Abstract
To evaluate PCR-generated artifacts (i.e., chimeras, mutations, and heteroduplexes) with the 16S ribosomal DNA (rDNA)-based cloning approach, a model community of four species was constructed from alpha, beta, and gamma subdivisions of the division Proteobacteria as well as gram-positive bacterium, all of which could be distinguished by HhaI restriction digestion patterns. The overall PCR artifacts were significantly different among the three Taq DNA polymerases examined: 20% for Z-Taq, with the highest processitivity; 15% for LA-Taq, with the highest fidelity and intermediate processitivity; and 7% for the conventionally used DNA polymerase, AmpliTaq. In contrast to the theoretical prediction, the frequency of chimeras for both Z-Taq (8.7%) and LA-Taq (6.2%) was higher than that for AmpliTaq (2.5%). The frequencies of chimeras and of heteroduplexes for Z-Taq were almost three times higher than those of AmpliTaq. The total PCR artifacts increased as PCR cycles and template concentrations increased and decreased as elongation time increased. Generally the frequency of chimeras was lower than that of mutations but higher than that of heteroduplexes. The total PCR artifacts as well as the frequency of heteroduplexes increased as the species diversity increased. PCR artifacts were significantly reduced by using AmpliTaq and fewer PCR cycles (fewer than 20 cycles), and the heteroduplexes could be effectively removed from PCR products prior to cloning by polyacrylamide gel purification or T7 endonuclease I digestion. Based upon these results, an optimal approach is proposed to minimize PCR artifacts in 16S rDNA-based microbial community studies.
The detection, identification, and characterization of microbial populations and their activities in environments are a great challenge to microbiologists. The application of culture-independent nucleic acid techniques has greatly advanced the detection and identification of microorganisms in natural habitats. In the last decade, the use of 16S rRNAs or ribosomal DNAs (rDNAs) as molecular markers has become routine for microbial ecologists. Several different rRNA-based approaches have been used to characterize microbial communities, such as cloning plus sequencing (31), amplified rDNA restriction analysis (23), terminal restriction fragment length polymorphism (RFLP) analysis (1, 21), RFLP analysis (25), denaturing gradient gel electrophoresis (DGGE) (16, 26), temperature gradient gel electrophoresis (TGGE) (27), single-strand conformation polymorphism analysis (19), and heteroduplex mobility assay (12). Nearly every study applying these approaches reveals novel microbial groups, and many of them are still undetectable by cultivation (2, 3, 7, 8, 11, 13–15, 17, 29, 39, 41). Use of these methods with PCR, however, can cause bias (32, 33) and artifacts that lead to overestimation of community diversity (38).
Although PCR-generated chimeras have received much attention (4, 17, 18, 20, 28, 34, 35, 38), PCR-generated heteroduplexes and mutations have largely been ignored. It is impossible to avoid the formation of heteroduplexes in the PCR products when a mixture of homologous genes is used as PCR templates. When a heteroduplex molecule is cloned and transformed, two homoduplex molecules of 16S rRNA genes will be produced and segregated as a result of plasmid propagation (30). When the mixed 16S rRNA genes are subjected to RFLP, DGGE or TGGE analysis, artificial RFLP patterns or DGGE or TGGE conformations will be generated.
PCR-generated mutations pose another potential problem. Compared to other DNA polymerases, Taq DNA polymerase has a higher intrinsic misincorporation rate during synthesis (6). Such errors can accumulate and be enlarged during PCR amplification (38). When an error occurs at the restriction enzyme-recognizing site, an artificial RFLP pattern will occur. Although the phenomenon of PCR-induced mutations is well known, their effects on community diversity studies have not been adequately addressed. In addition, when the amplified target gene has secondary structure, deletion mutations may exist (5). Since the 16S rRNA gene has a stable secondary structure, deletion mutations could be produced during PCR amplifications.
Theoretically, PCR-generated chimeras should be fewer in amplifications with DNA polymerases with higher processitivity and decrease as elongation time increases and cycle number decreases. The PCR-induced mutations should be lower for DNA polymerases with either higher fidelity (point mutation) (38) or higher processitivity (deletion mutation) (5) and decrease as the cycle number decrease. The frequency for formation of heteroduplexes in the PCR products should decrease when lesser amounts of PCR products are synthesized. In addition, there is a potential increase of total PCR artifacts as species diversity increases. To test these hypotheses and minimize the three types of PCR artifacts, we evaluated PCR artifacts under different amplification conditions, and here we provide a general approach for reducing artifacts in 16S rRNA gene-based cloning studies.
MATERIALS AND METHODS
Strains used.
The 16S rRNA genes were amplified from more than 40 different gram-negative and positive bacteria with the eubacterium-specific primers FD1 and R1540 (36, 40) and digested with HhaI. Ten strains that showed distinct HhaI digestion patterns were selected for constructing model microbial communities (Table 1).
TABLE 1.
Strains and model communities
| Strain (community)a | Species | Subdivision or group |
|---|---|---|
| C1-4 (4, 7, 10) | Unidentified alpha Proteobacterium | Alpha |
| C3-2 (7, 10) | Unidentified alpha Proteobacterium | Alpha |
| G-191 (7, 10) | Alcaligenes faecalis | Beta |
| Tol-4 (4, 7, 10) | Azoarcus tolulyticus | Beta |
| C10-5 (7, 10) | Marinobacter sp. | Gamma |
| ATCC 10145 (10) | Pseudomonas aeruginosa | Gamma |
| B9-12 (4, 7, 10) | Halomonas variabilis | Gamma |
| MW-31 (10) | Acetobacter sp. | Gamma |
| A2-1 (10) | Pseudomonas caricapapayae | Gamma |
| P39 (4, 7, 10) | Rhodococcus equi | Gram positive |
Numbers in parentheses indicate strains used in 4-, 7-, and 10-species communities, respectively. Sequence similarities of 16S genes were within the ranges of 76 to 89% in the 4 species community, 75 to 98% in the 7-species community, and 69 to 98% in the 10-species community.
The selected 16S rRNA genes were cloned into plasmid vector pCR II with the TA cloning kit (Invitrogen BV, San Diego, Calif.). The cloned 16S rRNA genes were amplified from plasmid DNAs using primers TA-F (5′GCCGCCAGTGTGCTGGAATT3′) and TA-R (5′TAGATGCATGCTCGAGCGGC3′), which are specific to the polylinker of the vector pCR II. The amplified 16S rDNA fragments were purified by using the Wizard PCR Preps DNA Purification System (Promega, Madison, Wis.) and quantified with PicoGreen dye (Molecular Probes, Inc. Eugene, Oreg.) by using a LB50 Luminescence Spectrometer (Perkin-Elmer, Branchburg, N.J.). The amounts of template were calculated to yield equivalent genomic DNA under the assumption of seven rRNA copies per genome (4.64 Mb) as in Escherichia coli.
Experimental design and PCR amplification conditions.
The effects of various PCR amplification parameters, including DNA polymerase, cycle number, elongation time, and template concentration, on the formation of PCR artifacts were examined in a four-species community, which contained C1-4, To1-4, B9-12, and P39, one each from the alpha, beta, and gamma subdivisions of the division Proteobacteria, and a gram-positive bacterium. All treatments were carried out in triplicate. For the amplifications above 20 cycles, three amplifications were combined, whereas for the amplifications below 20 cycles, 10 amplifications were combined.
To determine the effect of DNA polymerase on the formation of PCR artifacts, three Taq DNA polymerases were examined: TaKaRa Z-Taq (Pan Vera Corporation, Madison, Wis.), with the highest processitivity; LA-Taq (Pan Vera Corporation), with the highest accuracy and intermediate processitivity; and the conventionally used AmpliTaq DNA polymerase (Perkin-Elmer). PCR amplifications were carried out under the optimum conditions for each Taq as suggested by the manufacturers: in 1× Z-Taq buffer containing 3 mM Mg2+ and a 200 μM concentration of each deoxynucleoside triphosphate (dNTP) with Z-Taq; in 1× GC buffer I, including 2.5 mM Mg2+ and a 400 μM concentration of each dNTP with LA-Taq; and in a buffer of 10 mM Tris-HCl (pH 8.3 at 25°C)–50 mM KCl, containing 1.5 mM Mg2+ and a 200 μM concentration of each dNTP with AmpliTaq. The standard reaction mixture contained 20 pmol of each forward and reverse primer, 230 fg of cloned 16S rDNA (equivalent to 100 pg of genomic DNA) from each strain, bovine serum albumin (100 μg/ml), and 1 U of the Taq DNA polymerase in a final volume of 20 μl. Reaction mixtures were incubated in a thermocycler, (model Gene Amp PCR system 9700; PE Applied Biosystem, Branchburg, N.J.) at 95°C for 5 min, followed by 30 cycles at 95°C for 40 s, 58°C for 30 s, and 72°C for 4 min and then by a final extension at 72°C for 7 min.
To examine the effect of elongation time on the formation of PCR artifacts, the 16S rDNAs were amplified with Z-Taq as described above, except the template concentration from each strain was equivalent to 30 ng of genomic DNA. The extension times compared were 20 s, 2 min, and 4 min. To examine the effect of template concentration on the formation of PCR artifacts, the 16S rDNAs were mixed in equal ratios with an equivalent of 100 pg, 1 ng, or 10 ng of genomic DNA per strain and amplified with Z-Taq. In addition, the effect of PCR cycle number on the formation of PCR artifacts was evaluated with Z-Taq for 22, 25, and 28 cycles as well as with AmpliTaq for 15, 20, 25, and 30 cycles. Both amplifications used a mixture of four strains of an equivalent of 100 pg of genomic DNA per strain.
To examine the effect of species diversity on the formation of PCR artifacts, model communities were constructed consisting of 4, 7, and 10 species (Table 1). The total template concentration used in each community was equivalent to 500 pg of genomic DNA, with equal ratios of each strain. 16S rDNAs were amplified with AmpliTaq for 15 cycles.
16S rRNA gene cloning.
Multiple amplications 3 or 10 were combined and purified from low-melting-point agarose gel by using the Wizard PCR Preps DNA Purification System prior to ligation. For amplification with fewer than 20 cycles, PCR products were concentrated by ethanol precipitation. The ratio of 16S rDNAs to pCR II vector was 1:1. Two microliters of each ligation reaction mixture was transformed by heat pulse into E. coli Top10F' competent cells (Invitrogen).
16S rDNA RFLP analysis.
The 16S rDNA inserts were amplified directly from transformant cells in 20 μl with primers TA-F and TA-R (41). A 5-μl sample of the amplification mixture was digested with 1 U of HhaI (Gibco BRL, Gaithersburg, Md.) in a final volume of 15 μl at 37°C overnight. The resulting products were resolved by electrophoresis in 1.6% agarose gel in 1× Tris-borate-EDTA (TBE) buffer at 94 V for 4 h. The gel was stained with 0.5 μg of ethidium bromide per ml and visualized by UV excitation. PCR artifacts were detected by comparing the RFLP patterns to those of the reference strains using the Molecular Analyst Program (Bio-Rad, Hercules, Calif.).
Detection of chimeras and mutations.
DNA sequencing was carried out on an automated sequencer (model 373A; Applied Biosystems, Foster City, Calif.). Primers FD1 (E. coli 16S rRNA gene position 8 to 27), F925 (position 906 to 925), R529 (position 529 to 512), and R1392 (position 1406 to 1392) were used to obtain the sequences of both ends from both strands. About 200 bp from each individual primer was compared to the database containing the reference 16S rRNA gene sequences using the FASTA program (Genetics Computer Group Sequence Analysis Software Package; University of Wisconsin, Madison). If the sequences of the two ends showed the highest similarity to different reference strains but the sequences of both strands at the same end showed the highest similarity to the same reference strain, this clone was considered a chimera.
Primers F270 (position 246 to 261), F519 (position 512 to 529), F1099 (position 1099 to 1114), R350 (position 342 to 357), R925 (position 925 to 906), and R1540 (position 1541 to 1525) were used to obtain full 16S rRNA gene sequences. The sequences were assembled with PhredPhrap and Consed (University of Washington, Seattle) and compared to reference sequences using MAP, MAPSORT, and GAP (Genetics Computer Group). The error rate was the percentage of total examined clones that have a misincorporated nucleotide at the HhaI sites.
Detection and elimination of heteroduplexes.
To detect the heteroduplexes, an 8-μl aliquot of the PCR product from the clone with altered RFLP pattern was mixed with 2 μl of 50 mM EDTA (final concentration, 10 mM), denatured at 95°C for 5 min, and renatured at 25°C for 40 min (9, 35). The sample was separated on a 5% nondenatured polyacrylamide (49:1 ratio of acrylamide to bis) gel (16 by 20 cm) with a D Gene System (Bio-Rad) in 1× TBE buffer at 250 V at least for 3 h. The gel was stained with 0.5 μg of ethidium bromide per ml for 15 min, destained in 1× TBE for 30 min, and visualized by UV excitation. A heteroduplex was determined by comparing its banding pattern to those of reference homoduplex molecules. The clones showing extra bands that migrated more slowly than the homoduplex molecules but faster than single-stranded DNA molecules were considered heteroduplexes.
To remove heteroduplexes prior to cloning, 45 μl of amplified 16S rDNAs (Z-Taq) was mixed with 5 μl of 10× loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol, 25% Ficoll [type 400]), and separated on 5% nondenatured polyacrylamide gel. The bands corresponding to homoduplexes of 16S rDNAs were excised. The 16S rDNAs were recovered using a QIAEX II gel extraction kit (QIAGEN Inc., Valencia, Calif.) according to the manufacturer's instructions, except that the crushed gel strips were diffused twice with a 1.5× volume of diffuse buffer (0.5 M ammonium acetate 10 mM manganese acetate, 1 mM EDTA, and 0.1% sodium dodecyl sulfate) at 50°C for 30 min. 16S rDNAs were eluted in 40 μl of TE (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA) and concentrated to 20 μl by ethanol precipitation. The final concentration of the recovered 16S rDNAs was estimated on the 0.8% agarose gel, and about 20 ng was used for ligation.
To digest heteroduplexes, 60 μl of amplified 16S rDNAs (LA-Taq) was concentrated to 6 μl by ethanol precipitation, and incubated with 1 μl (30 U) of T7 endonuclease I in a volume of 20 μl in a buffer containing 50 mM Tris (pH 8.0), 50 mM potassium glutamate, 10 mM MgCl2, 5 mM dithiothreitol, and 5% glycerol at 37°C for 20 min and then purified with the QIAEX II gel extraction kit and eluted in 20 μl of TE buffer (10 mM Tris [pH 8.0], 0.1 mM EDTA). Addition of 3′-A overhangs postamplification was carried out in a 20-μl reaction mixture containing 1× PCR buffer, 1.5 mM MgCl2, 0.1 mM dATP, and 0.5 U of Taq DNA polymerase at 65°C for 20 min. The resulting 16S rDNAs (about 20 ng) were directly used for ligation.
RESULTS
Detection of heteroduplexes by PAGE.
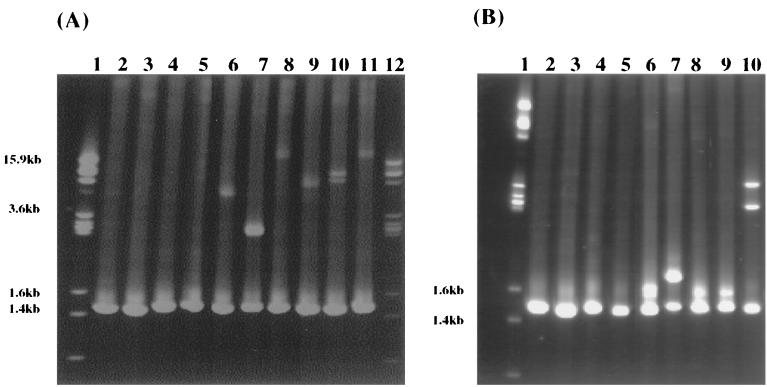
To test whether heteroduplex molecules of nearly entire 16S rRNA genes (1.5 kb) can be detected by polyacrylamide gel electrophoresis (PAGE) cloned 16S genes from the two strains were mixed equally and subjected to PCR amplification. The migration of heteroduplexes was retarded due to the “bubble” formation between mismatches (Fig. 1). The decrease of the heteroduplex mobility was inversely proportional to the sequence similarity of the two parental molecules (Fig. 1A, lanes 6 to 11). These results suggested that heteroduplexes, even those formed between distantly related 16S rRNA genes (76% similar), could be detected by PAGE. Some heteroduplexes of 16S rDNA formed between clones with misincorporated nucleotides and their parental strains were also detectable on the 5% nondenatured polyacrylamide gel (Fig. 1B, lanes 6 to 9).
FIG. 1.
Detection of 16S rDNA heteroduplex molecules by polyacrylamide gel electrophoresis. (A) Lanes 1 and 12, marker III; lanes 2 to 5, 16S rDNAs of C10-5, Tol-4, B9-12, and P-39, respectively; lanes 6 to 12, heteroduplexes formed between C10-5 and Tol-4 (84% similar), C10-5 and B9-12 (89% similar), C10-5 and P-39 (76% similar), Tol-4 and B9-12 (83% similar), Tol-4 and P-39 (79% similar), and B9-12 and P-39 (77% similar), respectively. (B) Lane 1, marker III; lanes 2 to 5, 16S rDNAs of C10-5, To-4, B9-12, and P-39, respectively; lanes 6 to 10, heteroduplexes formed between C10-5 and its mutation clone, Tol-4 and its mutation clone, B9-12 and its mutation clone, and P39 and its mutation clone, respectively; lane 10, positive control.
This PAGE method may fail to detect heteroduplex clones if the two 16S rDNAs are not equally abundant, which occurs very often due to the random partition of plasmids among daughter cells (30). To determine the effects of the ratios of two parental molecules on heteroduplex detection, the 16S rDNAs were coamplified from the mixture of C10-5 and B9-12 (similarity, 89%) and C10-5 and P-39 (similarity, 76%), respectively. The ratios of the two strains in the mixture were examined from 9:1 to 1:9, which caused the artificial RFLP patterns. The heteroduplexes formed from different ratios were all successfully detected by PAGE (data not shown).
Effects of DNA polymerases on formation of PCR artifacts.
The overall PCR-generated artifacts were enzyme dependent and significantly different among the three Taq DNA polymerases examined. The proportion of the overall PCR artifacts of LA-Taq was lower than that of Z-Taq but significantly higher than that of AmpliTaq (Table 2). No significant difference in chimera was observed between Z-Taq (8.7% ± 2.2%) and LA-Taq (6.2% ± 2.2%). However, the frequency of chimeras for AmpliTaq was significantly lower than those for Z-Taq and LA-Taq (Table 2).
TABLE 2.
Effects of PCR amplification conditions on formation of PCR artifacts
| Treatment | No. of clones examined | Mean % chimeras ± SD | Mean error ratea ± SD (%) | Mean % heteroduplexes ± SD | Mean % total PCR artifacts ± SD |
|---|---|---|---|---|---|
| DNA polymerase | |||||
| Z-Taq | 162 | 8.7 ± 2.2 | 9.3 ± 0.0d | 3.7 ± 1.1 | 20.4 ± 0.0d |
| LA-Taq | 162 | 6.2 ± 2.2 | 6.8 ± 0.6 | 1.2 ± 1.2 | 14.8 ± 3.2 |
| AmpliTaq | 162 | 2.5 ± 1.2 | 3.7 ± 1.1d | 1.2 ± 0.6 | 7.4 ± 1.1d |
| Elongation time | |||||
| 20 s | 162 | 10.3 ± 1.5d | 12.8 ± 2.1 | 1.8 ± 1.0 | 25.5 ± 3.9 |
| 2 min | 168 | 8.5 ± 2.6 | 10.6 ± 2.3 | 3.0 ± 1.6 | 22.1 ± 1.1 |
| 4 min | 165 | 3.7 ± 1.1d | 9.6 ± 2.1 | 3.0 ± 1.0 | 16.1 ± 3.2 |
| Template concn (ng/ml)b | |||||
| 0.1 | 162 | 6.8 ± 2.2 | 2.5 ± 1.2d | 1.3 ± 0.6 | 10.5 ± 2.2 |
| 1.0 | 162 | 8.6 ± 1.2 | 6.2 ± 1.6 | 1.2 ± 1.2 | 16.1 ± 2.7 |
| 10 | 162 | 6.2 ± 0.6 | 8.0 ± 0.6d | 3.1 ± 1.6 | 17.3 ± 1.2 |
| PCR cycle no. | |||||
| 22 | 162 | 2.5 ± 0.6d | 8.0 ± 1.6 | 2.5 ± 0.6 | 13.0 ± 1.9 |
| 25 | 162 | 4.3 ± 0.6 | 8.7 ± 1.6 | 3.1 ± 1.6 | 16.0 ± 1.2 |
| 28 | 155 | 8.2 ± 2.0d | 9.9 ± 3.3 | 2.7 ± 0.8 | 20.8 ± 3.0 |
| PCR cycle no.c | |||||
| 15 | 162 | NDd, f | 2.5 ± 1.6 | 0.6 ± 0.6 | 3.1 ± 1.6 |
| 20 | 162 | 0.6 ± 0.6a | 1.9 ± 1.1 | 0.6 ± 0.6 | 3.1 ± 1.6 |
| 25 | 162 | 3.7 ± 1.9 | 1.9 ± 1.1 | 2.5 ± 2.5 | 6.2 ± 3.3 |
| 30 | 162 | 4.3 ± 0.6d, e | 3.7 ± 1.1 | 1.9 ± 1.1 | 9.9 ± 1.6 |
Error rate is the percentage of total clones examined that had artificial RFLP patterns caused by the misincorporated base at the HhaI site.
Template concentration of each strain was equivalent to 30 ng of genomic DNA per ml.
Amplification was carried out with AmpliTaq.
Student t test shows significant difference at the 0.05 level.
Student t test shows significant difference at the 0.05 level.
ND, not detected.
Significant differences in the PCR-induced mutations were observed among the three enzymes. While the error rate for LA-Taq (6.8% ± 0.6%) was lower than that for Z-Taq (9.3% ± 0.0%), it was higher than that for AmpliTaq (3.7% ± 1.1%) (Table 2). (Results are given as means ± standard deviations.) No significant differences in the percentages of heteroduplexes were observed among three Taq DNA polymerases. The frequencies of heteroduplexes (1.2 to 3.7%) were lower than the frequencies of chimeras and mutations for all three Taq DNA polymerases (Table 2).
Effect of elongation time on formation of PCR artifacts.
The total PCR artifacts decreased as the elongation time increased (Table 2). The highest percentage of PCR artifacts (25.5% ± 3.9%) was observed at an elongation time of 20 s. When the extension time increased to 4 min, the total PCR artifacts decreased to 16.1% ± 3.2%. Among the three types of PCR artifacts, the percentage of chimeras decreased dramatically from 10.3 to 3.7 as the elongation time increased. While the PCR-induced mutations varied from 9.6 to 12.8%, they were not significantly different among the three treatments. In addition, no significant difference in the percentage of heteroduplexes was observed when elongation time changed. About 2 to 3% heteroduplexes was observed for all three treatments (Table 2).
Effect of template concentration on formation of PCR artifacts.
Since DNA template concentration will affect PCR amplification kinetics, it is expected that template concentration will have considerable effect on the formation of PCR artifacts. We found that the percentage of total PCR artifacts decreased from 17.3 to 10.5 as the template concentrations of each strain decreased from 10 ng to 100 pg (Table 2). This decrease was mainly due to the decrease in the error rates (from 8.0 to 2.5%). No significant difference in the percentages of heteroduplexes or chimeras was observed (Table 2).
Effect of PCR cycle number on formation of PCR artifacts.
As expected, a positive correlation was observed between the formation of PCR artifacts and the number of PCR cycles, with PCR artifacts increasing from 13.0 to 20.8% as the cycles increased from 22 to 28. While the percentage of chimeras increased significantly as the cycle number increased, there was no significant change in the frequency of heteroduplexes. A slight increase in mutations was observed (Table 2).
Since AmpliTaq from Perkin-Elmer gave the lowest proportion of PCR artifacts, the effect of PCR cycle number on the formation of PCR artifacts was also examined with this enzyme. No difference of PCR artifacts was observed when the number of PCR cycles was 20 or less, while the total PCR artifacts increased dramatically as the cycle number increased above 20. No chimera was detected when the cycle number was 15, whereas 4.3% chimeras was observed for 30 PCR cycles. Slight increases were observed in both mutations (1.9 to 3.7%) and heteroduplexes (0.6 to 2.5%). Similar to that described above, the overall PCR artifacts were significantly lower in the amplifications with AmpliTaq than those of Z-Taq (Table 2).
Effects of species diversity on formation of PCR artifacts.
Total PCR artifacts increased as the species diversity increased (Table 3). Among 162 clones examined, four PCR artifacts (2.5%) were detected in the four-species community. All these four clones were mutations of P39. Six artifacts (3.7%) were found in the seven-species community; three were mutations, and the other three were heteroduplexes. There were nine PCR artifacts (5.6%) in the 10-species community. Three were mutations, and six were heteroduplexes. No chimeric molecule was detected in all three model communities.
TABLE 3.
Effects of species diversity on the formation of PCR artifactsa
| No. of species in community | Mean % chimeras | Mean error rate ± SD (%) | Mean % heteroduplexes ± SD | Mean % total PCR artifact ± SD |
|---|---|---|---|---|
| 4 | NDb | 2.5 ± 1.6 | ND | 2.5 ± 1.6 |
| 7 | ND | 1.9 ± 1.9 | 1.9 ± 0.0 | 3.7 ± 1.8 |
| 10 | ND | 1.9 ± 0.0 | 3.7 ± 2.1 | 5.6 ± 2.1 |
For each community, 54 clones were examined in triplicate.
ND, not detected.
Elimination of heteroduplexes by polyacrylamide gel purification or T7 endonuclease I digestion.
Since heteroduplex molecules migrate more slowly than homoduplex molecules in polyacrylamide gel, the heteroduplex molecules should be separated from the PCR products by PAGE. To test the effectiveness of this purification method, a clone library was constructed using the 16S rDNAs purified from the polyacrylamide gel. No heteroduplexes were observed in this library, whereas 3.7% was detected in the control library (Table 4).
TABLE 4.
Elimination of heteroduplexes with polyacrylamide gel purification and T7 endonuclease I digestion
| DNA polymerase | Treatment | Mean % Chimeras ± SD | Mean Error rate ± SD (%) | Mean % heteroduplexes ± SD | Mean % total PCR artifacts ± SD |
|---|---|---|---|---|---|
| Z-Taq | Control | 8.7 ± 2.2 | 9.3 ± 0.0 | 3.7 ± 1.1 | 20.4 ± 0.0 |
| Gel purification | 8.0 ± 2.7 | 7.4 ± 1.9 | NDa | 15.4 ± 4.3 | |
| LA-Taq | Control | 1.9 ± 1.1 | 3.1 ± 0.6 | 3.1 ± 1.2 | 8.1 ± 2.5 |
| T7-endo Ib | 5.6 ± 1.1 | 3.1 ± 0.6 | ND | 8.7 ± 0.6 |
ND, not detected.
T-endoI, T7 endonuclease I.
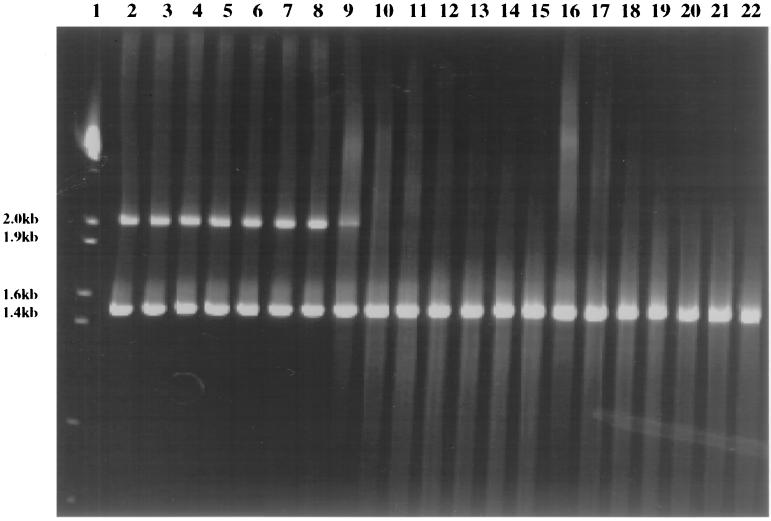
T7 endonuclease I can cut the single-stranded DNA bubbles in a heteroduplex molecule and chop them into small DNA fragments. The heteroduplexes formed from C10-5 and B9-12 were incubated with 30 and 60 U of T7 endonuclease I at 37°C for different time periods (Fig. 2). The intensity of the bands corresponding to heteroduplexes decreased for both samples incubated with 30 and 60 U of T7 endonuclease I at 37°C for 10 min and for the sample incubated with 60 U of enzyme on ice for 60 min (Fig. 2, lane 16). The smears resulting from the digested heteroduplex molecules were observed on PAGE; however, the intensity of the bands corresponding to 16S rDNA homoduplex molecules remained the same even after incubation with the enzyme at 37°C for 60 min (Fig. 2). These results suggested that T7 endonuclease I was effective in removing heteroduplexes, whereas the homoduplex molecules were protected under the conditions examined. The effectiveness of T7 endonuclease I in the elimination of heteroduplexes was further examined by cloning. A 16S rDNA library was constructed using the PCR products incubated with 30 U of T7 endonuclease I at 37°C for 20 min. No heteroduplexes were detected in this library, whereas 3.1% heteroduplexes was observed in the control library (Table 4).
FIG. 2.
Digestion of heteroduplexes with T7 endonuclease I. Lane 1, marker III; lanes 2 to 8, hetroduplexes of 16S rDNAs formed between C10-5 and B9-12 (89% similar) incubated with H2O on ice for 60 min (lane 2) or at 37°C from 10 to 60 min with 10-min intervals (lanes 3 to 8, respectively); lanes 10 to 16, heteroduplexes incubated with 30 U of T7 endonuclease I on ice for 60 min (lane 9) or at 37°C from 10 to 60 min with 10-min intervals (lanes 10 to 15, respectively); lanes 16 to 22, heteroduplexes treated with 60 U of T7 endonuclease on ice for 60 min (lane 16) or at 37°C from 10 to 60 min with 10-min intervals (lanes 17 to 22, respectively).
DISCUSSION
PCR-based cloning approaches are powerful tools for analyzing microbial community diversity despite intrinsic problems of bias and artifacts. With careful planning and experimental condition control, the artifacts can be minimized. Although the frequencies of artifacts observed in this study cannot be extrapolated to other studies due to differences in experimental conditions, they do provide valuable information for improving the methodologies of PCR-based cloning studies.
The effects of three types of PCR artifacts on the 16S-based cloning studies depend on experimental purpose. Single-base mutations may have little or no effect on the overall tree topology when the entire 16S rRNA gene sequences are compared. Clean sequences may not be obtained if the clone resulted from a heteroduplex; thus, it should not be a concern when the sequence is required for the analysis. However, all three types of PCR artifacts can have serious impacts when the RFLP or terminal RFLP analysis DGGE, or TGGE, etc., is used for microbial community analysis.
The existence of heteroduplexes in a cloned 16S rRNA gene library has been an unappreciated problem and can lead to overestimating the diversity of a microbial community. We found that the occurrence of heteroduplexes indirectly correlated with DNA polymerases. More heteroduplexes were observed in the amplifications with Z-Taq than with AmpliTaq. This could be due to the fact that more PCR products were synthesized by Z-Taq than by AmpliTaq. High PCR product production will favor heteroduplex formation. This explanation is further supported by the observed increase in proportion of heteroduplexes as the cycle number and template concentrations increased. Heteroduplex frequency also appeared to be a function of species diversity. The frequency of heteroduplexes in the 10-species community was about two to four times higher than those of 7- or 4-species communities. This could be due to the fact that the probability of annealing between the two strands from the same origin decreases as the number of the heterogeneous genes increases. The corresponding increase in heteroduplexes with species diversity is potentially a problem for analyzing natural microbial communities, which may have hundreds to thousands of phylotypes. Besides, there is usually more than one rrn copy in a genome. Theoretically, the frequency of forming heteroduplex molecules between different copies of the 16S rRNA gene in the same genome is higher than that between different 16S rRNA genes, because a heteroduplex should be more stable when the two parental genes have higher sequence similarity.
We proved that heteroduplex molecules of the entire 16S rRNA gene (1.5 kb) could be effectively detected by PAGE in a wide similarity range of 16S rRNA genes. However, the conditions for detecting such long fragments are different from those for DGGE, single-strand conformation polymorphism analysis, or heteroduplex mobility assay. We found that a gel with a lower cross-linking ratio (49:1), which yields a bigger pore size, should be used. Including 10% glycerol in the polyacrylamide gel helped to detect heteroduplexes, but a low concentration of urea in the gel did not. In addition, the conditions for forming heteroduplexes by denaturation-renaturation were also critical. We found that quickly cooling to 25°C for renaturation was much better than slowly cooling to 25°C (37) or quickly cooling on ice (9). Renaturation on ice resulted in more single-stranded DNA fragments. Including 10 mM EDTA in denaturation-renaturation buffer was helpful. However, we had difficulty forming heteroduplexes by denaturation at 98°C for 7 min and renaturation at 60°C for 40 min (12). We could detect the heteroduplexes formed between both closely (98.5% similar) and distantly (76% similar) related 16S rRNA genes by 5% nondenatured polyacrylamide gel.
We recommend the elimination heteroduplexes prior to cloning. It is possible to use an enzyme such as T7 endonuclease I to cut the bubble in a heteroduplex and further destroy it (22). However, the experimental conditions for this treatment are critical. Low concentrations of enzyme or short incubation time do not remove all heteroduplexes, whereas high concentrations of enzyme or long incubation time can digest the homoduplex molecules. Also, postamplification with Taq is required for this approach to generate an A overhang for TA cloning. PAGE was also effective in removing heteroduplex molecules; however, this approach may be difficult for separating the heteroduplexes formed between highly related strains or heteroduplexes having very close conformations to the parental homoduplex molecules.
PCR-generated mutation is another little-recognized problem for 16S rRNA gene-based cloning studies. In general, misincorporated nucleotides in the PCR products is not a big concern since the errors are distributed randomly over the amplified fragment. Theoretically, less than one misincorporated nucleotide is expected when the entire 16S rRNA gene is amplified using an enzyme with an average of fidelity such as 8 × 10−6 /base/replication. However, the error rate observed in this study was much higher than predicted. The highest error rate was observed for Z-Taq; however, the fidelity of Z-Taq (8.6 × 10−6 /base/duplication) is very close to that of AmpliTaq. These results indicated that the PCR-generated errors were not merely the consequences of infidelity of Taq polymerases. Since both Z-Taq and LA-Taq have higher processivity than AmpliTaq, we suspected that the higher error rate might be caused by a lack of PCR reagents, especially dNTPs. This explanation is supported by two observations: first, more PCR products were synthesized by Z-Taq and LA-Taq compared to AmpliTaq when equal units of enzymes were used; second, more PCR-induced mutations were observed when more templates were used (Table 2).
PCR amplification fidelity is affected by many factors—not only the enzyme used but also buffer conditions, divalent metal cations, and thermal cycling parameters. It was reported that Taq fidelity decreased when the concentration of Mg2+ was in great excess compared to total dNTPs (11). The Mg2+ and dNTP concentrations used in this study were the optimum concentrations recommended by the manufacturers. The rates of excess of Mg2+ over total dNTPs with Z-Taq, LA-Taq, and AmpliTaq were 2.2, 0.9, and 0.7 mM, respectively. All these assays were within the range of high-fidelity conditions described by Eckert et al. (11). Whether it has true impact on the high error rate needs to be further examined.
We also observed that certain types of artificial RFLP patterns that were caused by a misincorporated single nucleotide appeared in many independent amplifications with each of the three Taq DNA polymerases. To better understand this phenomenon, eight clones, each with a distinct artificial RFLP pattern that most frequently arose, were studied in detail. Clones 16, 6, 34, and 7 gained an HhaI site whereas clones 15, 40, 14, and 35 lost an HhaI site, all due to a base substitution (Table 5). Moreover, both error sites of clone 15 (mutation of C1-4) and clone 14 (mutation of B9-12) were at the E. coli position 1109, where C was in a loop (Table 5). The 16S rDNA sequences of the two clones were significantly different (79.4% similar). Hence, we suspect that the secondary structure of 16S rRNA gene contributed to the high error rate observed.
TABLE 5.
Nature of mutation in most frequently detected clones
| Clone no. | Reference strain | Error position in:
|
Base substitutiona | RFLP shift | |
|---|---|---|---|---|---|
| Reference strain | E. coli | ||||
| 15 | C1-4 | 1033 | 1109 | GCGC→GCGT | Lost HhaI |
| 16 | C1-4 | 976 | 1049 | GTGC→GCGC | Gained HhaI |
| 6 | Tol-4 | 994 | 1026 | GTGC→GCGC | Gained HhaI |
| 40 | Tol-4 | 1232 | 1265 | GCGC→TCGC | Lost HhaI |
| 14 | B9-12 | 1072 | 1109 | GCGC→GCGT | Lost HhaI |
| 34 | B9-12 | 540 | 576 | ACGC→GCGC | Gained HhaI |
| 7 | P39 | 854 | 878 | GCAC→GCGC | Gained HhaI |
| 35 | P39 | 739 | 763 | GCGC→GTGC | Lost HhaI |
HhaI restriction recognition sequence, GCGC.
Consistent with the results of Wang and Wang (34, 35), we found that the longer extension times and fewer PCR cycles decreased the frequency of chimeras. The percentages of chimeras found in this study, however, were more than three times lower than those observed by Wang and Wang (34, 35), probably due to different experimental systems and conditions. For example, they used pairs of cloned 16S rRNA genes with sequence similarity of 99.3, 86, and 82%, whereas sequence similarity varied from 76 to 89% in our four-species model community. Also, our chimera detection method could be less sensitive since we detected only an altered RFLP pattern. In theory, using Taq DNA polymerase with higher processitivity should lower the frequency of chimeras, since chimeric molecules are mainly caused by incomplete synthesis during the PCR cycle (24, 38). Conversely, the highest frequency of chimeras was observed for Z-Taq, which has the highest processitivity. Likely, other factors contributed to the formation of chimeras. For example, an undenatured region or secondary structure in the templates will make it difficult for DNA polymerase to read through, causing termination of DNA synthesis.
While some tools are available for detecting chimeras and heteroduplexes after cloning, no tools are available for identifying PCR-generated single-base mutations in natural samples. Thus, it is critical to minimize PCR artifact formation prior to cloning. PCR cycling is one key parameter to reducing all three types of PCR artifacts. We suggest that the PCR amplification for any cloning-based community studies be performed with as few cycles as possible (http://www.esd.ornl.gov/people/zhou/zhou.html). The appropriate cycle number will depend on the amount of template used, amplification efficiency, and existence and degree of inhibitory substances and thus should be determined experimentally. To minimize PCR artifacts, we suggest using the PCR products prior to or during the exponential period for cloning. To obtain enough products for cloning, we suggest combining multiple amplifications followed by concentration with ethanol precipitation. Mixing PCR products from independent amplifications can also help to minimize experimental errors and amplification bias. The concentrated sample can then be quantified and used for constructing 16S rRNA gene library. Because the extent of 16S rRNA gene artifacts can never be known in a natural sample, we suggest that interpretations be focused on comparative studies with replication and under identical PCR conditions.
ACKNOWLEDGMENTS
We thank Joe-Chang Cho for discussion and advice on heteroduplex detection.
This research was supported by the Natural and Accelerated Bioremediation Research and the Biotechnology Investigations-Ocean Margins Program, Office of Biological and Environmental Research, The United States Department of Energy. Oak Ridge National Laboratory is managed by University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725, and the Center for Microbial Ecology is funded by the National Science Foundation, DEB-9120006.
REFERENCES
- 1.Avaniss-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequence. BioTechniques. 1994;17:144–149. [PubMed] [Google Scholar]
- 2.Borneman J, Skroch P W, O'Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brakenhoff R H, Schoenmakers J G G, Lubsen N H. Chimeric cDNA clones: a novel PCR artifact. Nucleic Acids Res. 1991;19:1949–1950. doi: 10.1093/nar/19.8.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cariello N F, Thilly W G, Swenberg J A, Skopek T R. Deletion mutagenesis during polymerase chain reaction: dependence on DNA polymerase. Gene. 1990;99:105–108. doi: 10.1016/0378-1119(91)90040-i. [DOI] [PubMed] [Google Scholar]
- 6.Cline J, Braman J C, Hogrefe H H. PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerase. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong E F. Archaeal means and extremes. Science. 1998;280:542–543. doi: 10.1126/science.280.5363.542. [DOI] [PubMed] [Google Scholar]
- 9.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 10.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert K A, Kunkel T A. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerse. Nucleic Acids Res. 1990;18:3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espejo R T, Feijóo C G, Romero J, Vásquez M. PAGE analysis of the heteroduplexes formed between PCR-amplified 16S rRNA genes: estimation of sequence similarity and rDNA complexity. Microbiology. 1998;144:1611–1617. doi: 10.1099/00221287-144-6-1611. [DOI] [PubMed] [Google Scholar]
- 13.Felske A, Wolterink A, Van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (the Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Großkopf R, Janssen P H, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopczynski E D, Bateson M M, Ward D M. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl Environ Microbiol. 1994;60:746–748. doi: 10.1128/aem.60.2.746-748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee D-H, Zo Y-G, Kim S-J. Nonradioactive method to study genetic profiles of natural bacterial gommunities by PCR-single-strand-conformation polymorphism. Appl Environ Microbiol. 1996;62:3112–3120. doi: 10.1128/aem.62.9.3112-3120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by SSU rDNA analysis of a mixed culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 21.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowell J L, Klein D A. Heteroduplex resolution using T7 endonuclease I in microbial community analyses. BioTechniques. 2000;28:676–681. doi: 10.2144/00284st03. [DOI] [PubMed] [Google Scholar]
- 23.Massol-Deyá A, Weller R, Ríos-Hernández L, Zhou J-Z, Hickey R F, Tiedje J M. Succession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl Environ Microbiol. 1997;63:270–276. doi: 10.1128/aem.63.1.270-276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyerhans A, Vartanian J-P, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pääbo S, Irwin D M, Wilson A C. DNA damage promotes jumping between templates during enzymatic amplification. J Biol Chem. 1990;265:4718–4721. [PubMed] [Google Scholar]
- 29.Reysenbach A, Wickham G S, Pace N R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snyder L, Champness W. Molecular genetics of bacteria. Washington, D.C.: ASM Press; 1997. [Google Scholar]
- 31.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by SSU rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki M T, Rappe M S, Giovannoni S J. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G C-Y, Wang Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology. 1996;142:1107–1114. doi: 10.1099/13500872-142-5-1107. [DOI] [PubMed] [Google Scholar]
- 35.Wang G C-Y, Wang Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl Environ Microbiol. 1997;63:4645–4650. doi: 10.1128/aem.63.12.4645-4650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisburg W W, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White M B, Carvalho M, Derse D, O'Brien S J, Dean M. Detecting single base substitution as heteroduuplex polymorphisms. Genomics. 1992;12:301–306. doi: 10.1016/0888-7543(92)90377-5. [DOI] [PubMed] [Google Scholar]
- 38.Wintzingerode F V, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microb Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 39.Wise M G, McArthur J V, Shimkets L J. Bacterial diversity of a Carolina bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Appl Environ Microbiol. 1997;63:1505–1514. doi: 10.1128/aem.63.4.1505-1514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J-Z, Fries M R, Chee-Sanford J, Tiedje J M. Phylogenetic analysis of a new group of denitrifiers capable of anaerobic growth on toluene: description of Azoarcus tolulyticus sp. nov. Int J Syst Bacteriol. 1995;45:500–506. doi: 10.1099/00207713-45-3-500. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J-Z, Davey M E, Figueras J B, Rivkina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]