Abstract
A soil plot was inoculated with a mixture of Pseudomonas fluorescens Pf0-2, the wild type, and Pf0-5a, a Tn5 insertion mutant in adnA, at 7.84 log CFU/g of soil. Over a period of 231 days, culturable populations of both strains were measured at selected times below and away from the point of inoculation. Pf0-5a did not spread as fast and attained significantly lower populations than Pf0-2. At sample depths below the inoculation site, the adnA mutant showed a significant decrease in CFU/g of soil as compared to Pf0-2. Pf0-2 was first detected at the 1.5-cm annular site at 3 days after inoculation, whereas Pf0-5a required 7 days to travel the same distance. At this distance, the wild-type strain could be detected at a 21.5- to 25-cm depth, whereas Pf0-5a could be detected only as deep as 15.5 to 18 cm. At 4.5 cm from the site of inoculation and in soil fractions corresponding to 13 to 18 cm, Pf0-2 was the only strain detected. These results suggest that the transcription factor AdnA provides a fitness advantage in P. fluorescens, allowing it to spread and survive in soil under field conditions.
The effective use of microbial agents, such as the pseudomonads, to remedy plant diseases, decontaminate soil, improve texture, and enhance plant nutrition requires that such agents be ecologically adapted to the location where they are released. However, inoculation of selected microbes into soil or on seeds often results in rapid loss of their population within a short time (2). Such phenomena and the factors influencing this decline have been reviewed (23).
DeFlaun et al. (9) isolated two Tn5 insertion mutants of Pseudomonas fluorescens strain Pf0-1 that were deficient in attachment to soil particles and seeds and were nonmotile. The locus affected by these Tn5 insertions was identified as adnA. In complementation experiments, cloned wild-type adnA restored the wild-type phenotype described for the Tn5 insertion mutants (7a). Sequencing and expression experiments revealed that AdnA is a homologue of FleQ (7a), a transcriptional activator of the NtrC/NifA family in Pseudomonas aeruginosa which, in association with sigma 54, mediates transcription of structural genes for flagellar synthesis (4).
Other regulation factors have been linked to fitness in natural environments, e.g., RosR in Rhizobium etli and GacS in Pseudomonas syringae (6, 12, 13). RosR is likely to regulate exopolysaccharide production, whereas GacS has been implicated in regulation of several activities, such as swarming, antibiotic, toxin, and siderophore production in the genus Pseudomonas (11, 14, 18, 19, 24). The rosR mutant showed a reduced ability to grow in the rhizosphere and compete for nodule occupancy under laboratory conditions (3, 6). In field experiments, the gacS mutant attained significantly lower populations than those of the parent strain in bean foliage, but there was no effect on the populations in soil surrounding germinating seeds (12). These experiments were conducted in either laboratory conditions or for the short term and studied the effects of regulation factors on fitness of soil influenced by roots or bean foliage.
In this study, we examined the role of adnA on long-term survival, spread, and competence in soil under natural field conditions. We present evidence that adnA provides a fitness advantage in P. fluorescens in soil and discuss future research.
MATERIALS AND METHODS
Bacterial strains and semiselective media.
To determine how adnA affects soil persistence and spread of P. fluorescens under natural field conditions, we used rifampin-resistant (Rifr) strains Pf0-2 and Pf0-5a, derivatives of Pf0-1 and Pf0-5, respectively. Pf0-1 was first isolated by Compeau et al. (8) and was later used by DeFlaun et al. (9) in sand and seed attachment assays. Pf0-5 was generated by Tn5 mutagenesis and described as a nonflagellated, nonmotile, attachment-deficient mutant of Pf0-1 (9).
We generated the Rifr derivatives Pf0-2 and Pf0-5a by plating dense cultures of Pf0-1 and Pf0-5 on Luria-Bertani agar containing 50 μg of rifampin/ml. Rifr derivatives arose at a frequency of 3.5 × 10−7, and two of each strain were tested for their growth characteristics. There were no significant differences in growth rates in either minimal medium or Luria-Bertani broth between either of the Rifr derivatives and Pf0-1 and Pf0-5 (data not shown). P. fluorescens was quantified from soil by plating on MacConkey (Mac) agar, containing either rifampin (50 μg/ml) or rifampin and kanamycin (50 μg/ml; resistance mediated by Tn5 in Pf0-5), which selects preferentially for the test pseudomonads.
Inoculation site and procedure.
The experiment was conducted in an agricultural field situated at the University of Massachusetts Agricultural Experiment Station in Waltham. The experimental site, a fallow area which had not been cultivated for at least 10 years, was prepared with conventional tillage implements. An area of 20 by 10 m was fenced in chicken wire, and a plot of 1 m2 was demarked by wooden stakes. Prior to inoculation, soil samples were taken to determine soil characteristics (Table 1) and CFU of indigenous bacteria on Mac agar resistant to rifampin and kanamycin. Plating assays indicated that indigenous bacteria with these characteristics were below the limit of detection (12 CFU/g of fresh soil). Soil cores were extracted using 0.8- by 13-cm brass soil samplers (model GC-1; La Motte Co., Chesterton, Md.), which were washed, individually wrapped in aluminum foil, and autoclaved between samplings.
TABLE 1.
Soil characteristics of the plot used in this study
| Depth (cm) | OMa (%) | Texture
|
Bulk density (g/cm3) | pH | H2O holding capacity (%) | ||
|---|---|---|---|---|---|---|---|
| % Silt | % Sand | % Clay | |||||
| 1–15 | 4.9 | 49.8 | 43.9 | 6.3 | 1.004 | 6.3 | 42.3 |
| 16–30 | 2.4 | 56.3 | 42.6 | 1.1 | 1.142 | 6.2 | 39.5 |
Organic matter.
Inoculum strains Pf0-2 and Pf0-5a were grown in minimal media plus glucose (10) to dense cultures and were standardized to an optical density at 550 nm of 0.21. Bacterial suspensions were mixed with 50 g of γ-irradiated sterilized soil with similar physical and chemical properties (8) at 60% of water-holding capacity to a final density of 4.26 × 107 Pf0-2 (61%) and 2.72 × 107 Pf0-5a (39%) CFU/g of soil. By using a 250-ml conical centrifuge bottle (Corning catalog no. 25350-250), a conical depression 5.75 cm in diameter and 4.7 cm in depth was made in the soil at the site of inoculation, which corresponded to the volume occupied by the soil used as inoculum. The inoculum was carefully tapped into the depression to obtain a level soil surface.
Assay for soil populations of Pf0-2 and Pf0-5a and sampling strategy.
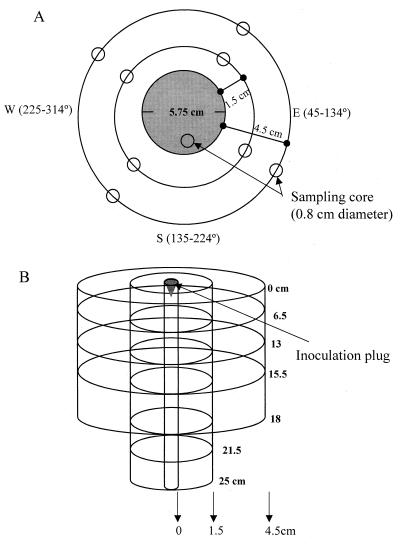
Figure 1 illustrates the soil plot and sampling plan used in this study. We slightly modified the design described by Krimsky et al. (17) to assess risk of microbial release. To measure spread of the strains, values for log CFU per gram of soil were determined at the inoculation plug (0 to 2 cm) and 1.5, 4.5, 13.5, and 40 cm from the inoculation plug (Fig. 1A). Soil fractions corresponding to depths of 0 to 6.5, 6.5 to 13, 13 to 15.5, 15.5 to 18, 18 to 21.5, and 21.5 to 25 cm below (Fig. 1B) were sampled. For measuring soil populations 1.5, 4.5, 13.5, and 40 cm from the inoculation plug, we established the four cardinal points in the outer ring of the soil plots and used a compass clockwise to determine angles randomly for sampling sites (Fig. 1A). Measurements at and below the inoculation plug were taken inside the circle demarked by the inoculation plug at different days after inoculation (dai). The inoculation plug was 5.75 cm in diameter, while the soil core samplers were 0.8 cm in diameter, which allowed ample room for several samplings. To measure persistence, the plot was sampled periodically from 0 to 231 dai and finally 365 dai. Soil corers were inserted to the desired depth and were then wrapped, labeled, and transported for analysis on the same day. For sampling at depths of 18 and 25 cm, two corers were bound end to end with duct tape. To maintain the integrity of the soil plot after sampling, the holes left by plug removal were filled with soil from a nearby uninoculated site. Because the bacterial reservoir was depleted any time that soil samples were taken, a new set of sites were generated for subsequent sampling times. A second independent plot with the same sampling strategy but with fewer time points and depths sampled was examined in parallel.
FIG. 1.
Diagram of sampling strategy and field plot inoculated with P. fluorescens strains Pf0-2 and Pf0-5a. (A) Top view of plot. The inoculation plug (shaded area) consisted of a cone with a diameter of 5.75 cm and a volume of 41 ml. The four cardinal points were established just beyond the outer ring of the plots. Sampling sites at test distances (1.5 and 4.5 cm) were determined by using a compass clockwise and randomly picking an angle between 315 and 44°, 45 and 134°, 135 and 224°, and 225 and 314° for north, east, south, and west, respectively. Rings indicating distances away from the site of inoculation were demarked with toothpicks. Distances of 13.5 and 40 cm from the point of inoculation were also tested, but no test organisms were detected. Plug is drawn to scale. (B) Side view of field plot. Movement of the strains below the site of inoculation was measured in soil fractions of depths (in centimeters) of 0 to 2, 2 to 6.5, 6.5 to 13, 13 to 15.5, 15.5 to 18, 18 to 21.5, and 21.5 to 25. Movement of the strains from the site of inoculation was measured at distances of 1.5 and 4.5 cm from the site and at depths of 0 to 6.5, 6.5 to 13, 13 to 15.5, 15.5 to 18, 18 to 21.5, and 21.5 to 25 cm below the inoculation site. Side view is not drawn to scale.
The soil plugs, weighing from 0.5 to 1.8 g, were carefully expelled, weighed, diluted in 6 ml of sterile water, and vortexed at high speed for 1 min. Serial dilutions (10-fold) were plated (either 500 or 100 μl) on Mac agar containing either rifampin alone or rifampin and kanamycin (50 μg/ml) and were incubated at 33°C for 42 h. We quantified populations of Pf0-2 by subtracting CFU on Mac rifampin-kanamycin plates from CFU on Mac rifampin plates.
Statistical analysis.
Plate counts were adjusted to CFU/gram of dry soil and log10 transformed. For measurements at and below the inoculation plug, we used a Student's t test of paired observations to determine whether the proportion of the strains at each depth remained similar to the nominal proportions in the inoculum (1.5:1, Pf0-2:Pf0-5a). At each timepoint tested, we paired values for log CFU/gram of soil from Pf0-2 with values for log CFU/gram of soil from Pf0-5a and calculated the difference in the pair. The number of pairs (replicates) corresponded to the number of times sampled. For example, for the 0 to 2-cm depth we measured populations at 7, 17, 31, 45, 73, and 231 dai. Then we determined the mean difference over six samplings and tested against the difference between the populations of the strains in the inoculum.
Values for log CFU/gram of soil taken 1.5 and 4.5 cm from the inoculation plug were treated by analysis of variance (ANOVA) as a randomized complete block design with four replicates. The treatments represented each of the strains and the replicates formed by each of the cardinal points. This analysis was performed for each dai sampled.
To test whether there was a soil gradient or any other bias affecting bacterial distribution in the experimental plot, we tested total populations (sum of Pf0-2 and Pf0-5a) that were 1.5 cm from and 6.5 to 13 cm below the inoculation plug for each cardinal point. We employed a randomized complete design using the cardinal points as treatments and 10 replicates, which corresponds to the number of times sampled.
RESULTS
Precipitation and soil water content.
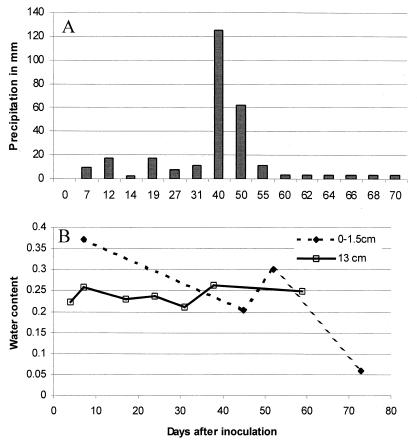
Figure 2A and B show precipitation in millimeters and soil water content of the plots up to 70 dai, respectively. Precipitation was registered after any appreciable rain occurred. Total rainfall on the plots was 287.6 mm with an average of 4.1 mm/day. Soil water content was determined by removing 2 to 10 g of soil at various depths and taking soil weight before and after drying the samples for 24 h at 60°C. Soil water content was more variable at the surface and ranged from 7 to 35%, compared to 22 to 27% at 13 cm, but the averages were 23% for both depths. The values for water content at 18 and 25 cm were very similar to those from 13 cm (data not shown). During the winter season (between 70 and 231 dai), precipitation and soil water content were not registered.
FIG. 2.
Precipitation in millimeters (A) and soil water content (B) for plots inoculated with P. fluorescens for 70 days at depths of 0 to 1.5 cm and 13 cm. Precipitation is reported as the average per day from 60 to 70 dai.
Effect of adnA on vertical movement in soil.
Mean representations and culturable populations of Pf0-2 and Pf0-5a (adnA mutant) in soil at the surface (depth 0 to 2 cm) and from fractions at depths of 2 to 6.5, 6.5 to 13, 13 to 15.5, 15.5 to 18, 18 to 21.5, and 21.5 to 25 cm below the inoculation plug were determined (Table 2). Pf0-2 significantly increased its representation in soil from 1.5:1 (Pf0-2:Pf0-5a) at the time of inoculation, to 6:1, 5:1, 49:1, 99:1, and 6:1 in soil fractions corresponding to depths of 2 to 6.5, 6.5 to 13, 13 to 15.5, 15.5 to 18, and 18 to 21.5 cm through 231 days, respectively. There were no significant differences between the representation of both strains at the surface (depth, 0 to 2 cm) and at the depth of 21.5 to 25 cm. Populations were highest at the surface and declined with depth.
TABLE 2.
Soil representation and populations of P. fluorescens strains Pf0-2 and Pf0-5a (adnA mutant) at various depths from the inoculation plug
| Strain or parameter | Populationa or parameter at following depth (cm)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Inoculum | 0–2 | 2–6.5 | 6.5–13 | 13–15.5 | 15.5–18 | 18–21.5 | 21.5–25 | |
| Pf0-2 | 7.62 | 6.6 ± 0.4 | 5.03 ± 0.41 | 4.4 ± 0.3 | 4.02 ± 0.3 | 4.05 ± 0.4 | 2.7 ± 0.35 | 1.38 ± 0.2 |
| Pf0-5a | 7.43 | 6.4 ± 0.4 | 4.2 ± 0.35 | 3.7 ± 0.28 | 2.3 ± 0.41 | 2.07 ± 0.6 | 1.9 ± 0.5 | 1.2 ± 0.2 |
| Ratio (Pf0-2:Pf0-5a)bc | 1.5:1 | 1.5:1 | 6:1∗∗ | 5:1∗ | 49:1∗∗ | 99:1∗ | 6:1∗ | 1.5:1 |
| n | 6 | 5 | 6 | 5 | 5 | 3 | 4 | |
Mean values for log CFU/gram of dry soil followed by standard error. Means of strains were determined using the values of each time sampled.
We used a Student's t test for paired observations to determine whether the ratio of the strains was significantly different from that at inoculation, i.e., 1.5:1. At each depth, the populations of both strains were paired and each time sampled was considered a replication. Results are representative of two experiments.
Significantly different ratios are indicated at P ≤ 0.1 (∗) and P ≤ 0.05 (∗∗).
Effect of adnA on movement from the inoculation site.
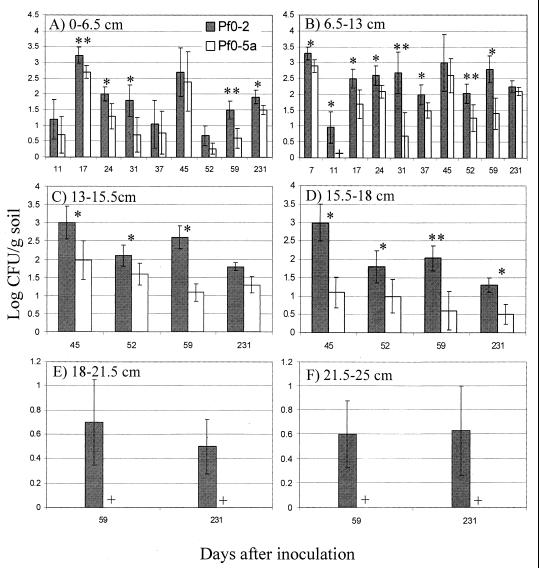
Populations for Pf0-2 and Pf0-5a in soil fractions below the 1.5-cm-depth test point were assayed (Fig. 3). At the total population level, values for log CFU/gram of soil reached 3.5 in soil fractions spanning from 0 to 18 cm (Fig. 3A to D), whereas numbers in soil fractions obtained from 18- to 25-cm depths did not exceed 0.8 log units (Fig. 3E and F). Values for log CFU/gram of soil appeared more variable in the 0- to 6.5-cm fraction (Fig. 3A). Pf0-2 titers were significantly higher than those of Pf0-5a for most of the dai sampled for soil fractions of 0 to 6.5, 6.5 to 13, and 13 to 15.5 cm (Fig. 3A to C) and for all dai sampled for the 15.5- to 18-cm fraction (Fig. 3D). Of note, in the soil fractions corresponding to 18 to 25 cm, only Pf0-2 was detected (Fig. 3E and F).
FIG. 3.
Soil populations of P. fluorescens strains Pf0-2 (wild type) and Pf0-5a (adnA mutant) at different depths 1.5 cm from the point of inoculation: 0 to 6.5 cm (A), 6.5 to 13 cm (B), 13 to 15.5 cm (C), 15.5 to 18 cm (D), 18 to 21.5 cm (E), and 21.5 to 25 cm (F). On day 0, strains were mixed and inoculated to 7.84 total log CFU/g of soil. At different times, 0.5- to 1.8-g samples were taken and strains were enumerated by plating serial 10-fold dilutions on Mac agar plates containing rifampin alone or rifampin and kanamycin. Means were determined from four replicates. Significant differences were determined by ANOVA. ∗, ∗∗, statistically significant differences between soil populations of the two strains at P ≤ 0.1 and 0.05, respectively; +, no Pf0-5a detected. Results are representative of two experiments.
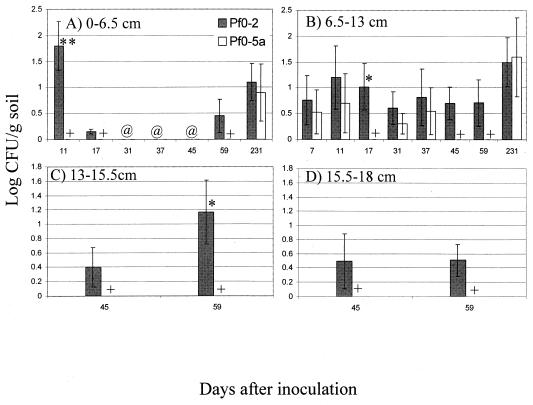
Soil populations in four soil fractions from the 4.5-cm radial test points were examined (Fig. 4). Values for log CFU/gram of soil did not exceed two and were at their lowest in the 13- to 15.5-cm and 15.5- to 18-cm fractions (Fig. 4). In the 0- to 6.5-cm soil fraction, Pf0-2 attained significantly higher populations than did Pf0-5a only at 11 dai and was undetected from 31 to 45 dai. Furthermore, Pf0-5a was not detected until 231 dai (Fig. 4A). In the 6.5- to 13-cm fraction, Pf0-2 was detected in all dai sampled and showed significant differences from Pf0-5a only at 17 dai (Fig. 4B). In the 13- to 15.5-cm and 15.5- to 18-cm soil fractions, Pf0-5a was not detected at either 45 or 59 dai (Fig. 4C and D). Pf0-2 attained significantly higher values for log CFU/gram of soil than Pf0-5a at 59 dai in the 13- to 15.5-cm soil fraction (Fig. 4C). Numbers of both strains fell below the detection limit in all soil fractions and were not detected in any samples taken at annular test points of 13.5 or 40 cm (data not shown).
FIG. 4.
Soil populations of P. fluorescens strains Pf0-2 (wild type) and Pf0-5a (adnA mutant) at different depths 4.5 cm from the point of inoculation: 0 to 6.5 cm (A), 6.5 to 13 cm (B), 13 to 15.5 cm (C), and 15.5 to 18 cm (D). Strains were inoculated and enumerated as described for Fig. 3. Means were determined from four replicates. Significant differences were determined by ANOVA. ∗, ∗∗, statistically significant differences between soil populations of the two strains at P ≤ 0.1 and 0.05, respectively; +, no Pf0-5a detected; @, both strains not detected. Results are representative of two experiments.
Distribution of total CFU of test strains from the point of inoculation.
There were no significant differences in soil total populations (sum of Pf0-2 and Pf0-5a) in terms of log CFU/gram of soil at four cardinal points (north, east, south, and west) at 1.5 cm from and 6.5 to 13 cm below the inoculation site. These data suggest that bacterial movement of P. fluorescens in the soil conditions tested proceeded in a radial fashion at similar rates (data not shown).
DISCUSSION
This study provides evidence that the transcriptional activator AdnA confers a long-term fitness advantage in P. fluorescens under field conditions. The mutant strain Pf0-5a has a single Tn5 insertion which is in the adnA gene (9). Complementation experiments with cloned adnA confirmed that its inactivation led to the adhesion-deficient and nonflagellated phenotypes (7a). In two separate, yearlong experiments, Pf0-5a showed a significant decrease in its survival and persistence in soil compared to the parent strain, Pf0-2 (Table 2). Moreover, Pf0-5a was not detected in the deeper soil fractions or as frequently or as numerously as Pf0-2 at 1.5 and 4.5 cm from the inoculation site (Fig. 3 and 4).
We determined soil populations by a standard plating technique, which has been shown to provide information for a small fraction of total and metabolically active cells (1, 7, 21). However, Johnsen et al. (16) recently showed that quantitative PCR correlated well with culturable counts of Pseudomonas in soil obtained on selective media.
As proposed previously (7a), the product of adnA has high similarity to FleQ, a transcriptional factor of the NtrC/NifA family of regulators. Such activators generally work as part of a two-component regulatory system which, under the right environmental conditions and in association with sigma 54, mediates nitrogen fixation in Rhizobium and formation of flagella in P. aeruginosa (4, 20). Although the adnA mutation may have pleiotropic effects, we speculate that its primary function is in flagellum synthesis because of its location in a region of flagellar genes and homology to FleQ, a transcription factor for flagellum synthesis. We favor the notion that the effects in soil are principally related to motility. DeFlaun et al. (9) showed that under water-saturated conditions the mutant strain moves down faster than the parent strain in soil columns; however, the adnA mutant strain was not detected in the deepest soil fractions (Fig. 3E and F). A major means of bacterial spread and transport in soil is by percolating water (22). In our case, the average soil water content was 23%, well below the threshold for percolating water. Perhaps motility is important for spread under non-water-saturated soil conditions. Similar results were reported for nonmotile variants of Rhizobium leguminosarum biovar phaseoli, which did not spread as well as the wild type from the point of inoculation in soils that were moderately wet (15).
Despite this evidence, we cannot rule out the possibility that adnA is influencing spread and persistence in soil of P. fluorescens by activating transcription of other systems. To address this, we have undertaken experiments that will identify genes regulated by adnA. As is the case with identifying genes affected by transcriptional factors (5), we foresee three functions regulated by adnA: genes important for soil fitness, other genes involved in regulation, and genes affecting other cellular functions. As adnA is predicted to be a transcriptional activator, this gene provides a handle to study environment-bacterium interactions and adaptation to soil environments.
ACKNOWLEDGMENTS
This work was supported by grants from the Department of Energy (DE-FG02-97ER62493) and the National Science Foundation (DEB 9120897).
B.M. and E.A.R. contributed equally to this work.
REFERENCES
- 1.Amann R I. Fluorescently labeled, ribosomal-RNA-targeted oligonucleotide probes in the study of microbial ecology. Mol Ecol. 1995;4:543–553. [Google Scholar]
- 2.Angle J S, Levin M A, McIntosh M, Glew J G. Pseudomonas aurofaciens in soil: survival and recovery efficiency. Microb Releases. 1994;2:247–254. [PubMed] [Google Scholar]
- 3.Araujo R A, Robleto E A, Handelsman J. A hydrophobic mutant of Rhizobium etli altered in nodulation competitiveness and growth in the rhizosphere. Appl Environ Microbiol. 1994;60:1430–1436. doi: 10.1128/aem.60.5.1430-1436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bittinger M A, Handelsman J. Identification of genes in the RosR regulon of Rhizobium etli. J Bacteriol. 2000;182:1706–1713. doi: 10.1128/jb.182.6.1706-1713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bittinger M A, Milner J, Saville B J, Handelsman J. RosR, a determinant of nodulation competitiveness in Rhizobium etli. Mol Plant-Microbe Interact. 1997;10:180–186. doi: 10.1094/MPMI.1997.10.2.180. [DOI] [PubMed] [Google Scholar]
- 7.Borneman J, Skroch P W, O'Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Casaz, P., A. Happel, J. Keithan, D. Read, S. R. Strain, and S. B. Levy. The Pseudomonas fluorescens transcription acitvator AdnA is required for adhesion and motility. Microbiology, in press. [DOI] [PubMed]
- 8.Compeau G, Al-Achi B J, Platsouka E, Levy S B. Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl Environ Microbiol. 1988;54:2432–2438. doi: 10.1128/aem.54.10.2432-2438.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeFlaun M, Marshall B M, Kulle E-P, Levy S B. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl Environ Microbiol. 1994;60:2637–2642. doi: 10.1128/aem.60.7.2637-2642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeFlaun M, Tanzer A S, McAteer A L, Marshall B, Levy S B. Development of an adhesion assay and characterization of an adhesion-deficient mutant of Pseudomonas fluorescens. Appl Environ Microbiol. 1990;56:112–119. doi: 10.1128/aem.56.1.112-119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grewal S, Han B, Johnstone K. Identification and characterization of a locus which regulates multiple functions in Pseudomonas tolaasii, the cause of brown blotch disease of Agaricus biporus. J Bacteriol. 1995;177:4658–4668. doi: 10.1128/jb.177.16.4658-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano S, Ostertag E M, Savage S A, Baker L S, Willis D K, Upper C D. Contribution of the regulatory gene lemA to field fitness of Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1997;63:4304–4312. doi: 10.1128/aem.63.11.4304-4312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hrabak E M, Rich J J, Barta T M, Lindow S E, Panopolous N J. Isolation and characterization of Pseudomonas syringae pv. syringae mutant deficient in lesion formation. Mol Plant-Microbe Interact. 1990;3:149–156. [Google Scholar]
- 14.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Issa S, Wood M, Simmons L. Active movement of chickpea and bean rhizobia in dry soil. Soil Biol Biochem. 1993;25:951–958. [Google Scholar]
- 16.Johnsen K, Enger O, Jacobsen C S, Thirup L, Torsvik V. Quantitative selective PCR of 16S ribosomal DNA correlates well with selective agar plating in describing population dynamics of indigenous Pseudomonas spp. in soil hot spots. Appl Environ Microbiol. 1999;65:1786–1788. doi: 10.1128/aem.65.4.1786-1788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krimsky S, Wrubel R P, Naess I G, Levy S B, Wetzler R E, Marshall B. Standardized microcosms in microbial risk assessment. BioScience. 1995;45:590–599. [Google Scholar]
- 18.Laville J C, Volstard C, Keel C, Maurhofer M, Defago G, Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci USA. 1992;89:1562–1566. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao C H, McCallus D, Fett W. Molecular characterization of two gene loci required for production of the key pathogenicity factor pectate lyase in Pseudomonas viridiflava. Mol Plant-Microbe Interact. 1994;7:391–400. doi: 10.1094/mpmi-7-0391. [DOI] [PubMed] [Google Scholar]
- 20.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trevors J, van Elsas J, van Overbeek L, Starboud M. Transport of a genetically engineered Pseudomonas fluorescens strain through a soil microcosm. Appl Environ Microbiol. 1990;56:401–408. doi: 10.1128/aem.56.2.401-408.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Veen J A, van Overbeek L S, van Elsas J D. Fate and activity of microorganisms introduced into soil. Microbiol Mol Biol Rev. 1997;61:121–135. doi: 10.1128/mmbr.61.2.121-135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J P, Normark S. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science. 1996;273:1234–1236. doi: 10.1126/science.273.5279.1234. [DOI] [PubMed] [Google Scholar]