A, Ikeuchi K, Imai Y, Kataoka A, Nukina N, Takahashi R, Chiba T. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene 2008;27:6002–11 [DOI] [PubMed] [Google Scholar]
- 12. Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, Janakiraman M, Schultz N, Hanrahan AJ, Pao W, Ladanyi M, Sander C, Heguy A, Holland EC, Paty PB, Mischel PS, Liau L, Cloughesy TF, Mellinghoff IK, Solit DB, Chan TA. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet 2010;42:77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berthier A, Navarro S, Jiménez-Sáinz J, Roglá I, Ripoll F, Cervera J, Pulido R. PINK1 displays tissue-specific subcellular location and regulates apoptosis and cell growth in breast cancer cells. Hum Pathol 2011;42:75–87 [DOI] [PubMed] [Google Scholar]
- 14. Yin K, Lee J, Liu Z, Kim H, Martin DR, Wu D, Liu M, Xue X. Mitophagy protein PINK1 suppresses colon tumor growth by metabolic reprogramming via p53 activation and reducing acetyl-CoA production. Cell Death Differ 2021;28:2421–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kung-Chun Chiu D, Pui-Wah Tse A, Law CT, Ming-Jing Xu I, Lee D, Chen M, Kit-Ho Lai R, Wai-Hin Yuen V, Wing-Sum Cheu J, Wai-Hung Ho D, Wong CM, Zhang H, Oi-Lin Ng I, Chak-Lui Wong C. Hypoxia regulates the mitochondrial activity of hepatocellular carcinoma cells through HIF/HEY1/PINK1 pathway. Cell Death Dis 2019;10:934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Agnihotri S, Golbourn B, Huang X, Remke M, Younger S, Cairns RA, Chalil A, Smith CA, Krumholtz SL, Mackenzie D, Rakopoulos P, Ramaswamy V, Taccone MS, Mischel PS, Fuller GN, Hawkins C, Stanford WL, Taylor MD, Zadeh G, Rutka JT. PINK1 is a negative regulator of growth and the Warburg effect in glioblastoma. Cancer Res 2016;76:4708–19 [DOI] [PubMed] [Google Scholar]
- 17. Martin SA, Hewish M, Sims D, Lord CJ, Ashworth A. Parallel high-throughput RNA interference screens identify PINK1 as a potential therapeutic target for the treatment of DNA mismatch repair-deficient cancers. Cancer Res 2011;71:1836–48 [DOI] [PubMed] [Google Scholar]
- 18. Lee HJ, Jang SH, Kim H, Yoon JH, Chung KC. PINK1 stimulates interleukin-1β-mediated inflammatory signaling via the positive regulation of TRAF6 and TAK1. Cell Mol Life Sci 2012;69:3301–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kang R, Xie Y, Zeh HJ, Klionsky DJ, Tang D. Mitochondrial quality control mediated by PINK1 and PRKN: links to iron metabolism and tumor immunity. Autophagy 2019;15:172–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao Y, Li J, Li J, Hu C, Zhang L, Yan J, Li L, Zhang L. Tetrahydroxy stilbene glycoside alleviated inflammatory damage by mitophagy via AMPK related PINK1/Parkin signaling pathway. Biochem Pharmacol 2020;177:113997. [DOI] [PubMed] [Google Scholar]
- 21. O’Flanagan CH, Morais VA, Wurst W, De Strooper B, O’Neill C. The Parkinson’s gene PINK1 regulates cell cycle progression and promotes cancer-associated phenotypes. Oncogene 2015;34:1363–74 [DOI] [PubMed] [Google Scholar]
- 22. Miyahara K, Takano N, Yamada Y, Kazama H, Tokuhisa M, Hino H, Fujita K, Barroga E, Hiramoto M, Handa H, Kuroda M, Ishikawa T, Miyazawa K. BRCA1 degradation in response to mitochondrial damage in breast cancer cells. Sci Rep 2021;11:8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ, Panel members. Strategies for subtypes–dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 2011;22:1736–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Subik K, Lee JF, Baxter L, Strzepek T, Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG, Tang P. The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer 2010;4:35–41 [PMC free article] [PubMed] [Google Scholar]
- 25. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008;7:11–20 [DOI] [PubMed] [Google Scholar]
- 26. Tang Z, Li L, Tang Y, Xie D, Wu K, Wei W, Xiao Q. CDK2 positively regulates aerobic glycolysis by suppressing SIRT5 in gastric cancer. Cancer Sci 2018;109:2590–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song W, Wang Z, Gu X, Wang A, Chen X, Miao H, Chu J, Tian Y. TRIM11 promotes proliferation and glycolysis of breast cancer cells via targeting AKT/GLUT1 pathway. Onco Targets Ther 2019;12:4975–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao W, Li W, Dai W, Huang N, Qiu J. LINK-A promotes cell proliferation through the regulation of aerobic glycolysis in non-small-cell lung cancer. Onco Targets Ther 2018;11:6071–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo Y, Wei L, Zhou Y, Lu N, Tang X, Li Z, Wang X. Flavonoid GL-V9 induces apoptosis and inhibits glycolysis of breast cancer via disrupting GSK-3beta- modulated mitochondrial binding of HKII. Free Radic Biol Med 2020;146:119–29 [DOI] [PubMed] [Google Scholar]
- 30. Lee M, Yoon JH. Metabolic interplay between glycolysis and mitochondrial oxidation: the reverse Warburg effect and its therapeutic implication. World J Biol Chem 2015;6:148–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yonashiro R, Eguchi K, Wake M, Takeda N, Nakayama K. Pyruvate dehydrogenase PDH-E1beta controls tumor progression by altering the metabolic status of cancer cells. Cancer Res 2018;78:1592–603 [DOI] [PubMed] [Google Scholar]
- 32. Rabas N, Palmer S, Mitchell L, Ismail S, Gohlke A, Riley JS, Tait SWG, Gammage P, Soares LL, Macpherson IR, Norman JC. PINK1 drives production of mtDNA-containing extracellular vesicles to promote invasiveness. J Cell Biol 2021;220:e202006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu L, Wu W, Jiang S, Yu S, Yan Y, Wang K, He J, Ren Y, Wang B. Pan-cancer analysis of the mitophagy-related protein PINK1 as a biomarker for the immunological and prognostic role. Front Oncol 2020;10:569887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221082613 for Pink1 promotes cell proliferation and affects glycolysis in breast cancer by Jing Li, Xuting Xu, Huilian Huang, Liqin Li, Jing Chen, Yunfeng Ding and Jinliang Ping in Experimental Biology and Medicine