Abstract
Chia seed peptides (CSP) can be a source of multifunctional biopeptides to treat non-communicable diseases. However, interactions and binding affinity involved in targeting specific receptors remains unexplored. In this study, molecular simulation techniques were used as virtual screening of CSP to determine drug-like candidates using a multi-target-directed ligand approach. CSP fraction with the best bioactivities in vitro was sequenced. Then, a prediction model was built using physicochemical descriptors (hydrophobicity, hydrophilicity, intestinal stability, antiangiogenic, antihypertensive, and anti-inflammatory) to calculate potential scores and rank possible biopeptides. Furthermore, molecular dynamics simulations (MDS) and ensemble molecular docking analysis were carried out using four human protein targets (ACE, angiotensin converting enzyme; VEGF, vascular endothelial growth factor; GLUC, glucocorticoid and MINC, mineralocorticoid receptors). Five known-sequence peptides (NNVFYPF, FNIVFPG, SRPWPIDY, QLQRWFR, GSRFDWTR) and five de novo peptides (DFKF, DLRF, FKAF, FRSF, QFRF) had the lowest energy score and higher affinity for ACE and VEGF. The therapeutic effects of these selected peptides can be related to the inhibition of the enzymes involved in angiogenesis and hypertension, due to formation of stable complexes with VEGF and ACE binding sites, respectively. The application of MDS is a good resource for identifying bioactive peptides for future experimental validation.
Keywords: bioactive peptides, ensemble docking, multifunctional bioactivities, molecular dynamics simulations, chronic diseases
1. Introduction
In recent years, there has been an increased interest in finding bioactive peptides that can prevent the risk of chronic diseases and/or boost the immune system. The advancement of peptidomics and bioinformatics in food science has enabled integrated studies to be carried out for the rapid development of food-derived bioactive peptides [1]. Moreover, the application of an integrated approach in the study of bioactive peptides allows for their production to be optimized, leads to the discovery of peptides of specific interest, and contributes to the understanding of the mechanism of action between bioactive peptides and human target receptors [2]. Accordingly, this integrated approach is more cost-effective and time-saving compared with the standard approach used, which involves more laborious experimental tests to study bioactive peptides [2,3]. In view of the above, these advancements have provided important tools for efficient discovery of novel peptides with profitable biological activities, for the analysis of peptide datasets, and an understanding of their structure-activity relationships [3]. The latter will allow researchers to explore the complete picture and/or entire spectrum of bioactivities from any protein source.
Chia (Salvia hispanica L.) seeds are recognized as an emerging protein source. Aside from its role as dietary nutrients, it is being progressively recognized for its bioactive properties due to the presence of essential and non-essential amino acids, ω-3 fatty acids, phytosterol, and dietary fiber [4,5,6]. Recently, our research group demonstrated that chia seed peptides, obtained by sequential and microwave-assisted proteolysis, exhibited prominent chemical and cellular antioxidant [7], antimicrobial [8], and anti-aging [9] properties. Similarly, other studies have reported that chia seed peptides possess in vitro antioxidant [10], antibacterial [11], and anti-inflammatory [12] activities. Therefore, our aim was to apply bioinformatics and in silico methodologies (i.e., physicochemical properties prediction, molecular dynamics and molecular docking simulations) to predict potential bioactivities associated with chia seed peptides. The outcomes of this study will generate new knowledge into their mechanism of action associated with intermolecular interactions within enzymes involved in well-recognized chronic diseases such as hypertension and inflammation.
2. Results and Discussion
Chia seed is an interesting protein source that can be studied to identify peptides and explore their potential bioactivities. Since chia seed peptides (CSP) obtained by sequential and microwave-assisted proteolysis have demonstrated, in our laboratory, to have good chemical and cellular antioxidant [7], antimicrobial [8], and anti-aging [9] in vitro activities, they were selected for peptidomics sequencing and bioinformatic analysis. Many traditional in silico methods are challenging to adapt to certain classes of biomolecules, mostly when the biomolecules consist of large and highly flexible chemical moieties (like peptides, toxins, and antibodies). For example, generating libraries of large peptides is currently feasible, due to massive mass spectrometry techniques and/or peptidomics approaches. However, the prediction of properties, such as those related to the bioactivity of these biomolecules enables either simplifying assumptions to be made or the application of new approaches for predicting bioactivity [13]. Here, we conducted an in silico prediction of physicochemical properties (hydrophobicity, hydrophilicity, intestinal stability, antiangiogenic, antihypertensive, and anti-inflammatory, using molecular dynamics simulations (MDS) and ensemble docking–virtual screening (VS) methods, with potential bioactive peptides from chia seed, and correlated these properties with human molecular (protein) targets (angiotensin converting enzyme (ACE), vascular endothelial growth factor (VEGF), glucocorticoid (GLUC), and mineralocorticoid (MINC) receptors).
CSP library was initially built to reflect differences in physicochemical properties of selected peptides, and subsequentially those properties were used as descriptors. Several studies have reinforced the use of descriptors, showing the effectiveness of predicting a potential pharmaco-kinetic behavior based on ligand virtual screening techniques [14,15]. However, very few examples of peptides have shown the expected effectiveness [15,16]. Overall, a total of 1954 peptides were identified from the <3 kDa CSP fraction obtained from controlled enzymatic proteolysis. After the list of peptides were screened using Peptide Ranker, a total of 83 peptides were predicted as being bioactive (49 database peptides and 34 de novo peptides) according to their score (>0.8 threshold) (Table 1). This web server allowed us to predict the likehood of all the peptides for being bioactive using a neural network based on amino acid sequences and specific structure feature analysis [17]. Interestingly, it is important to note that the shortlisted database-peptides had a length between 7 and 17 amino acid residues, while shortlisted de novo peptides had a length between 4 and 6 amino acid residues. Scientific literature reports that bioactive peptides tend to be between 2–20 amino acids in length [18], with cores (hydrophobic or hydrophilic) of 4–5 amino acids that guide the interaction within the binding sites. Furthermore, several in silico bioactivity predictors have incorporated “peptide sequence length” as a descriptor into their quantitative structure relationship models [19,20]. Our de novo peptides fullfilled the necessary length to be considered as bioactive by different in silico predictors.
Table 1.
In silico analysis results of selected peptide sequences from chia seed.
| No. | Peptide Sequence | Peptide Ranker Score | PreAIP | AntiAngio-Pred | AHTpin | |||
|---|---|---|---|---|---|---|---|---|
| Score | Prediction | Score | Prediction | Score | Prediction | |||
| Database peptides | ||||||||
| 1 | FNLVFFLL | 0.951 | 0.416 | AIP | −0.21 | Non-AAP | −0.58 | Non-AHT |
| 2 | EGDVFWIPRF | 0.940 | 0.418 | AIP | −0.5 | Non-AAP | −0.12 | Non-AHT |
| 3 | DHFPFIY | 0.933 | 0.518 | AIP | 0.04 | AAP | 0.47 | AHT |
| 4 | EGGIWPF | 0.929 | 0.344 | AIP | 0.48 | AAP | −0.1 | Non-AHT |
| 5 | GFEWITF | 0.922 | 0.57 | AIP | 0.47 | AAP | −0.89 | Non-AHT |
| 6 | GLDFPELPLGM | 0.919 | 0.481 | AIP | −0.61 | Non-AAP | 1.31 | AHT |
| 7 | GQTPLFPRIF | 0.912 | 0.412 | AIP | 0.41 | AAP | 0.65 | AHT |
| 8 | GDAHYDPLFPF | 0.909 | 0.323 | Non-AIP | −1.23 | Non-AAP | 0.95 | AHT |
| 9 | NNVFYPF | 0.903 | 0.344 | AIP | −0.34 | Non-AAP | 0.22 | AHT |
| 10 | EYPPLGRF | 0.901 | 0.395 | AIP | 1.01 | AAP | 1.04 | AHT |
| 11 | KPLPFELF | 0.898 | 0.409 | AIP | 0.6 | AAP | 0.55 | AHT |
| 12 | DVWDPFQDFPL | 0.895 | 0.461 | AIP | 0.11 | AAP | 0.41 | AHT |
| 13 | SDKNGYFF | 0.883 | 0.418 | AIP | −0.82 | Non-AAP | −1.16 | Non-AHT |
| 14 | VPIPVPLPF | 0.883 | 0.318 | Non-AIP | −0.24 | Non-AAP | 2.29 | AHT |
| 15 | SNVFDPF | 0.876 | 0.338 | Non-AIP | −0.87 | Non-AAP | −0.06 | Non-AHT |
| 16 | TPLFPRIF | 0.876 | 0.393 | AIP | 1.03 | AAP | 0.58 | AHT |
| 17 | DQNPRSFFL | 0.873 | 0.444 | AIP | 1 | AAP | −0.67 | Non-AHT |
| 18 | QLQRWFR | 0.871 | 0.519 | AIP | 2.22 | AAP | −0.62 | Non-AHT |
| 19 | GFEWVAF | 0.868 | 0.59 | AIP | −0.94 | Non-AAP | 0.3 | AHT |
| 20 | SFNLPIL | 0.867 | 0.408 | AIP | −0.4 | Non-AAP | −0.15 | Non-AHT |
| 21 | QEGGIWPF | 0.863 | 0.37 | AIP | 0.39 | AAP | −0.52 | Non-AHT |
| 22 | GSRFDWTR | 0.858 | 0.488 | AIP | 2.17 | AAP | −1.36 | Non-AHT |
| 23 | ADFYNPR | 0.853 | 0.303 | Non-AIP | 0.92 | AAP | 0.44 | AHT |
| 24 | APSKDAPMF | 0.851 | 0.452 | AIP | −0.16 | Non-AAP | −0.24 | Non-AHT |
| 25 | GFEWITFK | 0.847 | 0.578 | AIP | 0.28 | AAP | −0.66 | Non-AHT |
| 26 | NGFEWITF | 0.842 | 0.549 | AIP | 0.4 | AAP | −1.03 | Non-AHT |
| 27 | VNEGDVFWIPRF | 0.841 | 0.414 | AIP | −1.1 | Non-AAP | −0.52 | Non-AHT |
| 28 | SSNVFDPF | 0.841 | 0.304 | Non-AIP | −0.93 | Non-AAP | −0.17 | Non-AHT |
| 29 | FNIVFPG | 0.839 | 0.385 | AIP | −1.68 | Non-AAP | 0.76 | AHT |
| 30 | VPVFPPPLN | 0.837 | 0.435 | Non-AIP | −0.26 | Non-AAP | 2 | AHT |
| 31 | GIDIPPPR | 0.835 | 0.316 | Non-AIP | 0.55 | AAP | 0.47 | AHT |
| 32 | APAEKGFAGF | 0.832 | 0.402 | AIP | −1.39 | Non-AAP | 0.23 | AHT |
| 33 | DQNPRSFF | 0.830 | 0.433 | AIP | 1.05 | AAP | −0.92 | Non-AHT |
| 34 | SRPWPIDY | 0.827 | 0.486 | AIP | 2.26 | AAP | −0.04 | Non-AHT |
| 35 | QNGFEWITF | 0.825 | 0.573 | AIP | 0.46 | AAP | −1.47 | Non-AHT |
| 36 | RPGDVFVFPR | 0.825 | 0.383 | AIP | −0.98 | Non-AAP | 0.22 | AHT |
| 37 | DNGIIYPW | 0.823 | 0.32 | Non-AIP | −0.36 | Non-AAP | 0.15 | AHT |
| 38 | NPQAGRF | 0.822 | 0.376 | AIP | −0.43 | Non-AAP | 0.03 | AHT |
| 39 | APVGSPVGSTGGNFGVF | 0.817 | 0.476 | AIP | −1.1 | Non-AAP | 0.39 | AHT |
| 40 | APPPVLAL | 0.816 | 0.396 | AIP | 0.61 | AAP | 0.07 | AHT |
| 41 | FPLLNYL | 0.813 | 0.554 | AIP | −0.18 | Non-AAP | 1.31 | AHT |
| 42 | RNNVFYPF | 0.811 | 0.378 | AIP | 0.61 | AAP | 0.49 | AHT |
| 43 | GNIFRGL | 0.811 | 0.452 | AIP | −0.34 | Non-AAP | −0.6 | Non-AHT |
| 44 | FPGLADRM | 0.810 | 0.333 | Non-AIP | −1.01 | Non-AAP | 0.67 | AHT |
| 45 | SNEWDPSFR | 0.806 | 0.393 | AIP | 0.81 | AAP | −1.34 | Non-AHT |
| 46 | SMLSPHW | 0.806 | 0.42 | AIP | 0.22 | AAP | −0.24 | Non-AHT |
| 47 | SLDVWDPFQDFPL | 0.804 | 0.464 | AIP | 0.44 | AAP | 0.59 | AHT |
| 48 | SPDLIRRM | 0.803 | 0.399 | AIP | 1.12 | AAP | −0.74 | Non-AHT |
| 49 | FGNVFKGM | 0.803 | 0.32 | Non-AIP | −1.68 | Non-AAP | −0.8 | Non-AHT |
| De novo peptides | ||||||||
| 1 | QFRF | 0.980 | 0.361 | AIP | ND | - | −0.05 | Non-AHT |
| 2 | FDRF | 0.978 | 0.301 | Non-AIP | ND | - | −0.79 | Non-AHT |
| 3 | GRPW | 0.971 | 0.266 | Non-AIP | ND | - | 0.84 | AHT |
| 4 | FWDR | 0.964 | 0.347 | AIP | ND | - | −0.74 | Non-AHT |
| 5 | FRSF | 0.962 | 0.339 | Non-AIP | ND | - | −0.76 | Non-AHT |
| 6 | GPHW | 0.958 | 0.354 | AIP | ND | - | 1 | AHT |
| 7 | KPPF | 0.955 | 0.294 | Non-AIP | ND | - | 2.09 | AHT |
| 8 | WLPR | 0.943 | 0.306 | Non-AIP | ND | - | 1.13 | AHT |
| 9 | FWDH | 0.938 | 0.318 | Non-AIP | ND | - | −0.61 | Non-AHT |
| 10 | FDKF | 0.937 | 0.32 | Non-AIP | ND | - | −0.87 | Non-AHT |
| 11 | FRGL | 0.937 | 0.318 | Non-AIP | ND | - | −0.38 | Non-AHT |
| 12 | DFKF | 0.932 | 0.336 | Non-AIP | ND | - | −0.87 | Non-AHT |
| 13 | KDFLFP | 0.929 | 0.352 | AIP | −0.03 | Non-AAP | −0.79 | Non-AHT |
| 14 | EFRF | 0.922 | 0.289 | Non-AIP | ND | - | 0.99 | AHT |
| 15 | APHW | 0.918 | 0.389 | AIP | ND | - | 0.03 | AHT |
| 16 | RPAF | 0.909 | 0.28 | Non-AIP | ND | - | 0.89 | AHT |
| 17 | ARGW | 0.908 | 0.317 | Non-AIP | ND | - | −0.76 | Non-AHT |
| 18 | FKAF | 0.907 | 0.31 | Non-AIP | ND | - | −0.67 | Non-AHT |
| 19 | WEFLTF | 0.907 | 0.329 | Non-AIP | 0.43 | AAP | −0.89 | Non-AHT |
| 20 | HVFF | 0.878 | 0.349 | AIP | ND | - | −0.21 | Non-AHT |
| 21 | WAPH | 0.873 | 0.313 | Non-AIP | ND | - | 0.99 | AHT |
| 22 | RPSF | 0.872 | 0.334 | Non-AIP | ND | - | 0.7 | AHT |
| 23 | HPAYW | 0.871 | 0.392 | AIP | 0.1 | AAP | 1.81 | AHT |
| 24 | DLRF | 0.863 | 0.319 | Non-AIP | ND | - | −0.56 | Non-AHT |
| 25 | QLRF | 0.863 | 0.344 | AIP | ND | - | 0.21 | AHT |
| 26 | GKFL | 0.850 | 0.316 | Non-AIP | ND | - | −0.48 | Non-AHT |
| 27 | QRYF | 0.848 | 0.327 | Non-AIP | ND | - | 0.89 | AHT |
| 28 | FWDNH | 0.834 | 0.345 | AIP | −0.63 | Non-AAP | 0.05 | AHT |
| 29 | FPLK | 0.834 | 0.336 | Non-AIP | ND | - | 1.07 | AHT |
| 30 | RAFL | 0.831 | 0.349 | AIP | ND | - | −0.42 | Non-AHT |
| 31 | FPLLN | 0.820 | 0.483 | AIP | −0.1 | Non-AAP | 0.22 | AHT |
| 32 | WDPSYR | 0.816 | 0.325 | Non-AIP | 2.67 | AAP | 0.11 | AHT |
| 33 | GLKF | 0.810 | 0.348 | AIP | ND | - | −0.48 | Non-AHT |
| 34 | HPNPRL | 0.808 | 0.358 | AIP | 0.51 | AAP | 0.98 | AHT |
ND: Not determined due to server limitations; PreAIP: Peptides with a score > 0.342 are predicted as anti-inflammatory peptides (AIP); AngioPred: a positive score is predicted as an antiangiogenic peptide (AAP); AHTpin: a positive score is predicted as an antihypertensive peptide (AHT); a score > 1 is predicted as high stability, while a score < 1 is predicted as normal stability.
The descriptors of the whole library were used to filter out the peptides with PreAIP, AHTpin, and AntioAngioPred servers for the prediction of their anti-inflammatory, antihypertensive, and antiangiogenic potentials before the ensemble docking analyses. In this scenario, physicochemical properties such as hydrophilicity, hydropathy, hydrophobicity, intestinal absorption (Table 2), and potential bioactivities (e.g., anti-angiogenic, anti-inflammatory, and antihypertensive) were prioritized, expecting that, among the screened peptide library, peptides with potential activity towards previously described protein targets could be identified [21]. Overall, it was observed that 52 and 42 of the 83 selected peptides were predicted as anti-inflamatoy and antihypertensive, respectively. This suggests that most of the selected peptides contained amino acids and structural features that contributed to those bioactivities.
Table 2.
Physicochemical chracteristics of selected chia seed peptide sequences.
| No. | Peptide Sequence | Intestinal Stability | Hydrophobicity | Hydropathicity | Hydrophilicity | Charge | Molecular Weight |
|---|---|---|---|---|---|---|---|
| Database peptides | |||||||
| 1 | FNLVFFLL | 0.73 | 0.42 | 2.56 | −1.78 | 0 | 1012.35 |
| 2 | EGDVFWIPRF | 2.391 | −0.02 | −0.01 | −0.27 | −1 | 1265.57 |
| 3 | DHFPFIY | 1.094 | 0.11 | 0.07 | −0.94 | −1 | 938.13 |
| 4 | EGGIWPF | 0.945 | 0.19 | 0.07 | −0.67 | −1 | 805 |
| 5 | GFEWITF | 2.512 | 0.24 | 0.66 | −1.09 | −1 | 899.11 |
| 6 | GLDFPELPLGM | 2.79 | 0.12 | 0.46 | −0.29 | −2 | 1188.54 |
| 7 | GQTPLFPRIF | 1.079 | −0.01 | 0.16 | −0.58 | 1 | 1175.52 |
| 8 | GDAHYDPLFPF | 2.144 | 0.02 | −0.35 | −0.37 | −2 | 1278.52 |
| 9 | NNVFYPF | 0.798 | 0.06 | −0.01 | −1.2 | 0 | 900.09 |
| 10 | EYPPLGRF | 0.392 | −0.15 | −0.79 | −0.07 | 0 | 978.21 |
| 11 | KPLPFELF | 1.859 | 0.05 | 0.32 | −0.33 | 0 | 990.3 |
| 12 | DVWDPFQDFPL | 1.608 | −0.03 | −0.41 | −0.23 | −3 | 1378.65 |
| 13 | SDKNGYFF | 1.183 | −0.17 | −0.98 | −0.1 | 0 | 977.14 |
| 14 | VPIPVPLPF | 2.283 | 0.3 | 1.46 | −1.01 | 0 | 978.35 |
| 15 | SNVFDPF | 0.585 | 0.01 | 0.06 | −0.43 | −1 | 824.97 |
| 16 | TPLFPRIF | 1.143 | 0.05 | 0.69 | −0.75 | 1 | 990.3 |
| 17 | DQNPRSFFL | 1.023 | −0.27 | −0.89 | −0.01 | 0 | 1123.34 |
| 18 | QLQRWFR | 1.066 | −0.48 | −1.47 | −0.19 | 2 | 1033.3 |
| 19 | GFEWVAF | 2.466 | 0.27 | 0.97 | −1.06 | −1 | 855.06 |
| 20 | SFNLPIL | 2.342 | 0.2 | 1.29 | −1.06 | 0 | 803.04 |
| 21 | QEGGIWPF | 1.213 | 0.08 | −0.38 | −0.56 | −1 | 933.15 |
| 22 | GSRFDWTR | 0.667 | −0.44 | −1.56 | 0.38 | 1 | 1024.21 |
| 23 | ADFYNPR | 1.132 | −0.33 | −1.4 | 0.13 | 0 | 882.02 |
| 24 | APSKDAPMF | 1.538 | −0.09 | −0.34 | 0.17 | 0 | 963.22 |
| 25 | GFEWITFK | 2.682 | 0.07 | 0.09 | −0.58 | 0 | 1027.3 |
| 26 | NGFEWITF | 2.499 | 0.13 | 0.14 | −0.93 | −1 | 1013.23 |
| 27 | VNEGDVFWIPRF | 2.24 | −0.02 | 0.05 | −0.33 | −1 | 1478.84 |
| 28 | SSNVFDPF | 0.2 | −0.02 | −0.05 | −0.34 | −1 | 912.06 |
| 29 | FNIVFPG | 2.136 | 0.28 | 1.26 | −1.16 | 0 | 793.02 |
| 30 | VPVFPPPLN | 1.974 | 0.14 | 0.57 | −0.79 | 0 | 979.3 |
| 31 | GIDIPPPR | 0.964 | −0.13 | −0.53 | 0.3 | 0 | 864.1 |
| 32 | APAEKGFAGF | 3.43 | 0.05 | 0.12 | −0.05 | 0 | 994.24 |
| 33 | DQNPRSFF | 1.044 | −0.36 | −1.48 | 0.21 | 0 | 1010.17 |
| 34 | SRPWPIDY | 1.413 | −0.22 | −1.21 | −0.15 | 0 | 1033.25 |
| 35 | QNGFEWITF | 2.539 | 0.04 | −0.27 | −0.8 | −1 | 1141.38 |
| 36 | RPGDVFVFPR | 3.379 | −0.19 | −0.21 | 0.1 | 1 | 1189.51 |
| 37 | DNGIIYPW | 0.959 | 0.07 | −0.28 | −0.76 | −1 | 977.19 |
| 38 | NPQAGRF | 0.929 | −0.31 | −1.27 | 0.06 | 1 | 788.95 |
| 39 | APVGSPVGSTGGNFGVF | 2.935 | 0.14 | 0.53 | −0.56 | 0 | 1549.95 |
| 40 | APPPVLAL | 1.457 | 0.24 | 1.32 | −0.76 | 0 | 777.06 |
| 41 | FPLLNYL | 1.841 | 0.22 | 1.11 | −1.43 | 0 | 879.14 |
| 42 | RNNVFYPF | 0.769 | −0.17 | −0.58 | −0.67 | 1 | 1056.29 |
| 43 | GNIFRGL | 1.294 | −0.03 | 0.33 | −0.41 | 1 | 775.99 |
| 44 | FPGLADRM | 2.43 | −0.09 | 0.04 | −0.01 | 0 | 906.16 |
| 45 | SNEWDPSFR | 0.992 | −0.37 | −1.81 | 0.43 | −1 | 1137.29 |
| 46 | SMLSPHW | 1.339 | 0.02 | −0.23 | −0.91 | 0 | 857.09 |
| 47 | SLDVWDPFQDFPL | 1.558 | 0 | −0.12 | −0.31 | −3 | 1578.91 |
| 48 | SPDLIRRM | 1.23 | −0.38 | −0.59 | 0.55 | 1 | 987.27 |
| 49 | FGNVFKGM | 1.854 | 0.07 | 0.44 | −0.57 | 1 | 899.19 |
| De novo peptides | |||||||
| 1 | QFRF | ND | −0.31 | −0.6 | −0.45 | 1 | 596.73 |
| 2 | FDRF | ND | −0.32 | −0.6 | 0.25 | 0 | 583.68 |
| 3 | GRPW | ND | −0.33 | −1.85 | −0.1 | 1 | 514.64 |
| 4 | FWDR | ND | −0.38 | −1.52 | 0.02 | 0 | 622.73 |
| 5 | FRSF | ND | −0.2 | 0.07 | −0.42 | 1 | 555.67 |
| 6 | GPHW | ND | 0.01 | −1.53 | −0.97 | 0 | 495.6 |
| 7 | KPPF | ND | −0.16 | −1.07 | 0.12 | 1 | 487.64 |
| 8 | WLPR | ND | −0.23 | −0.8 | −0.55 | 1 | 570.74 |
| 9 | FWDH | ND | −0.04 | −1.2 | −0.85 | −1 | 603.69 |
| 10 | FDKF | ND | −0.15 | −0.45 | 0.25 | 0 | 555.67 |
| 11 | FRGL | ND | −0.11 | 0.42 | −0.33 | 1 | 491.63 |
| 12 | DFKF | ND | −0.15 | −0.45 | 0.25 | 0 | 555.67 |
| 13 | KDFLFP | 1.208 | −0.29 | −0.6 | 0.25 | 0 | 597.71 |
| 14 | EFRF | ND | 0.04 | −0.97 | −1.1 | 0 | 509.62 |
| 15 | APHW | ND | −0.02 | 0.07 | −0.13 | 0 | 765.97 |
| 16 | RPAF | ND | −0.24 | −0.38 | 0 | 1 | 489.61 |
| 17 | ARGW | ND | −0.25 | −1 | −0.22 | 1 | 488.6 |
| 18 | FKAF | ND | 0.09 | 0.88 | −0.62 | 1 | 511.66 |
| 19 | WEFLTF | 1.16 | 0.22 | 0.72 | −1.27 | −1 | 842.04 |
| 20 | HVFF | ND | 0.34 | 1.65 | −1.75 | 0 | 548.69 |
| 21 | WAPH | ND | 0.04 | −0.98 | −1.1 | 0 | 509.62 |
| 22 | RPSF | ND | −0.37 | −1.02 | 0.2 | 1 | 505.61 |
| 23 | HPAYW | 1.398 | 0.03 | −1.04 | −1.34 | 0 | 672.81 |
| 24 | DLRF | ND | −0.33 | −0.35 | 0.43 | 0 | 549.66 |
| 25 | QLRF | ND | −0.33 | −0.35 | −0.28 | 1 | 562.71 |
| 26 | GKFL | ND | 0.05 | 0.57 | −0.33 | 1 | 463.62 |
| 27 | QRYF | ND | −0.46 | −1.63 | −0.4 | 1 | 612.73 |
| 28 | FWDNH | 1.425 | −0.16 | −1.66 | −0.64 | −1 | 717.81 |
| 29 | FPLK | ND | −0.01 | 0.28 | −0.32 | 1 | 503.68 |
| 30 | RAFL | ND | −0.09 | 0.97 | −0.45 | 1 | 505.65 |
| 31 | FPLLN | 1.991 | 0.19 | 1.06 | −1.18 | 0 | 602.78 |
| 32 | WDPSYR | 1.486 | −0.4 | −2.1 | 0.1 | 0 | 822.95 |
| 33 | GLKF | ND | 0.05 | 0.57 | −0.33 | 1 | 463.62 |
| 34 | HPNPRL | 1.534 | −0.4 | −1.77 | 0.15 | 1 | 732.91 |
ND: Not determined due to server limitations (peptides ≤ 4 amino acid residues).
Particularly, in relation to the prediction of antiangiogenic peptides, due to the software limitation for processing peptides with lengths of at least of 5 amino acid residues, the majority of de novo peptides (i.e., 27) were not subjected to such analysis because of their short amino acid length. From the rest, 29 peptides were predicted as antiangiogenic, while 27 were considered as non-angiogenic. The process of angiogenesis is a vital step towards the formation of malignant cancer tumors [22]. Antiangiogenic peptides could therefore be promising candidates in the treatment for cancer. Thus, most of the peptides showed potential antiangiogenic (anti-tumor), anti-inflammatory, and antihypertensive properties. Other studies have applied in silico analysis of select peptides obtained from different protein sources to identify potential bioactivies such as amaranth [23], milk protein [24], chicken breast [17], and kerfir milk [25], with promising results.
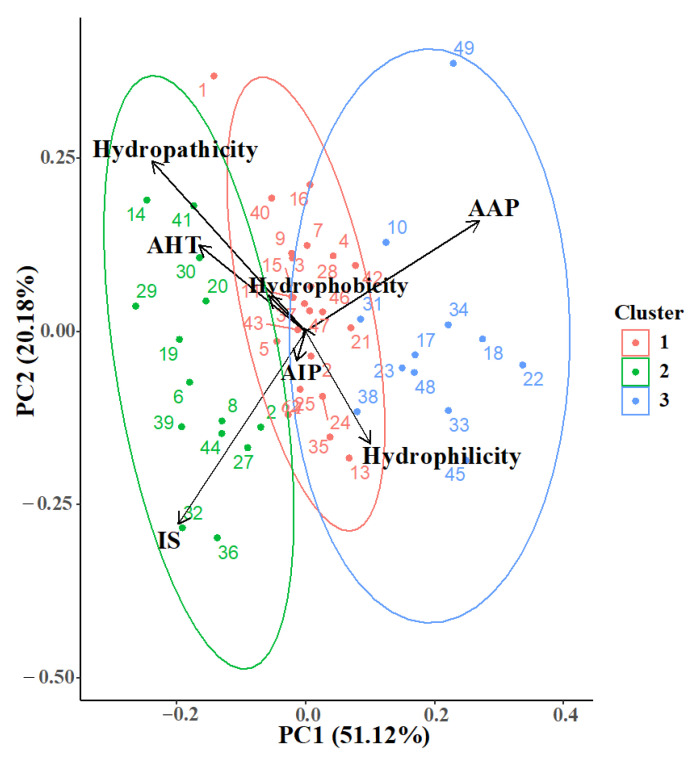
Dimensional reduction in calculated physicochemical properties was evaluated using Principal Component Analysis (PCA), which displayed a clustering hypothesis against three potential bioactivies, e.g., antiangiogenic (AAP), anti-inflamatory (AIP), and anti-hypertensive (AHT). Surprinsigly, physicochemical properties such as hydrophilicity, hydrophobicity and hydropathicity, clustered together with the aforementioned bioactivites, respectively (Figure 1). However, due to the high complexity of the analyzed biomolecules (e.g., tautomeric states, rotable bonds and side-chain diversity), the variance coverage was limited to nearly 72% (PC1-PC2). Furthermore, in chemoinformatics approaches toward natural products, these methodologies gave insights into the relationship between the calculated properties and possible bioactivity [26]. Acording to PCA and clustering analysis, four different molecular targets were selected; those targets were related to hypertension (angiotensin converting enzyme), inflammation (glucocorticoid and mineralocorticoid receptors), and angiogenesis (vascular endothelial growth factor).
Figure 1.
Principal Component Analysis of calculated physicochemical and bioactivity properties. AAP, antiangiogenic; AHT, antihypertensive; AIP, anti-inflammatory; IS, Intestinal Stability. Each dot corresponds to a peptide in the library (Table 1).
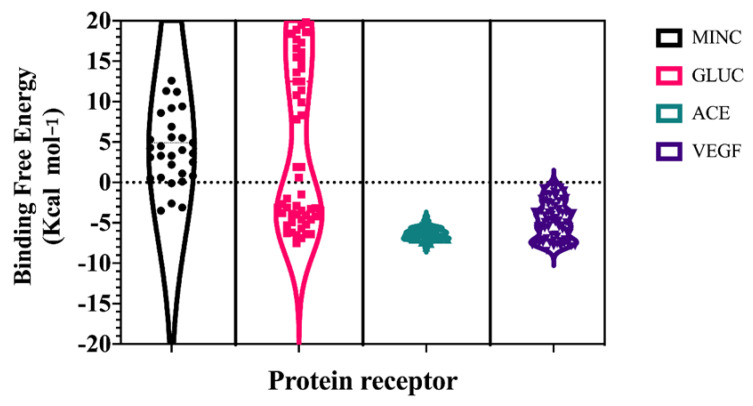
A hybrid all-atom Molecular Dynamics Simulations and ensemble docking-virtual screening workflow was applied to each target; 300 ns of Gaussian Accelerated Molecular Dynamics simulations were performed on each target, in order to recover a major conformational diversity of protein structure that stems into more robust molecular docking analysis [27,28]. Three diverse conformations of each target were used as initial docking coordinates for the peptide library, and the score (Kcal mol−1) was averaged with the Vinardo Scoring Function. As shown in Figure 2, the avegare scoring values of targets and peptide library complexes showed lower energy values for VEGF and ACE (~−3 Kcal mol−1 and −5 Kcal mol−1, respectively) and higher values for mineralocortidoid and glucocorticoid receptors (~3 Kcal mol−1 and 12 Kcal mol−1, respectively). This difference may arise from more constrained binding sites on mineralocorticoid and glucocorticoid receptors, which generally evolve around the structure of phenanthrene core ligands [21,29].
Figure 2.
Ensemble docking-virtual screening scores of chia seed peptides towards select human protein targets: MINC (mineralocorticoid), GLUC (glucocorticoid), ACE (angiotensin converting enzyme), and VEGF (vascular endothelial growth factor).
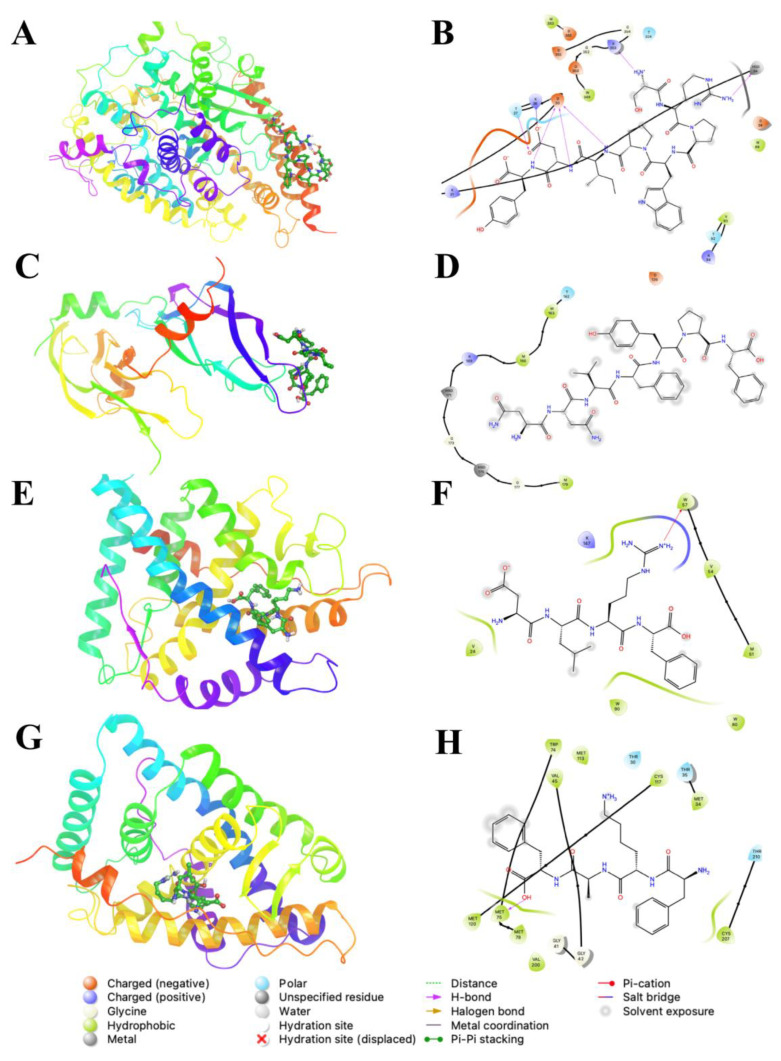
Figure 3 shows the binding modes and ligand interaction diagrams of selected peptides from library vs. selected molecular targets. In the case of ACE (Figure 3A,B) and VEGF (Figure 3C,D), these targets displayed superficial binding sites (interfacial regions), which are highly dependent on the conformation of receptor proteins. Furthermore, the resulting binding modes for VEGF and ACE were more stable (lower energy) and in good agreement with those previously described [30]. Additionally, for ACE, the lowest energy peptide (SRPWPIDY) is posed on the same binding site of the spike protein for the SARS-CoV-2 virus (Figure 3A), displaying electrostatic interactions with amino acid residues (D30 and E37); whereas basic residues K26, H34 and R393 mediate atractions to D7 and COO- terminal group of the Y8 residue. Moreover, the indole moiety of peptides was estabilized by aliphatic sidechains of V93 and backbone of T92 and K94. ACE was able to bind peptides through a flexible loop comprising A386–R393, which displays amphiphatic propierties, since it includes hydrophobic sidechains (A386, A387, F390) and backbone and amide sidechain from N388.
Figure 3.
Binding modes (left) and ligand interaction diagrams (right) of selected peptides from library vs. selected molecular targets. (A,B): Angiotensin converting enzyme (ACE) in complex with WPRSPIDY; (C,D): Vascular Endothelial growth factor (VEGF) in complex with FVNYPF; (E,F): Glucocorticoid receptor (GLUC) in complex wit KFAF; (G,H): Mineralocorticoid receptor (MINC) in complex with RLDF. Ligand interactions were calculated around a 5 Å distance cutoff.
The resulting peptide library for VEGF binding modes corresponds to the previously described binding sites of natural products and pharmaceutical drugs [22]. The proposed binding sites of VEGF are located on the C-terminal and comprise residues such as Y124, P125, Y130, and M179, which mainly impose short-range van der Waals (vdW) interactions with ligands; whereas D126, E129 and R167 are involved in long range electrostatic interactions. All the displayed interactions contribute to the recognition of de novo peptides that contain positively charged residues such as R and K, along with aliphatic residues L, A and F (Figure 3C,D). However, certain short length peptides, as de novo, resemble the binding modes of natural ligands of nuclear receptors; those peptides displayed lower energy scores due to hydrogen bond interactions with W31 and T210 of the GLUC receptor, and hydrophobic interactions L39, I132, and M212 (Figure 3E,F). MINC showed the same pattern of hydrogen bonding through W57 and K147; whereas hydrophobic interactions with aliphatic sidechains of peptides were stabilized with A47, L46, F103, M51, V54, and L83 (Figure 3G,H).
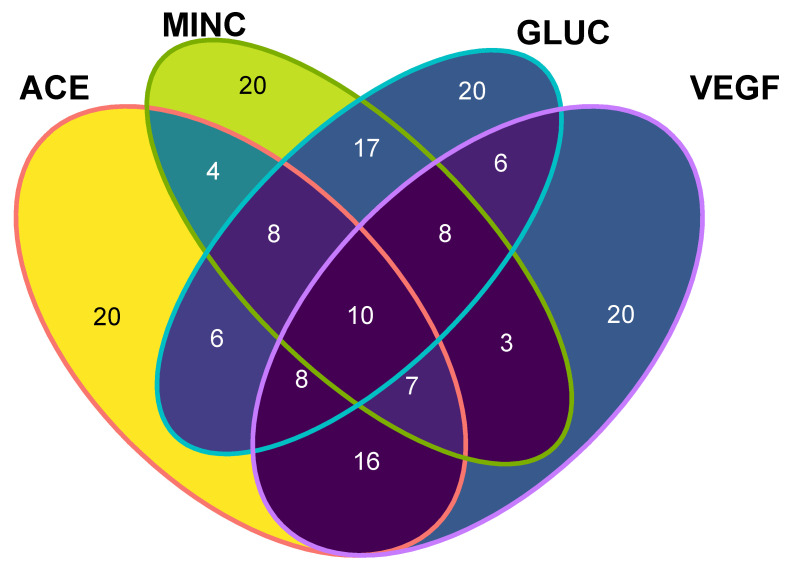
The aim of this study was to integrate a multi-target-directed ligand approach, where peptides with lower binding energies can be shared as ligands towards the four evaluated human target proteins. To achieve this goal, the 20 lowest-binding, free energy peptides for each target protein were plotted and those that rely on the intersection of a Venn diagram (five known-sequence peptides (NNVFYPF, FNIVFPG, SRPWPIDY, QLQRWFR, GSRFDWTR) and five de novo peptides (DFKF, DLRF, FKAF, FRSF, QFRF)) were selected as potential multi-target-directed ligands, and turned into candidates for a future synthesis and in vitro evaluation (Figure 4). Those peptides can be considered as multifunctional peptides, which may be preferred over single-activity peptides, as they can simultaneously trigger, modulate, or inhibit multiple physiological pathways. The subset of selected peptides contained five know-sequence peptides (already described in databases and mass spectra data) and five de novo peptides (non previously described), which demonstrate that the in silico methods may assist in the identification of possible ligand hits.
Figure 4.
Venn diagram of identified peptides with their human target protein interactions. The numbers indicate independent subsets of shared peptides with the lowest binding free energy among molecular targets. Ten different peptides comprise the subset of multi-target directed ligands. Angiotensin converting enzyme (ACE), Mineralocorticoid receptor (MINC), Glucocorticoid receptor (GLUC), Vascular Endothelial growth factor (VEGF).
Finally, we applied an absorption, distribution, metabolism, and excresion (ADME) prediction to evaluate drug-likeness parameters on the subset of selected multi-target directed peptides. The pharmaco-kinetic profiles of the complete subset assessed 25 different properties within the acceptable range for 95% of known drugs [31]. Table 3 lists the criteria used for each descriptor, as well as the calculated parameters and chemical similarity to approved drugs.
Table 3.
Calculated drug-likeness parameter for subset of 10 selected peptide sequences.
| Parameters | NNVFYPF | FNIVFPG | SRPWPIDY | QLQRWFR | GSRFDWTR | DFKF | DLRF | FKAF | FRSF | QFRF |
|---|---|---|---|---|---|---|---|---|---|---|
| MW (g/mol) | 899.9 | 792.93 | 899.07 | 10.33.19 | 1024.1 | 555.63 | 549.62 | 511.62 | 555.63 | 596.68 |
| H-bond donors | 8 | 6 | 8.5 | 17.5 | 14.5 | 5.75 | 7.75 | 5.75 | 7.75 | 9.75 |
| H-bond acceptors | 19.25 | 17 | 19 | 22.5 | 22.4 | 10.25 | 11.25 | 9.25 | 10.95 | 12.75 |
| logP o/w a | −3.56 | −1.83 | −2.79 | −5.68 | −4.64 | −1.06 | −1.83 | −1.43 | −1.28 | −3.14 |
| logS wat b | −1.87 | −3.77 | −0.49 | −1.18 | −0.24 | −0.68 | −0.64 | −0.79 | −2.72 | −0.96 |
| NlogK has Serum Protein Binding c | −2.39 | −1.88 | −2.53 | −3.35 | −3.18 | −1.23 | −1.54 | −1.02 | −1.26 | -1.75 |
| Apparent Caco-2 Permeability (nm/s) d | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Apparent MDCK Permeability (nm/s) e | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| logKp for skin permeability f | −9.04 | −6.92 | −8.53 | −14.29 | −13.53 | −8.69 | −9.95 | −8.26 | −8.62 | −10.60 |
| Qualitative Model for Human Oral Absorption | Low | Low | Low | Low | Low | Low | Low | Low | Low | Low |
| Most similar pharmaceutical drugs | Troxerutin, Voglibose, Monoxerutin | Lymecycline, Troxerutin, Proglumetacin | Razoxane, Hexoprenaline, Dihydralazine | Everolimus, Amiodarone, Fenethylline | Everolimus, Droxidopa, Polaprezinc | Hexoprenaline, Lisinopril, Lymecycline | Hexoprenaline, Voglibose, Lymecycline | Hexoprenaline, Lisinopril, Lymecycline | Aminopterin, Lymecycline, Hexobendine | Hexobendine, Hexoprenaline, Lymecycline |
a Predicted logarithm of partitioning coefficient for octanol/water phases (range for 95% of drugs: −2.0 to 6.0). b Predicted logarithm of aqueous solubility in mol/dm3 (range for 95% of drugs: −6.0 to 0.5). c Predicted logarithm of serum protein binding (range for 95% of drugs: −1.5 to 1.5). d Predicted apparent Caco-2 cell rate permeability in nm/s (range for 95% of drugs: <25, >500). e Predicted apparent MDCK cells rate permeability in nm/s (range for 95% of drugs: <25, >500). f Predicted apparent for skin permeability rate permeability Kp in cm/h).
As shown in Table 3, the subset of selected peptides displayed lower calculated logP o/w partition coefficients that can be associated with increased permeability across membranes, despite having high molecular weight (>500 Da) and several hydrogen bond interactions. Similarly, different in silico techniques have addressed that the prediction of skin permeability by complex molecules is difficult to achieve; however, those predictions are based on comparisons to experimental data or complex algorithms based on quantitative structure–activity relationships (QSAR) [32,33]. Additonally, the calculated coefficient for binding to serum proteins and skin permeability are in the range of 95% of approved drugs, suggesting a positive interaction with epidermal tissues. The above obervations confirm that chia seed peptides could contribute in the improvement of the skin health and can be used as potential functional ingredients for development of cosmeceutical skin products, as previously demonstrated in vitro [9]. An interesting finding was that the entire subset of multi-target peptides showed structural similarity to antihypertensive and antiangiogenic drugs, such as lymecycline, everolimus and lisinopril, among others.
3. Materials and Methods
3.1. Materials
Alcalase (E.C. 3.4.21.62) and flavourzyme (E.C. 232-752-2) were acquired from Sigma Aldrich (St. Luis, MO, USA). Ultrafiltration units were purchased from Millipore (Bedford, MA, USA). Chia (Salvia hispanica L.) seeds were obtained from Healthworks (pesticide-free, Scottsdale, AZ, USA). All chemical reagents used in this study were analytical grade or HPLC grade.
3.2. Preparation of Chia Seed Peptides
Peptides from chia seed protein were obtained as described in detail by Urbizo-Reyes, San Martin-González, Garcia-Bravo, López Malo Vigil and Liceaga [7]. Briefly, chia seeds were devoid of mucilage using a combined treatment of ultrasound and vacuum-assisted filtration. Afterwards, chia seeds were defatted by a mechanical oil press and the resulting meal was proteolyzed using sequential enzymatic (alcalase followed by flavourzyme) microwave-assisted hydrolysis. The subsequent protein hydrolysate had a yield of 78.2% extracted protein. Resulting chia seed peptides (CSP) were freeze-dried and stored at −20 °C until used.
3.3. Peptidomics Analysis of Low Molecular Weight Peptides by Liquid Chromatography-Mass Spectrometry
Following proteolysis, low molecular weight peptides were obtained by ultrafiltration using centrifugal filters of 3 kDa cut-off membrane (regenerated cellulose, Amicon® Ultra-4). Peptides present in the <3 kDa fraction were identified by liquid chromatography-mass spectrometry (LC-MS/MS) in the Proteomics Core Facility at the Indiana University School of Medicine (Indianapolis, Indiana, USA). Briefly, lyophilized samples of the <3 kDa fraction were resuspended in 0.1% of formic acid and reverse phase LC-MS was performed on the QE-Plus mass spectrometer. Peptide sequences were obtained from mass spectrometry data (raw files) using a database of all Salvia spp. proteins in the UniProt database and were also searched for de novo sequencing of peptides [8,9]. A total of 1954 peptides were identified (i.e., 1565 peptides using UniProt database and 389 de novo peptides).
3.4. In Silico Analysis of Identified Peptides for their Potential Bioactivities
First, each identified peptide was screened for their bioactive likelihood using the PeptideRanker bioinformatic tool (http://distilldeep.ucd.ie/PeptideRanker/, accessed on 6 March 2022), which estimates a bioactivity potential score based on a novel N-to-1 neural network. This server ranked all assessed peptides by giving scores ranging from 0 to 1. Values near 1 indicate peptides with high likelihood to be bioactive. Therefore, if the probabilities are ≥0.8, the peptide is said to be predicted as bioactive [34].
Selected peptides were subjected to in silico prediction and physicochemical characterization through different online servers. The predictions were made using the tools PreAIP (http://kurata14.bio.kyutech.ac.jp/PreAIP/, accessed on 7 March 2022) for their anti-inflammatory potential, AHTPIN (http://crdd.osdd.net/raghava/ahtpin/, accessed on 7 March 2022) for their antihypertensive potential, AntiAngioPred (http://webs.iiitd.edu.in/raghava/antiangiopred/, accessed on 8 March 2022) for their anti-angiogenic potential, HLP (http://crdd.osdd.net/raghava/hlp/, accessed on 8 March 2022) for their intestinal stability, and PlifePred (https://webs.iiitd.edu.in/raghava/plifepred/, accessed on 9 March 2022) for their plasma stability. Additionally, some physicochemical properties (i.e., hydrophobicity, hydropathicity, hydrophilicity, charge, and molecular weight) were determined using the AHTPIN tool.
3.5. Construction of Human Molecular Targets and Identified Peptides
For this analysis, the peptides identified in the previous step and some human targets were constructed. The three-dimensional structures of identified peptides sequences were modeled using the OPLS-AA forcefield with an alpha helix template (Maestro Suite v 2.12). The resulting structures were minimized using a conjugated gradient and Steepest Descent algorithms with the biomolecular simulation program, Amber20 package (https://ambermd.org/, accessed on 3 March 2022). The target crystallographic structures were Human Angiotensin Converting Enzymes, ACE-1 (PDB code: 1O8A), Vascular Endothelial Growth Factor, VEGF (PDB code: 2VPF), and Glucocorticoid and Mineralocorticoid Nuclear Receptors (PDB code: 1P93 and 2AA2, respectively). Modeling of missing or incomplete residues and disulfide bonds were carried out with Maestro Suite v 2.12, (Schrödinger, LLC, New York, NY, USA) using OPLS-AA force field to guide the structure completion. The tritiable residue protonation was calculated using the PROPKA web-server at pH 7 (e.g., histidine was protonated on the delta/epsilon nitrogen) depending on the polar environment.
All targets were prepared in octahedral water boxes 15 Å buffered with TIP3P explicit water molecules and neutralized with Na+ or Cl- ions using Tleap routine from AmberTools 20. The systems were neutralized with a salt solution of 0.15 M NaCl. The resulting systems comprised between 100,000–172,000 atoms.
3.6. Molecular Dynamics and Ensemble Docking Calculations
All Molecular Dynamics calculations were carried out in Amber20 using the amberff14 and GAFF force fields with PMEMD.cuda software. Initial MD stages (minimization and thermalization) were performed at 1 atm of pressure and 298 °K. The minimization consisted of 5000 steps using the steepest-descendent method, followed by 10,000 with the conjugate gradient method. Covalent bonds between hydrogen and heavy atoms were constrained using the SHAKE algorithm. Initial simulations steps applied an NVT ensemble, coupled with a Langevin thermostat, to reach and maintain a constant temperature (298 °K). Linear increases in the temperature from 0 to 298 °K, in intervals of 50 °K, were applied along 5,000,000 steps. The atoms were restricted in protein alpha carbons atoms with 5 Kcal/mol/A3 and 2.5 Kcal/mol/A3. An isothermal-semi isobaric ensemble (NPT) simulation, using the Berendsen barostat, was coupled to maintain 1 atm of pressure keeping restrains. Afterwards, restrains were reduced by 2 Kcal/mol/A3 units for proteins 250,000 steps until restriction disappeared. The systems continued for 10 ns more in simulation in the anisotropic ensemble at 1 atm, keeping track of Cα root-mean-square deviation (RMSD), potential energy and system density as markers of stable behavior. Long-range electrostatic interactions were calculated, periodic boundary conditions, and particle mesh Ewald (PME) methods using a 12 Å cutoff and force switch on a 10 Å radius. Time steps of 2.0 fs were set for production simulations. Production calculations were performed along 300 ns for each target using Gaussian Accelerated Molecular Dynamics (GaMD) to enhance conformational sampling. After each GaMD production, the carbon alpha root mean square deviation (A) was calculated and projected onto first two principal components using Bio3D package for selection of three representative conformations for ensemble docking.
Binding modes and score calculations (Autodock Vina scoring function) were performed with Smina v 1.0 package for Ensemble docking—Virtual screening. All representative conformation for each previously selected target was used for docking calculations. The search parameters were 0.375 Å mesh step, 64 as exhaustiveness value, and a maximum value of 25,000,000 evaluations. The octahedral search space dimensions for all targets were set to 25 × 25 × 40 Å centered on each identified active site or interaction regions, as described for ACE enzymes. All scores were approximated using consensus scoring approximation.
4. Conclusions
In this work, we presented a pipeline based on in silico analysis upon a set of chia seed peptides identified using a peptidomics and bioinformatics approach that sought to screen potential bioactivities and correlate them with pharmacological (drug) targets. Structural analysis and physicochemical prediction of descriptors allowed us to select probable molecular targets and virtually screen their intermolecular interactions through molecular dynamics simulations and ensemble docking. With this approach, we were able to identify 10 multifunctional chia seed peptides (NNVFYPF, FNIVFPG, SRPWPIDY, QLQRWFR, GSRFDWTR, DFKF, DLRF, FKAF, FRSF, QFRF) with low binding free energy (ca. −5 Kcal mol−1) and stable intermolecular interactions during formation of a ligand-receptor complex. Furthermore, this information corroborates the high in vitro biological activity previously reported for chia seed peptides and foresees sequences found in the peptide fractions that can provide multifunctional bioactivities towards human protein receptors involved in chronic diseases such as hypertension and inflammation. However, further research will be required to test the specified peptide sequences using in vivo models. Nevertheless, this study provides promising use of bioinformatics and in silico analyses to establish ligand-receptor interactions from new functional ingredients and emerging protein sources.
Acknowledgments
The authors also acknowledge the technical support of Emanuel Villafán (INECOL A.C.); Eng. Jesús Alaniz (Thubat Kaal 2.0 at CNS-IPICyT, A.C) and Héctor Manuel Olivier Hernández (Xiuhcoatl Hybrid Cluster at CINVESTAV).
Author Contributions
Conceptualization, J.E.A.-T., A.V.-L. and A.M.L.; Formal analysis, J.E.A.-T. and A.V.-L.; Writing—original draft preparation, J.E.A.-T. and A.V.-L.; Writing—review and editing, A.M.L.; Supervision, A.M.L.; funding acquisition, A.V.-L. and A.M.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by National Supercomputing Center—IPICYT for the computational resources through grant TKII-AMVL001 and by LANCAD—Xiuhcoatl Hybrid Cluster supercomputing resources granted to AVL, as well as to the resources of High Performance Computing at INECOL A.C, and supercomputing resources at Miztli through LANCAD-UNAM—DGTIC-347. This work was also supported by the USDA National Institute of Food and Agriculture, Hatch Act formula funds project 1019794.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vidal-Limon A., Aguilar-Toalá J.E., Liceaga A.M. Integration of molecular docking analysis and molecular dynamics simulations for studying food proteins and bioactive peptides. J. Agric. Food Chem. 2022;70:934–943. doi: 10.1021/acs.jafc.1c06110. [DOI] [PubMed] [Google Scholar]
- 2.Tu M., Cheng S., Lu W., Du M. Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: Sequence, structure, and functions. TrAC Trends Anal. Chem. 2018;105:7–17. doi: 10.1016/j.trac.2018.04.005. [DOI] [Google Scholar]
- 3.Agyei D., Tsopmo A., Udenigwe C.C. Bioinformatics and peptidomics approaches to the discovery and analysis of food-derived bioactive peptides. Anal. Bioanal. Chem. 2018;410:3463–3472. doi: 10.1007/s00216-018-0974-1. [DOI] [PubMed] [Google Scholar]
- 4.Zettel V., Hitzmann B. Applications of chia (Salvia hispanica L.) in food products. Trends Food Sci. Technol. 2018;80:43–50. doi: 10.1016/j.tifs.2018.07.011. [DOI] [Google Scholar]
- 5.Kulczyński B., Kobus-Cisowska J., Taczanowski M., Kmiecik D., Gramza-Michałowska A. The chemical composition and ntritional value of Chia seeds-Current state of knowledge. Nutrients. 2019;11:1242. doi: 10.3390/nu11061242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melo D., Machado T.B., Oliveira M.B.P.P. Chia seeds: An ancient grain trending in modern human diets. Food Funct. 2019;10:3068–3089. doi: 10.1039/C9FO00239A. [DOI] [PubMed] [Google Scholar]
- 7.Urbizo-Reyes U., San Martin-González M.F., Garcia-Bravo J., López Malo Vigil A., Liceaga A.M. Physicochemical characteristics of chia seed (Salvia hispanica) protein hydrolysates produced using ultrasonication followed by microwave-assisted hydrolysis. Food Hydrocoll. 2019;97:105187. doi: 10.1016/j.foodhyd.2019.105187. [DOI] [Google Scholar]
- 8.Aguilar-Toalá J.E., Deering A.J., Liceaga A.M. New insights into the antimicrobial properties of hydrolysates and peptide fractions derived from Chia seed (Salvia hispanica L.) Probiotics Antimicrob. Proteins. 2020;12:1571–1581. doi: 10.1007/s12602-020-09653-8. [DOI] [PubMed] [Google Scholar]
- 9.Aguilar-Toalá J.E., Liceaga A.M. Identification of chia seed (Salvia hispanica L.) peptides with enzyme inhibition activity towards skin-aging enzymes. Amino Acids. 2020;52:1149–1159. doi: 10.1007/s00726-020-02879-4. [DOI] [PubMed] [Google Scholar]
- 10.Cotabarren J., Rosso A.M., Tellechea M., García-Pardo J., Rivera J.L., Obregón W.D., Parisi M.G. Adding value to the chia (Salvia hispanica L.) expeller: Production of bioactive peptides with antioxidant properties by enzymatic hydrolysis with Papain. Food Chem. 2019;274:848–856. doi: 10.1016/j.foodchem.2018.09.061. [DOI] [PubMed] [Google Scholar]
- 11.Coelho M.S., Soares-Freitas R.A.M., Arêas J.A.G., Gandra E.A., Salas-Mellado M.d.l.M. Peptides from Chia present atibacterial ativity and inhibit colesterol synthesis. Plant Foods Hum. Nutr. 2018;73:101–107. doi: 10.1007/s11130-018-0668-z. [DOI] [PubMed] [Google Scholar]
- 12.Grancieri M., Martino H.S.D., Gonzalez de Mejia E. Digested total protein and protein fractions from chia seed (Salvia hispanica L.) had high scavenging capacity and inhibited 5-LOX, COX-1-2, and iNOS enzymes. Food Chem. 2019;289:204–214. doi: 10.1016/j.foodchem.2019.03.036. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q., Yang C., Xie Y., Wang Y., Li X., Wang K., Huang J., Yan W. GM-Pep: A high efficiency strategy to de novo design functional peptide sequences. J. Chem. Inf. Model. 2022;62:2617–2629. doi: 10.1021/acs.jcim.2c00089. [DOI] [PubMed] [Google Scholar]
- 14.Panyayai T., Ngamphiw C., Tongsima S., Mhuantong W., Limsripraphan W., Choowongkomon K., Sawatdichaikul O. FeptideDB: A web application for new bioactive peptides from food protein. Heliyon. 2019;5:e02076. doi: 10.1016/j.heliyon.2019.e02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kartal C., Kaplan Türköz B., Otles S. Prediction, identification and evaluation of bioactive peptides from tomato seed proteins using in silico approach. J. Food Meas. Charact. 2020;14:1865–1883. doi: 10.1007/s11694-020-00434-z. [DOI] [Google Scholar]
- 16.Coscueta E.R., Batista P., Gomes J.E., da Silva R., Pintado M.M. Screening of novel bioactive peptides from goat casein: In silico to in vitro validation. Int. J. Mol. Sci. 2022;23:2439. doi: 10.3390/ijms23052439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao C., Zhao M., Zhou F., Gallego M., Toldrá F., Mora L. Data on bioactive peptides derived from chicken hydrolysate with potential alcohol dehydrogenase stabilizing activity and in silico analysis of their potential activity and applicability. Data Brief. 2020;29:105163. doi: 10.1016/j.dib.2020.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitts D.D., Weiler K. Bioactive proteins and peptides from food sources. Applications of bioprocesses used in isolation and recovery. Curr. Pharm. Des. 2003;9:1309–1323. doi: 10.2174/1381612033454883. [DOI] [PubMed] [Google Scholar]
- 19.Udenigwe C.C., Aluko R.E. Food protein-derived bioactive peptides: Production, processing, and potential health benefits. J. Food Sci. 2012;77:R11–R24. doi: 10.1111/j.1750-3841.2011.02455.x. [DOI] [PubMed] [Google Scholar]
- 20.Pripp A.H., Isaksson T., Stepaniak L., Sørhaug T., Ardö Y. Quantitative structure activity relationship modelling of peptides and proteins as a tool in food science. Trends Food Sci. Technol. 2005;16:484–494. doi: 10.1016/j.tifs.2005.07.003. [DOI] [Google Scholar]
- 21.Stockner T., Sterk H., Kaptein R., Bonvin A.M.J.J. Molecular dynamics studies of a molecular switch in the glucocorticoid receptor. J. Mol. Biol. 2003;328:325–334. doi: 10.1016/S0022-2836(03)00316-4. [DOI] [PubMed] [Google Scholar]
- 22.Yadav M., Khandelwal R., Mudgal U., Srinitha S., Khandekar N., Nayarisseri A., Vuree S., Singh S.K. Identification of potent VEGF inhibitors for the clinical treatment of glioblastoma, a virtual screening approach. Asian Pac. J. Cancer Prev. 2019;20:2681–2692. doi: 10.31557/APJCP.2019.20.9.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamal H., Mudgil P., Bhaskar B., Fisayo A.F., Gan C.-Y., Maqsood S. Amaranth proteins as potential source of bioactive peptides with enhanced inhibition of enzymatic markers linked with hypertension and diabetes. J. Cereal Sci. 2021;101:103308. doi: 10.1016/j.jcs.2021.103308. [DOI] [Google Scholar]
- 24.Mudgil P., Baby B., Ngoh Y.-Y., Kamal H., Vijayan R., Gan C.-Y., Maqsood S. Molecular binding mechanism and identification of novel anti-hypertensive and anti-inflammatory bioactive peptides from camel milk protein hydrolysates. LWT Food Sci. Technol. 2019;112:108193. doi: 10.1016/j.lwt.2019.05.091. [DOI] [Google Scholar]
- 25.Amorim F.G., Coitinho L.B., Dias A.T., Friques A.G.F., Monteiro B.L., Rezende L.C.D.d., Pereira T.d.M.C., Campagnaro B.P., De Pauw E., Vasquez E.C., et al. Identification of new bioactive peptides from Kefir milk through proteopeptidomics: Bioprospection of antihypertensive molecules. Food Chem. 2019;282:109–119. doi: 10.1016/j.foodchem.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Medina-Franco J.L., Saldívar-González F.I. Cheminformatics to characterize pharmacologically active natural products. Biomolecules. 2020;10:1566. doi: 10.3390/biom10111566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao Y., McCammon J.A. Chapter Six—Gaussian Accelerated Molecular Dynamics: Theory, Implementation, and Applications. In: Dixon D.A., editor. Annual Reports in Computational Chemistry. Volume 13. Elsevier; Amsterdam, The Netherlands: 2017. pp. 231–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao Y., Bhattarai A., Wang J. Ligand Gaussian accelerated molecular dynamics (LiGaMD): Characterization of ligand binding thermodynamics and kinetics. J. Chem. Theory Comput. 2020;16:5526–5547. doi: 10.1021/acs.jctc.0c00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edman K., Hosseini A., Bjursell M.K., Aagaard A., Wissler L., Gunnarsson A., Kaminski T., Köhler C., Bäckström S., Jensen T.J., et al. Ligand binding mechanism in steroid receptors: From conserved plasticity to differential evolutionary constraints. Structure. 2015;23:2280–2290. doi: 10.1016/j.str.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Wrapp D., Wang N., Corbett Kizzmekia S., Goldsmith Jory A., Hsieh C.-L., Abiona O., Graham Barney S., McLellan Jason S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schrödinger . QikProp v 2022-2. LLC; New York, NY, USA: 2021. [Google Scholar]
- 32.Chang Y.-C., Chen C.-P., Chen C.-C. Predicting skin permeability of chemical substances using a quantitative structure-activity relationship. Procedia Eng. 2012;45:875–879. doi: 10.1016/j.proeng.2012.08.252. [DOI] [Google Scholar]
- 33.Zeng R., Deng J., Dang L., Yu X. Correlation between the structure and skin permeability of compounds. Sci. Rep. 2021;11:10076. doi: 10.1038/s41598-021-89587-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mudgil P., Kamal H., Priya Kilari B., Mohd Salim M.A.S., Gan C.-Y., Maqsood S. Simulated gastrointestinal digestion of camel and bovine casein hydrolysates: Identification and characterization of novel anti-diabetic bioactive peptides. Food Chem. 2021;353:129374. doi: 10.1016/j.foodchem.2021.129374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.