Abstract
During sclerotial infection of Sclerotinia sclerotiorum the mycoparasite Coniothyrium minitans penetrates through the host cell wall, which contains β-1,3-glucan as its major component. A PCR-based strategy was used to clone a β-1,3-glucanase-encoding gene, designated cmg1, from a cDNA library of the fungus. The nucleotide and deduced amino acid sequences of this gene showed high levels of similarity to the sequences of other fungal exo-β-1,3-glucanase genes. The calculated molecular mass of the deduced protein (without the predicted 24-amino-acid N-terminal secretion signal peptide) was 83,346 Da, and the estimated pI was 4.73. Saccharomyces cerevisiae INVSc1 expressing the cmg1 gene secreted a ∼100-kDa β-1,3-glucanase enzyme (as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis) into the culture medium. N-terminal sequence analysis of the purified recombinant enzyme revealed that the secreted enzyme starts at Ala-32, seven amino acids downstream from the predicted signal peptidase cleavage site. The purified recombinant glucanase inhibited in vitro mycelial growth of S. sclerotiorum by 35 and 85% at concentrations of 300 and 600 μg ml−1, respectively. A single copy of the cmg1 gene is present in the genome of C. minitans. Northern analyses indicated increases in the transcript levels of cmg1 due to both carbon starvation and the presence of ground sclerotia of S. sclerotiorum; only slight repression was observed in the presence of 2% glucose. Expression of cmg1 increased during parasitic interaction with S. sclerotiorum.
Coniothyrium minitans Campbell is a destructive mycoparasite highly specialized on Sclerotinia spp. (50). It has been used successfully in field and glasshouse experiments to control Sclerotinia diseases of a number of crop plants (7, 20, 36, 48). Long-term field studies of agricultural and horticultural crops indicated that C. minitans is more efficient against Sclerotinia sclerotiorum than the other mycoparasites tested (14, 20). In a 5-year crop rotation experiment C. minitans was found to be superior to Trichoderma strains isolated from infected sclerotia in reducing the contamination of soil with S. sclerotiorum sclerotia (14). Two years after the last application of the biocontrol agents, plots treated with C. minitans contained many fewer sclerotia than plots treated with Trichoderma, indicating the outstanding long-term effect of the Coniothyrium treatments (14).
Despite the powerful potential of C. minitans to control Sclerotinia diseases, knowledge concerning the mechanisms accounting for its parasitic activity is rather limited. Light and electron microscopy studies have demonstrated that C. minitans penetrates through the external rind cells and cortex of the sclerotia of S. sclerotiorum (49). This process is followed by both inter- and intracellular growth of the parasite within subcortical layers. According to Philips and Price (43), penetration is achieved merely by physical pressure exerted by the mycoparasite. However, other workers have claimed that penetration is facilitated by the production of cell wall-degrading enzymes by the fungus (21, 23).
β-1,3-Glucan, a major component of the cell walls and resting structures of most fungi, is degraded by β-1,3-glucanases, which are grouped according to their mechanisms of hydrolysis (44). Endo-β-1,3-glucanases (EC 3.2.1.39) cleave at random sites along the glucan chain. Exo-β-1,3-glucanases (EC 3.2.1.58) release glucose monomers from the nonreducing end of the glucan chain. Fungal noncellulolytic glucanases have been implicated as important factors in the mycoparasitic activities of various fungal species (2, 4, 8, 11, 29).
The role of extracellular hydrolases in sclerotial parasitism has been poorly characterized. The cell walls of sclerotia of S. sclerotiorum, the main target organism of C. minitans, contain β-1,3-glucan as their major component (22); therefore, genes coding for glucan-degrading enzymes could be important parasitic traits of this fungus. The primary aims of the present work were to clone and characterize a β-1,3-glucanase-encoding gene from C. minitans. In order to begin to assess the role of this enzyme in the parasitic activity of the fungus, the expression patterns of the gene, as well as the inhibitory effect of the gene product on mycelial growth of S. sclerotiorum, were also investigated.
MATERIALS AND METHODS
Fungal strains and culture conditions.
C. minitans Cm-2 isolated from sclerotia of S. sclerotiorum and S. sclerotiorum Sc-1 were obtained from the culture collection of the Plant Protection Institute, Budapest, Hungary. Both fungi were maintained on potato dextrose agar (PDA) (Difco). For mycelium production, conidiospores of C. minitans washed with distilled water from 12- to 14-day-old PDA plates were used to inoculate a synthetic medium (SM) at a final concentration of 106 spores ml−1. SM contained (per liter) 3.0 g of NaNO3, 1.0 g of KH2PO4, 0.5 g of KCl, 0.5 g of MgSO4 · 7H2O, 1.0 g of peptone, 0.01 g of FeSO4 · 7H2O, 0.003 g of ZnSO4 · 7H2O, and 0.003 g of CoCl2 · 6H2O. It was supplemented with glucose (1.0 or 20.0 g liter−1), ground sclerotia of S. sclerotiorum (5.0 g liter−1), or cell wall extract from sclerotia (2.0 g liter−1) as a carbon source. Liquid cultures were incubated in a rotary shaker at 180 to 200 rpm at 23 ± 2°C. Cell wall extract was prepared from sclerotia of S. sclerotiorum collected from PDA plates. Sclerotia were freeze-dried, ground to a powder, suspended in distilled water (0.2 g ml−1), and centrifuged at 16,000 × g for 10 min. This extraction procedure was repeated several times until no protein could be detected in the supernatant. The suspension was then sonicated twice for 5 min with a VirSonic 300 ultrasonic disintegrator (Virtis). The suspension was centrifuged again at 16,000 × g for 10 min, and the sediment was washed with distilled water twice. After the final centrifugation step the upper part of the pellet, a homogeneous, gellike black substance, was gently scraped off, freeze-dried, ground in a mortar to a fine powder, and stored at room temperature. Saccharomyces cerevisiae INVSc1 (Invitrogen, San Diego, Calif.) was cultured in SC medium as recommended by the manufacturer.
Isolation and manipulation of nucleic acids.
Fungal genomic DNA was isolated as described previously (15). Total RNA was extracted from the mycelium by the LiCl precipitation method (47). DNA electrophoresis, RNA electrophoresis, blotting, hybridization, and general recombinant DNA techniques were carried out by using standard protocols (46). DNA sequencing was performed by the dideoxy chain termination method using a Sequenase kit (version 2.0; U.S. Biochemicals, Cleveland, Ohio). The two strands were sequenced independently. Nucleotide sequence analyses and comparisons were carried out by using the GCG software (12) and the BLAST method (basic local alignment search tool) (1).
Reverse transcription-PCR experiments.
Degenerate oligonucleotide primers GP1 (5′-AA[A/G]GG[A/C/G/T]GA[C/T]GG[A/C/G/T]GT[A/C/G/T]AC[A/C/ G/T]GA[C/T]GA-3′) and GP2 (5′-TG[A/G][A/T]A[A/G]TA[A/C/G/T]GG[A/C/G/T]GT[C/T]TC[A/C/G/T]GT[C/T]TG-3′) were constructed on the basis of conserved regions of known fungal β-1,3-glucanase sequences. First-strand cDNA was synthesized by standard protocols (46) using 10 μg of total RNA as the template; this RNA was extracted from C. minitans mycelium grown for 12 days in liquid SM containing 0.2% (wt/vol) Sclerotinia cell wall preparation as the sole carbon source. First-strand cDNA was used as the template in 50-μl PCR mixtures containing each degenerate primer at a concentration of 0.5 μM, 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 100 μM, and 1 U of Taq polymerase (Promega, Madison, Wis.). Amplifications were performed with a Perkin-Elmer DNA thermal cycler by using the following program: one cycle of 95°C for 2 min, 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, one cycle of 72°C for 10 min, and storage at 4°C. The PCR product was electrophoresed, purified from the agarose gel with a DNA extraction kit from Fermentas (Vilnius, Lithuania), and cloned into plasmid pKS(+) (Stratagene, La Jolla, Calif.).
Preparation and screening of the cDNA library.
Total RNA was isolated from mycelium of C. minitans grown as described above. Poly(A)+ mRNA was purified from the total RNA by using the Oligotex mRNA purification system (Qiagen, Chatsworth, Calif.). Five micrograms of poly(A)+ mRNA was used to prepare a cDNA library in the Lambda ZAP Express vector (Stratagene) by following the manufacturer's recommendations. Plaque hybridization was performed as recommended in the supplier's protocol by using the radiolabelled PCR product (see above) as the probe. Phagemids were excised from positive lambda clones as recommended by the manufacturer. The resulting plasmid containing the putative glucanase gene was designated pCMGL.
Expression of cmg1 in yeast.
A 2.8-kb MluI-XhoI fragment of pCMGL containing the entire coding region was cloned into plasmid pYES2 (Invitrogen), yielding the vector pYGL. This strategy generated a transcriptional fusion of the cDNA with a galactose-inducible yeast promoter. Plasmids pYGL and pYES2 were transformed independently into S. cerevisiae INVSc1 as described by Gietz et al. (16). Transformants were selected for uracil prototrophy. Galactose-induced protein expression was performed as described in Invitrogen's instruction manual. After 9 h of galactose induction, yeast cultures were pelleted by centrifugation at 1,500 × g for 5 min at 4°C. Extracellular protein samples were prepared from the culture supernatants following dialysis, freeze-drying, and resuspension in 20 mM Tris-HCl (pH 6.8). Intracellular proteins were extracted from the pelleted yeast cells by vortexing them vigorously with acid-washed glass beads in 50 mM sodium phosphate (pH 7.4)–1 mM EDTA–5% (vol/vol) glycerol–1 mM phenylmethylsulfonyl fluoride. Both intra- and extracellular protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 4% stacking gels and 8% separating gels by using standard protocols (46), except that the reducing agent (dithiothreitol) was omitted and the samples were not boiled. Renaturation of the separated proteins was carried out by rinsing the gel twice for 10 min in 50 mM phosphate buffer (pH 5.0) containing 25% (vol/vol) propanol and twice in the same buffer without propanol. β-1,3-Glucanase activity was then detected in the gel as described by Pan et al. (41). The protein concentration was determined as described by Bradford (6) by using ovalbumin as the standard.
Characterization of the recombinant enzyme.
Extracellular proteins of strain INVSc1 transformed with pYGL were fractionated on a Mono Q HR 5/5 anion-exchange column (Pharmacia, Uppsala, Sweden) with linear NaCl gradient elution. The β-1,3-glucanase activities of the fractions were assayed by using laminarin (5 mg ml−1) as the substrate. Release of reducing sugars in 66 mM phosphate buffer (pH 5.0) at 40°C for 20 min was measured as described by Miller (38). The fraction showing the highest activity was concentrated with a Vivaspin 4 concentrator (Vivascience, Binbrook, United Kingdom). The protein homogeneity of the fraction was determined by standard SDS-PAGE followed by silver staining, as described by Wray et al. (52). The substrate specificity of the enzyme was evaluated by measuring the reducing sugars released from laminarin, carboxymethyl cellulose (medium viscosity), arabinoxylan, and lichenin (all obtained from Sigma Chemical Co., St. Louis, Mo.); pustulan (Calbiochem, San Diego, Calif.); and Avicel (Merck, Darmstadt, Germany). The exoglucanase nature of the enzyme encoded by cmg1 was determined by monitoring the amounts and rates of release of both glucose and glucose equivalent reducing sugars from laminarin. A glucose oxidase kit obtained from Sigma (catalog no. 510-A) was used for glucose detection, whereas reducing sugars were detected as described by Miller (38). N-terminal amino acid sequence analysis was performed at the Analysis-Synthesis Laboratory of the Agricultural Biotechnology Center, Gödöllo", Hungary. Proteins from the glucanase-active fraction were electrophoretically transferred to a polyvinylidene difluoride membrane after standard SDS-PAGE by using a Mini Trans-Blot Cell (Bio-Rad Laboratories, Hercules, Calif.). The membrane was stained with Coomassie brilliant blue R250 as described by Matsudaira (35). The single protein band detected after staining was excised and subjected to sequence analysis with an ABI 471A protein sequencer (Applied Biosystems, Foster City, Calif.).
Mycelial growth inhibition.
The inhibitory effect of the purified, recombinant enzyme on mycelial growth of S. sclerotiorum was determined as described by Woo et al. (51), with some modifications. Mycelial discs (1 mm in diameter) of S. sclerotiorum Sc-1 were cut from a 3-day-old PDA plate and were placed into wells of a 24-well cell culture plate containing 400 μl of water agar (1%) medium. The plate was incubated at 25°C for 12 h under humid conditions. Fifty microliters of sodium citrate buffer (50 mM, pH 4.8) containing 0, 300, or 600 μg of recombinant glucanase per ml were then sprayed onto the inocula, and mycelial growth was measured with an inverted microscope at zero time and after 1 and 7 h of incubation at 25°C. Heat-inactivated enzyme obtained by boiling the sample for 5 min was used as a control. The experiment was done twice, and four replicates were used each time.
Dual-culture plate assay.
A total of 200 ± 20 sclerotia of S. sclerotiorum were placed on cellophane-covered SM agar plates containing 0.1% glucose and inoculated with 200 μl of a C. minitans Cm-2 conidium suspension (106 conidia ml−1). The plates were incubated at 22°C in the dark. Control plates were inoculated separately with C. minitans or S. sclerotiorum. After 3, 5, and 10 days of incubation mycelia were collected, frozen in liquid nitrogen, and stored at −70°C until total RNA was extracted and hybridized to the radiolabelled cmg1 sequence. Relative expression levels were quantified by comparing scanned images of photographs taken from ethidium bromide-stained gels and the corresponding autoradiographs by using the analySIS software (Soft Imaging Systems Gmbh, Münster, Germany).
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the GenBank database under accession number AF247649.
RESULTS
Isolation and sequence analysis of the putative glucanase-encoding cDNA.
When degenerate oligonucleotide primers GP1 and GP2 were used in reverse transcription-PCR, a ∼700-bp DNA fragment was amplified from mRNA extracted from C. minitans Cm-2 after growth for 12 days in SM containing an S. sclerotiorum cell wall preparation as the sole carbon source. The PCR product was cloned into plasmid pKS(+) and partially sequenced. A BLAST search of this sequence indicated that the cloned DNA was part of a β-1,3-glucanase gene; the fragment was therefore radiolabelled and used to screen a λ ZAP II cDNA library prepared from C. minitans grown on Sclerotinia cell wall extract. Plasmid pCMGL obtained after in vivo excision from a positive plaque was characterized further.
The nucleotide sequence of the inserted cDNA of plasmid pCMGL, designated cmg1, was determined and found to consist of 2,966 bp, including a 13-mer poly(A) tail. The first translation initiation codon was at positions 58 to 60 on the cDNA. It was followed by an in-frame stop codon at positions 76 to 78. The actual translation initiation point was therefore assumed to be the second ATG at positions 233 to 235. This phenomenon, known as translation reinitiation, has been observed in a number of eukaryotic genes (26). The sequence context of the second ATG, TCATCATGG, fits the consensus sequence surrounding translation initiation sites of filamentous fungal genes (17).
Southern analysis of genomic DNA of C. minitans Cm-2 showed that cmg1 is a single-copy gene (data not shown).
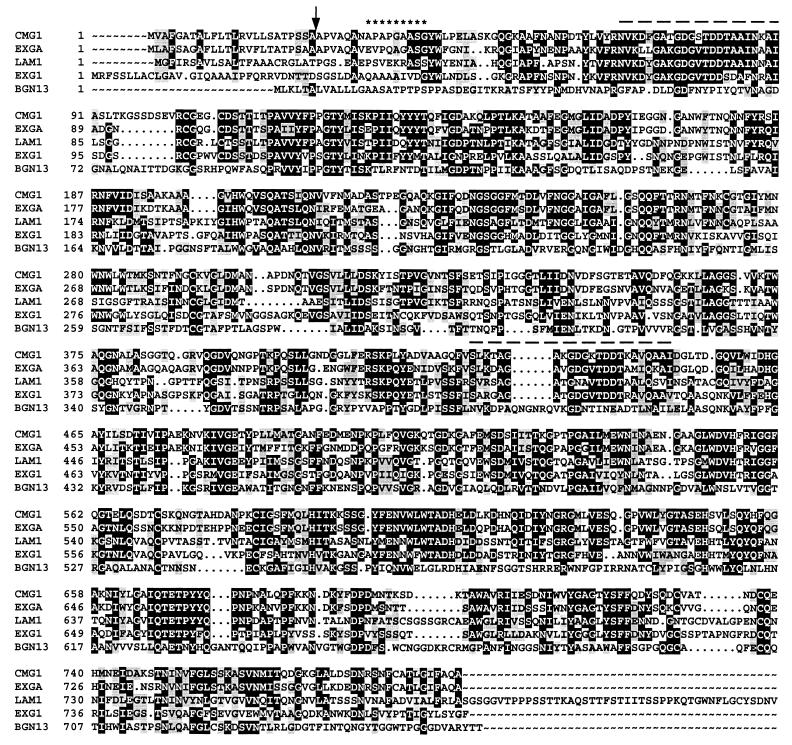
The cDNA contains an open reading frame encoding a putative 792-amino-acid protein. The open reading frame is preceded by a relatively large, 232-bp 5′ noncoding region and is followed by a 338-bp 3′ noncoding region. A signal peptide cleavage site was predicted between residues 24 and 25 (Fig. 1) by the SignalP program (39). Six potential N-glycosylation sites (NXT/S) typical of secreted fungal enzymes (3) were identified in the protein sequence (data not shown). These results collectively suggest that pCMGL encodes an extracellular enzyme. The calculated molecular mass of the secreted protein is 82,346 Da, and its estimated pI is 4.73. A comparison of its deduced amino acid sequence with the amino acid sequences of other proteins deposited in databases revealed high levels of homology to various fungal exo-β-1,3-glucanases (Fig. 1). The overall amino acid sequence of the deduced protein showed 69, 42, and 41% identities with exoglucanase sequences from Ampelomyces quisqualis, Trichoderma harzianum, and Cochliobolus carbonum, respectively (9, 40, 45); 27% overall similarity was also found with BGN13.1, an endo-β-1,3-glucanase from T. harzianum (11). The BLAST search revealed two short regions, stretching from residues 68 to 90 and from residues 428 to 450, that showed homologies to motifs present in various proteins and enzymes that interact with glucans (Fig. 1). Nikolskaya et al. (40) suggested that these regions might be involved in substrate binding.
FIG. 1.
Alignment of CMG1 of C. minitans with EXGA of A. quisqualis, LAM1.3 (LAM1) of T. harzianum, EXG1 of C. carbonum, and BGN13.1 (BGN13) of T. harzianum. Identical and similar residues are indicated by black and grey backgrounds, respectively. The C terminus of LAM1.3 of T. harzianum shows no homology to the other sequences presented and therefore is not shown. The arrow indicates the predicted signal peptide cleavage site. Asterisks indicate the N-terminal residues of the yeast-expressed enzyme, as determined by protein sequencing. The dashed lines indicate the putative substrate binding regions.
Characterization of the recombinant enzyme.
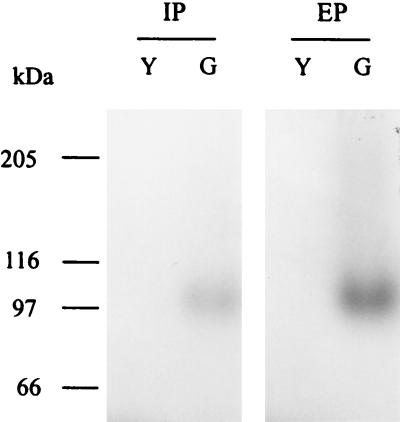
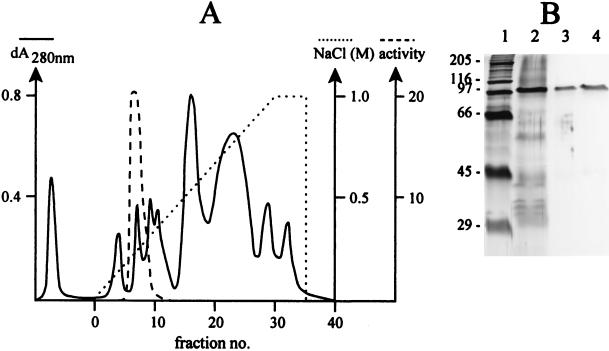
When intra- and extracellular proteins from S. cerevisiae INVSc1 transformed with pYGL (containing the entire coding region of cmg1) were separated by SDS-PAGE and stained for β-1,3-glucanase activity, a distinct band at ∼100 kDa was detected, whereas no activity was observed in the control yeast transformed with pYES2 (Fig. 2). The recombinant CMG1 enzyme was then purified from 1,000-fold-concentrated culture supernatant of INVSc1(pYGL) by anion-exchange chromatography. A fraction that eluted at 250 mM NaCl showed the greatest β-1,3-glucanase activity. This fraction was desalted, concentrated, and checked for homogeneity by SDS-PAGE (Fig. 3). A single protein band appeared at ∼100 kDa (Fig. 3B, lanes 3 and 4). N-terminal sequence analysis of this protein yielded the sequence APAPGAASG, which corresponds to residues 32 to 40 deduced from the nucleotide sequence of cmg1 (Fig. 1).
FIG. 2.
Detection of β-1,3-glucanase activity in polyacrylamide gels. S. cerevisiae INVSc1 transformed either with pYES2 (lanes Y) or with pYGL (lanes G) was induced by galactose, and 100 μg of intracellular protein (IP) or 15 μl of 10×-concentrated culture supernatant (extracellular protein [EP]) was separated by SDS-PAGE, renatured, and screened for β-1,3-glucanase activity. The positions of molecular mass markers are indicated on the left.
FIG. 3.
Purification of the recombinant glucanase. (A) Ion-exchange chromatography. (B) Silver-stained protein in SDS-PAGE gel. Lane 1, molecular mass standards; lane 2, concentrated culture supernatant of S. cerevisiae INVScl(pYGL); lanes 3 and 4, samples from the glucanase active fraction.
The substrate specificity of CMG1 was tested with a number of polysaccharide substrates containing different β-glycosyl linkages, such as laminarin, pustulan, lichenin, microcrystalline cellulose, carboxymethyl cellulose, and xylan. Sclerotial cell wall extract was also included in this test. The recombinant enzyme exhibited high activity only with laminarin, 20-fold-lower activity with the cell wall extract, and no activity with any other substrate tested. To prove that cmg1 codes for an exo-acting glucanase, the release of glucose and the release of glucose equivalent reducing sugars from laminarin were compared. Both the amounts and the rates of liberation of glucose and glucose equivalent reducing sugars appeared to be the same (data not shown), suggesting that the sole hydrolysis product is glucose; thus, CMG1 acts as an exoglucanase.
Inhibition of mycelial growth of S. sclerotiorum by CMG1.
The inhibitory activity of the purified enzyme was tested with S. sclerotiorum mycelial cultures in cell culture plates. We found that CMG1 applied at concentrations of 300 and 600 μg ml−1 caused 35 and 85% growth inhibition, respectively. No inhibition was observed when the enzyme was inactivated by boiling prior to application.
Expression of the cmg1 gene.
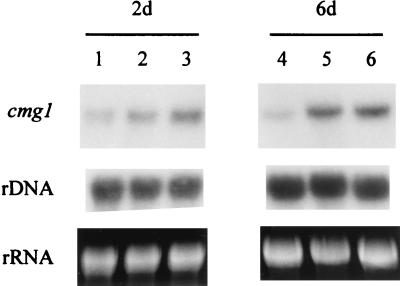
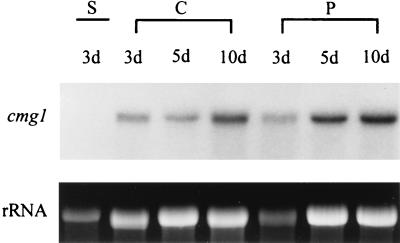
Expression of cmg1 under different growth conditions was studied by performing Northern analyses. C. minitans Cm-2 was grown as a shaken culture in SM containing different carbon sources. Total-RNA samples extracted from the mycelia after 2 and 6 days of culturing were electrophoresed, blotted, and hybridized to the radiolabelled cmg1 gene. As shown in Fig. 4, cmg1 transcripts were present in each sample. Hybridization signals having similar intensities were detected with an initial glucose concentration of 0.1%, representing starvation conditions (Fig. 4, lanes 2 and 5), and with 0.5% freeze-dried, ground sclerotia of S. sclerotiorum (lanes 3 and 6). Lower levels of expression were observed with an initial glucose concentration of 2% (lanes 1 and 4), especially after 6 days of culturing. The glucose content of the medium was still 1% after 6 days of growth, indicating that the carbon supply was not limiting. We also tested expression of cmg1 during parasitism in a dual-culture plate assay on SM containing 0.1% glucose. Two hundred sclerotia of S. sclerotiorum placed on this medium were inoculated with 200 μl of a C. minitans Cm-2 conidium suspension (106 conidia ml−1); control plates were inoculated only with C. minitans conidia or S. sclerotiorum sclerotia. Northern hybridization to RNA samples extracted after 3, 5, and 10 days of incubation clearly indicated that expression of cmg1 increased in the presence of sclerotia, the natural targets of C. minitans (Fig. 5). A quantitative comparison of the relative expression levels, performed by analyzing scanned images of photographs taken from ethidium bromide-stained gels and the corresponding autoradiographs, revealed a twofold increase in samples taken on days 3 and 5 and a 1.5-fold increase in the case of the 10-day-old samples. We determined by both Northern hybridization (Fig. 5, lane 1) and low-stringency Southern hybridization (data not shown) that S. sclerotiorum contained no sequences homologous to cmg1.
FIG. 4.
Northern blot analysis of cmg1. C. minitans Cm-2 was grown as a shaken culture in SM supplemented with 2% glucose (lanes 1 and 4), 0.1% glucose (lanes 2 and 5), or 0.5% freeze-dried, ground sclerotia of S. sclerotiorum (lanes 3 and 6). Five micrograms of total RNA extracted after 2 days (2d) (lanes 1 to 3) or 6 days (6d) (lanes 4 to 6) of growth was electrophoresed on a formaldehyde gel, blotted, and hybridized to the radiolabelled cmg1 gene (upper panels). The middle panels show control hybridizations with a ribosomal DNA (rDNA) probe. The bottom panels show ethidium bromide-stained rRNA. Representative results of two independent experiments are shown.
FIG. 5.
Expression of cmg1 during sclerotial parasitism. RNA was extracted from dual cultures of C. minitans and S. sclerotiorum (P) and from mycelium of S. sclerotiorum (S) or C. minitans (C) grown alone, as controls. Five micrograms of RNA was loaded onto a formaldehyde gel and hybridized to the radiolabelled cmg1 gene. The cultures were 3, 5, and 10 days old (3d, 5d, and 10d, respectively) when samples were removed. The bottom panel shows ethidium bromide-stained rRNA as an indication of the amounts of RNA loaded. Representative results of three independent experiments are shown.
DISCUSSION
It has been suggested that β-1,3-glucanases contribute to the mycoparasitic activities of several fungal species by facilitating penetration through host cell wall structures (2, 4, 8, 11, 29). However, information on the cell wall-degrading hydrolases of C. minitans is sparse. Although earlier studies (23) showed the presence of two synergistically interacting glucanases in the culture filtrate of this fungus, no data on the physicochemical properties, inducibility, and genetic background of these enzymes have been available.
In this study we characterized an exo-β-1,3-glucanase gene of C. minitans, designated cmg1. The degenerate oligonucleotide primers which we used to clone this gene were constructed on the basis of conserved sequence motifs found in three filamentous fungal exoglucanases (9, 40, 45). Our results suggest that these primers may be useful in cloning β-1,3-glucanase genes from other filamentous fungi.
Sequence analysis of the deduced protein product of cmg1 revealed 61, 42, and 41% overall similarities with EXGA of A. quisqualis, LAM1.3 of T. harzianum, and EXG1 of C. carbonum, respectively. All three of these are exo-β-1,3-glucanase sequences. A comparison of the CMG1 sequence with the sequence of BGN13.1 of T. harzianum, the only endo-acting β-1,3-glucanase cloned from filamentous fungi (11), showed a lower level of overall homology (27%), indicating that CMG1 is more closely related to the exo-acting glucanases. We confirmed the exoglucanase nature of this protein when we analyzed the amounts and the rates of liberation of glucose and glucose equivalent reducing sugars liberated from laminarin by the recombinant enzyme. Substrate specificity tests revealed that CMG1 is highly specific for 1,3-β-glycosyl linkages, as no reducing sugars were released by the enzyme from polymeric substrates containing 1,4-, 1,3(4)-, or 1,6-β-glycosyl linkages. The molecular mass of the secreted recombinant enzyme as estimated by SDS-PAGE (100 kDa) appeared to be somewhat larger than that calculated from the nucleotide sequence of the gene (82.3 kDa). The difference may be due to glycosylation of the enzyme since we identified the presence of potential N-glycosylation sites in the deduced protein sequence. Penttilä et al. (42) observed that overglycosylated cellulases of filamentous fungi expressed in S. cerevisiae were larger than the original proteins secreted by the host organisms.
Expression of the cmg1 cDNA clone in S. cerevisiae provided evidence that its protein product is an extracellular enzyme secreted into the culture medium. Nucleotide sequence analysis predicted a signal peptide cleavage site between amino acids Ala-24 and Ala-25. However, N-terminal sequence analysis of the recombinant enzyme revealed that the mature glucanase starts at Ala-32, seven amino acids downstream of the predicted signal peptidase cleavage site. This difference might be explained by further proteolytic cleavage, a step that occurs frequently during processing of eukaryotic extracellular enzymes (18). The predicted signal peptidase cleavage site in CMG1 is immediately followed by a characteristic stretch of dipeptides (either X-Ala or X-Pro) (Fig. 1) which are known to be substrates of dipeptidyl aminopeptidase A. This enzyme is known to be involved in proteolytic processing of a range of secreted yeast proteins (e.g., the α mating pheromone of S. cerevisiae and AEP, an alkaline extracellular protease of Yarrowia lipolytica) (25, 34).
Expression of the majority of the genes encoding cell wall-degrading enzymes in mycoparasitic fungi is repressed by glucose and can be induced by autoclaved mycelium, fungal cell wall extracts, or polymers, such as laminarin and chitin (9, 19, 28, 45). Interactions between a parasite and its host have also been reported to trigger expression of chitinase and proteinase genes of T. harzianum (10, 53). Small diffusible molecules derived from host cell walls have been shown to elevate expression of these genes. Physiological stress and carbon starvation also appear to be involved in upregulation of some of the genes encoding mycolytic enzymes (28, 31, 33). However, we found that cmg1 is not entirely repressed by glucose. Compared to the levels detected on 2% glucose, expression of cmg1 transcripts increased under starvation conditions (0.1% glucose), as well as in the presence of autoclaved, ground sclerotia of S. sclerotiorum (Fig. 4). C. minitans cultures grown under these conditions were examined microscopically to check whether autolysis contributed to the elevated transcription of cmg1. No sign of autolysis was detected. The only change found in these cultures was an increase in sporulation incited by carbon limitation. Moreover, during parasitic attack of sclerotia of S. sclerotiorum by C. minitans we found greater expression of cmg1 in the mycelium parasitizing the host than in the controls. These results indicate that multiple regulatory mechanisms are involved in expression of cmg1 and also suggest that the gene plays an important role in sclerotial parasitism.
Various strategies have been suggested to utilize genes encoding cell wall-degrading enzymes of mycoparasitic fungi. First, superior fungal strains that exhibit greater biocontrol activities have been produced by genetic transformation (13, 27, 37), an approach that became especially promising after the recent development of a transformation system for C. minitans (24). Second, transgenic plants that exhibit increased resistance to fungal pathogens have been obtained by introducing genes of fungal origin (5, 30). Finally, expression of these genes in heterologous hosts may yield enzyme production at a commercially reasonable scale (32). The cmg1 gene of C. minitans, the first described gene cloned from this mycoparasitic fungus, appears to be a good candidate for use in any of these strategies.
ACKNOWLEDGMENTS
This work was supported by grants from OTKA (grant F 029999) and FKFP (grant 0315/99). G.G. was partially supported by a Bolyai Research Fellowship from the Hungarian Academy of Sciences.
We thank László Vajna (Plant Protection Institute, Budapest, Hungary) for providing the fungal strains C. minitans Cm-2 and S. sclerotiorum Sc-1 and for helpful advice on culturing practices. We are also indebted to Zsuzsanna Buzás and András Patthy for protein sequencing. We are grateful to Kurt Zeller for comments and linguistic improvement of the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D P. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Archambault C, Coloccia G, Kermashe S, Jabai-Hare S. Characterization of an endo-1,3-β-d-glucanase produced during the interaction between Stachybotrys elegans and its host Rhizoctonia solani. Can J Microbiol. 1998;44:989–997. [PubMed] [Google Scholar]
- 3.Archer D B, Peberdy J F. The molecular biology of secreted enzyme production by fungi. Crit Rev Biotechnol. 1997;17:273–306. doi: 10.3109/07388559709146616. [DOI] [PubMed] [Google Scholar]
- 4.Benhamou N, Chet I. Cellular and molecular mechanisms involved in the interaction between Trichoderma harzianum and Pythium ultimum. Appl Environ Microbiol. 1997;63:2095–2099. doi: 10.1128/aem.63.5.2095-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolar J P, Norelli J L, Wong K-W, Hayes C K, Harman G E, Aldwinckle H S. Expression of endochitinase from Trichoderma harzianum in transgenic apple increases resistance to apple scab and reduces vigor. Phytopathology. 2000;90:72–77. doi: 10.1094/PHYTO.2000.90.1.72. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Budge S P, Whipps J M. Glasshouse trials of Coniothyrium minitans and Trichoderma species for the biological control of Sclerotinia sclerotiorum in celery and lettuce. Plant Pathol (London) 1991;40:59–66. [Google Scholar]
- 8.Chiu S C, Tzean S S. Glucanolytic enzyme production by Schizophyllum commune Fr. during mycoparasitism. Physiol Mol Plant Pathol. 1995;46:83–94. [Google Scholar]
- 9.Cohen-Kupiec R, Broglie K E, Friesem D, Broglie R M, Chet I. Molecular characterization of a novel β-1,3-exoglucanase related to mycoparasitism of Trichoderma harzianum. Gene. 1999;226:147–154. doi: 10.1016/s0378-1119(98)00583-6. [DOI] [PubMed] [Google Scholar]
- 10.Cortes C, Gutierrez A, Olmedo V, Inbar J, Chet I, Herrera-Estrella A. The expression of genes involved in parasitism by Trichoderma harzianum is triggered by a diffusible factor. Mol Gen Genet. 1998;260:218–225. doi: 10.1007/s004380050889. [DOI] [PubMed] [Google Scholar]
- 11.de la Cruz J, Pintor-Toro J A, Benítez T, Llobell A, Romero R C. A novel endo-β-1,3-glucanase, BGN13.1, involved in the mycoparasitism of Trichoderma harzianum. J Bacteriol. 1995;177:6937–6945. doi: 10.1128/jb.177.23.6937-6945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores A, Chet I, Herrera-Estrella A. Improved biocontrol activity of Trichoderma harzianum by over-expression of the proteinase-encoding gene prb1. Curr Genet. 1997;31:30–7. doi: 10.1007/s002940050173. [DOI] [PubMed] [Google Scholar]
- 14.Gerlagh M, Goossen-van de Geijn H M, Fokkema N J, Vereijken P F G. Long-term biosanitation by application of Coniothyrium minitans on Sclerotinia sclerotiorum-infected crops. Phytopathology. 1999;89:141–147. doi: 10.1094/PHYTO.1999.89.2.141. [DOI] [PubMed] [Google Scholar]
- 15.Giczey G, Kerényi Z, Dallmann G, Hornok L. Homologous transformation of Trichoderma hamatum with an endochitinase encoding gene, resulting in increased levels of chitinase activity. FEMS Microbiol Lett. 1998;165:247–252. doi: 10.1111/j.1574-6968.1998.tb13153.x. [DOI] [PubMed] [Google Scholar]
- 16.Gietz R D, Jean A S, Woods R A, Schiestl R H. Improved method for high-efficiency transformation of intact yeast cell. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, United Kingdom: IRL Press; 1988. pp. 93–139. [Google Scholar]
- 18.Halban P A, Irminger J C. Sorting and processing of secretory proteins. Biochem J. 1994;299:1–18. doi: 10.1042/bj2990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haran S, Schickler H, Chet I. Molecular mechanisms of lytic enzymes involved in the biocontrol activity of Trichoderma harzianum. Microbiology. 1996;142:2321–2331. [Google Scholar]
- 20.Huang H C. Control of sclerotinia wilt of sunflower by hyperparasites. Can J Plant Pathol. 1980;2:26–32. [Google Scholar]
- 21.Huang H C, Kokko E G. Ultrastructure of hyperparasitism of Coniothyrium minitans on sclerotia of Sclerotinia sclerotiorum. Can J Bot. 1987;65:2483–2489. [Google Scholar]
- 22.Jones D. Ultrastructure and composition of the cell walls of Sclerotinia sclerotiorum. Trans Br Mycol Soc. 1970;53:351–360. [Google Scholar]
- 23.Jones D, Gordon A H, Bacon J S D. Co-operative action by endo- and exo-β-(1,3)glucanases from parasitic fungi in the degradation of cell-wall glucans of Sclerotinia sclerotiorum (lib.) de Bary. Biochem J. 1974;140:47–55. doi: 10.1042/bj1400047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones E, Carpenter M, Fong D, Goldstein A, Thrush A, Crowhurst, Stewart A. Co-transformation of the sclerotial mycoparasite Coniothyrium minitans with hygromycin B resistance and β-glucuronidase markers. Mycol Res. 1999;103:929–937. [Google Scholar]
- 25.Julius D, Blair L, Brake A, Sprague G, Thorner J. Yeast alpha factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell. 1983;32:839–852. doi: 10.1016/0092-8674(83)90070-3. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 27.Limón M C, Pintor-Toro J A, Benítez T. Increased antifungal activity of Trichoderma harzianum transformants that overexpress a 33-kDa chitinase. Phytopathology. 1999;89:254–261. doi: 10.1094/PHYTO.1999.89.3.254. [DOI] [PubMed] [Google Scholar]
- 28.Lora J M, de la Cruz J, Llobell A, Benítez T, Pintor-Toro J A. Molecular characterization and heterologous expression of an endo-beta-1,6-glucanase gene from the mycoparasitic fungus Trichoderma harzianum. Mol Gen Genet. 1995;247:639–645. doi: 10.1007/BF00290356. [DOI] [PubMed] [Google Scholar]
- 29.Lorito M, Hayes C K, Di Pietro A, Woo S L, Harman G E. Purification, characterization, and synergistic activity of a glucan 1,3-β-glucosidase and an N-acetyl-β-glucosaminidase from Trichoderma harzianum. Phytopathology. 1994;84:398–405. [Google Scholar]
- 30.Lorito M, Woo S L, Garcia I, Colucci G, Harman G E, Pintor-Toro J A, Filippone E, Muccifora S, Lawrence C B, Zoina A, Tuzun S, Scala F. Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proc Natl Acad Sci USA. 1998;95:7860–7865. doi: 10.1073/pnas.95.14.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mach R L, Peterbauer C K, Payer K, Jaksits S, Woo S L, Zeilinger S, Kullnig C M, Lorito M, Kubicek C P. Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl Environ Microbiol. 1999;65:1858–1863. doi: 10.1128/aem.65.5.1858-1863.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margolles-Clark E, Hayes C K, Harman G E, Penttilä M E. Improved production of Trichoderma harzianum endochitinase by expression in Trichoderma reesei. Appl Environ Microbiol. 1996;62:2145–2151. doi: 10.1128/aem.62.6.2145-2151.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margolles-Clark E, Harman G E, Penttilä M E. Enhanced expression of endochitinase in Trichoderma harzianum with the cbhl promoter of Trichoderma reesei. Appl Environ Microbiol. 1996;62:2152–2155. doi: 10.1128/aem.62.6.2152-2155.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matoba S, Morano K A, Klionsky D J, Kim K, Ogrydziak D M. Dipeptidyl aminopeptidase processing and biosynthesis of alkaline extracellular protease from Yarrowia lipolytica. Microbiology. 1997;143:3263–3272. doi: 10.1099/00221287-143-10-3263. [DOI] [PubMed] [Google Scholar]
- 35.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 36.McQuilken M P, Mitchell S J, Budge S P, Whipps J M, Fenlon J S, Archer S A. Effect of Coniothyrium minitans on sclerotial survival and apothecial production of Sclerotinia sclerotiorum in field-grown oilseed rape. Plant Pathol (London) 1995;44:883–896. [Google Scholar]
- 37.Migheli Q, González-Candelas L, Dealessi L, Camponogara A, Ramón-Vidal D. Transformants of Trichoderma longibrachiatum overexpressing the β-1,4-glucanase gene egl1 show enhanced biocontrol of Pythium ultimum on cucumber. Phytopathology. 1998;88:673–677. doi: 10.1094/PHYTO.1998.88.7.673. [DOI] [PubMed] [Google Scholar]
- 38.Miller G L. Use of dinitrosalicylic reagent for determination of reducing sugars. Anal Biochem. 1959;31:426–428. [Google Scholar]
- 39.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 40.Nikolskaya A N, Pitkin J W, Schaeffer H J, Ahn J H, Walton J D. EXG1p, a novel exo-β1,3-glucanase from the fungus Cochliobolus carbonum, contains a repeated motif present in other proteins that interact with polysaccharides. Biochim Biophys Acta. 1998;1425:632–636. doi: 10.1016/s0304-4165(98)00117-2. [DOI] [PubMed] [Google Scholar]
- 41.Pan S Q, Ye X S, Kuc J. Direct detection of β-1,3-glucanase isozymes on polyacrylamide electrophoresis and isoelectrofocusing gels. Anal Biochem. 1989;182:136–140. doi: 10.1016/0003-2697(89)90730-6. [DOI] [PubMed] [Google Scholar]
- 42.Penttilä M E, André L, Lehtovara P, Bailey M, Teeri T, Knowles J K C. Efficient secretion of two fungal cellobiohydrolases by Saccharomyces cerevisiae. Gene. 1988;63:103–112. doi: 10.1016/0378-1119(88)90549-5. [DOI] [PubMed] [Google Scholar]
- 43.Philips A J L, Price K. Structural aspects of the parasitism of sclerotia of Sclerotinia sclerotiorum (Lib.) de Bary by Coniothyrium minitans Campb. Phytopathol Z. 1983;107:193–203. [Google Scholar]
- 44.Pitson S M, Seviour R J, McDougall R J. Noncellulolytic fungal β-glucanases: their physiology and regulation. Enzyme Microb Technol. 1993;15:178–192. doi: 10.1016/0141-0229(93)90136-p. [DOI] [PubMed] [Google Scholar]
- 45.Rotem Y, Yarden O, Sztejnberg A. The mycoparasite Ampelomyces quisqualis expresses exgA encoding an exo-β-1,3-glucanase in culture and during mycoparasitism. Phytopathology. 1999;89:631–638. doi: 10.1094/PHYTO.1999.89.8.631. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 47.Stiekema W, Heidekamp F, Dirkse W, van Beckum J, de Haan P, ten Bosch C, Louwerse J. Molecular cloning and analysis of four potato tuber mRNAs. Plant Mol Biol. 1988;11:255–269. doi: 10.1007/BF00027383. [DOI] [PubMed] [Google Scholar]
- 48.Trutmann P, Kean P J, Merriman P R. Biological control of Sclerotinia sclerotiorum on aerial parts of plants by Coniothyrium minitans. Trans Br Mycol Soc. 1982;78:521–529. [Google Scholar]
- 49.Tu J C. Mycoparasitism by Coniothyrium minitans on Sclerotinia sclerotiorum and its effect on sclerotial germination. Phytopathol Z. 1984;109:261–268. [Google Scholar]
- 50.Whipps J M, Gerlagh M. Biology of Coniothyrium minitans and its potential for use in disease biocontrol. Mycol Res. 1992;96:897–907. [Google Scholar]
- 51.Woo S L, Donzelli B, Scala F, Mach R, Harman G E, Kubicek C P, Del Sorbo G, Lorito M. Disruption of the ech42 (endochitinase-encoding) gene affects biocontrol activity in Trichoderma harzianum. Mol Plant-Microbe Interact. 1999;12:419–429. [Google Scholar]
- 52.Wray W, Boulikas T, Wray V P, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- 53.Zeilinger S, Galhaup C, Payer K, Woo S L, Mach R L, Fekete C, Lorito M, Kubicek C P. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet Biol. 1999;26:131–140. doi: 10.1006/fgbi.1998.1111. [DOI] [PubMed] [Google Scholar]