Abstract
Strains of Xylella fastidiosa isolated from grape, almond, maple, and oleander were characterized by enterobacterial repetitive intergenic consensus sequence-, repetitive extragenic palindromic element (REP)-, and random amplified polymorphic DNA (RAPD)-PCR; contour-clamped homogeneous electric field (CHEF) gel electrophoresis; plasmid content; and sequencing of the 16S-23S rRNA spacer region. Combining methods gave greater resolution of strain groupings than any single method. Strains isolated from grape with Pierce's disease (PD) from California, Florida, and Georgia showed greater than previously reported genetic variability, including plasmid contents, but formed a cluster based on analysis of RAPD-PCR products, NotI and SpeI genomic DNA fingerprints, and 16S-23S rRNA spacer region sequence. Two groupings of almond leaf scorch (ALS) strains were distinguished by RAPD-PCR and CHEF gel electrophoresis, but some ALS isolates were clustered within the PD group. RAPD-PCR, CHEF gel electrophoresis, and 16S-23S rRNA sequence analysis produced the same groupings of strains, with RAPD-PCR resolving the greatest genetic differences. Oleander strains, phony peach disease (PP), and oak leaf scorch (OLS) strains were distinct from other strains. DNA profiles constructed by REP-PCR analysis were the same or very similar among all grape strains and most almond strains but different among some almond strains and all other strains tested. Eight of 12 ALS strains and 4 of 14 PD strains of X. fastidiosa isolated in California contained plasmids. All oleander strains carried the same-sized plasmid; all OLS strains carried the same-sized plasmid. A plum leaf scald strain contained three plasmids, two of which were the same sizes as those found in PP strains. These findings support a division of X. fastidiosa at the subspecies or pathovar level.
Xylella fastidiosa (Wells et al.) (35) is a gram-negative, xylem-inhabiting bacterium that causes Pierce's disease (PD) of grape, phony peach disease (PP), periwinkle wilt, citrus variegated chlorosis, and leaf scorch diseases of almond (almond leaf scorch [ALS]), plum (PLS), elm, maple, oak (OLS), and sycamore (16, 27). All strains of X. fastidiosa are currently classified as one species but differ in important respects in plant host range and pathogenicity. The geographic isolation of the plant diseases caused by X. fastidiosa restricts the easy comparison of the pathological characteristics of the various strains. Previous studies differentiated X. fastidiosa strains on the basis of pathogenicity, nutritional requirements (16), DNA homology (19), structural protein analysis (3), restriction fragment length polymorphisms (RFLPs) (4), and random amplified polymorphic DNA (RAPD)-PCR (1, 6, 9, 25, 27). RFLP and DNA-DNA hybridization studies revealed distinct differences between PD strains and strains that cause PP, PLS, and periwinkle wilt (4, 19). Pooler et al. (25) distinguished five groups of X. fastidiosa using RAPD-PCR: the citrus group, plum-elm group, grape-ragweed group, almond group, and mulberry group. Strains of X. fastidiosa causing OLS formed a separate cluster from PD strains (6).
The high levels of DNA homology (>85%) (35) among X. fastidiosa strains indicate that differences among strains would primarily lie in the linear arrangement of cistrons within the genome. Genetic analyses using repetitive extragenic palindromic element (REP)-PCR, which utilizes conserved PCR primer sequences that reside within repetitive elements distributed throughout the prokaryote genome, have been widely used to identify and assess the genetic diversity of prokaryotes (34). Differences revealed by RAPD-PCR (36) and REP-PCR (34) have also been useful in characterizing differences among closely related strains (6, 25). Contour-clamped homogeneous electric field (CHEF) electrophoresis (7) allows the separation of large DNA molecules by electrophoresis. By digesting the genome with enzymes which cut infrequently, DNA fingerprints which consist of a comparatively small number (generally fewer than 30 bands) of large fragments can be generated. Fewer fragments allow easier comparison of strains than the hundreds of fragments that are typically produced using conventional restriction enzyme digestion. Significant sequence heterogeneity within the 16S-23S rRNA spacer region of Proteobacteria has been used to discriminate between different species of Proteobacteria and to detect heterogeneity within species (2, 18), but closely related microorganisms often have very similar spacer regions. RAPD analysis distinguished coffee strains from citrus strains of X. fastidiosa (9, 29). Plasmids have been found in some strains of X. fastidiosa (4, 24) but have not been extensively studied.
The objective of this study was to analyze the genetic relatedness of X. fastidiosa strains in California that were isolated from grape, almond, and oleander. For comparison, strains of X. fastidiosa which cause PD, PP, PLS, and OLS isolated from the southeastern United States were also characterized. X. fastidiosa strains were analyzed by RAPD-PCR (36), REP-PCR (34), CHEF electrophoresis (7, 8) of genomic DNA digested with rare-cutting restriction enzymes, 16S-23S rRNA spacer sequences, and plasmid content.
MATERIALS AND METHODS
Strains tested.
Forty-six strains of X. fastidiosa that were analyzed in this study are listed in Table 1. In addition, we analyzed over 80 other strains isolated from grapevines from widespread regions of California by REP-and enterobacterial repetitive intergenic consensus sequence (ERIC)-PCR. Each strain was isolated from the indicated host plants (Table 1) with disease symptoms. All strains were grown by plating 100 μl of 1010-CFU/ml culture on PW agar medium (10) or the modified PWG medium (15) and incubating at 28°C for 7 to 10 days.
TABLE 1.
Strains of X. fastidiosa used in this study
| Strain | Host of origin | County in California or state from which strain was isolated | GenBank accession no. of 16S-23S rRNA sequence |
|---|---|---|---|
| ALS1 | Almond | San Joaquin | AF073240 |
| ALS2 | Almond | San Joaquin | AF073243 |
| ALS3 | Almond | San Joaquin | AF073244 |
| ALS4 | Almond | San Joaquin | AF073245 |
| ALS5 | Almond | San Joaquin | AF073246 |
| ALS6 | Almond | San Joaquin | AF073247 |
| ALS7 | Almond | San Joaquin | AF073248 |
| ALS9 | Almond | San Joaquin | AF073249 |
| Manteca | Almond | San Joaquin | AF073241 |
| Contra Costa | Almond | Contra Costa | AF073250 |
| Dixon | Almond | Solano | AF073251 |
| Tulare | Almond | Tulare | AF073242 |
| Conn Creek | Grape | Napa | AF073225 |
| Stags Leap | Grape | Napa | AF073226 |
| Fetzer | Grape | Napa | AF073227 |
| STL | Grape | Napa | AF073228 |
| Santa Cruz | Grape | Santa Cruz | AF073229 |
| Meyley | Grape | Santa Cruz | AF073230 |
| UCLA | Grape | Los Angeles | AF073231 |
| Preston Ranch | Grape | Sonoma | AF073232 |
| VinoF | Grape | Sonoma | AF073233 |
| Medeiros | Grape | Fresno | AF073234 |
| Traver | Grape | Tulare | AF073235 |
| Moore Park | Grape | Ventura | AF073236 |
| Douglas | Grape | San Luis Obispo | AF073237 |
| Oxford | Grape | Alameda | AF073238 |
| Hopland | Grape | Mendocino | AF073239 |
| Maple | Maple | Alameda | AF073219 |
| Ann1 | Oleander | Palm Springs | AF073215 |
| PF1 | Oleander | Palm Springs | AF073216 |
| T1c | Oleander | Orange | AF073217 |
| TR1 | Oleander | Orange | AF073218 |
| PD 95-2 | Grape | Florida | AF073220 |
| PD 95-4 | Grape | Florida | AF073221 |
| PD 95-9 | Grape | Florida | AF073222 |
| R116V3 | Grape | Georgia | AF073223 |
| R118V3-4 | Grape | Georgia | AF073224 |
| 5S2 | Peach | Georgia | AF073206 |
| 5R1 | Peach | Georgia | AF073207 |
| 4S3 | Peach | Georgia | AF073208 |
| Plum 2#4 | Plum | Georgia | AF073209 |
| Oak 88-9 | Oak | Florida | AF073210 |
| Oak 92-3 | Oak | Florida | AF073211 |
| Oak 92-10 | Oak | Florida | AF073212 |
| OLS#2 | Oak | Georgia | AF073213 |
| Stucky | Oak | Georgia | AF073214 |
DNA extraction for PCR.
Bacterial DNA was extracted using the CTAB minipreparation method described by Wilson (37). Aerosol-resistant tips were used in procedures for the extraction of DNA that was used in PCRs.
PCR conditions and primers.
In PCRs for the specific detection of X. fastidiosa strains, primers RST31 (5′-GCGTTAATTTTCGAAGTGATTCGATTGC-3′) and RST33 (5′-CACCATTCGTATCCCGGTG-3′) were used as described by Minsavage et al. (21).
RAPD-PCRs were performed using 10-base primers (Kit AA; Operon Technologies, Inc., Alameda, Calif.). Amplifications were performed in a 25-μl reaction volume containing 25 ng of genomic DNA; 25 pmol of a single primer; 2 mM MgCl2; 100 μM (each) dATP, dCTP, dGTP, and dTTP; 10 mM Tris HCl [pH 9]; 50 mM KCl; 0.1% Triton X-100; and 0.5 U of Amplitaq DNA polymerase (Perkin-Elmer, Foster City, Calif.). RAPD-PCRs were done with a DNA Thermal Cycler (Perkin-Elmer) using 1 cycle of 1 min at 94°C; 45 cycles of 1 min at 94°C, 1 min at 34°C, and 2 min at 72°C; and a final extension of 10 min at 72°C.
Eighty X. fastidiosa grape strains that were isolated from separate grapevines showing typical symptoms of Pierce's disease were analyzed by ERIC- and REP-PCR. A subset of strains shown in Table 1 was also analyzed by ERIC-PCR. Individual colonies growing on PW agar medium were transferred directly into the REP-PCR mixture, as previously described by Opgenorth et al. (23). PCR conditions using ERIC- and REP-PCR primers and agarose gel electrophoresis conditions of the PCR products were the same as described by Opgenorth et al. (23). Similarities and differences among the strains were compared qualitatively.
The 16S-23S rRNA intergenic spacer regions of X. fastidiosa strains were PCR amplified using primers G1 and L1, which are located in highly conserved regions within the 16S and 23S rRNA genes, respectively (18). Primer G1 (5′-GAAGTCGTAACAAGG-3′) is located at the 3′ end of the 16S rRNA gene 30 to 40 nucleotides upstream of the spacer region, and L1 (5′-CAAGGCATCCACCGT-3′) is located at the 5′ end of the 23S rRNA gene, approximately 20 bases downstream from the spacer region. PCR mixtures contained 25 ng of genomic DNA; 25 pmol of G1; 25 pmol of L1; 10 mM Tris [pH 8.0]; 50 mM KCl; 2.5 mM MgCl2; 200 μM (each) dATP, dCTP, dGTP, and dTTP, and 0.5 U of Amplitaq DNA polymerase (Perkin-Elmer). A DNA Thermal Cycler (Perkin-Elmer) was used, with 1 cycle of 5 min at 94°C; 35 cycles of 94°C for 40 s, 55°C for 1 min, and 72°C for 2 min; and a final extension of 10 min at 72°C.
Except for REP-PCR products, gel electrophoresis of PCR amplification products was performed with 1.4% agarose gels in TAE buffer (20) at 3.5 V/cm. After the gels were stained with ethidium bromide (0.5 μg/ml), PCR products were visualized on a UV transilluminator and photographed.
Sequencing of the 16S-23S rRNA intergenic spacer region.
PCR-amplified intergenic spacer regions were purified using Ultrafree tubes (Millipore, Bedford, Mass.). Both strands of the spacer regions were sequenced by the dideoxy chain termination method (30) using a cycle-sequencing format (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Sequencing reaction mixtures were electrophoresed, and nucleotide sequences were recorded using the ABI 377 automated sequencer (Perkin-Elmer Applied Biosystems). Phylogenetic analysis of the 16S-23S rRNA sequences was resolved using the PAUP Software program, version 3.1 (33).
Analysis of RAPD-PCR product profiles.
RAPD-PCR product profiles of all isolates were compared, and the similarities of PCR products between pairs of strains were scored with the Jaccard similarity coefficient (32). The Jaccard coefficient, S, is the proportion of shared DNA fragments in two isolates and is calculated with the formula S = 2nxy/(nx + ny), where nx is the total number of fragments in isolate x, ny is the total number in isolate y, and nxy is the number of fragments common to the two isolates. The distance (d) between two strains is calculated with the formula d = 1 − S. A d value of 0 indicates that the two isolates have identical RAPD-PCR products, and a value of 1.0 indicates that the two isolates have no PCR products in common. A distance matrix of pairwise comparison between strains was constructed. The relationship between strains was analyzed with the neighbor-joining program of PHYLIP (12).
CHEF gel electrophoresis.
DNA for CHEF electrophoresis was prepared by a modification of the method of Cooksey and Graham (8). Cells grown on plates were harvested and washed with SE buffer (75 mM NaCl; 25 mM EDTA [pH 7.5]), and the cell density was adjusted to 109 CFU/ml in SE buffer. A 0.5-ml sample of the bacterial suspension was mixed with 0.5 ml of 2% agarose (pulse-field certified; Bio-Rad, Richmond, Calif.) in 10 mM Tris, 10 mM MgCl2, and 0.1 mM EDTA (pH 7.5). Plugs were cast in plug molds (1 by 0.5 cm; Bio-Rad) and placed at 4°C for 5 min to allow the agarose to solidify. Ten plugs were placed in 5 ml of lysis solution consisting of proteinase K (0.5 mg/ml; Bethesda Research Laboratories, Gaithersburg, Md.), 1% N-lauroylsarcosine, and 0.5 M EDTA (pH 9.5). After overnight incubation at 50°C, the plugs were washed four times with 20 mM Tris (pH 8.0) and 50 mM EDTA, 1 h each time, at room temperature with gentle agitation. In the second wash, 1 mM phenylmethylsulfonyl fluoride (Sigma, St. Louis, Mo.) was added to inactivate the residual proteinase K. Plugs were then washed five times with TE (10 mM Tris, 10 mM EDTA, [pH 8.0]), 10 min for each wash, with gentle agitation at room temperature. The genomic DNA immobilized in the plugs was incubated with the appropriate restriction buffer at room temperature for 1 h prior to the addition of the restriction enzyme. Digestions were carried out overnight at 37°C in 1× restriction buffer, 1 mg of bovine serum albumin per ml, and one of the following enzymes: NheI (10 U), NotI (10 U), SacII (10 U), SpeI (6 U), or XbaI (6 U) (New England Biolabs, Beverly, Mass.). Agarose gel electrophoresis of digested DNA fragments was performed with a 1% agarose gel (13 by 14 cm) using 0.5× TBE buffer (20). One-third of the agarose plug was inserted into each well in the gel. Wells in the gel were filled with agarose to seal the wells. Agarose plugs containing concatemers of λ DNA (Bio-Rad) were used as molecular size standards. Electrophoresis was carried out with 0.5× TBE at 14°C for 20 h at 6 V/cm with a 1- to 12-s switch time ramp at an included angle of 120° using a CHEF-DR III pulsed-field electrophoresis system (Bio-Rad). After electrophoresis, the gel was stained in 0.5 μg of ethidium bromide per ml for 30 min. DNA was visualized and photographed as described above.
The similarities in restriction fragment length patterns generated by CHEF electrophoresis between pairs of strains were scored with the Jaccard coefficient as described above. A distance matrix was obtained by pairwise comparison between strains. The relatedness of strains was determined by neighbor-joining analysis using the PHYLIP computer package (12).
Plasmid isolation and analysis.
Strains of X. fastidiosa were grown by plating 100 μl of 1010 CFU/ml culture on PWG medium and incubation at 28°C for 7 days. Cells from one plate were resuspended in 1 ml of TE buffer, washed once in TE buffer, and finally resuspended in 100 μl of TE buffer. Plasmids were isolated by the alkaline minipreparation method (20). Plasmid DNA was resuspended in 10 μl of TE buffer. Five microliters of native plasmid DNA was electrophoresed to determine the number of plasmids present in the strains. Total plasmid DNA in the remaining 5 μl was digested with HindIII by standard procedures (20), and the resulting fragments were analyzed by agarose gel electrophoresis with TAE buffer (20).
RESULTS
Analysis of PCR products using primers specific for X. fastidiosa.
All strains gave the same-sized PCR product (0.75 kb) (results not shown) in reactions using primers RST31 and RST33. PCR products of X. fastidiosa strains isolated from oak, oleander, peach, and plum and all ALS strains excluding ALS1, Manteca, and Tulare were digested into two fragments of approximately 0.55 and 0.2 kb using RsaI (results not illustrated). In contrast, the 0.75-kb PCR products from the three ALS strains, all PD strains, and the strain isolated from maple were not digested with RsaI.
Sequence analysis of the 16S-23S rRNA intergenic spacer region.
A region of approximately 500 bases containing the 16S-23S rRNA intergenic spacer region of all strains listed in Table 1 was sequenced. The intergenic spacer region of all X. fastidiosa strains contained tRNAs for alanine (positions 133 to 191) and isoleucine (positions 225 to 272). Analysis of this region revealed four groupings of strains: (i) most almond strains, all peach strains, and the plum strain, (ii) oak strains, (iii) oleander strains, and (iv) grape strains, a few almond strains, and the maple strain. With the exception of almond strains ALS1, Manteca, and Tulare, all strains of X. fastidiosa isolated from the same host had identical sequences within the 16S-23S rRNA region. Strains ALS1, Manteca, and Tulare and the strain isolated from maple had the same sequence as strains isolated from grape. Strains isolated from grape and maple and almond strains ALS1, Manteca, and Tulare differed from the other strains at positions 56 and 485. The sequence of strains isolated from oleander (28) was distinguished from all other strains at two base positions (213 and 328). Strains of X. fastidiosa isolated from oak differed from all other strains at position 351. The 16S-23S sequences of strains isolated from almond (except for ALS1, Manteca, and Tulare), plum, and peach were identical and differed from other strains at positions 327, 337, 408, 419, and 431.
ERIC- and REP-PCR fingerprinting of X. fastidiosa strains.
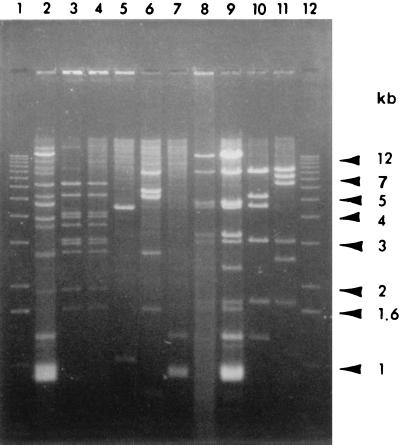
Grape strains of X. fastidiosa from plants from the southeastern United States had the same ERIC-PCR profiles as grape strains from California (Fig. 1, lanes 1 to 5). All 80 strains from widely distributed locations in California had the same ERIC- and REP-PCR profiles (data not shown). The same profile was also observed in a California isolate from maple exhibiting widespread leaf scorching and one of the almond strains (ALS1). Two other almond strains (ALS2 and ALS3) were identical to each other but possessed three or four DNA bands differing from those in the grape strains. The two oak strains from the southeastern United States were similar to each other and different from all the other strains. Similarly, the oleander strains were identical to each other but different from the other strains. Despite numerous attempts, we were not able to amplify an ERIC-PCR profile from either the plum or peach strains from the southeastern United States.
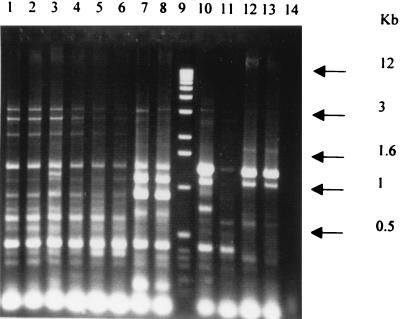
FIG. 1.
REP-PCR analysis of X. fastidiosa strains. ERIC-PCR profiles of X. fastidiosa strains from diverse plant hosts or geographical locations are shown. Lane 1, grape isolate from Georgia (strain R116V3); lane 2, grape from Florida (PD95-2); lane 3, grape from California (Stags Leap); lane 4, maple from California; lane 5, grape from California (Conn Creek); lane 6, almond (ALS1); lane 7, almond (ALS2); lane 8, almond (ALS3); lane 9, 1-kb size standard; lane 10, oak from Florida (88-9); lane 11, oak from Georgia (OLS#2); lane 12, oleander (Ann1); lane 13, oleander (TR1); lane 14, water control.
RAPD-PCR analysis.
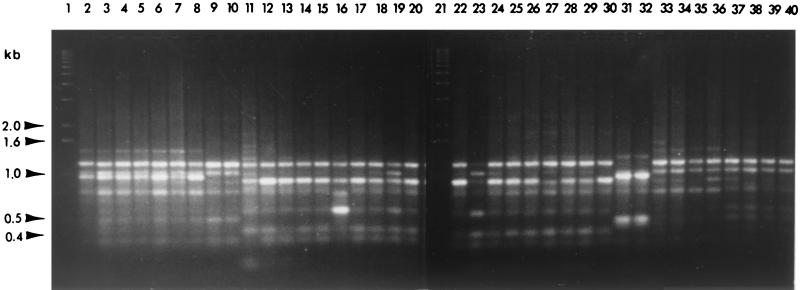
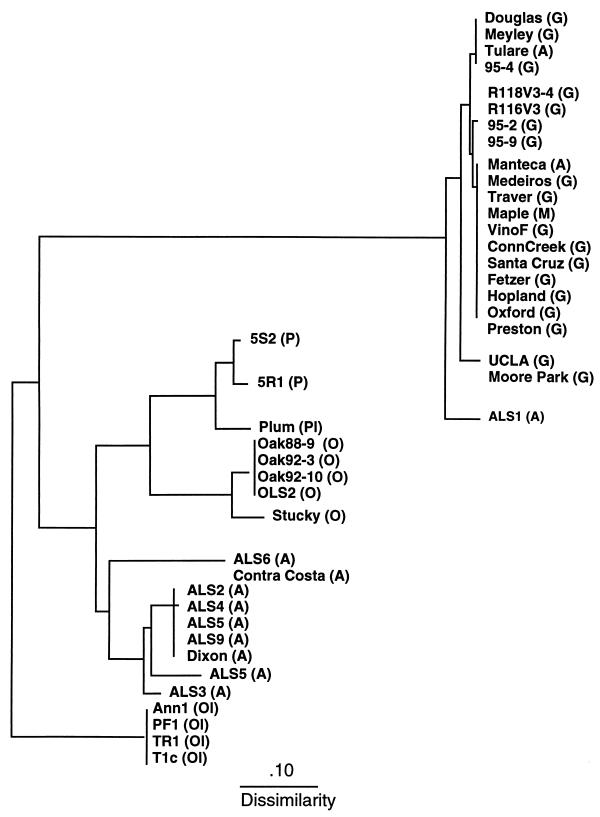
Of the 20 individual 10-base oligomers used as primers in RAPD-PCR analysis of X. fastidiosa strains listed in Table 1, 4 primers (OP-AA-05, OP-AA-15, OP-AA-18, and OP-AA-19) yielded no PCR product. Six primers (OP-AA-07, OP-AA-13, OP-AA-14, OP-AA-16, OP-AA-17, and OP-AA-19) yielded one or two PCR products (results not shown) that were uniform among all strains tested. Ten primers (OP-AA-01, OP-AA-02, OP-AA-03, OP-AA-04, OP-AA-06, OP-AA-08, OP-AA-09, OP-AA-10, OP-AA-11, and OP-AA-12) yielded complex RAPD fingerprints that allowed the distinction of strains isolated from different hosts. The PCR products of selected strains using primer OP-AA-01 are shown in Fig. 1. PCR products of all strains were compared, and a distance matrix was constructed. A dendrogram produced by the neighbor-joining algorithm of the phylogeny inference package, PHYLIP, is shown (see Fig. 3). As depicted in Fig. 2 and 3, all strains of X. fastidiosa causing PD were similar to ALS strains ALS1, Manteca, and Tulare. This included PD strains from California, Florida, and Georgia. Strains causing ALS were further divided into second and third groupings; the latter consisted of strains ALS6 and Contra Costa (Fig. 2 and 3). Strains isolated from oleander, oak, peach, and plum each formed separate clusters. Strains of X. fastidiosa isolated from the plum and peach clusters were more similar to OLS strains than to other groupings (Fig. 2 and 3).
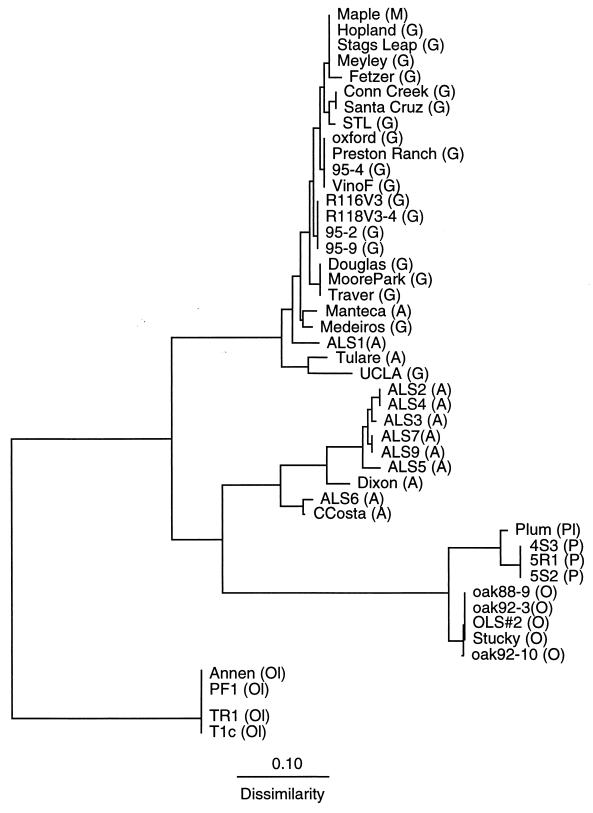
FIG. 3.
Phylogram based on distance data obtained by RAPD-PCR analysis. A distance matrix was constructed by pairwise comparison of the RAPD PCR products among strains. The Jaccard similarity coefficient (32) was used (S = 2nxy/nx + ny), where nxy is the number of PCR products common to strains x and y, nx is the number of bands present in strain x, and ny is the number of bands present in strain y. The scale bar indicates relative dissimilarity between strains. A, almond; G, grape; M, maple; O, oak; Ol, oleander; P, peach; Pl, plum.
FIG. 2.
Agarose gel electrophoresis of RAPD-PCR products (primer OP-AA-01) synthesized using the template DNA of various strains. Lane 1, 1-kb ladder; lane 2, strain ALS2; lane 3, ALS3; lane 4, ALS4; lane 5, ALS5; lane 6, ALS7; lane 7, ALS9; lane 8, Dixon; lane 9, ALS6; lane 10, Contra Costa; lane 11, ALS1; lane 12, Manteca; lane 13, Tulare; lane 14, Conn Creek; lane 15, Douglas; lane 16, Hopland; lane 17, Medeiros; lane 18, Meyley; lane 19, Moore Park; lane 20, Oxford; lane 21, 1-kb ladder; lane 22, Santa Cruz; lane 23, Traver; lane 24, UCLA; lane 25, 95-2; lane 26, 95-4; lane 27, 95–9; lane 28, R116V3; lane 29, R118V3-4; lane 30, Maple; lane 31, T1c; lane 32, TR1; lane 33, Plum 2#4; lane 34, 4S3; lane 35, 5R1; lane 36, 5S2; lane 37, Oak 88-9; lane 38, Oak 92–3; lane 39, Oak 92–10; lane 40, Stucky.
CHEF electrophoresis of genomic DNA.
Restriction fragments of genomic DNA of X. fastidiosa strains produced by digestion with enzymes NheI, SacII, and XbaI were less than 50 kb in size and too numerous to make good comparisons between strains. Fragments of genomic DNA digested with NotI and SpeI were generally larger than 50 kb and proved to be useful for comparing strains. Figure 4 shows a CHEF gel of DNAs from representative X. fastidiosa strains digested by NotI and SpeI. The groupings based on SpeI digestion of genomic DNA corresponded to groupings established by NotI digestion of genomic DNA.
FIG. 4.
CHEF-agarose gel electrophoresis of the genomic DNA of various strains digested with enzyme NotI digests. Lane 1, one concatemer; lane 2, UCLA; lane 3, Douglas; lane 4, 95-4; lane 5, R118V3-4; lane 6, ALS1; lane 7, Manteca; lane 8, Maple; lane 9, ALS2; lane 10, ALS3; lane 11, ALS5; lane 12, ALS6; lane 13, 5S2; lane 14, 5R1; lane 15, Plum 2#4; lane 16, Oak 92-3; lane 17, Stucky; lane 18, Ann1; lane 19, TR1; lane 20, one concatemer.
In general, CHEF and RFLP patterns of strains isolated from the same plant host were similar or identical (Fig. 4). The exceptions were the almond strains ALS1, Manteca, and Tulare, which had fingerprints similar to those of strains isolated from grape from California, Florida, and Georgia and of the strain isolated from maple. These groupings agreed with those established by ERIC-PCR fingerprinting. Thus, almond strains formed three distinct RFLP groups. Group 1 included strains ALS1, Manteca, and Tulare, which had restriction fingerprints that were similar to those of grape strains. The second RFLP group consisted of strains ALS2, ALS3, ALS4, ALS5, ALS7, ALS9, and Dixon. The third group, consisting of strains ALS6 and Contra Costa, differed from strains of almond groups 1 and 2. A distance matrix constructed by pairwise comparison of restriction fragments among strains with the Jaccard coefficient and analyzed by neighbor-joining analysis resulted in the tree shown in Fig. 5. Groupings of strains elucidated by CHEF electrophoresis agreed with groupings established by REP- and RAPD-PCR analysis.
FIG. 5.
Phylogram based on distance data obtained by CHEF-gel electrophoresis of NotI- and SpeI-restricted genomic DNA. A distance matrix was constructed by pairwise comparison of the RAPD-PCR products among strains. The Jaccard similarity coefficient (32) was used. The scale bar indicates relative dissimilarity between strains. A, almond; G, grape; M, maple; O, oak; Ol, oleander; P, peach; and Pl, plum.
Plasmid analysis.
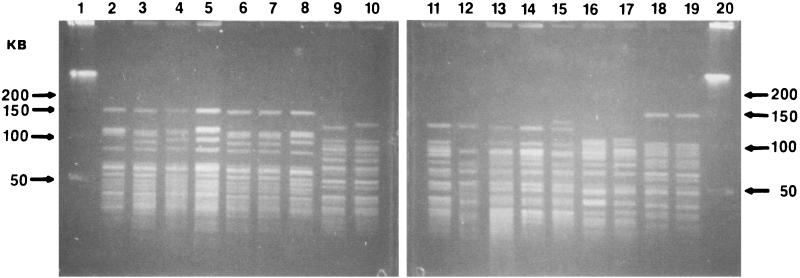
Plasmids were detected in 27 of 44 strains tested. Native plasmid DNA was electrophoresed to determine the number of plasmids in the 27 strains (results not shown; Table 2). When total plasmid DNA was digested with HindIII, nine different RFLP patterns (Fig. 6; Table 2) were observed. Figure 6 shows HindIII digests of the various plasmids observed among the X. fastidiosa strains tested. HindIII-digested plasmid DNAs of all OLS strains (Stucky, OLS#2, Oak 88-9, Oak 92-3, and Oak 92-10) were identical (plasmid profile B, Table 2). The same plasmid profile was observed in all oleander leaf scorch strains (Ann1, PF1, TR1, and T1c) (plasmid profile H). The strain causing PLS contained three plasmids (results not shown; plasmid profiles E, F, and G), two of which appeared to have the same HindIII patterns as the plasmids isolated from PP strains 5S3 and 5R1 (plasmid profiles E and F). Peach strain 4S3 was not evaluated for plasmid content. ALS strains ALS1 and ALS3 did not harbor any plasmids. The restriction profile of the plasmid present in strain ALS6 (plasmid I profile) differed from that found in other almond strains. Contra Costa (almond group 3) contained no plasmids, whereas ALS6 (almond group 3) contained one plasmid. ALS strains Dixon, Tulare, ALS2, ALS4, ALS5, ALS7, and ALS9 and PD strains Medeiros, Moore Park, and Traver carried the same plasmid (plasmid profile A). Strain UCLA contained four plasmids (results not shown; plasmid profiles J through M). HindIII digestion of plasmid DNA isolated from strain UCLA indicated that a subset of the fragments were the same size as those found in plasmid profile A. The HindIII profiles of plasmid DNA of Florida grape strains 95-2 and 95-10 were identical (plasmid profile C). The restriction pattern of the plasmid present in strain 95-4 differed from those found in 95-2 and 95-10 (plasmid profile D).
TABLE 2.
Plasmids found among strains of X. fastidiosaa
| Plasmid profile | Host/disease | Strain(s) | Sizes of HindIII fragments (kb) |
|---|---|---|---|
| A | Almond/ALS | ALS2, ALS4, ALS5, ALS7, ALS9, Dixon, Tulare | 6.2, 5.2, 4.03, 3.9, 3.6, 3.1, 2.96, 2.8, 1.95, 1.65 |
| A | Grape/PD | Medeiros, Moore Park, Traver | 6.2, 5.2, 4.03, 3.9, 3.6, 3.1, 2.96, 2.8, 1.95, 1.65 |
| B | Oak/OLS | Oak 88-9, Oak 92-3, Oak 92-10, OLS#2, Stucky | 8.5, 5.2, 4.6, 3.1, 1.7, 1.3 |
| C | Grape/PD | PD 95-2, PD 95-9 | 7.6, 5.4, 5.0, 2.7, 1.6 |
| E + F | Peach/PP | 5S3, 5R1 | 12.4, 8.6, 4.9, 4.8, 3.2, 3.1, 2.3, 1.7, 1.6, 1.3 |
| E + F + G | Plum | Plum 2#4 | 12.4, 8.6, 4.9, 4.8, 3.2, 3.1, 2.3, 1.7, 1.6, 1.3, 1.2, 0.89 |
| H | Oleander/leaf scorch | Ann1, PF1, T1c, TR1 | 9.5, 7.5, 6.5, 3.1, 2.57, 1.7 |
| I | Almond/ALS | ALS6 | 4.375, 1.05 |
| J + K + L + M | Grape/PD | UCLA | 12.2, 10.2, 8.43, 7.4, 6.2, 5.2, 4.7, 3.9, 3.6, 2.8, 1.95, 1.3, 0.97, 0.89 |
Strains ALS1, ALS3, Conn Creek, Contra Costa, Douglas, Fetzer, Hopland, Manteca, Maple, Meyley, Oxford, Preston Ranch, R116V3, R118V3-4, Santa Cruz, Stags Leap, STL, and VinoF did not carry plasmids.
FIG. 6.
Agarose gel electrophoresis of HindIII-digested plasmid DNA from various X. fastidiosa strains. Lane 1, 1-kb ladder; lane 2, strain UCLA; lane 3, Tulare; lane 4, ALS4; lane 5, ALS6; lane 6, 95-2; lane 7, 95-4; lane 8, peach; lane 9, Plum 2#4; lane 10, Oak 88-9; lane 11, Ann1; lane 12, 1-kb ladder.
DISCUSSION
All of the methods of genetic analysis used in this study produced the same host-associated groupings of X. fastidiosa strains, but RAPD and CHEF methods distinguished the greatest degree of genetic heterogeneity. The 16S-23S spacer region sequences distinguished four major groups: (i) 9 of 12 almond strains, plum strains, and all peach strains; (ii) oak strains; (iii) oleander strains; and (iv) all grape strains, a maple strain, and 3 almond strains. REP-PCR and RAPD analyses delineated five groups by distinguishing almond strains from peach and plum strains from the above four groups and separating almond strains into two groups: one within the grape group and another almond strain group distinct from grape. CHEF analysis identified the same five groups as did REP-PCR and RAPD analyses but revealed greater differences between grape and almond strains. CHEF analysis identified six groupings among the strains studied by distinguishing differences in groupings among most almond strains; however, CHEF analysis gave less resolution of genetic heterogeneity than RAPD analysis.
ALS and PD are thought to be caused by the same or similar strains of X. fastidiosa (11, 22), but anomalies have been noted. For example, in southern central California, ALS was absent from almond orchards that were adjacent to vineyards with a high incidence of PD (26). The reverse situation was noted in northern central California, where the incidence of PD was low in vineyards that were adjacent to almond orchards with a very high incidence of ALS (26). Our results suggest that some almond strains (e.g., ALS1, Manteca, and Tulare) may cause either PD or ALS under natural conditions, but other strains (almond groups 2 and 3) probably cause disease mostly in field-grown almonds. Unlike oak, oleander, peach, and plum groups, which appear to be genetically uniform, ALS strains (groups 1, 2, and 3) differ considerably from each other in REP- and RAPD-PCR and genomic DNA restriction fingerprints. However, strains from both distinctive groups of ALS strains produced ALS symptoms after mechanical inoculation (R. P. Almeida and A. H. Purcell, unpublished data). It would be interesting to determine whether the three groups of ALS strains differ in virulence in almond and grape.
Strains that cause PD or alfalfa dwarf disease under greenhouse conditions occur naturally among an unusually broad range of host plants (13, 17). Hewitt (14) speculated that the PD pathogen was introduced into California via the importation of plants (such as wild grapes used for grape rootstocks) from southeastern North America, where PD is endemic. The similarity of RAPD-PCR products and genomic DNA restriction patterns among PD strains from California, Florida, and Georgia supports this hypothesis. Grape isolates from California were genetically uniform when both ERIC and REP-PCR primers were used. Because REP-PCR analyzes the distribution of repetitive sequences in bacterial genomes, it is less likely to detect minor genetic changes that can be detected using select RAPD primers. The results obtained with REP-PCR suggest that California PD strains form a coherent cluster compared to Xylella strains that infect other woody plant species growing in diverse geographical areas.
The genetic variation among PD strains of X. fastidiosa strains revealed by RAPD-PCR suggests either that these differences among PD strains have evolved over the past 130-plus years or that genetically different strains were introduced into the state or that some PD strains are indigenous to western North America. Among the California PD strains that were analyzed by RAPD-PCR, the north coastal strains (found in Alameda, Napa, Santa Cruz, and Sonoma counties) were more similar to each other than to southern California strains (found in Ventura, San Luis Obispo, and Los Angeles counties) or to central California strains (found in Fresno, Tulare, and Contra Costa counties) (Table 1). Additional strains will have to be analyzed to determine if these putative geographical groupings consistently occur in these regions of California. Alternatively, some of the differences that were revealed by RAPD analyses could be due to the presence of extrachromosomal DNA, although there was no consistent association between a particular RAPD pattern and the presence or absence of plasmid DNA.
In contrast to the results reported by Chen et al. (4), we found plasmids in most of the X. fastidiosa strains analyzed. It is interesting to note that, although grape strains from the east and west coasts of the United States have similar chromosomal DNA, they differed in plasmid DNA content. Furthermore, the same plasmid profile (plasmid profile A) was common to both Californian PD and ALS strains whose chromosomal DNAs were genetically different, suggesting that this plasmid might be transferable between almond and grape strains.
All of the molecular genetic analyses indicate that the strains of X. fastidiosa causing oleander leaf scorch (28) are distinct from PD, ALS, OLS, PP, and PLS strains. We did not detect any genetic differences among the four oleander strains we examined. Oleander leaf scorch was observed in California in the mid-1990s, and the origin of the oleander leaf scorch strains is unknown. Our results best support the hypothesis that the current outbreak of oleander disease is the result of a single introduction or limited number of introductions of a genetically uniform X. fastidiosa strain. Additional oleander strains will have to be analyzed in order to substantiate this hypothesis.
The low variability of 16S-23S rRNA intergenic spacer region sequences among all X. fastidiosa strains tested indicates that they are phylogenetically closely related. CHEF electrophoresis of large genomic restriction fragments, REP- and RAPD-PCR products, and RsaI restriction of the genomic region specific to X. fastidiosa and flanked by oligonucleotide primers RST 31 and 33 (21) grouped oleander leaf scorch strains with ALS, OLS, PP, and PLS strains. But the sequence of the 16S-23S rRNA spacer region suggests that oleander leaf scorch strains are more closely related to PD strains. This implies potential independent evolution of these particular genetic loci.
RFLP studies by Chen et al. (4) indicated that strains of PD, ALS, and alfalfa dwarf disease comprise a PD RFLP group. The PLS cluster was distantly related to the Pierce's disease group. Our results show that most ALS strains differ from PD strains and that the plum group is closer to the almond group than to the PD group.
If our results are supported by additional DNA-DNA hybridization analyses, these genetic groups of X. fastidiosa may represent distinct species. The availability of a complete sequence from a citrus variegated chlorosis strain (31) and several other X. fastidiosa strains in the near future (http: //www.jgi.doe.gov/tempweb/jgi_microbial/html/index.html) should further clarify species differences among strains of Xylella. At the least, our findings support the suggestion of other groups (6, 9, 19, 24, 29) that X. fastidiosa should be taxonomically differentiated at the subspecies or pathovar level. However, the abilities of various strains to infect and cause disease in the various plant host need to be examined before pathovars can be designated.
ACKNOWLEDGMENTS
This work was supported by grants from the Napa Valley Pierce's Disease Task Force and the American Vineyard Foundation.
We thank Stuart Saunders for help in maintaining cultures and other technical assistance.
REFERENCES
- 1.Banks D, Albibi R, Chen J, Lamikanra O, Jarret R L, Smith B J. Specific detection of Xylella fastidiosa Pierce's disease strains. Curr Microbiol. 1999;39:85–88. doi: 10.1007/s002849900423. [DOI] [PubMed] [Google Scholar]
- 2.Barry T, Colleran G, Glennon M, Dunican L K, Gannon F. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991;1:233–236. doi: 10.1101/gr.1.1.51. [DOI] [PubMed] [Google Scholar]
- 3.Chang C J, Schaad N W. Electrophoretic protein profiles of total cell envelopes of xylem-limited plant pathogenic rickettsia-like bacteria (RLB) Phytopathology. 1982;72:935–936. [Google Scholar]
- 4.Chen J, Chang C J, Jarret R L. Plasmids from Xylella fastidiosa. Can J Microbiol. 1992;38:993–995. [Google Scholar]
- 5.Chen J, Chang C J, Jarret R L, Gawel N. Genetic variation among Xylella fastidiosa strains. Phytopathology. 1992;82:973–977. [Google Scholar]
- 6.Chen J, Lamikanra O, Chang C J, Hopkins D L. Randomly amplified polymorphic DNA analysis of Xylella fastidiosa Pierce's disease and oak leaf scorch pathotypes. Appl Environ Microbiol. 1995;61:1688–1690. doi: 10.1128/aem.61.5.1688-1690.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu G, Vollrath D, Davis R W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 8.Cooksey D A, Graham J H. Genomic fingerprinting of two pathovars of phytopathogenic bacteria by rare-cutting restriction enzymes and field inversion gel electrophoresis. Phytopathology. 1989;79:745–750. [Google Scholar]
- 9.da Costa P I, Franco C F, Miranda V S, Teixeira D C, Hartung J S. Strains of Xylella fastidiosa rapidly distinguished by arbitrarily primed PCR. Curr Microbiol. 2000;40:279–282. doi: 10.1007/s002849910055. [DOI] [PubMed] [Google Scholar]
- 10.Davis M J, French W J, Schaad N W. Axenic culture of the bacteria associated with phony peach disease and plum leaf scald. Curr Microbiol. 1981;6:309–314. [Google Scholar]
- 11.Davis M J, Thompson S W, Purcell A H. Etiological role of the xylem-limited bacterium causing Pierce's disease in almond leaf scorch. Phytopathology. 1979;70:472–475. [Google Scholar]
- 12.Felsenstein J. PHYLIP (phylogeny inference package) version 3.5c. Seattle, Wash: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 13.Freitag J H. Host range of the Pierce's disease virus of grapes as determined by insect transmission. Phytopathology. 1951;41:920–934. [Google Scholar]
- 14.Hewitt W B. The probable home of Pierce's disease virus. Plant Dis Rep. 1958;42:211–215. [Google Scholar]
- 15.Hill B L, Purcell A H. Acquisition and retention of Xylella fastidiosa by an efficient vector, Graphocephala atropunctata. Phytopathology. 1995;85:209–212. [Google Scholar]
- 16.Hopkins D L. Xylella fastidiosa: xylem-limited bacterial pathogen of plants. Annu Rev Phytopathol. 1989;27:271–290. [Google Scholar]
- 17.Hopkins D L, Adlerz W C. Natural hosts of Xylella fastidiosa in Florida. Plant Dis. 1988;72:429–431. [Google Scholar]
- 18.Jensen M A, Webster J A, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamper S M, French W J, DeKloet S R. Genetic relationships of some fastidious xylem-limited bacteria. Int J Syst Bacteriol. 1985;35:185–188. [Google Scholar]
- 20.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning—a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 368–369. [Google Scholar]
- 21.Minsavage G V, Thompson C M, Hopkins D L, Leite R M V B C, Stall R E. Development of a polymerase chain reaction protocol for detection of Xylella fastidiosa in plant tissue. Phytopathology. 1994;84:456–461. [Google Scholar]
- 22.Mircetich S M, Lowe S L, Moller W J, Nyland G. Etiology of almond leaf scorch disease and transmission of the casual agent. Phytopathology. 1976;66:17–24. [Google Scholar]
- 23.Opgenorth D C, Smart C D, Louws F J, deBruijn F J, Kirkpatrick B C. Identification of Xanthomonas fragariae field isolates by rep-PCR genomic fingerprinting. Plant Dis. 1996;80:868–873. [Google Scholar]
- 24.Pooler M R, Hartung J S. Sequence analysis of a 1296-nucleotide plasmid from Xylella fastidiosa. FEMS Microbiol Lett. 1997;155:217–222. doi: 10.1016/s0378-1097(97)00391-1. [DOI] [PubMed] [Google Scholar]
- 25.Pooler M R, Hartung J S, Fenton R G. Genetic relationships among strains of Xylella fastidiosa from RAPD-PCR data. Curr Microbiol. 1995;31:134–137. doi: 10.1007/BF00294290. [DOI] [PubMed] [Google Scholar]
- 26.Purcell A H. Almond leaf scorch: leafhopper and spittle bug vectors. J Econ Entomol. 1980;73:834–838. [Google Scholar]
- 27.Purcell A H. Xylella fastidiosa, a regional problem or global threat? J Plant Pathol. 1997;79:99–105. [Google Scholar]
- 28.Purcell A H, Saunders S R, Hendson M, Grebus M E, Henry M J. Causal role of Xylella fastidiosa in oleander leaf scorch. Phytopathology. 1999;89:53–58. doi: 10.1094/PHYTO.1999.89.1.53. [DOI] [PubMed] [Google Scholar]
- 29.Rosato Y E, Neto J R, Miranda V S, Carlos E F, Manfio G P. Diversity of a Xylella fastidiosa population isolated from Citrus sinensis affected by citrus variegated chlorosis in Brazil. Syst Appl Microbiol. 1998;21:593–598. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson A A J G, Reinach F C, Arruda P, Abreu F A, Acencio M, Alvarenga R, Alves L M, Araya J E, Baia G S, Baptista C S, et al. The genome sequence of the plant pathogen Xylella fastidiosa. Nature. 2000;406:151–157. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- 32.Sneath P H A, Sokal R R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman and Co.; 1973. [Google Scholar]
- 33.Swofford D L. PAUP (phylogenic analysis using parsimony). Washington, D.C.: Laboratory of Molecular Systematics, National Museum of Natural History, Smithsonian Institution; 1993. [Google Scholar]
- 34.Versalovic J, Schneider M, deBruijn F J, Lupski J R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 35.Wells J M, Raju B C, Hung H-Y, Weisburg W G, Mandelco-Paul L, Brenner D J. Xylella fastidiosa gen. nov., sp. nov.: gram-negative, xylem-limited, fastidious plant bacteria related to Xanthomonas spp. Int J Syst Bacteriol. 1987;37:136–143. [Google Scholar]
- 36.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1987. pp. 2.4.1–2.4.2. [Google Scholar]