Abstract
Combined pituitary hormone deficiency (CPHD) is characterized by deficiency of growth hormone and at least one other pituitary hormone. Pathogenic variants in more than 30 genes expressed during the development of the head, hypothalamus, and/or pituitary have been identified so far to cause genetic forms of CPHD. However, the etiology of around 85% of the cases remains unknown. The aim of this study was to unveil the genetic etiology of CPHD due to congenital hypopituitarism employing whole exome sequencing (WES) in two newborn patients, initially tested and found to be negative for PROP1, LHX3, LHX4 and HESX1 pathogenic variants by Sanger sequencing and for copy number variations by MLPA. In this study, the application of WES in these CPHD newborns revealed the presence of three different heterozygous gene variants in each patient. Specifically in patient 1, the variants BMP4; p.Ala42Pro, GNRH1; p.Arg73Ter and SRA1; p.Gln32Glu, and in patient 2, the SOX9; p.Val95Ile, HS6ST1; p.Arg306Gln, and IL17RD; p.Pro566Ser were identified as candidate gene variants. These findings further support the hypothesis that CPHD constitutes an oligogenic rather than a monogenic disease and that there is a genetic overlap between CPHD and congenital hypogonadotropic hypogonadism.
Keywords: combined pituitary hormone deficiency, congenital hypopituitarism, hypogonadotropic hypogonadism, whole exome sequencing, pathogenic gene variants
1. Introduction
Combined pituitary hormone deficiency (CPHD) is characterized by deficiencies of growth hormone (GH) and at least one more anterior pituitary hormone. The most frequent hormone deficiencies besides that of GH are deficiencies of thyroid stimulating hormone (TSH), the gonadotrophins, luteinizing hormone (LH), follicle stimulating hormone (FSH), and lastly, adrenocorticotropic hormone (ACTH). The normal secretion of LH and FSH is under the control of the gonadotropin releasing hormone (GnRH) produced by the hypothalamus that binds to GnRH receptors in the anterior pituitary. Reduced production and secretion of gonadotrophins leads to hypogonadotropic hypogonadism (HH) [1]. CPHD may either present already in the neonatal period, representing cases of congenital hypopituitarism, or later during life secondary to insults of the hypothalamic–pituitary region, such as tumors or irradiation, leading to the acquired forms of CPHD.
Cases of congenital hypopituitarism are, therefore, diagnosed during the neonatal period, most commonly with the clinical picture of hypoglycemia, due to GH and ACTH/cortisol deficiency, protracted jaundice, due to TSH/thyroxine and cortisol deficiency and, especially in males, micropenis, due to GH and gonadotrophin deficiencies.
In less severe cases, if left undiagnosed beyond the neonatal period, the patient with congenital hypopituitarism may be diagnosed later on with short stature as the first clinical manifestation accompanied by other features such as delayed puberty or metabolic abnormalities.
Neuroimaging may reveal abnormalities of the hypothalamic–pituitary region, mainly hypoplasia of the anterior pituitary, ectopic posterior pituitary, and hypoplastic or absent pituitary stalk resulting in impaired pituitary hormone secretion, while some cases are further characterized by craniofacial malformations [2]. Thus, depending on the hormone deficiencies identified and the underlying etiology, there is a wide spectrum of phenotypic characteristics ranging from mild to more severe. The incidence of congenital forms of CPHD is estimated at 1:8000 live births worldwide [3].
As stated above, the etiology of CPHD might be either genetic or acquired due to trauma, tumor, infection, infiltration or autoimmune diseases affecting the hypothalamic–pituitary region [3,4,5]. As far as the genetic etiology is concerned, several genes, mostly transcription factors, have been identified to be associated with congenital forms of CPHD and participate in the Bmp, Shh, Fgf, Wnt or Notch signaling pathways [6]. Pathogenic variants of the transcription factor genes PROP1, POU1F1, HESX1, LHX3, and LHX4 are the most frequent causes of CPHD [7].
In recent years, the application of next generation sequencing methodologies (NGS), including the screening of large gene panels in many patients simultaneously, and whole exome sequencing have revealed new variants of known CPHD genes and new genes associated with CPHD [8,9,10]. However, the etiology of approximately 85% of the cases still remains unknown [3]. Furthermore, various studies demonstrated that the genetic causes of CPHD and pituitary stalk interruption syndrome (PSIS) are oligogenic rather than monogenic [11,12,13]. Similarly, congenital hypogonadotropic hypogonadism (CHH) is now considered as an oligogenic disease, and it has been shown by various studies that mutations in genes associated with CHH may also be related with CPHD, PSIS and holoprosencephaly, indicating a genetic overlap between these syndromes [14,15,16].
The aim of this study was to unveil the genetic etiology in two newborn patients with congenital hypopituitarism, employing WES.
2. Materials and Methods
2.1. Patients
Two male (46, XY) newborns, diagnosed with CPHD, were referred to the Laboratory of Molecular Endocrinology, Division of Endocrinology, Diabetes and Metabolism, Center for Rare Pediatric Endocrine Diseases, First Department of Pediatrics, National and Kapodistrian University of Athens, School of Medicine, “Aghia Sophia” Children’s Hospital, for genetic testing. The parents of the patients provided written informed consent.
Patient 1 was delivered by caesarean section due to intrauterine growth restriction (IUGR) with a birth weight of 2200 g. Shortly after birth, he presented with refractory hypoglycemia and mild hypotonia. On physical examination, he had micropenis (1 cm) with bilaterally palpable small testes (testes volume: 1 mL measured by orchidometer). Endocrinological workup revealed secondary hypothyroidism, secondary adrenal insufficiency, and hypogonadotropic hypogonadism (HH) (Table 1).
Table 1.
Laboratory hormone tests of the two newborns diagnosed with CPHD.
| Hormones | Patients | Normal Ranges | |
|---|---|---|---|
| 1 | 2 | ||
| TSH | 5.66 | <0.004 | 0.5–5 μUI/mL |
| FT4 | 0.784 | 0.776 | 0.8–1.8 ng/dL |
| ACTH | <1 | 3.49 | 7–63 pg/mL |
| Cortisol | 0.523 | 2.87 | 6.24–18 μg/dL |
| D4-Andostenedione | <0.30 | NA | 0.05–0.45 ng/mL |
| DHEA-S | <15 | 29.4 | 6–21 μg/dL |
| LH | <0.1 | <0.1 | 0–1.3 mIU/mL |
| FSH | 0.123 | 0.226 | 0.1–2.4 mIU/mL |
| Testosterone | <20 | 22.5 | 75–400 ng/dL |
| GH | 4.54 | 1.19 | 5–40 ng/mL |
| IGF-1 | <15 | <0.25 | 15–129 ng/mL |
| PRL | 291.5 | 27.06 | 5–20 ng/mL |
| Aldosterone | 1185 | NA | 50–900 pg/mL |
| Renin | 27.6 | NA | <37 ng/mL/h |
Notes: TSH: Thyroid Stimulating Hormone; FT4: Free Thyroxine; ACTH: Adrenocorticotropic Hormone; DHEA-S: Dehydroepiandrosterone-sulfate; LH: Luteinizing Hormone; FSH: Follicle Stimulating Hormone; GH: Growth Hormone; IGF-1: Insulin-like Growth Factor-1; PRL: Prolactin; NA: Not Available.
MRI scan of the hypothalamic–pituitary region depicted hypoplastic anterior pituitary and ectopic posterior pituitary with absence of the pituitary stalk. The patient was put on substitutive treatment with hydrocortisone, testosterone during the mini-puberty period, levothyroxine and somatropin.
Patient 2 presented with protracted jaundice, direct hyperbilirubinemia, severe episodes of hypoglycemia and severe hypertransaminasemia. Clinical examination revealed a hypoplastic and thin penis (1.5 cm). Ultrasound of the scrotum depicted hypoplastic testicles bilaterally with hydrocele on the right. Hormonal assessment revealed multiple pituitary hormone deficiencies with central hypothyroidism, secondary adrenal insufficiency, hypogonadotropic hypogonadism, and growth hormone deficiency (Table 1). MRI scan depicted a hypoplastic anterior pituitary, an ectopic posterior pituitary lobe and thinning of the pituitary stalk. The patient was treated with substitution of hydrocortisone, thyroxine, testosterone during the mini-puberty period, and somatropin.
Both patients were tested by direct sequencing of the PROP1, LHX3, LHX4 and HESX1 genes, with no pathogenic variant identified. They were also found to be negative for copy number variants (CNV) of the genes GH1, POU1F1, GHRHR, LHX3, LHX4, HESX1 and PROP1 employing multiplex ligation-dependent probe amplification (MLPA) (SALSA MLPA Probemix P216 Growth Hormone Deficiency mix-1; MRC-Holland, Amsterdam, The Netherlands).
The study was approved by the Institutional Scientific and Bioethics Committee and is in accordance with the Declaration of Helsinki.
2.2. DNA Isolation
DNA was extracted from 400 μL peripheral blood leucocytes employing the Maxwell 16 Blood DNA Purification Kit (Promega, Madison, WI, USA), an automated method that uses paramagnetic particles to bind DNA, according to the manufacturer’s instructions.
2.3. Whole Exome Sequencing, Filtering and Bioinformatic Analysis
WES was carried out on an Ion S5™ XL System (ThermoFisher Scientific, Waltham, MA, USA) using the Ion AmpliSeq™ Exome RDY Kit (ThermoFisher Scientific, Waltham, MA, USA). The amount of DNA used for WES was ≥50 ng.
Base calling, alignment to the human genome assembly hg19 and variant calling were performed on Torrent SuiteTM Server according to the manufacturer’s instructions. The variants were annotated by: (1) Ion Reporter (v5.2.0.66; https://ionreporter.thermofisher.com, accessed 17 May 2019) and (2) ANNOVAR (http://annovar.openbioinformatics.org accessed 17 May 2019) through VarAFT software (Variant Annotation and Filter Tool, Version 2.15; https://varaft.eu/, accessed 16 September 2019 and 9 February 2021).
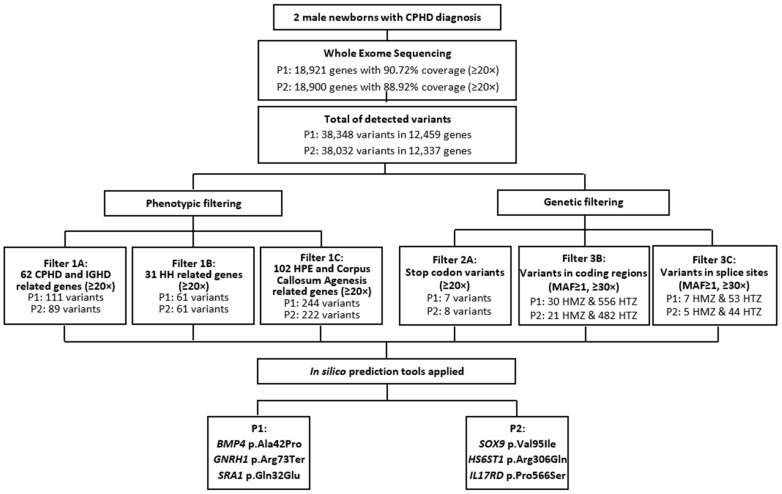
Initial filtering was carried out employing an in silico panel of 194 genes related to CPHD and isolated growth hormone deficiency (IGHD): ARNT2, BMP2, BMP4, CDON, CHD7, FGF10, FGF18, FGF8, FGFR1, GATA2, GHRH, GHRHR, GLI2, GLI3, GLI4, GPR161, HESX1, HHIP, HNRNPU, IFT172, IGSF1, LHX3, LHX4, NEUROD1, NFKB2, NR5A1, OTX2, PAX6, PCSK1, PITX1, PITX2, PNPLA6, POLR3A, POU1F1, PROK1, PROKR2, PROP1, RBM28, ROBO1, SHH, SIX1, SIX2, SIX3, SIX4, SIX5, SIX6, SOX1, SOX2, SOX3, TBX19, TCF7L1, TGIF, TGIF1, TRH, TRHR, TSHB, WDR11, WNT4, WNT5A, ZFHX3, ZSWIM6; HH: ANOS1, AXL, CCDC141, DCAF17, DMXL2, DUSP6, FEZF1, FGF17, FLRT3, GNRH1, GNRHR, HS6ST1, IL17RD, KAL1, KISS1, LEPR, LEP, LHB, NR0B1, NSMF, PROK2, SEMA3A, SOX10, TAC3, TACR3, OTUD4, RNF216, SEMA3E, SEMA7A, SRA1; as well as holoprosencephaly (HPE) and Corpus Callosum Agenesis: AHI1, AKT3, AMPD2, ANOP1, ARID1B, ARL13B, ARX, ASPM, ATR, ATRX, B9D2, BCOR, CASK, CC2D2A, CENPJ, CEP152, CEP290, CEP41, CEP63, CREBBP, CTBP1, DCX, DHCR24, DHCR7, DIS3L2, DISC1, DISP1, DLL1, EFNB1, EOMES, EP300, EPG5, FGFR2, FH, FKRP, FKTN, FLNA, FOXH1, GAS1, GPSM2, GTDC2, HCCS, HHAT, HYLS1, IGBP1, IGF1, INPP5E, ISPD, KAT6B, KCC3, KIF7, L1CAM, LRP2, MED1, MED12, MID1, MKS1, NDE1, NFIX, NIN, NODAL, NPHP1, NPHP3, NSD1, OFD1, OTX1, PDHA1, PDHB, POMGNT1, POMT1, POMT2, PTCH, PYCR1, RAB18, RAB3GAP1, RAB3GAP2, RBBP8, RBM10, RELN, RNU4ATAC, RPGRIP1L, RPS6KA3, SCKL3, SLC12A6, SPG11, SPRY4, STRA6, SUFU, TCTN1, TCTN2, TCTN3, TMEM138, TMEM216, TMEM237, TMEM67, TUBA1A, TUBB2B, TUBB3, VAX1, ZIC2, to select variants with minor allele frequency (MAF) values < 2% and depth ≥ 20× (filter 1). Next, variants resulting in premature stop codons with depth ≥ 20× (filter 2) as well as homozygous and heterozygous variants in coding regions and splice sites with MAF values < 1% and depth ≥ 30× (filter 3) were also checked (Figure 1).
Figure 1.
NGS analysis pipeline and filtering. Abbreviations: P1 = Patient 1; P2 = Patient 2; MAF = Minor Allele Frequency; HMZ = homozygous; HTZ = heterozygous; CPHD = Combined Pituitary Hormone Deficiency, IGHD = Isolated Growth Hormone Deficiency, HH = Hypogonadotropic Hypogonadism, HPE = Holoprosencephaly.
Another 233 genes found after relevant literature search were checked for pathogenic variants: ACVRL1, AES, AGAP1, AHR, AKAP2, AKT1S1, ALK, ALMS1, AMN1, ANK1, ANOS1, ARNT2, ASCL1, ATF2, ATP13A1, AXIN1, BAMBI, BMP1, BMPER, BMPR1A, BMPR1B, BOD1L1, CARTPT, CCKBR, CCN4, CDY1, CEBPB, CGA, CHRD, CPE, CRIM1, CRY1, CSNK1A1, CSNK1D, CSNK2A1, CTNNB1, CXCR4, DAAM1, DEK, DHH, DISP3, DKK3, DLK1, DLL3, DNER, DR1, DVL1, DVL2, E2F4, EID1, ENG, EYA4, EZH2, FAAH2, FANCB, FANCC, FANCD2, FBXW2, FGF1, FGF13, FGF14, FGF4, FGFR1OP, FGFR1OP2, FIBP, FKBP8, FRAT2, FRS3, FSTL1, FSTL5, FUS, FZD2, FZD4, FZD5, FZD6, GAP43, GDNF, GH, GH1, GLG1, GLI1, GLIPR2, GRK1, HES6, HEY1, HIF1A, HMGA2, HMX2, HMX3, ID1, ID2, ID3, ID4, IGSF10, IL6ST, IRS4, ISL1, ITGA2B, JAG1, JUN, KISS1R, KLF4, LAMB2, LDB1, LHX1, LHX2, LHX8, LTBP3, LTBP4, MAMLD1, MASTL, MECP2, MITOL, MLH3, MYO9B, NAALADL1, NBL1, NEUROG2, NKX2-1, NLE1, NOBOX, NOS1, NOTCH1, NOTCH2, NPC1, NPFFR1, NPVF, NR4A1, NRP2, OTP, OVOL1, PALLD, PAX2, PDE3A, PIAS4, PIK3CA, PJA1, PLEKHA5, PLPP3, PLXND1, POFUT1, POLR3B, POLR3D, POU3F2, PPAP2B, PPHLN1, PPM1L, PPRC1, PRKAG1, PRKAR1A, PTCH1, PTCHD2, PTTG1, RARA, RB1, RBPJ, RD3, RNPC3, ROR2, RORA, RXRA, SENP2, SFRP1, SIM1, SIRT1, SLC15A4, SLC20A1, SLC9A3R1, SLX4, SMAD1, SMAD2, SMAD3, SMAD4, SMAD6, SMAD7, SMARCA4, SMURF1, SNIP1, SOSTDC1, SP1, SP3, SPIN2A, SPRED2, SPRY1, SPRY2, SRF, SRY, STAT3, STAT5B, STIL, STRAP, STUB1, SYNPO2, TAB1, TANC2, TBX3, TCF12, TCF3, TCF4, TDGF1, TET2, TGFB2, TGFBI, TGFBR1, TGFBR2, TLE4, TLE5, TMEM231, TNRC6A, TP53, TRAPPC9, TRIP11, TSHZ1, TSPAN11, TSPYL2, TWSG1, TXNDC5, WDR12, WIF1, WISP1, WNT5B, WNT6, WNT9A, YBX1, ZBTB20, ZEB1, ZFY, ZFYVE9, ZIC1, ZNF488, ZNF8, ZSWIM6.
Ion Reporter and VarAFT software gathered the pathogenicity scores from 15 predictive bioinformatic tools. The following in silico tools were further employed for the interpretation of the filtered variants: protein analysis through evolutionary relationships (Panther; http://www.pantherdb.org), and Human Splicing Finder (HSF; http://www.umd.be/HSF3/). The variants were also searched in the VarSome (https://varsome.com/), Ensembl (http://www.ensembl.org), Human Gene Mutation Database (HGMD; www.hgmd.cf.ac.uk) and Single Nucleotide Polymorphism Database (dbSNP; www.ncbi.nlm.nih.gov/snp). These in silico tools were accessed various times over a 2 years period (2019–2021). The classification of the variants’ pathogenicity was performed taking into consideration the information gathered from the above-mentioned databases and in silico tools for the application of the American College of Medical Genetics (ACMG) criteria. The nucleotide sequences were visualized using the software IGV 2.3.91 (Broad Institute; https://software.broadinstitute.org/software/igv/, September 2019 and February 2021).
To verify candidate variants, Sanger sequencing was carried out for the patients and their parents. All variants reported herein were confirmed by Sanger sequencing the corresponding exon of each gene in the patients and their parents.
The frequencies of the candidate variants identified in our patients were also searched in a 500 Greek WES in-house dataset.
3. Results
3.1. Patient 1
Employing WES, 18,921 genes were sequenced with 90.72% coverage (>20×), and 38,348 variants in 12,459 genes were detected.
Three heterozygous variants in the genes, BMP4, GNRH1 and SRA1 related to patient’s phenotype, were identified (Table 2).
Table 2.
Pathogenic variants detected by WES and their interpretation by some of the used bioinformatic tools.
| Patient | Gene | Transcript ID | Nucleotide Variant | Protein Variant | dbSNP | Inheri Tance |
Frequency | ACMG Classification | Prediction Tools | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gnomAD Exomes | 500 Greek Exomes | SIFT | PolyPhen2 | Mutation Taster | CADD | GERP | ||||||||
| 1 | BMP4 | NM_001202.4 | c.124G > C | p.[Ala42Pro;=] | rs140920120 | M | 0.000283 | HTZ:0.002 | VUS | B | LP | B | 22.6 | 5.19 |
| HMZ: 0 | (BP4, PS3) | |||||||||||||
| GNRH1 | NM_000825.3 | c.217C > T | p.[Arg73Ter;=] | rs375970738 | P | 0.00000805 | 0 | LP | - | - | - | 14.98 | 4.91 | |
| (PVS1, PM2) | ||||||||||||||
| SRA1 | NM_001035235.4 | c.94C > G | p.[Gln32Glu;=] | rs35610885 | M | 0.00715 | HTZ: 0.008 | VUS | P | LP | DC | 27.6 | 5.01 | |
| HMZ: 0 | (BP4, PP5) | |||||||||||||
| 2 | SOX9 | NM_000346.4 | c.283G > A | p.[Val95Ile;=] | Novel | M | NA | 0 | VUS | B | LP | DC | 23.3 | 4.24 |
| (PM2, BP4) | ||||||||||||||
| HS6ST1 | NM_004807.3 | c.917G > A | p.[Arg306Gln;=] | rs201307896 | M | 0.000706 | 0 | P | B | LP | DC | 29.9 | 4.78 | |
| (PS3, PM2, PM5, PP5, PP3) | ||||||||||||||
| IL17RD | NM_017563.5 | c.1696C > T | p.[Pro566Ser;=] | rs61742267 | M | 0.0144 | HTZ: 0.028 | VUS | B | LP | DC | 22.6 | 5.64 | |
| HMZ: 0 | (PM5, BP4) | |||||||||||||
Abbreviations: dbSNP = database Single Nucleotide Polymorphism, gnomAD = Genome Aggregation Database, ACMG = American College of Medical Genetics, SIFT = Sorting Intolerant From Tolerant, Polyphen2 = Polymorphism Phenotyping v2, CADD = Combined Annotation-Dependent Depletion, GERP = Genomic Evolutionary Rate Profiling, M = Maternal, P = Paternal, NA: not annotated, VUS = Variant of Uncertain Significance, B:Benign, P = Pathogenic, LP = Likely Pathogenic, DC = Disease Causing, PD = Probably Damaging, HTZ = Heterozygous, HMZ = Homozygous.
Notes: CADD: value > 20 = Deleterious, GERP: −12.3 = less conserved to 6.17 = more conserved, ACMG classification: P = Pathogenic, LP = Likely Pathogenic, and VUS = Variant of Uncertain Significance. The BMP4 variant c.124G > C, p.Ala42Pro (193 ×), of maternal inheritance, was identified by the CPHD gene panel employed. Residue Ala42 is conserved and the p.Ala42Pro is a rare variant (f = 0.000283). According to ACMG criteria, it is classified as Variant of Unknown Significance (VUS).
The GNRH1 variant c.217C > T, p.Arg73Ter (58×), of paternal inheritance, was identified by the CHH gene panel. It is a nonsense variant resulting in protein truncation by 19 amino acids. It is an ultrarare variant with frequency in gnomAD Exomes 0.00000805. According to ACMG criteria, it is classified as likely pathogenic.
The SRA1 variant c.94C > G, p.Gln32Glu (200×), of maternal inheritance, was identified by the CHH gene panel. Residue Gln32 is highly conserved among species, and the in silico conservation tools Panther and GERP classified this variant as pathogenic. According to ACMG criteria, it is classified as VUS.
3.2. Patient 2
Employing WES, sequencing was achieved for 18,900 genes with 88.92% coverage (>20×), and 38,032 variants in 12,337 genes were detected.
Three heterozygous variants in the genes SOX9, HS6ST1 and IL17RD, related to patient’s phenotype, were identified (Table 2).
The novel SOX9 gene variant c.283G > A, p.Val95Ile (82×), of maternal inheritance, was identified by the CHH gene panel. It was found to be pathogenic by PolyPhen2, Mutation Taster, DANN and CADD bioinformatic tools. Residue Val95 is conserved, and the in silico conservation tools Panther and GERP classified it as pathogenic. According to ACMG criteria, it is classified as VUS.
The HS6ST1 gene variant c.917G > A, p.Arg306Gln (143×), of maternal inheritance, was identified by the CHH gene panel employed. According to the ACMG criteria, it is classified as pathogenic.
The IL17RD gene variant c.1696C > T, p.Pro566Ser (37×), of maternal inheritance, was identified by the CHH gene panel. Its gnomAD Exomes frequency is 0.0144, and it is classified as VUS according to ACMG criteria.
4. Discussion
The genetic etiology of CPHD is heterogeneous, and patients present complex phenotypes. Despite the wide application of WES in large cohorts of patients, most cases still remain without genetic identification. In recent years, various studies revealed a possible oligogenic or multigenic basis for CPHD.
In this study, the application of WES in two CPHD newborns revealed the presence of several gene variants mostly associated with CHH and the identification of a novel SOX9 variant.
Patient 1 was found to be heterozygote for variants in three genes, the BMP4, GNRH1 and SRA1.
The bone morphogenic protein-4 (BMP4), a member of the BMP family that belongs to the transforming growth factor-beta (TGF-β) family, plays a significant role in early organogenesis, pituitary development and function [17]. BMP4 activity is required for initial organ commitment and development of the pituitary anlage. Particularly BMP4 expression in early development contributes to the formation of Rathke’s pouch in the mouse, while BMP4 together with BMP2, and 7 later secreted by surrounding tissues, contribute to the polarization of the pouch [18]. Inhibition of Bmp signaling in the oral ectoderm of mouse embryos disrupts pituitary cell specification [19].
The BMP4 gene encodes a 408 amino acid protein that consists of an N-terminal signal peptide, which directs the protein to the secretory pathway, a prodomain that ensures proper folding, and the C-terminal mature peptide [20].
The p.Ala42Pro BMP4 gene variant identified in our patient is located in the highly conserved prodomain part of the protein and up to now has been classified as a VUS according to the ACMG criteria. Various BMP4 variants have been reported in patients with tooth agenesis, amongst them the BMP4 variant p.Ala42Pro identified in our patient and a female patient and her mother with tooth agenesis [21,22]. Other variants also found in the prodomain region have been reported in patients with a wide spectrum of phenotypes, such as Stickler syndrome, anophthalmia, ocular malformation, cleft palate and scleroma, as well as colorectal cancer predisposition [23,24,25,26].
BMP4 gene variants have been associated with a wide range of phenotypic characteristics, including ocular, craniofacial, finger and urogenital abnormalities, and psychomotor retardation, as well as CPHD [23,27,28,29,30]. Specifically, the BMP4 variants p.Arg300Pro (VUS) and de novo p.Trp265Ter have been identified in heterozygosity in two patients with CPHD; on neuroimaging, these patients presented hypoplastic anterior pituitary gland, ectopic posterior pituitary, and absent pituitary stalk, as is the case in our patient, while clinically they presented sclerosed nodules at the hands, short metacarpals IV, azoospermia [29] and, GH and TSH deficiencies [30]. After exome sequencing in 20 patients (trio analysis) with isolated pituitary stalk interruption syndrome (PSIS), the BMP4 variant p.Ala334Asp was identified together with a SIX6 gene variant p.Glu129Lys (and DCHS1 gene variants) in a patient who presented with growth retardation, pituitary hormone deficiencies and MRI findings of ectopic posterior pituitary, small stalk and anterior pituitary [11].
The GNRH1 gene encodes a preproprotein that generates the GnRH decapeptide, a member of the gonadotropin-releasing hormone (GnRH) family of peptides. It consists of four exons, three of which are translated, and encodes a 92 amino acid preprohormone that is organized to a signal sequence, followed by the GnRH decapeptide, a conserved GKR cleavage site, and the GnRH-associated peptide (GAP). The decapeptide is secreted in a pulsatile manner by the GnRH neurons in the hypothalamus and triggers the synthesis and secretion of LH and FSH by the anterior pituitary gonadotrophs [31].
The GNRH1 pathogenic variant p.Arg73Ter was identified in heterozygosity in our patient 1, inherited from his father. Residue R73 resides in the GAP region of the protein and results in its truncation by 19 amino acids, and although a nonsense variant, it is considered as VUS since the resulting protein truncation leaves the GnRH decapeptide intact.
This variant has been previously reported in heterozygosity in a male patient investigated for absent puberty at the age of 17 6/12 years [32]. Chan and coworkers speculated that, since the premature stop codon is close to a splice junction, it may escape nonsense-mediated decay and, if this is the case, an abnormal protein is produced. Although, the effect of this variant on GNRH1 synthesis is unclear, the authors suggested that the CHH of this family with the p.Arg73Ter might be of autosomal dominant inheritance with variable expressivity since the father, heterozygous for the variant, exhibited a mild phenotype of delayed puberty [32].
Furthermore, recently a homozygous splice site variant of exon 2 (c.142-2A > C), damaging the natural acceptor site and activating a cryptic acceptor site, was described in a patient with lack of pubertal development and bilateral cryptorchidism, surgically corrected in childhood. In the patient’s lymphocytes, an aberrant GNRH1 transcript was detected, harboring a four bp deletion resulting in a premature stop codon at amino acid 74. The authors speculated that this may lead to mRNA nonsense mediated decay indicating a probable functional importance of the GAP region of GNRH1 [33].
Only a few GNRH1 gene pathogenic variants have been detected to date in patients with CHH, even though GnRH is the central regulator in reproductive function and sexual development. Thus far, four variants have been identified in the decapeptide encoding sequence, the p.Gly29GlyfsTer12 and p.Arg31His in homozygosity, and the p.Trp26Ter and p.Arg31Cys in heterozygosity [32,34,35]. The remaining variants have been found in heterozygosity either in the highly conserved signal peptide (p.Val18Met) or in the GAP (p.Thr58Ser and p.Arg73Ter) [32].
Our patient was very young (infant), and his severe micropenis and hypoplastic testicles along with the absence of LH and FSH surge during his mini-puberty suggest impaired future reproductive function However, since this variant is paternally inherited, we can assume that this variant without the synergistic effect of other reported variants identified in our patient, cannot lead to impaired reproductive capacity, if isolated.
The CHH phenotype of our patient, consisting of micropenis, small testes and low LH, FSH and testosterone, cannot be attributed solely to the GNRH1 p.Arg73Ter heterozygous variant, since hypogonadotropic hypogonadism 12 with or without anosmia (MIM Number 614841) is dominantly inherited. However, we may speculate that our patient’s phenotype might be due to a synergistic effect with the second CHH variant identified, the SRA1 p.Gln32Glu. Although heterozygous variants of the GNRH1 gene have been reported in the literature, no oligogenic case of GNRH1 has been reported so far, whereas this is not the case for GNRHR [36]. Furthermore, in a recent study applying WES in a large cohort of GnRH deficient Greek patients, no GNRH1 variant was identified either as a monogenic or as an oligogenic cause of CHH [37].
The third variant identified in our patient 1 was the SRA1 gene variant p.Gln32Glu, which has been previously reported in compound heterozygosity in an IHH Turkish male patient in trans with the p.Ile179Thr variant [38]. It has also been identified after WES in a heterozygous 46,XY NR5A1 patient, together with six other gene variants, supporting the concept of an oligogenic basis for DSD phenotypes [39].
The SRA1 gene plays a dual role, encoding a long non-coding RNA and a conserved steroid receptor RNA activator protein (SRAP), which, among its other functions, regulates steroid receptor-dependent gene expression [40]. SRA1 as a steroid receptor coactivates thyroid hormone, androgen, estrogen, progesterone, glucocorticoid and retinoic acid receptors [41,42,43,44]. It has also been shown that DAX-1 functions by binding to the RNA coactivator SRA and p160 coactivator proteins and that SRA1 is important to stabilize complexes of SF-1 and DAX-1 for the recruitment of p160 coactivators in order to regulate target gene expression during steroidogenesis [45].
To date, various SRA1 gene variants, p.Pro20Leu, p.Tyr35Asn, p.Arg126His and p.Ile179Thr, have been identified in homozygosity, compound heterozygosity, heterozygosity or as digenically inherited with PNPLA6, RNF216 and SEMA7A gene variants in patients with CHH [38,46,47]. All but one of the patients reported with SRA1 variants are of Turkish, Cypriot, and Greek (this report) origin, indicating the possibility of a founder effect in the East Mediterranean area.
In Patient 2, three heterozygous variants in the genes SOX9, HS6ST1 and IL17RD were identified.
The variant p.Val95Ile of the SOX9 gene identified in patient 2 is a novel variant. SOX9 gene encodes the transcription factor SOX9, which is a member of the SRY-related HMG box (SOX) family of transcription factors. It is essential for both sex and skeletal development and is expressed in various tissues during embryogenesis, such as the cartilage, brain, pituitary, heart, lungs, pancreas, testes, etc. SOX proteins play an important role in the differentiation, establishment or maintenance of stem and progenitor cells of various tissues [48]. Various studies have reported the presence of potential populations of stem cells in the pituitary. In particular, it has been shown by in vitro studies that a small population of progenitor cells in the adult pituitary gland express SOX2 and SOX9 [49]. Furthermore, it was demonstrated that SOX2 and SOX9 expressing progenitor cells can self-renew and give rise to endocrine cells in vivo, suggesting that they are tissue stem cells, playing a role in the physiological maintenance of the pituitary gland as well as in the induction of pituitary tumors [50,51].
In mammals, during embryonic gonadal development, the factors SRY and SF-1 induce the expression of the SOX9 gene. SOX9 itself then suppresses the expression of the SRY gene, while it takes over the differentiation of the bipotential gonad to testis. Finally, SOX9, together with SF-1, induces the differentiation of Sertoli cells in the testes [52]. The role of SOX9 in testis formation and sex determination was first recognized in patients with campomelic dysplasia, of whom around 75% with a 46, XY male karyotype bearing one mutant SOX gene exhibit male-to-female sex reversal [53].
During embryonic development in mice (e13.5–14.5), the SOX9 protein is expressed in the migratory nerve cells of the brain, which are distributed in the posterior and mainly in the middle of the brain, while later on, SOX9 is expressed in cartilage and bone [54]. Thus, the SOX9 protein has been found to be involved in the formation of craniofacial structures, and patients with gene variants present with micrognathia, cleft lip and palate, and craniofacial malformations [55].
The HS6ST1 gene encodes the enzyme heparan sulfate 6-O-sulfotransferase 1 (HS6ST1), which participates in the biosynthesis of heparan sulfates (HSs). HSs are expressed in different developmental stages of several tissues and are involved in cell communication regulating the binding of many ligands–receptors, such as FGF8–FGFR1 [56,57]. The impaired interaction of the FGF8 ligand with the FGFR1 receptor is known to be involved in the pathogenesis of HH [58].
Pathogenic variants in the KAL1 gene, also involved in the FGFR signaling pathway, are the main genetic cause of CHH. However, pathogenic variants of the HS6ST1 gene are also common in patients with normosmic/anosmic IHH (approximately 2%), resembling that of KAL1-HH phenotype, but also as an oligogenic cause of CHH together with FGFR1 and NELF genes [36,59,60].
The HS6ST1 gene pathogenic variant, c.917G > A, p.Arg306Gln, was detected in patient 2 and his mother. A variant in the same codon resulting in a different amino acid, c.917G > A, p.Arg306Trp (reported as p.Arg296Trp), has been previously reported in a male patient with Kallmann syndrome and small pituitary gland [59], as was the case in our patient. The patient’s mother presented delayed puberty without anosmia, indicating an incomplete penetrance of this variant [59]. It was also shown by in vitro experiments that the activity of the p.Arg306Gln HS6ST1 mutated enzyme is significantly reduced compared to the wild type enzyme [59].
The IL17RD gene (Interleukin 17 Receptor D) encodes a 739 amino acid membrane protein, member of the interleukin 17 receptor protein family, that also regulates the signaling pathway of FGF8/FGFR1, and its pathogenic variants have been associated with CHH with or without anosmia and delayed puberty and in some cases with deafness [59,61].
The IL17RD p.Pro566Ser identified herein has not been reported previously in CHH patients. It has been reported in a patient with evening fatigue [62], which might be due to subnormal cortisol secretion as a plausible explanation for his fatigue, although cortisol secretion has not been investigated in the patient. However, a homozygous c.1697C > T variant that results in a different amino acid change of the same codon, p.Pro566Leu, classified as probably pathogenic, has been reported in a normosmic CHH patient [33]. Two patients carrying other probably pathogenic variants of the IL17RD gene were identified after exome sequencing in 37 PSIS families. In these patients, the IL17RD variants were co-inherited with variants in other genes; specifically, one carried a GLI2 and an IL17RD variant, whereas the other carried variants in three genes, PROP1, IL17RD and SMARCA2 [56]. Although it is highly probable that the combination of several gene variants may lead to various clinical phenotypes, the contribution of each gene variant to the patient’s phenotype was not investigated in detail and, therefore, could not be established.
5. Conclusions
In this study, each patient was found to harbor three possible causative gene variants, after WES. Although to date numerous studies have shown that CPHD as well as PSIS have an oligogenic rather than a monogenic etiology [11,12,63,64,65], such a conclusion could not be reached in this work.
It is of interest that most of the variants detected in this study were related to CHH, indicating that there is a significant phenotypic and genetic overlap between CHH and CPHD that has also been observed in other studies [66], further supporting the notion that CPHD and CHH share common genetic background [66] and allowing us to speculate that a synergistic action of these gene variants may underlie our patient’s phenotype.
Among the six genes identified, only two (BMP4 and SOX9) were expressed in the pituitary gland, and four genes (GNRH1, SRA1, HS6ST1 and IL17RD), two for each patient, are known CHH genes. However, it is worth stating that two of these genes, HS6ST1 and IL17RD, belong to the FGF signaling network, which plays a major role in the hypothalamic–pituitary axis. Thus far, FGFR1 and FGF8 gene variants have been identified to be causative genes not only for Kallmann syndrome, but also for CPHD, septo-optic dysplasia and holoprosencephaly [14,15,67]. Furthermore, the fact that the HS6ST1 variant, p.Arg306Gln, identified in our patient has also been previously reported in a CHH patient with small pituitary gland [16] strengthens the possible role of this gene in our patient’s CPHD phenotype.
None of the identified variants herein was a de novo one. Patient 1 inherited two CHH variants, one from each parent, and a CPHD variant (BMP4), inherited from his mother. Patient 2 inherited the three variants identified from his mother, who to our knowledge does not present any of her son’s phenotypic characteristics, as is the case for patient’s 1 parents. Therefore, we can speculate that this is due either to incomplete penetrance of the gene variants, as has been previously reported for CPHD [3,11,63], or to the involvement of other etiologic factors, genetic or environmental.
Furthermore, it is highly probable that apart from the variants reported in this work, there might also exist other gene variants acting synergistically. These variants could have escaped identification either because of the WES methodological limitations, or because they are variants of genes yet unknown to be related with CPHD.
This work has several limitations, the main one being the small number of patients studied and the lack of functional studies that could possibly delineate the impact of each variant identified in the patients’ phenotypes. There are also methodological drawbacks: WES has a diagnostic turnover of 35–50%, which might be due either to technical limitations (incomplete coverage of all exons, short length reads, sequencing of GC-rich regions, repetitive sequences, and sequences with high homology), or to methodological limitations such as inability to detect intronic or structural variants, as well as the limitations of data analysis with respect to the VUS, particularly in genes with unknown function. In this study, four variants were classified as VUS; however, considering their frequency and the in silico predictions, their possible contribution to an oligogenic model of CPHD inheritance cannot be overlooked.
Further work is required to fully delineate the genetic basis of CPHD, for example, with large cohorts of patients as well as other methodologies such as whole genome sequencing, a more comprehensive sequencing option, since it provides analysis of the entire genome or the long-read WES methodology. However, the presentation of the identified gene variants in the international literature may provide further evidence to researchers for the critical role of oligogenic combinations in the pathogenesis of complex human disorders of varied penetrance and severity.
As the mystery of the genetic basis of congenital hypopituitarism is slowly unraveled, many new variants and complex combinations of variants of different genes are expected to be added to the still incomplete list of genes involved in pituitary ontogenesis and function.
Acknowledgments
We would like to express our thanks to the patients’ families and to N. Marinakis, Laboratory of Medical Genetics, National and Kapodistrian University of Athens, Athens, Greece for providing the Greek exome data.
Author Contributions
Conceptualization, A.S. and C.K.-G.; methodology, A.S. and E.B.T.; software, E.B.T. and I.F.; validation, E.B.T. and I.F.; formal analysis, A.S. and E.B.T.; clinical investigation, I.A.V., E.N., N.I., T.S. and C.K.-G.; resources, C.K.-G.; writing—original draft preparation, E.B.T. and A.S.; writing—review and editing, E.B.T., A.S. and C.K.-G.; visualization, E.B.T., A.S. and C.K.-G.; supervision, C.K.-G.; project administration, A.S.; funding acquisition, E.B.T. and C.K.-G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of “Agia Sofia” Children’s Hospital, Athens Greece (protocol code 15026/24-6-16 and date of approval 24 June 2016).
Informed Consent Statement
Informed consent was obtained from the parents of patients involved in the study.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
E.B.T. was supported by the Hellenic Foundation for Research and Innovation (HFRI) and the General Secretariat for Research and Technology (GSRT), under the HFRI PhD Fellowship grant (GA. no. 74156/2017).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Turkyilmaz A., Cayir A., Yarali O., Kurnaz E., Baykan E.K., Ates E.A., Demirbilek H. Clinical characteristics and molecular genetic analysis of a cohort with idiopathic congenital hypogonadism. J. Pediatr. Endocrinol. Metab. 2021;34:771–780. doi: 10.1515/jpem-2020-0590. [DOI] [PubMed] [Google Scholar]
- 2.Mehta A., Dattani M.T. Developmental disorders of the hypothalamus and pituitary gland associated with congenital hypopituitarism. Best Pract. Res. Clin. Endocrinol. Metab. 2008;22:191–206. doi: 10.1016/j.beem.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Fang Q., George A.S., Brinkmeier M.L., Mortensen A.H., Gergics P., Cheung L.Y.M., Daly A., Ajmal A., Millan M.P., Ozel A.B., et al. Genetics of Combined Pituitary Hormone Deficiency: Roadmap into the Genome Era. Endocr. Rev. 2016;37:636–675. doi: 10.1210/er.2016-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz A., Urban R. Neuroendocrine consequences of traumatic brain injury. Curr. Opin. Endocrinol. Diabetes Obes. 2013;20:354–358. doi: 10.1097/MED.0b013e32836318ba. [DOI] [PubMed] [Google Scholar]
- 5.de Bree R., Lips P., Leemans C.R. The need for patients endocrine function vigilance following treatment of head and neck cancer. Curr. Opin. Otolaryngol. Head Neck Surg. 2008;16:154–157. doi: 10.1097/MOO.0b013e3282f4479d. [DOI] [PubMed] [Google Scholar]
- 6.Simm F., Griesbeck A., Choukair D., Weiß B., Paramasivam N., Klammt J., Schlesner M., Wiemann S., Martinez C., Hoffmann G.F., et al. Identification of SLC20A1 and SLC15A4 among other genes as potential risk factors for combined pituitary hormone deficiency. Genet. Med. 2018;20:728–736. doi: 10.1038/gim.2017.165. [DOI] [PubMed] [Google Scholar]
- 7.Larson A., Nokoff N.J., Meeks N.J.L. Genetic causes of pituitary hormone deficiencies. Discov. Med. 2015;19:175–183. [PubMed] [Google Scholar]
- 8.Budny B., Zemojtel T., Kaluzna M., Gut P., Niedziela M., Obara-Moszynska M., Rabska-Pietrzak B., Karmelita-Katulska K., Stajgis M., Ambroziak U., et al. SEMA3A and IGSF10 Are Novel Contributors to Combined Pituitary Hormone Deficiency (CPHD) Front. Endocrinol. 2020;11:368. doi: 10.3389/fendo.2020.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinjo K., Nagasaki K., Muroya K., Suzuki E., Ishiwata K., Nakabayashi K., Hattori A., Nagao K., Nozawa R.-S., Obuse C., et al. Rare variant of the epigenetic regulator SMCHD1 in a patient with pituitary hormone deficiency. Sci. Rep. 2020;10:10985. doi: 10.1038/s41598-020-67715-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elizabeth M.S., Verkerk A.J., Hokken-Koelega A.C., Verlouw J.A., Argente J., Pfaeffle R., Visser T.J., Peeters R.P., De Graaff L.C. Unique near-complete deletion of GLI2 in a patient with combined pituitary hormone deficiency and post-axial polydactyly. Growth Horm. IGF Res. 2020;50:35–41. doi: 10.1016/j.ghir.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Zwaveling-Soonawala N., Alders M., Jongejan A., Kovačič L., Duijkers F.A., Maas S.M., Fliers E., Van Trotsenburg A.S.P., Hennekam R.C. Clues for Polygenic Inheritance of Pituitary Stalk Interruption Syndrome From Exome Sequencing in 20 Patients. J. Clin. Endocrinol. Metab. 2018;103:415–428. doi: 10.1210/jc.2017-01660. [DOI] [PubMed] [Google Scholar]
- 12.Jee Y.H., Gangat M., Yeliosof O., Temnycky A.G., Vanapruks S., Whalen P., Gourgari E., Bleach C., Yu C.H., Marshall I., et al. Evidence That the Etiology of Congenital Hypopituitarism Has a Major Genetic Component but Is Infrequently Monogenic. Front. Genet. 2021;12:697549. doi: 10.3389/fgene.2021.697549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brauner R., Bignon-Topalovic J., Bashamboo A., McElreavey K. Pituitary stalk interruption syndrome is characterized by genetic heterogeneity. PLoS ONE. 2020;15:e0242358. doi: 10.1371/journal.pone.0242358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCabe M.J., Gaston-Massuet C., Tziaferi V., Gregory L.C., Alatzoglou K.S., Signore M., Puelles E., Gerrelli D., Farooqi I.S., Raza J., et al. Novel FGF8 Mutations Associated with Recessive Holoprosencephaly, Craniofacial Defects, and Hypothalamo-Pituitary Dysfunction. J. Clin. Endocrinol. Metab. 2011;96:E1709–E1718. doi: 10.1210/jc.2011-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raivio T., Avbelj M., McCabe M.J., Romero C.J., Dwyer A., Tommiska J., Sykiotis G., Gregory L.C., Diaczok D., Tziaferi V., et al. Genetic Overlap in Kallmann Syndrome, Combined Pituitary Hormone Deficiency, and Septo-Optic Dysplasia. J. Clin. Endocrinol. Metab. 2012;97:E694–E699. doi: 10.1210/jc.2011-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kałużna M., Budny B., Rabijewski M., Kałużny J., Dubiel A., Trofimiuk-Müldner M., Wrotkowska E., Hubalewska-Dydejczyk A., Ruchała M., Ziemnicka K. Defects in GnRH Neuron Migration/Development and Hypothalamic-Pituitary Signaling Impact Clinical Variability of Kallmann Syndrome. Genes. 2021;12:868. doi: 10.3390/genes12060868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wozney J.M., Rosen V., Celeste A.J., Mitsock L.M., Whitters M.J., Kriz R.W., Hewick R.M., Wang E.A. Novel regulators of bone formation: Molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 18.Ericson J., Norlin S., Jessell T., Edlund T. Integrated FGF and BMP signaling controls the progression of progenitor cell differentiation and the emergence of pattern in the embryonic anterior pituitary. Development. 1998;125:1005–1015. doi: 10.1242/dev.125.6.1005. [DOI] [PubMed] [Google Scholar]
- 19.Treier M., Gleiberman A.S., O’Connell S.M., Szeto D.P., McMahon J.A., McMahon A.P., Rosenfeld M.G. Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev. 1998;12:1691–1704. doi: 10.1101/gad.12.11.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sieber C., Kopf J., Hiepen C., Knaus P. Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. 2009;20:343–355. doi: 10.1016/j.cytogfr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y., Lu Y., Mues G., Wang S., Bonds J., D’Souza R. Functional evaluation of a novel tooth agenesis-associated bone morphogenetic protein 4 prodomain mutation. Eur. J. Oral Sci. 2013;121:313–318. doi: 10.1111/eos.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu M., Wang H., Fan Z., Xie C., Liu H., Liu Y., Han D., Wong S.-W., Feng H. BMP4 mutations in tooth agenesis and low bone mass. Arch. Oral Biol. 2019;103:40–46. doi: 10.1016/j.archoralbio.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki S., Marazita M.L., Cooper M.E., Miwa N., Hing A., Jugessur A., Natsume N., Shimozato K., Ohbayashi N., Suzuki Y., et al. Mutations in BMP4 Are Associated with Subepithelial, Microform, and Overt Cleft Lip. Am. J. Hum. Genet. 2009;84:406–411. doi: 10.1016/j.ajhg.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nixon T.R.W., Richards A., Towns L.K., Fuller G., Abbs S., Alexander P., McNinch A., Sandford R.N., Snead M.P. Bone morphogenetic protein 4 (BMP4) loss-of-function variant associated with autosomal dominant Stickler syndrome and renal dysplasia. Eur. J. Hum. Genet. 2019;27:369–377. doi: 10.1038/s41431-018-0316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lubbe S.J., Pittman A., Matijssen C., Twiss P., Olver B., Lloyd A., Qureshi M., Brown N., Nye E., Stamp G., et al. Evaluation of germline BMP4 mutation as a cause of colorectal cancer. Hum. Mutat. 2011;32:E1928–E1938. doi: 10.1002/humu.21376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R.N., Green J., Wang Z., Deng Y., Qiao M., Peabody M., Zhang Q., Ye J., Yan Z., Denduluri S., et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014;1:87–105. doi: 10.1016/j.gendis.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reis L.M., Tyler R.C., Schilter K.F., Abdul-Rahman O., Innis J.W., Kozel B.A., Schneider A.S., Bardakjian T.M., Lose E.J., Martin D.M., et al. BMP4 loss-of-function mutations in developmental eye disorders including SHORT syndrome. Qual. Life Res. 2011;130:495–504. doi: 10.1007/s00439-011-0968-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bakrania P., Efthymiou M., Klein J.C., Salt A., Bunyan D.J., Wyatt A., Ponting C.P., Martin A., Williams S., Lindley V., et al. Mutations in BMP4 Cause Eye, Brain, and Digit Developmental Anomalies: Overlap between the BMP4 and Hedgehog Signaling Pathways. Am. J. Hum. Genet. 2008;82:304–319. doi: 10.1016/j.ajhg.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitfeld J., Martens S., Klammt J., Schlicke M., Pfäffle R., Krause K., Weidle K., Schleinitz D., Stumvoll M., Führer D., et al. Genetic analyses of bone morphogenetic protein 2, 4 and 7 in congenital combined pituitary hormone deficiency. BMC Endocr. Disord. 2013;13:56. doi: 10.1186/1472-6823-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez-Contreras F.J., Marbán-Calzón M., Vallespín E., del Pozo Á., Solís-López M., Lobato-Vidal N., Fernández-Elvira M., Rex-Romero M.D.V., Heath K.E., González-Casado I., et al. Loss of function BMP4 mutation supports the implication of the BMP/TGF-β pathway in the etiology of combined pituitary hormone deficiency. Am. J. Med. Genet. Part A. 2019;179:1591–1597. doi: 10.1002/ajmg.a.61201. [DOI] [PubMed] [Google Scholar]
- 31.Boehm U., Bouloux P.-M., Dattani M.T., de Roux N., Dodé C., Dunkel L., Dwyer A., Giacobini P., Hardelin J.-P., Juul A., et al. European Consensus Statement on congenital hypogonadotropic hypogonadism—pathogenesis, diagnosis and treatment. Nat. Rev. Endocrinol. 2015;11:547–564. doi: 10.1038/nrendo.2015.112. [DOI] [PubMed] [Google Scholar]
- 32.Chan Y.-M., de Guillebon A., Lang-Muritano M., Plummer L., Cerrato F., Tsiaras S., Gaspert A., Lavoie H.B., Wu C.-H., Crowley W.F., et al. GNRH1 mutations in patients with idiopathic hypogonadotropic hypogonadism. Proc. Natl. Acad. Sci. USA. 2009;106:11703–11708. doi: 10.1073/pnas.0903449106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amato L.G.L., Montenegro L.R., Lerario A.M., Jorge A.A.L., Junior G.G., Schnoll C., Renck A.C., Trarbach E.B., Costa E.M.F., Mendonca B.B., et al. New genetic findings in a large cohort of congenital hypogonadotropic hypogonadism. Eur. J. Endocrinol. 2019;181:103–119. doi: 10.1530/EJE-18-0764. [DOI] [PubMed] [Google Scholar]
- 34.Francou B., Paul C., Amazit L., Cartes A., Bouvattier C., Albarel F., Maiter D., Chanson P., Trabado S., Brailly-Tabard S., et al. Prevalence of KISS1 Receptor mutations in a series of 603 patients with normosmic congenital hypogonadotrophic hypogonadism and characterization of novel mutations: A single-centre study. Hum. Reprod. 2016;31:1363–1374. doi: 10.1093/humrep/dew073. [DOI] [PubMed] [Google Scholar]
- 35.Brachet C., Gernay C., Boros E., Soblet J., Vilain C., Heinrichs C. Homozygous p.R31H GNRH1 mutation and normosmic congenital hypogonadotropic hypogonadism in a patient and self-limited delayed puberty in his relatives. J. Pediatr. Endocrinol. Metab. 2020;33:1237–1240. doi: 10.1515/jpem-2020-0207. [DOI] [PubMed] [Google Scholar]
- 36.Sykiotis G.P., Plummer L., Hughes V.A., Au M., Durrani S., Nayak-Young S., Dwyer A.A., Quinton R., Hall J.E., Gusella J.F., et al. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc. Natl. Acad. Sci. USA. 2010;107:15140–15144. doi: 10.1073/pnas.1009622107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.I Stamou M., Varnavas P., Plummer L., Koika V., Georgopoulos N.A. Next-generation sequencing refines the genetic architecture of Greek GnRH-deficient patients. Endocr. Connect. 2019;8:468–480. doi: 10.1530/EC-19-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotan L.D., Cooper C., Darcan Ş., Carr I.M., Özen S., Yan Y., Hamedani M.K., Gürbüz F., Mengen E., Turan I., et al. Idiopathic Hypogonadotropic Hypogonadism Caused by Inactivating Mutations in SRA. J. Clin. Res. Pediatr. Endocrinol. 2016;8:125–134. doi: 10.4274/jcrpe.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camats N., Fernández-Cancio M., Audí L., Schaller A., Flück C.E. Broad phenotypes in heterozygous NR5A1 46,XY patients with a disorder of sex development: An oligogenic origin? Eur. J. Hum. Genet. 2018;26:1329–1338. doi: 10.1038/s41431-018-0202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanz R.B., Razani B., Goldberg A.D., OMalley B.W. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA) Proc. Natl. Acad. Sci. USA. 2002;99:16081–16086. doi: 10.1073/pnas.192571399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawashima H., Takano H., Sugita S., Takahara Y., Sugimura K., Nakatani T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: Expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem. J. 2003;369:163–171. doi: 10.1042/bj20020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chooniedass-Kothari S., Vincett D., Yan Y., Cooper C., Hamedani M.K., Myal Y., Leygue E. The protein encoded by the functional steroid receptor RNA activator is a new modulator of ER alpha transcriptional activity. FEBS Lett. 2010;584:1174–1180. doi: 10.1016/j.febslet.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 43.Xu B., Koenig R.J. An RNA-binding Domain in the Thyroid Hormone Receptor Enhances Transcriptional Activation. J. Biol. Chem. 2004;279:33051–33056. doi: 10.1074/jbc.M404930200. [DOI] [PubMed] [Google Scholar]
- 44.Liu C., Wu H.-T., Zhu N., Shi Y.-N., Liu Z., Ao B., Liao D.-F., Zheng X.-L., Qin L. Steroid receptor RNA activator: Biologic function and role in disease. Clin. Chim. Acta. 2016;459:137–146. doi: 10.1016/j.cca.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Xu B., Yang W.-H., Gerin I., Hu C.-D., Hammer G.D., Koenig R.J. Dax-1 and Steroid Receptor RNA Activator (SRA) Function as Transcriptional Coactivators for Steroidogenic Factor 1 in Steroidogenesis. Mol. Cell. Biol. 2009;29:1719–1734. doi: 10.1128/MCB.01010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neocleous V., Fanis P., Toumba M., Tanteles G.A., Schiza M., Cinarli F., Nicolaides N.C., Oulas A., Spyrou G.M., Mantzoros C.S., et al. GnRH Deficient Patients With Congenital Hypogonadotropic Hypogonadism: Novel Genetic Findings in ANOS1, RNF216, WDR11, FGFR1, CHD7, and POLR3A Genes in a Case Series and Review of the Literature. Front. Endocrinol. 2020;11:626. doi: 10.3389/fendo.2020.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gach A., Pinkier I., Szarras-Czapnik M., Sakowicz A., Jakubowski L. Expanding the mutational spectrum of monogenic hypogonadotropic hypogonadism: Novel mutations in ANOS1 and FGFR1 genes. Reprod. Biol. Endocrinol. 2020;18:8. doi: 10.1186/s12958-020-0568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jo A., Denduluri S., Zhang B., Wang Z., Yin L., Yan Z., Kang R., Shi L.L., Mok J., Lee M.J., et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014;1:149–161. doi: 10.1016/j.gendis.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fauquier T., Rizzoti K., Dattani M., Lovell-Badge R., Robinson I.C.A.F. SOX2-expressing progenitor cells generate all of the major cell types in the adult mouse pituitary gland. Proc. Natl. Acad. Sci. USA. 2008;105:2907–2912. doi: 10.1073/pnas.0707886105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rizzoti K., Akiyama H., Lovell-Badge R. Mobilized Adult Pituitary Stem Cells Contribute to Endocrine Regeneration in Response to Physiological Demand. Cell Stem Cell. 2013;13:419–432. doi: 10.1016/j.stem.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andoniadou C.L., Matsushima D., Gharavy S.N.M., Signore M., Mackintosh A.I., Schaeffer M., Gaston-Massuet C., Mollard P., Jacques T.S., Le Tissier P., et al. Sox2+ Stem/Progenitor Cells in the Adult Mouse Pituitary Support Organ Homeostasis and Have Tumor-Inducing Potential. Cell Stem Cell. 2013;13:433–445. doi: 10.1016/j.stem.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 52.Sekido R., Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 53.Foster J.W., Dominguez-Steglich M.A., Guioli S., Kwok C., Weller P.A., Stevanovic M., Weissenbach J., Mansour S., Young I.D., Goodfellow P.N., et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 54.Cela P., Hampl M., Shylo N., Christopher K., Kavkova M., Landova M., Zikmund T., Weatherbee S., Kaiser J., Buchtova M. Ciliopathy Protein Tmem107 Plays Multiple Roles in Craniofacial Development. J. Dent. Res. 2018;97:108–117. doi: 10.1177/0022034517732538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee Y.-H., Saint-Jeannet J.-P. Sox9 function in craniofacial development and disease. Genesis. 2011;49:200–208. doi: 10.1002/dvg.20717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang F., Zheng L., Cheng S., Peng Y., Fu L., Zhang X., Linhardt R. Comparison of the Interactions of Different Growth Factors and Glycosaminoglycans. Molecules. 2019;24:3360. doi: 10.3390/molecules24183360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kreuger J., Spillmann D., Li J.-P., Lindahl U. Interactions between heparan sulfate and proteins: The concept of specificity. J. Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loo B.-M., Salmivirta M. Heparin/Heparan Sulfate Domains in Binding and Signaling of Fibroblast Growth Factor 8b. J. Biol. Chem. 2002;277:32616–32623. doi: 10.1074/jbc.M204961200. [DOI] [PubMed] [Google Scholar]
- 59.Tornberg J., Sykiotis G.P., Keefe K., Plummer L., Hoang X., Hall J.E., Quinton R., Seminara S.B., Hughes V., Van Vliet G., et al. Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. Proc. Natl. Acad. Sci. USA. 2011;108:11524–11529. doi: 10.1073/pnas.1102284108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Danda V.S.R., Paidipelly S.R., Verepula M., Lodha P., Thaduri K.R., Konda C., Ruhi A. Exploring the Genetic Diversity of Isolated Hypogonadotropic Hypogonadism and Its Phenotypic Spectrum: A Case Series. J. Reprod. Infertil. 2020;22:38–46. doi: 10.18502/jri.v22i1.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu J., Choa R., Guo M.H., Plummer L., Buck C., Palmert M.R., Hirschhorn J.N., Seminara S.B., Chan Y.-M. A shared genetic basis for self-limited delayed puberty and idiopathic hypogonadotropic hypogonadism. J. Clin. Endocrinol. Metab. 2015;100:E646–E654. doi: 10.1210/jc.2015-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wright F., Hammer M., Paul S.M., Aouizerat B.E., Kober K.M., Conley Y.P., Cooper B.A., Dunn L.B., Levine J.D., Melkus G.D., et al. Inflammatory pathway genes associated with inter-individual variability in the trajectories of morning and evening fatigue in patients receiving chemotherapy. Cytokine. 2017;91:187–210. doi: 10.1016/j.cyto.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gregory L.C., Dattani M.T. The Molecular Basis of Congenital Hypopituitarism and Related Disorders. J. Clin. Endocrinol. Metab. 2020;105:e2103–e2120. doi: 10.1210/clinem/dgz184. [DOI] [PubMed] [Google Scholar]
- 64.McCormack S.E., Li N., Kim Y.J., Lee J.Y., Kim S.-H., Rapaport R., Levine M. Digenic Inheritance of PROKR2 and WDR11 Mutations in Pituitary Stalk Interruption Syndrome. J. Clin. Endocrinol. Metab. 2017;102:2501–2507. doi: 10.1210/jc.2017-00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fang X., Zhang Y., Cai J., Lu T., Hu J., Yuan F., Chen P. Identification of novel candidate pathogenic genes in pituitary stalk interruption syndrome by whole-exome sequencing. J. Cell. Mol. Med. 2020;24:11703–11717. doi: 10.1111/jcmm.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cangiano B., Swee D.S., Quinton R., Bonomi M. Genetics of congenital hypogonadotropic hypogonadism: Peculiarities and phenotype of an oligogenic disease. Qual. Life Res. 2021;140:77–111. doi: 10.1007/s00439-020-02147-1. [DOI] [PubMed] [Google Scholar]
- 67.Vaaralahti K., Raivio T., Koivu R., Valanne L., Laitinen E.-M., Tommiska J. Genetic Overlap between Holoprosencephaly and Kallmann Syndrome. Mol. Syndr. 2012;3:1–5. doi: 10.1159/000338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.