Abstract
A total of 26 strains of Vibrio cholerae, including members of the O1, O139, and non-O1, non-O139 serogroups from both clinical and environmental sources, were examined for the presence of genes encoding cholera toxin (ctxA), zonula occludens toxin (zot), accessory cholera enterotoxin (ace), hemolysin (hlyA), NAG-specific heat-stable toxin (st), toxin-coregulated pilus (tcpA), and outer membrane protein (ompU), for genomic organization, and for the presence of the regulatory protein genes tcpI and toxR in order to determine relationships between epidemic serotypes and sources of isolation. While 22 of the 26 strains were hemolytic on 5% sheep blood nutrient agar, all strains were PCR positive for hlyA, the hemolysin gene. When multiplex PCR was used, all serogroup O1 and O139 strains were positive for tcpA, ompU, and tcpI. All O1 and O139 strains except one O1 strain and one O139 strain were positive for the ctxA, zot, and ace genes. Also, O1 strain VO3 was negative for the zot gene. All of the non-O1, non-O139 strains were negative for the ctxA, zot, ace, tcpA, and tcpI genes, and all of the non-O1, non-O139 strains except strain VO26 were negative for ompU. All of the strains except non-O1, non-O139 strain VO22 were PCR positive for the gene encoding the central regulatory protein, toxR. All V. cholerae strains were negative for the NAG-specific st gene. Of the nine non-ctx-producing strains of V. cholerae, only one, non-O1, non-O139 strain VO24, caused fluid accumulation in the rabbit ileal loop assay. The other eight strains, including an O1 strain, an O139 strain, and six non-O1, non-O139 strains, regardless of the source of isolation, caused fluid accumulation after two to five serial passages through the rabbit gut. Culture filtrates of all non-cholera-toxigenic strains grown in AKI media also caused fluid accumulation, suggesting that a new toxin was produced in AKI medium by these strains. Studies of clonality performed by using enterobacterial repetitive intergenic consensus sequence PCR, Box element PCR, amplified fragment length polymorphism (AFLP), and pulsed-field gel electrophoresis (PFGE) collectively indicated that the V. cholerae O1 and O139 strains had a clonal origin, whereas the non-O1, non-O139 strains belonged to different clones. The clinical isolates closely resembled environmental isolates in their genomic patterns. Overall, there was an excellent correlation among the results of the PCR, AFLP, and PFGE analyses, and individual strains derived from clinical and environmental sources produced similar fingerprint patterns. From the results of this study, we concluded that the non-cholera-toxin-producing strains of V. cholerae, whether of clinical or environmental origin, possess the ability to produce a new secretogenic toxin that is entirely different from the toxin produced by toxigenic V. cholerae O1 and O139 strains. We also concluded that the aquatic environment is a reservoir for V. cholerae O1, O139, non-O1, and non-O139 serogroup strains.
Vibrio cholerae is a natural inhabitant of the aquatic environment (9, 12, 15, 21, 24, 31, 37). This vibrio species not only survives in riverine, estuarine, and coastal waters around the world but also lives in association with crustacean copepods, such as Acartia tonsa and Gerris spinolae, and with aquatic plants, either in the viable and culturable state or in the viable but nonculturable state (13, 25, 30, 32, 33, 36, 49, 53, 66, 70). Since this human pathogen is primarily an inhabitant of the aquatic environment, water plays an important role in the transmission and epidemiology of cholera (19, 38, 72). However, the epidemiological impact of environmental V. cholerae strains is not clearly understood, since the majority of strains isolated from the environment do not produce cholera toxin and lack not only the virulence gene cassette (45, 64) for cholera toxin but also zonula occludens toxin and accessory cholera enterotoxin. In addition, environmental vibrios lack the toxin-coregulated pilus (TCP) that is recognized as a significant factor in the pathogenicity of toxigenic strains of V. cholerae serogroups O1 and O139.
Strains of V. cholerae belonging to serogroup O1 biotype El Tor and serogroup O139 have been described as causative agents of diarrhea (1, 10, 34, 42, 55). Furthermore, results of genetic and phenotypic characterizations of such strains indicate that in order to cause diarrhea, they require genes for cholera toxin, the colonization TCP, and the central regulatory protein ToxR (27, 29, 46, 50, 76). However, nontoxigenic strains isolated from the environment and lacking the virulence gene cassette have been reported to evoke the secretory response in the ligated rabbit ileal loop assay (62, 63). Nontoxigenic O1 biotype El Tor strains have also been isolated from cases of diarrhea (11, 60).
V. cholerae non-O1, non-O139 strains are recognized as causative agents of sporadic and localized outbreaks, and the diarrhea caused by these strains is sometimes characterized by blood and mucous (8, 61). However, these strains continue to be considered of negligible significance, since they have been associated with illness only in a low percentage of patients hospitalized with secretory diarrhea (52). While analyzing the nucleotide sequence of asd genes in V. cholerae strains, Karaolis et al. (43) demonstrated that the sixth- and seventh-pandemic strains and United States Gulf Coast V. cholerae O1 isolates may have been derived from nontoxigenic strains and postulated that horizontal gene transfer occurred in V. cholerae, which resulted in the emergence of a new pathogenic strain. Furthermore, V. cholerae non-O1 serogroups have been reported to be involved in the emergence of a newer variant of V. cholerae; this hypothesis is supported by the genesis of V. cholerae O139, a serogroup believed to have evolved by horizontal gene transfer from serogroup O1 to a non-O1 serogroup (7).
Amplified fragment length polymorphism (AFLP), pulsed-field gel electrophoresis (PFGE), and PCR methods have been used in epidemiological investigations and also to study relatedness among bacterial strains in order to trace the origin and geographical distribution of V. cholerae (40, 69, 73, 75). It is accepted that cholera is a waterborne disease, but there is no molecular evidence to prove that strains causing human infections belong to the same clones as those found in aquatic environments. To answer this vexing question, we examined a set of strains belonging to O1, O139, and non-O1, non-O139 serogroups, including both clinical and environmental isolates, to determine whether virulence and regulatory genes were present and to compare their genomic organizations.
MATERIALS AND METHODS
Bacterial strains.
A total of 26 strains of V. cholerae were included in this study. Seven strains were serogroup O1 strains (four clinical strains and three environmental strains), eight strains were serogroup O139 strains (four strains each from clinical and environmental sources), and seven strains were non-O1, non-O139 strains (two clinical isolates and five environmental isolates) from laboratory stocks and had been identified previously by using standard bacteriological methods (77). Four additional isolates of V. cholerae O139, including reference strain ATCC 51394 (= MO45), provided by G. B. Nair, National Institute of Cholera and Enteric Diseases, Calcutta, India, were also included in the study. All isolates were examined for the oxidase reaction, and the identities of the V. cholerae O1 strains were confirmed by serogrouping by using growth from triple sugar iron agar slants with polyvalent O1 and monospecific Inaba and Ogawa antisera. V. cholerae strains which did not agglutinate with O1 antiserum were checked with monoclonal O139 antiserum supplied by the WHO Regional Office of South-East Asia, New Delhi, India. V. cholerae strains which did not agglutinate with either O1 or O139 antisera were assumed to belong to non-O1, non-O139 serogroups. The sources, years of isolation, and geographical sites of isolation of the strains are listed in Table 1. All strains were maintained in peptone agar stab cultures and did not undergo more than two subcultures prior to testing in this study.
TABLE 1.
V. cholerae strains of the O1, O139, and non-O1, non-O139 serogroups included in this study
| Strain | Place of isolation | Source | Year of isolation |
|---|---|---|---|
| V. cholerae O1 biotype E1 Tor strains | |||
| VO1 | Orissa, India | Diarrheal patient | 1969 |
| VO2 | Varanasi, India | Diarrheal patient | 1976 |
| VO13 | Varanasi, India | Diarrheal patient | 1989 |
| VO3 | Varanasi, India | Diarrheal patient | 1990 |
| VO4 | Varanasi, India | River water | 1986 |
| VO5 | Varanasi, India | River water | 1988 |
| VO7 | Varanasi, India | River water | 1988 |
| V. cholerae O139 strains | |||
| MO45a | Madras, India | Diarrheal patient | 1992 |
| CO594 | Calcutta, India | Diarrheal patient | 1992 |
| CO766 | Calcutta, India | Diarrheal patient | 1992 |
| CO788 | Calcutta, India | Diarrheal patient | 1992 |
| VO12 | Varanasi, India | Diarrheal patient | 1994 |
| VO14 | Varanasi, India | Diarrheal patient | 1993 |
| VO15 | Varanasi, India | Diarrheal patient | 1993 |
| VO16 | Varanasi, India | Diarrheal patient | 1994 |
| VO17 | Varanasi, India | Eichhornia crassipes | 1992 |
| VO18 | Varanasi, India | Eichhornia crassipes | 1992 |
| VO19 | Varanasi, India | River water | 1992 |
| VO20 | Varanasi, India | Hand pump | 1992 |
| V. cholerae non-O1, non-O139 strains | |||
| VO22 | Varanasi, India | Diarrheal patient | 1979 |
| VO23 | Varanasi, India | Diarrheal patient | 1979 |
| VO24 | Varanasi, India | River water | 1986 |
| VO25 | Varanasi, India | River water | 1988 |
| VO26 | Varanasi, India | River water | 1988 |
| VO27 | Varanasi, India | River water | 1986 |
| VO28 | Varanasi, India | River water | 1986 |
V. cholerae O139 reference strain MO45 (= ATCC 51394).
Hemolysis.
Strains of V. cholerae were grown in brain heart infusion broth (Difco) for 3 h and were subsequently streaked on 5% sheep blood nutrient agar plates. After overnight incubation at 37°C, colonies were examined for hemolysis on blood agar.
Preparation of culture filtrates.
Culture filtrates of all nine non-cholera-toxin-producing strains, including an O1 strain and an O139 serogroup representative, were prepared by the method of Sanyal et al. (63). Briefly, 10-ml portions of AKI medium or brain heart infusion broth in 50-ml conical flasks were inoculated with approximately five colonies from an overnight nutrient agar plate culture. The flasks were incubated at 37°C in a water bath for 16 to 18 h with shaking at 80 to 120 oscillations per min. The cultures were centrifuged at 22,000 × g for 20 min, the supernatants were filtered through membrane filters (pore size, 0.22 μm; Millipore), and the filtrates were stored in small volumes at 4°C.
Ileal loop test and passage through rabbit ileal loops.
Cultures and culture filtrates of non-cholera-toxigenic V. cholerae strains were tested for enterotoxin production in adult New Zealand and albino rabbits by the method of De and Chatterje (17). The strains were grown in peptone water for 3 to 4 h, diluted 10-fold in the same medium, and inoculated into rabbit ileal loops by using 1-ml portions of preparations containing 105 to 106 CFU/ml. V. cholerae toxigenic strain 569B grown in peptone water and uninoculated, sterile peptone water served as positive and negative controls, respectively. Culture filtrates of all strains prepared in AKI medium were tested by the same method by using 1-ml inocula. Each test was done in two rabbits; six to eight 8- to 10-cm loops were ligated in each rabbit, and the interloop length was 2 to 4 cm. The rabbits were sacrificed after 18 h.
V. cholerae strains that caused little or no fluid accumulation in the initial tests were passaged through the rabbit gut by using the method of Sanyal et al. (63). Briefly, each strain was recovered from the ileal loops on nutrient agar plates, and following overnight incubation approximately five colonies were inoculated into peptone water and incubated at 37°C for 3 to 4 h. Approximately 1 ml of diluted culture was again inoculated into rabbit ileal loops. This process was continued until unequivocal positive responses were obtained.
Ouchterlony immunodiffusion.
Ouchterlony immunodiffusion tests were performed by using concentrated (10×) culture filtrates of test strains prepared in AKI medium, and the new anti-cholera toxin serum. Cholera toxin gene-negative V. cholerae strain X-392, which has been reported to produce the new cholera toxin (62, 63), was obtained from J. B. Kaper of the Center for Vaccine Development, University of Maryland, College Park, and was used to prepare antiserum against the new cholera toxin in purified form as described previously (67).
DNA isolation.
Chromosomal DNA was extracted from each of the V. cholerae strains by the cetyltrimethylammonium bromide method, as described by Ausubel et al. (4). The purity of the DNA was assayed with a Beckman DU 640 spectrophotometer by using automatic calculation of the ratio of optical densities at 260 and 280 nm.
Multiplex PCR assay.
A multiplex PCR assay was performed (22, 65, 74; G. R. Matte, M. H. Matte, J. Chun, A. Huq, and R. R. Colwell, submitted for publication) in order to determine the presence of toxin genes ctxA, zot, ace, st, hlyA, tcpA, and ompU. In addition, all strains were examined for the presence of regulatory genes for TCP expression (tcpI) and the central regulatory protein (toxR). The primers used in this study and the expected amplicon sizes are listed in Table 2. PCR was performed with a PTC-200 thermal cycler (MJ Research, Inc., Waltham, Mass.). The instrument was programmed as follows: initial denaturation at 94°C for 2 min, followed by 30 cycles consisting of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min and a final extension step of 72°C for 10 min. For the tcpI gene, extension was at 72°C for 3 min and all of the other steps were the same as those described above. The amplification program for the st and ace genes began with denaturation at 94°C for 1 min, which was followed by 29 cycles consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension step of 72°C for 5 min. V. cholerae O139 strain MO45 (= ATCC 51394) was used as the PCR positive control for ctxA, zot, ace, tcpA, ompU, tcpI, and toxR. ST+ V. cholerae non-O1, non-O139 strain 618, provided by A. C. P. Vicente, Department of Genetics, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, was used as a positive control for the st gene. Amplified products were separated on a 1% agarose gel, stained with ethidium bromide, and photographed.
TABLE 2.
Sequences of primers used for detection of selected virulence and regulatory genes
| Target | Nucleotide sequence (5′-3′) | Directiona | Amplicon size (bp) | Reference, biotype, and/or serotype |
|---|---|---|---|---|
| ctxA | CGGGCAGATTCTAGACCTCCTG | F | 564 | 22 |
| CGATGATCTTGGAGCATTCCCAC | R | |||
| zot | TCGCTTAACGATGGCGCGTTTT | F | 947 | Matte et al.b |
| AACCCCGTTTCACTTCTACCCA | R | |||
| ace | TAAGGATGTGCTTATGATGGACACCC | F | 289 | 65 |
| CGTGATGAATAAAGATACTCATAGG | R | |||
| st | GAGAAACCTATTCATTGC | F | 216 | 74 |
| GCAAGCTGGATTGCAAC | R | |||
| ompU | ACGCTGACGGAATCAACCAAAG | F | 869 | Matte et al.b |
| GCGGAAGTTTGGCTTGAAGTAG | R | |||
| tcpA | CACGATAAGAAAACCGGTCAAGAG | F | ||
| TTACCAAATGCAACGCCGAATG | R | 620 | Matte et al.; classicalbc | |
| CGAAAGCACCTTCTTTCACACGTTG | R | 453 | Matte et al.; E1 Tor or O139bc | |
| tcpI | TAGCCTTAGTTCTCAGCAGGCA | F | 862 | Matte et al.b |
| GGCAATAGTGTCGAGCTCGTTA | ||||
| hlyA | GGCAAACAGCGAAACAAATACC | F | 738 | Matte et al.; classical |
| GAGCCGGCATTCATCTGAAT | F | 481 | Matte et al.; nonclassicalbc | |
| CTCAGCGGGCTAATACGGTTTA | R | |||
| toxR | CCTTCGATCCCCTAAGCAATAC | F | 779 | Matte et al.b |
| AGGGTTAGCAACGATGCGTAAG | R |
F, forward; R, reverse. ∗
Matte et al., submitted.
Biotype- and/or serotype-specific amplification occurs.
Genomic fingerprinting by ERIC- and BOX-PCR.
Enterobacterial repetitive intergenic consensus sequence PCR (ERIC-PCR) was performed as described by Rivera et al. (56) by using two oligonucleotide primers (5′-ATGTAAGCTCCTGGGGATTCAC-3′ and 5′-AAGTAAGTGACTGGGGTGAGCG-3′). Box elements PCR (BOX-PCR) was performed as described by Martin et al. (48) by using a single oligonucleotide primer (5′-CTACGGCAAGGCGACGCTGACG-3′). The amplicons were electrophoresed in 1.8% agarose at 60 V for 6 h and stained with SYBR Green I (FMC BioProducts, Rockland, Maine). The DNA band patterns were digitized with a Bio-Imager (Molecular Dynamics) and subjected to a fingerprint analysis. A 1-kb molecular size ladder (GIBCO-BRL, Gaithersburg, Md.) was included on each gel.
AFLP fingerprinting.
All AFLP procedures were performed as described by Janssen et al. (40), with the minor modifications for V. cholerae described by Jiang et al. (41). Briefly, 1 μg of DNA was double digested with restriction enzymes TaqI and ApaI. Following digestion, adapters were added to final concentrations of 0.4 μM for TaqI adapters (5′-GACGATGAGTCCTGAC-3′ and 3′-TACTCAGGACTGGC-5′) and 0.04 μM for ApaI adapters (5′-TCGTAGACTGCGTACAGGCC-3′ and 3′-CATCTGACGCATGT-5′). Ligation reactions were performed at 16°C overnight. Ligated template DNA was purified, stored in Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA [pH 8.0]) at −20°C, and used for PCR amplification within 48 h.
PCR was performed as described by Janssen et al. (40) by using primers A01 (5′-GACTGCGTACAGGCCCA-3′) and T01 (5′-CGATGAGTCCTGACCGAA-3′). Prior to PCR, primer T01 was end labeled with [γ-33P]ATP. Two microliters of template DNA was used for amplification in a total reaction volume of 25 μl. Amplification products were separated on 5% denaturing polyacrylamide and exposed to X-ray film (Kodak Inc.). Autoradiographs were digitized for fingerprint analysis.
PFGE analysis.
Intact genomic DNA was prepared by the method of Majumder et al. (47), with modifications. Briefly, vibrios were grown in Luria broth (Difco) overnight at 37°C. Cells were harvested and washed twice with 10 mM Tris-HCl (pH 7.6) buffer containing 1 M NaCl and suspended at a concentration of 108 CFU/ml in the same buffer. Agarose plugs were prepared by mixing equal volumes of the cell suspension and molten 1% agar (In Cert agarose; FMC BioProducts). Cells were lysed in the plugs and stored in 5 ml of buffer containing 10 mM Tris-HCl (pH 8.0) and 50 mM EDTA (pH 8.0) at 4°C until they were used.
Prior to restriction digestion, plugs containing genomic DNA were washed twice (30 min each) in 10 volumes of buffer containing 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA. Digestion with SfiI (GIBCO-BRL) was carried out at 50°C for 16 h in a solution containing 100 μg of bovine serum albumin per ml by using 40 U of enzyme per plug.
PFGE was carried out with a CHEF Mapper System (Bio-Rad, Hercules, Calif.) in 0.5× TBE buffer containing 0.1% (vol/vol) 100 mM thiourea. Electrophoresis was performed at 6 V/cm and 14°C at a field angle of 120° by using 1.2% SeaKem GTG agarose gels (FMC BioProducts). The pulse time was ramped from 2.98 to 54.17 s over 26.56 h. A lambda DNA ladder (Bio-Rad) was used as the molecular weight standard. Following electrophoresis, the gels were stained with SYBR Green I (Molecular Probes Inc., Eugene, Oreg.), the DNA band patterns were imaged with a Bio-Image analyzer (Molecular Dynamics), and the fingerprints were analyzed.
Analysis of fingerprint patterns.
Digital images of ERIC-PCR, BOX-PCR, AFLP, and PFGE fingerprint patterns were analyzed by using Molecular Fingerprinting Analyst (Bio-Rad) as recommended by the manufacturer and were graphically represented as dendrograms.
RESULTS
Twenty-two V. cholerae strains, including both clinical and environmental strains, were hemolytic on 5% sheep blood nutrient agar (Table 3). Although two O1 strains (VO3 and VO5) and two O139 strains (CO594 and VO17) were negative for hemolysis, they were PCR positive, like all of the other strains, for the genes representing the El Tor fragment for hemolysin (hlyA). In addition, except for two of the non-O1, non-O139 isolates (VO23 and VO28), all strains were also PCR positive for the genes encoding the classical fragment of hemolysin (Table 3).
TABLE 3.
Selected characteristics of V. cholerae O1, O139, and non-O1, non-O139 strains included in this study
| Strain | Hemolysis | Presence of the following genes as determined by PCR
|
NCT productionb | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ctxA | zot | ace | st | hlyAa | tcpA | ompU | tcpI | toxR | |||
| V. cholerae O1 biotype E1 Tor strains | |||||||||||
| VO1 | Beta | − | − | − | − | + | + | + | + | + | NTc |
| VO2 | Beta | + | + | + | − | + | + | + | + | + | NT |
| VO13 | Beta | + | + | + | − | + | + | + | + | + | NT |
| VO3 | None | + | − | + | − | + | + | + | + | + | NT |
| VO4 | Beta | + | + | + | − | + | + | + | + | + | NT |
| VO5 | None | + | + | + | − | + | + | + | + | + | NT |
| VO7 | Beta | + | + | + | − | + | + | + | + | + | NT |
| V. cholerae O139 strains | |||||||||||
| MO45d | Beta | + | + | + | − | + | + | + | + | + | NT |
| CO594 | None | + | + | + | − | + | + | + | + | + | NT |
| CO766 | Beta | + | + | + | − | + | + | + | + | + | NT |
| CO788 | Beta | − | − | − | − | + | + | + | + | + | NT |
| VO12 | Beta | + | + | + | − | + | + | + | + | + | NT |
| VO14 | Beta | + | + | + | − | + | + | + | + | + | NT |
| VO15 | Beta | + | + | + | − | + | + | + | + | + | NT |
| VO16 | Beta | + | + | + | − | + | + | + | + | + | NT |
| VO17 | None | + | + | + | − | + | + | + | + | + | NT |
| VO18 | Beta | + | + | + | − | + | + | + | + | + | NT |
| VO19 | Beta | + | + | + | − | + | + | + | + | + | NT |
| VO20 | Beta | + | + | + | − | + | + | + | + | + | NT |
| V. cholerae non-O1, non-O139 strains | |||||||||||
| VO22 | Beta | − | − | − | − | + | − | − | − | − | + |
| VO23 | Beta | − | − | − | − | − | − | − | − | + | + |
| VO24 | Beta | − | − | − | − | + | − | − | − | + | + |
| VO25 | Beta | − | − | − | − | + | − | − | − | + | + |
| VO26 | Beta | − | − | − | − | + | − | + | − | + | + |
| VO27 | Beta | − | − | − | − | + | − | − | − | + | + |
| VO28 | Beta | − | − | − | − | − | − | − | − | + | + |
Specific for V. cholerae classical hlyA. All V. cholerae strains were positive for nonclassical specific hlyA.
NCT, new cholera toxin detected by immunodiffusion assay.
NT, not tested.
Reference strain MO45 (= ATCC 51394).
Of the nine non-cholera-toxin-producing strains, only one environmental isolate belonging to the non-O1, non-O139 serogroup (VO24) caused fluid accumulation (range, 0.5 to 0.9 ml/cm) in the initial rabbit ileal loop test. However, other strains, including an O1 strain, an O139 strain, and six non-O1, non-O139 isolates from clinical sources, caused fluid accumulation after two to five serial passages through the rabbit gut, and the amount of fluid accumulated ranged from 0.5 to 1.1 ml/cm. Culture filtrates of all strains prepared in brain heart infusion broth and AKI medium also caused accumulation of fluid at volumes similar to those caused by the whole cells.
When concentrated culture filtrates of cholera toxin gene-negative V. cholerae O1 strain X-392 and non-cholera-toxin-producing V. cholerae non-O1, non-O139 strains were tested by gel diffusion, one strain (VO26) produced a precipitin band against new cholera toxin antiserum, showing a reaction of identity, whereas the other strains showed reactions of partial identity (data not shown) (Table 3).
Based on the results of the multiplex PCR, all V. cholerae O1 and O139 strains yielded positive results for tcpA, tcpI, and ompU, and one isolate each of V. cholerae O1 (VO1) and V. cholerae O139 (CO788) gave negative results for the ctxA, zot, and ace genes. Moreover, O1 strain VO3 was negative for the zot gene. However, all V. cholerae non-O1, non-O139 strains yielded negative results for ctxA, zot, ace, and tcpA, and VO26 gave a positive result for ompU. Further analysis showed that all non-O1, non-O139 strains were also PCR negative for the gene encoding tcpI, which negatively regulates expression of tcp (Table 3). All V. cholerae strains were also negative for the NAG-specific st gene. However, except for one non-O1, non-O139 strain (VO22), all of the isolates were positive as determined by PCR for the gene encoding the central regulatory protein, ToxR, which coordinates regulation of expression of the dynamic core genetic element, including TCP and outer membrane protein (OMP).
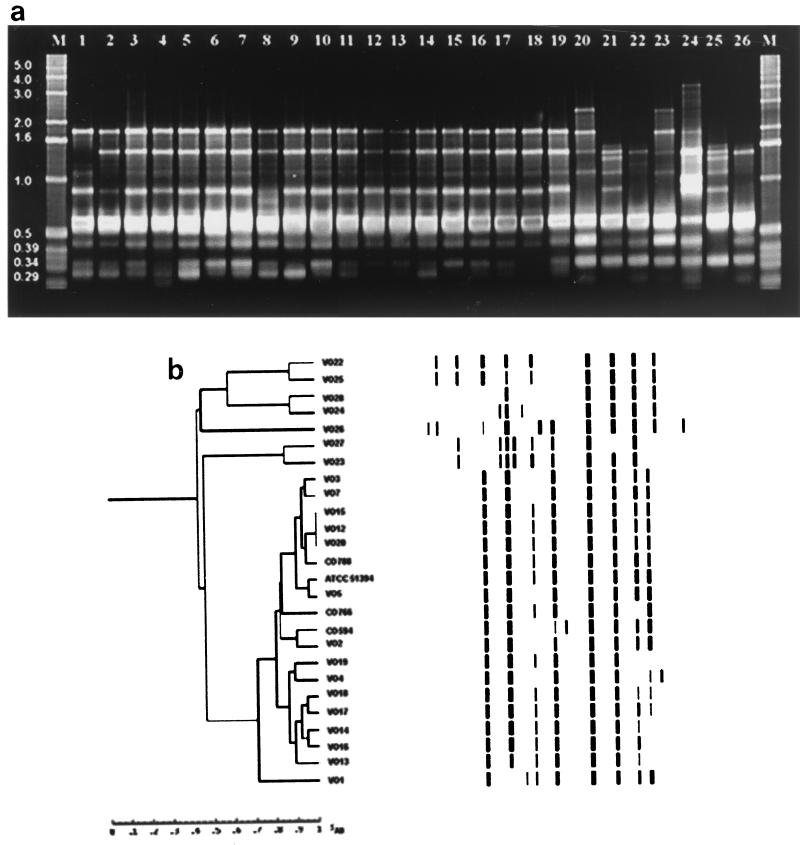
Genetic relatedness analyses of the clinical and environmental strains of V. cholerae performed by ERIC-PCR, BOX-PCR, AFLP, and PFGE methods proved to be interesting. ERIC-PCR of genomic DNA from the various V. cholerae strains yielded a total of 16 fingerprints with sizes ranging between 0.18 and 4.0 kb, as shown in Fig. 1. Two of the seven O1 isolates, one clinical strain (VO3) and one environmental strain (VO7), yielded similar fingerprints. Similarly, 3 of the 12 O139 serogroup strains, both clinical (VO12 and VO15) and environmental (VO20), yielded identical PCR profiles; a non-ctx-producing strain, CO788, yielded closely related fingerprints. The other O1 and O139 serogroup strains produced different fingerprints. A number of V. cholerae O1 and O139 strains had similar PCR profiles. However, they differed from each other by the presence of an analogous fragment that was approximately 0.29 kb long. When the seven V. cholerae non-O1, non-O139 strains were examined, clinical isolate VO22 and environmental isolate VO25 produced similar PCR profiles, clinical isolate VO23 and environmental isolate VO27 produced similar PCR profiles, and environmental isolates VO24 and VO28 strains produced similar PCR profiles.
FIG. 1.
(a) DNA fingerprints of clinical and environmental isolates of V. cholerae O1, O139, and non-O1, non-O139 strains generated by ERIC-PCR amplification. Lanes M, 1-kb molecular weight ladder; lanes 1 to 3 and 12, O1 strains VO1, VO2, VO3, and VO13, respectively; lanes 4 to 6, O1 strains VO4, VO5, and VO7, respectively; lanes 7 to 11 and 13 to 15, O139 strains MO45 (= ATCC 51394), CO594, CO766, CO788, VO12, VO14, VO15, and VO16, respectively; lanes 16 to 19, O139 strains VO17, VO18, VO19, and VO20, respectively; lanes 20 and 21, non-O1, non-O139 strains VO22, and VO23, respectively; lanes 22 to 26, non-O1, non-O139 strains VO24, VO25, VO26, VO27, and VO28, respectively. (b) Digitized ERIC-PCR profiles obtained from genomic DNA of clinical and environmental V. cholerae O1, O139, and non-O1, non-O139 isolates. The dendrogram was constructed by using the Molecular Fingerprinting Analyst (Bio-Rad) software with a simple-match similarity matrix, and data were clustered by the unweighted pair group method with arithmetic means.
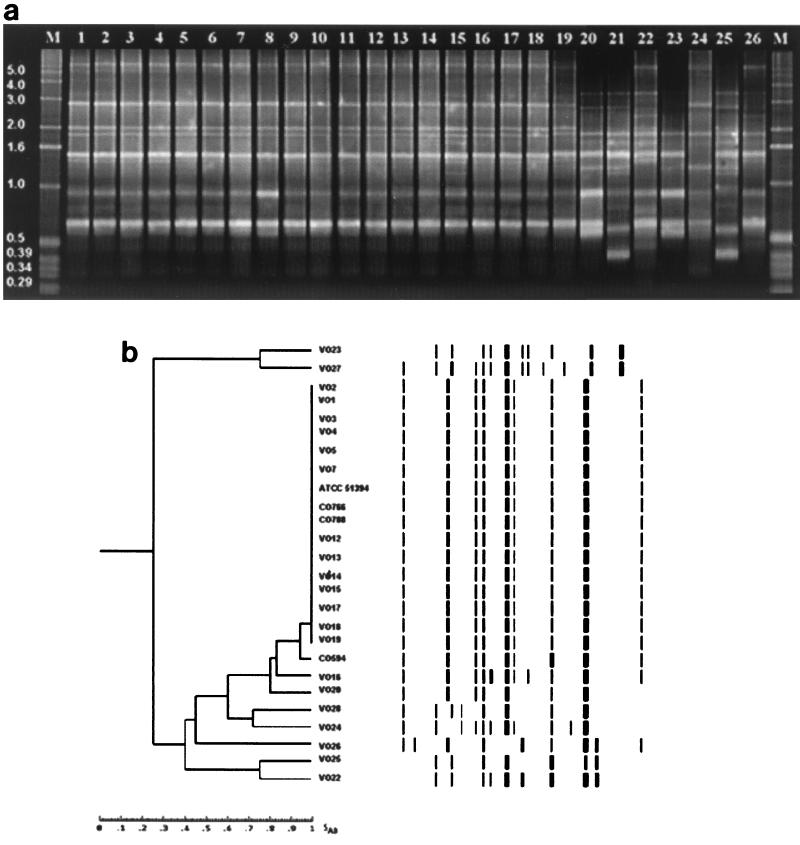
As shown in Fig. 2, BOX-PCR of genomic DNA from various V. cholerae strains resulted in amplification of multiple fragments of DNA that ranged from 0.29 to 6.0 kb long. All of the O1 serogroup strains and most of the O139 serogroup strains yielded identical fingerprint patterns, regardless of source of isolation; the only exception was O139 strain CO594, which produced a closely related PCR profile. Two isolates of serogroup O139, one clinical isolate (VO16) and one environmental isolate (VO20), did not group with the large O1-O139 group. However, non-cholera-toxigenic O1 and O139 strains could not be differentiated from their toxigenic counterparts based on BOX-PCR fingerprints. Like the ERIC-PCR results, when clinical and environmental V. cholerae non-O1, non-O139 strains were examined, strains VO22 and VO25, strains VO23 and VO27, and strains VO24 and VO28 also produced different but closely related PCR profiles.
FIG. 2.
(a) BOX-PCR profiles obtained by using DNA from clinical and environmental V. cholerae O1, O139, and non-O1, non-O139 isolates. Lanes M, 1-kb molecular weight ladder; lanes 1 to 3 and 12, O1 strains VO1, VO2, VO3, and VO13, respectively; lanes 4 to 6, O1 strains VO4, VO5, and VO7, respectively; lanes 7 to 11 and 13 to 15, O139 strains MO45 (= ATCC 51394), CO594, CO766, CO788, VO12, VO14, VO15, and VO16, respectively; lanes 16 to 19, O139 strains VO17, VO18, VO19, and VO20, respectively; lanes 20 and 21, non-O1, non-O139 strains VO22, and VO23, respectively; lanes 22 to 26, non-O1, non-O139 strains VO24, VO25, VO26, VO27, and VO28, respectively. (b) Digitized fingerprints of clinical and environmental V. cholerae O1, O139, and non-O1, non-O139 isolates generated by BOX-PCR amplification. The dendrogram was constructed by using the Molecular Fingerprinting Analyst (Bio-Rad) software as described in the legend to Fig. 1.
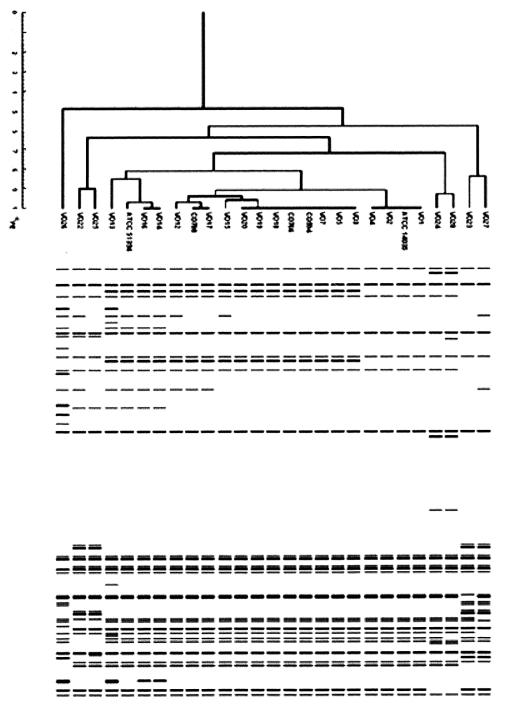
AFLP fragment analysis of V. cholerae non-O1, non-O139 strains and of O1 and O139 strains revealed that the band patterns of the non-O1, non-O139 strains tended to be more diverse, regardless of the source of isolation, while the patterns of the O1 and O139 strains were conserved (Fig. 3). However, differences in the band pattern were detected within and between the O1 and O139 serogroups.
FIG. 3.
Digitized images of AFLP patterns obtained from ApaI-TaqI template DNA by using the selective PCR primers A01 and T01 for clinical and environmental V. cholerae O1, O139, and non-O1, non-O139 isolates. The dendrogram was constructed by using the Molecular Fingerprinting Analyst (Bio-Rad) software as described in the legend to Fig. 1.
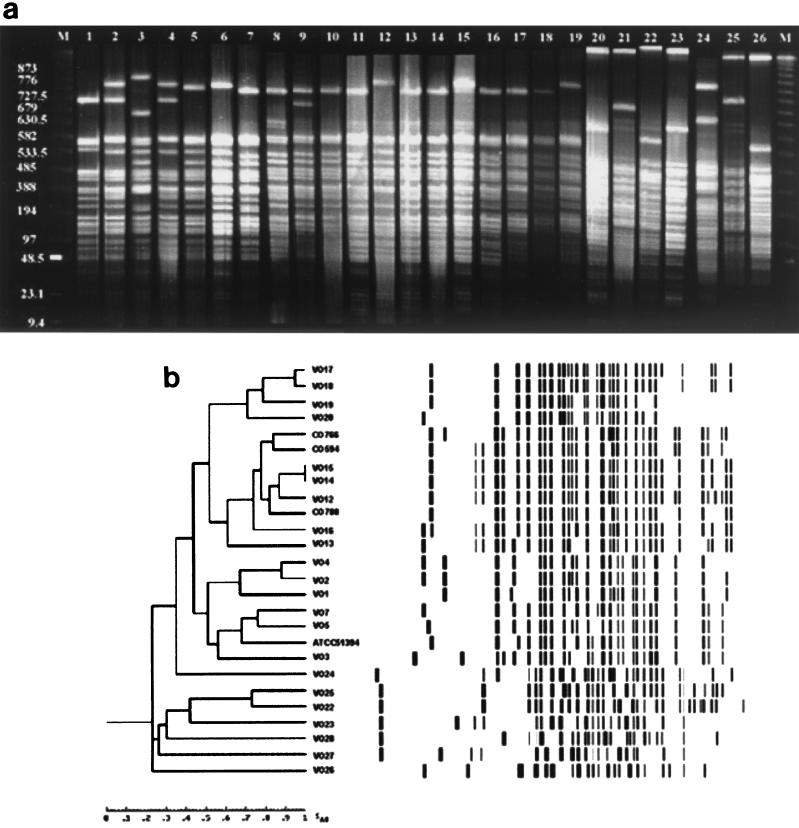
The PFGE profiles of the V. cholerae strains obtained with SfiI showed a variety of patterns. For example, clinical and environmental serogroup O1 isolates had similar restriction patterns. Similarly, strains of V. cholerae O139, whether clinical or environmental, displayed different but closely related restriction profiles (Fig. 4a). Non-cholera-toxigenic O1 and O139 strains were distinguishable from their toxigenic counterparts by the fingerprints obtained from SfiI-digested genomic DNA. The PFGE profiles of non-O1, non-O139 strains differed from each other and from the patterns displayed by the strains belonging to the O1 and O139 serogroups. However, two non-O1, non-O139 isolates, a clinical isolate (VO22) and an environmental isolate (VO25), displayed closely related restriction fingerprint patterns. These data are presented in dendrogram format in Fig. 4b.
FIG. 4.
(a) Fingerprint patterns obtained from PFGE of SfiI-digested DNA of clinical and environmental V. cholerae O1, O139, and non-O1, non-O139 isolates. Lanes M, 1-kb molecular weight ladder; lanes 1 to 3 and 12, O1 strains VO1, VO2, VO3, and VO13, respectively; lanes 4 to 6, O1 strains VO4, VO5, and VO7, respectively; lanes 7 to 11 and 13 to 15, O139 strains MO45 (= ATCC 51394), CO594, CO766, CO788, VO12, VO14, VO15, and VO16, respectively; lanes 16 to 19, O139 strains VO17, VO18, VO19, and VO20, respectively; lanes 20, and 21, non-O1, non-O139 strains VO22, and VO23, respectively; lanes 22 to 26, non-O1, non-O139 strains VO24, VO25, VO26, VO27, and VO28, respectively; (b) Digitized PFGE analysis of SfiI-digested profiles obtained from genomic DNA of clinical and environmental V. cholerae O1, O139, and non-O1, non-O139 isolates. The Dendrogram was generated by using the average percentages of matched bands summarizing the degrees of similarity of the SfiI restriction patterns of genomic DNA of V. cholerae O1, O139, and non-O1, non-O139 strains.
DISCUSSION
Unlike strains of the classical biotype, which are nonhemolytic, strains of the El Tor biotype produce and secrete a hemolysin into the culture medium. Historically, this feature has been used to distinguish the two biotypes. However, hemolytic activity does not always correlate with El Tor biotype, and many El Tor strains that do not produce hemolysin have been reported (6, 18, 58). The results of this study also indicate that a few strains of O1 biotype El Tor and O139, which were isolated from diarrheal and environmental samples, did not show hemolysis in initial tests but were hemolytic on 5% sheep blood nutrient agar plates after serial passage through rabbit ileal loops (data not shown). All strains, whether hemolytic or nonhemolytic on 5% sheep blood nutrient agar plates, were positive for the hlyA gene as determined by PCR. Thus, it is clear that sequences homologous to the hemolysin gene are present in both hemolytic and nonhemolytic strains of V. cholerae O1 biotype El Tor and O139 but may not be expressed in the nonhemolytic isolates.
The hemolysin produced by V. cholerae non-O1, non-O139 strains is identical to the El Tor hemolysin (78) and is considered to be a virulence factor that causes diarrhea (2, 35). However, we found that an El Tor-like hemolysin is not responsible for fluid production in the rabbit gut, because the nonhemolytic strains of V. cholerae also cause fluid accumulation. Furthermore, a number of hemolytic strains did not elicit a secretory response in the initial experiments (68, 71). All V. cholerae non-O1, non-O139 strains examined in this study were hemolytic on 5% sheep blood nutrient agar and had the genetic potential to produce hemolysin. However, the majority of the strains did not show a secretory response in initial tests, strongly suggesting that hemolysin does not play a role in initiating the secretory response.
The V. cholerae O1 biotype El Tor and O139 strains isolated from both diarrheal and environmental samples were characterized in detail to obtain an understanding of the role of the virulence traits in cholera. The environmental isolates, like the clinical isolates, were positive for ctxA, zot, ace, tcpA, and ompU amplicons, indicating that the genes comprising the virulence gene cassette and the genes encoding surface organelles required for intestinal adherence and colonization were intact. Thus, both the O1 biotype El Tor strains and the O139 strains retained the core of the CTX genetic element and also TCP and OMP both of which are recognized as important components in pathogenicity, even in the aquatic environment. These observations are in contrast to reports indicating that environmental isolates of V. cholerae O1 biotype El Tor are nontoxigenic (57) and that the hemolysin produced is most likely responsible for the enterotoxic activity (2, 35).
It is known that some V. cholerae non-O1, non-O139 strains have the ability to cause diarrhea-like symptoms. Recently, several localized outbreaks caused by V. cholerae non-O1, non-O139 have been reported (5, 16, 26, 64). Although some of these strains produce cholera or cholera-like toxin, the majority lack the virulence gene cassette but produce several other extracellular products, such as NAG-specific heat-stable toxin, a thermostable direct hemolysin, Shiga-like toxin, and hemagglutinin, which play some role in the disease process (3, 5, 23, 28, 54, 79).
We concluded from results of this study that in the absence of cholera toxin, NAG-specific heat-stable toxin, and/or TCP and OMP, V. cholerae O1, O139, and non-O1, non-O139 strains that have clinical or environmental origins have the ability to cause diarrhea by a mechanism entirely different from that of the toxigenic V. cholerae O1 and O139 strains. When culture filtrates from all of the non-cholera-toxigenic V. cholerae strains grown in AKI medium (39), mimicking hemolysin (59), were examined in rabbit guts, they caused fluid accumulation. The non-O1, non-O139 strains examined produced an enterotoxin that was antigenetically identifiable as a new V. cholerae toxin. This conclusion is supported by the fact that enterotoxic activity was completely neutralized (67) by the previously reported new cholera toxin antiserum (62, 63). Thus, we concluded that a new secretogenic toxin is the factor most likely responsible for enterotoxic activity in non-cholera-toxin-producing V. cholerae O1, O139, and non-O1, non-O139 strains.
Four different genetic fingerprinting methods, ERIC-PCR, BOX-PCR, AFLP analysis, and PFGE, were used to measure the relatedness of V. cholerae strains isolated from both clinical and environmental sources and the relatedness of O1 biotype El Tor, O139, and non-O1, non-O139 strains. Overall, the correlation of the results of all methods was excellent, and strains belonging to O1 biotype El Tor and O139 yielded fingerprint patterns that were closely related but distinct from those of the non-O1, non-O139 strains. The observed differences in the band patterns within and between the O1 and O139 serogroups suggested that there was divergence in genomic organization which may have arisen from genetic reassortment that took place in the environment over time. No significant difference between the clinical and environmental isolates of V. cholerae was observed. The most interesting finding of the BOX-PCR, AFLP, and PFGE analyses was that environmental isolates (VO24 and/or VO28) of the non-O1, non-O139 serogroups were closely related to the toxigenic V. cholerae O1 and O139 strains.
Collectively, the data indicate that strains of V. cholerae that cause diarrhea have a clonal origin in the aquatic environment. Furthermore, clinical V. cholerae strains showed fingerprint patterns similar to those of strains isolated from the environment. Although minor differences in band patterns were observed among the strains, we concluded that there is a clonal relationship between clinical and environmental isolates, which supports the hypothesis of Colwell et al. (14) that V. cholerae strains are autochthonous to the aquatic environment and that aquatic environments serve as multiple reservoirs of toxigenic and non-cholera-toxigenic V. cholerae strains belonging to the O1, O139, and non-O1, non-O139 serogroups (12–14, 32, 66). The first strain of V. cholerae O139 isolated in our laboratory belonged to the same clone and was isolated in January 1992, when it was originally isolated from an hydrophytic plant, Eichhornia crassipes, in the River Ganga in Varanasi, India; this was almost 8 months before V. cholerae was isolated from diarrheal patients in Madras, India.
It has been reported that non-cholera-toxin-producing V. cholerae non-O1, non-O139 strains possess toxR, the central regulatory protein gene, and can acquire the tcp gene from toxigenic V. cholerae O1 by horizontal gene transfer (44) when they are exposed to the filamentous bacteriophage VPIφ. The ctx element can be acquired by exposure to the toxinoferous phage CTXφ (20, 76). Data obtained in the present study indicate that there is not a significant difference in the genomic organizations of toxigenic V. cholerae O1 and O139 strains, whether they are of clinical or environmental origin. Both toxigenic V. cholerae O1 strains and non-cholera-toxin-producing V. cholerae non-O1, non-O139 strains possess toxR, the central regulatory protein gene, and these strains are present in the aquatic environment. Thus, it can be hypothesized that under the selective pressures of the biological and physicochemical conditions of the aquatic environment, events like acquisition of tcp and ctx genetic elements by non-O1, non-O139 strains from O1 V. cholerae strains can occur after exposure to bacteriophages and thereby favor emergence of a novel strain. However, given the rare occurrence of non-O1, non-O139 strains in the environment that possess both the tcpA and ctx genetic elements (52), it appears that when and if such events occur, there are attendant changes in the somatic antigen or in the cell surface properties (7, 51). A similar suggestion has recently been made concerning the asd gene sequences of O1 and non-O1, non-O139 strains in transition for the somatic O antigen in V. cholerae (43).
ACKNOWLEDGMENTS
A UNESCO Short-Term Fellowship in Biotechnology, funded by UNESCO and provided to D. V. Singh, is gratefully acknowledged. Fundação de Amparo à Pesquisa do Estado de São Paulo, FAPESP, is also gratefully acknowledged for postdoctoral fellowships awarded to Glavur R. Matté and Maria H. Matté. The Department of Biotechnology, Government of India, and the Rajiv Gandhi Centre for Biotechnology are also gratefully acknowledged for funds contributed to this study and for providing a fellowship to F. Sabeena. NIH grant 1RO1A139129-01 is also gratefully acknowledged.
REFERENCES
- 1.Albert M J, Siddique A K, Islam M S, Faruque A S G, Ansaruzzaman M, Faruque S M, Sack R B. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. doi: 10.1016/0140-6736(93)90481-u. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Mayerhofer G, Kotlarski I, Manning P A. Amino-terminal domain of the El Tor hemolysin of Vibrio cholerae O1 is expressed in classical strain and is cytotoxic. Vaccine. 1991;9:588–594. doi: 10.1016/0264-410x(91)90247-4. [DOI] [PubMed] [Google Scholar]
- 3.Arita M, Takeda T, Honda T, Miwatani T. Purification and characterization of Vibrio cholerae non-O1 heat stable enterotoxin. Infect Immun. 1986;52:45–49. doi: 10.1128/iai.52.1.45-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley and Sons Inc.; 1995. [Google Scholar]
- 5.Bagachi K, Echeverria P, Arther J D, Sethabutr O, Serichantalergs O, Hoge C W. Epidemic diarrhoea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp, Thailand. J Clin Microbiol. 1993;31:1315–1317. doi: 10.1128/jcm.31.5.1315-1317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrette T J, Blake P A. Epidemiological usefulness of changes in hemolytic activity of Vibrio cholerae biotype El Tor during the seventh pandemic. J Clin Microbiol. 1981;13:126–129. doi: 10.1128/jcm.13.1.126-129.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. Genesis of the novel epidemic Vibrio cholerae O139 strains: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blake P A, Weaver R W, Hollins D G. Diseases of humans (other than cholera) caused by vibrios. Annu Rev Microbiol. 1980;34:341–367. doi: 10.1146/annurev.mi.34.100180.002013. [DOI] [PubMed] [Google Scholar]
- 9.Chan E G S, McManus E A. Distribution, characterization, and nutrition of marine microorganisms from ten algae, Polysiphonium ianosa, and Ascophylum nodosum. Can J Microbiol. 1969;15:409–420. doi: 10.1139/m69-073. [DOI] [PubMed] [Google Scholar]
- 10.Cholera Working Group. Large epidemic of cholera like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet. 1993;342:387–390. [PubMed] [Google Scholar]
- 11.Coelho A, Andrade J R C, Vicente A C P, Salles C A. New variant of Vibrio cholerae O1 from clinical isolates in Amazonia. J Clin Microbiol. 1995;33:114–118. doi: 10.1128/jcm.33.1.114-118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colwell R R, Kaper J, Joseph S W. Vibrio cholerae, Vibrio parahaemolyticus, and other vibrios: occurrence and distribution in Chesapeake Bay. Science. 1977;198:394–396. [PubMed] [Google Scholar]
- 13.Colwell R R, Huq A. Vibrios in the environment: viable but non-culturable Vibrio cholerae. In: Wachsmuth I K, Olsvik Ø, Blake P A, Olsvik R, editors. Vibrio cholerae and cholera—molecular to global perspectives. Washington, D.C.: American Society for Microbiology; 1994. pp. 117–133. [Google Scholar]
- 14.Colwell R R, Huq A. Environmental reservoir of Vibrio cholerae: the causative agent of cholera. Ann NY Acad Sci. 1994;740:44–54. doi: 10.1111/j.1749-6632.1994.tb19852.x. [DOI] [PubMed] [Google Scholar]
- 15.Colwell R R, Seidler R J, Kaper J, Joseph S W, Garyes S, Lockman H, Maneval D, Bradford H, Roberts N, Huq I, Huq A. Occurrence of Vibrio cholerae serotype O1 in Maryland, and Louisiana estuaries. Appl Environ Microbiol. 1981;41:555–558. doi: 10.1128/aem.41.2.555-558.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalsgaard A, Albert M J, Taylor D N, Shimada T, Meza R, Serichantalergs O, Echeverria P. Characterization of Vibrio cholerae non-O1 serogroup obtained from an outbreak of diarrhea in Lima, Peru. J Clin Microbiol. 1995;33:2715–2722. doi: 10.1128/jcm.33.10.2715-2722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De S N, Chatterje D N. Experimental studies of the mechanism of action of Vibrio cholerae on the intestinal mucous membrane. J Pathol Bacteriol. 1953;66:559–562. doi: 10.1002/path.1700660228. [DOI] [PubMed] [Google Scholar]
- 18.de Moore C E. A non-hemolytic vibrio. Trop Geogr Med. 1963;15:97–107. [PubMed] [Google Scholar]
- 19.Faruque S M, Albert M J, Mekalanos J J. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faruque S, Asadulghani M, Saha M N, Abdul Alim A R M, Albert M J, Nasirul Islam K M, Mekalanos J J. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXφ: molecular basis for origination of new strains with epidemic potential. Infect Immun. 1998;66:5819–5825. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feachem R, Miller C, Drasar B. Environmental aspects of cholera epidemiology. II. Occurrence and survival of V. cholerae in the environment. Trop Dis Bull. 1981;78:865–880. [PubMed] [Google Scholar]
- 22.Fields P I, Popovic T, Wachsmuth K, Olsvik Ø. Use of polymerase chain reaction for detection of toxigenic Vibrio cholerae O1 strains from the Latin American cholera epidemic. J Clin Microbiol. 1992;30:2118–2121. doi: 10.1128/jcm.30.8.2118-2121.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finkelstein R A, Mukerjee S. Haemagglutination: a rapid method for differentiating Vibrio cholerae and El Tor vibrios. Proc Soc Exp Biol Med. 1963;112:355–359. [Google Scholar]
- 24.Ghosh A R, Koley H, De D, Garg S, Bhattacharya M K, Bhattacharya S K, Manna B, Nair G B, Shimada T, Takeda T, Takeda Y. Incidence and toxigenicity of Vibrio cholerae in freshwater lake during the epidemic of cholera caused by serogroup O139 Bengal in Calcutta, India. FEMS Microbiol Ecol. 1994;14:285–291. [Google Scholar]
- 25.Grimes D J, Atwell R A, Brayton P R, Palmer L M, Rollins D M, Roszak D B, Singleton F L, Tamplin M L, Colwell R R. The fate of enteric bacteria in estuarine and marine environment. Microb Sci. 1986;3:324–329. [PubMed] [Google Scholar]
- 26.Gupta N P, Gupta S P, Manglik V S, Prasad B G, Yajnik B S. Investigation into the nature of Vibrio cholerae strains isolated from the epidemic of gastroenteritis in Kumph fair at Allahabad in 1956. Indian J Med Sci. 1956;10:781–791. [Google Scholar]
- 27.Hall R H, Khambaty F M, Kothary M H, Keasler S P, Tall B D. Vibrio cholerae non-O1 serogroup associated with cholera gravis genetically and physiologically resembles O1 El Tor cholera strains. Infect Immun. 1994;62:3859–3863. doi: 10.1128/iai.62.9.3859-3863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanne H F, Finkelstein R A. Characterization and distribution of hemagglutinin produced by Vibrio cholerae. Infect Immun. 1979;36:209–214. doi: 10.1128/iai.36.1.209-214.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. Toxin, toxin co-regulated pili, and the tox R regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hood M A, Ness G E, Roderick G E. Isolation of Vibrio cholerae O1 from the eastern oyster Crassostrea viginea. Appl Environ Microbiol. 1981;41:559–560. doi: 10.1128/aem.41.2.559-560.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huq A, Colwell R R. Vibrios in the marine and estuarine environment: tracking of Vibrio cholerae. J Ecosyst Health. 1996;2:198–214. [Google Scholar]
- 32.Huq A, Colwell R R, Chowdhury M A R, Xu B, Moniruzzaman S M, Islam M S, Yunus M, Albert M J. Co-existence of Vibrio cholerae O1 and O139 Bengal in plankton in Bangladesh. Lancet. 1995;345:1245. doi: 10.1016/s0140-6736(95)92038-2. [DOI] [PubMed] [Google Scholar]
- 33.Huq A, Colwell R R, Rahman R, Ali A, Chowdhury M A R, Parveen S, Sack D A, Russek-Cohen E. Detection of Vibrio cholerae in the aquatic environment measured by fluorescent antibody and culture method. Appl Environ Microbiol. 1990;56:2370–2373. doi: 10.1128/aem.56.8.2370-2373.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huq A, Parveen S, Qadri F, Colwell R R. Comparison of Vibrio cholerae serotype O1 isolated from patient and aquatic environment. J Trop Med Hyg. 1993;96:86–92. [PubMed] [Google Scholar]
- 35.Ichinose Y, Yamamoto K, Nakasone N, Tanabe M J, Takeda T, Miwatani T, Iwanaga M. Enterotoxicity of El Tor-like hemolysin of non-O1 Vibrio cholerae. Infect Immun. 1987;55:1090–1093. doi: 10.1128/iai.55.5.1090-1093.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Islam M S, Drasar B S, Bradly D J. Survival of toxigenic Vibrio cholerae O1 with comman duck weed, Lemna minor, in artificial aquatic ecosystem. Trans R Soc Trop Med Hyg. 1990;84:422–424. doi: 10.1016/0035-9203(90)90345-f. [DOI] [PubMed] [Google Scholar]
- 37.Islam M S, Hasan M K, Miah M A, Yunus M, Zaman K, Albert M J. Isolation of Vibrio cholerae O139 synonym Bengal from the aquatic environment in Bangladesh: implications for disease transmission. Appl Environ Microbiol. 1994;60:1684–1686. doi: 10.1128/aem.60.5.1684-1686.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Islam M S, Drasar B S, Sack R B. The aquatic flora and fauna as reservoir of Vibrio cholerae: a review. J Diarrh Dis Res. 1994;12:87–96. [PubMed] [Google Scholar]
- 39.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol. 1986;30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 40.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 41.Jiang S C, Matte M, Matte G, Huq A, Colwell R R. Genetic diversity of clinical and environmental isolates of Vibrio cholerae determined by amplified fragment length polymorphism. Appl Environ Microbiol. 2000;66:148–153. doi: 10.1128/aem.66.1.148-153.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamal A M. The seventh pandemic of cholera. In: Barua D, Burros W, editors. Cholera. Philadelphia, Pa: The W. B. Saunders Company; 1974. pp. 1–14. [Google Scholar]
- 43.Karaolis D K R, Lan R, Reeves P R. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karaolis D K R, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 45.Kurazono H, Pal A, Bag P K, Nair G B, Karasawa T, Mihara T, Takeda Y. Distribution of genes encoding cholera toxin, zonula occludens toxin, accessory cholera toxin, and El Tor hemolysin in Vibrio cholerae of diverse origins. Microb Pathog. 1995;18:231–235. doi: 10.1016/s0882-4010(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 46.Levine M M, Kaper J B, Herrington D, Losonsky G, Morris J G, Clements M L, Black R E, Tall B, Hall R. Volunteer studies of cholera deletion mutants of Vibrio cholerae O1 prepared by recombinant techniques. Infect Immun. 1988;56:161–167. doi: 10.1128/iai.56.1.161-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Majumder R, Sengupta S, Khetawat G, Bhadra R K, Roychoudhury S, Das J. Physiological map of the genome of Vibrio cholerae 569B and localization of genetic markers. J Bacteriol. 1996;178:1105–1112. doi: 10.1128/jb.178.4.1105-1112.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin B, Humbert O, Camara M, Guenzi E, Walker J, Mitchell T, Andrew P, Prudhomme M, Alloing G, Hakenbeck H, Morrison D A, Boulnois G J, Claverys J P. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 1992;20:3479–3483. doi: 10.1093/nar/20.13.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller C J, Feachem R G, Drasar B S. Cholera epidemiology in developed and developing countries: new thoughts on transmission, seasonality, and control. Lancet. 1985;i:261–263. doi: 10.1016/s0140-6736(85)91036-0. [DOI] [PubMed] [Google Scholar]
- 50.Miller J F, Mekalanos J J, Falkow S. Coordinated regulation and sensory transduction in the control of bacterial virulence. Science. 1989;243:916–922. doi: 10.1126/science.2537530. [DOI] [PubMed] [Google Scholar]
- 51.Montila R, Chowdhury M A R, Huq A, Xu B, Colwell R R. Serogroup conversion of Vibrio cholerae non-O1 to Vibrio cholerae O1: effect of growth state of cells, temperature, and salinity. Can J Microbiol. 1996;42:87–93. doi: 10.1139/m96-014. [DOI] [PubMed] [Google Scholar]
- 52.Mukhopadhyay A K, Garg S, Mitra R, Basu A, Dutta D, Bhattacharya S K, Shimada T, Takeda T, Takeda Y, Nair G B. Temporal shift in traits of Vibrio cholerae strains isolated from hospitalized patients in Calcutta: 3 years (1993-1996) analysis. J Clin Microbiol. 1996;34:2537–2543. doi: 10.1128/jcm.34.10.2537-2543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nalin D R. Cholera, copepods and chitinase. Lancet. 1976;ii:958. doi: 10.1016/s0140-6736(76)90915-6. [DOI] [PubMed] [Google Scholar]
- 54.O'Brien A D, Chen M E, Holmes R K, Kaper J, Levine M M. Environmental and human isolates of Vibrio cholerae and Vibrio parahaemolyticus produce a Shigella dysenteriae type 1 (Shiga)-like toxin. Lancet. 1984;i:77–78. doi: 10.1016/s0140-6736(84)90006-0. [DOI] [PubMed] [Google Scholar]
- 55.Ramamurthy T, Garg S, Sharma R, Bhattacharya S K, Nair G B, Shimada T, Takeda T, Karasawa T, Kurazono H, Pal A, Takeda Y. Emergence of a novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 56.Rivera I G, Chowdhury M A R, Huq A, Jacobs D, Martins M T, Colwell R R. Enterobacterial repetitive intergenic consensus sequence and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139 and non-O1 strains. Appl Environ Microbiol. 1995;61:2898–2904. doi: 10.1128/aem.61.8.2898-2904.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodrigue D C, Popovic T, Wachsmuth I K. Non-toxigenic Vibrio cholerae O1 infection in the Unites States. In: Wachsmuth I K, Blake P A, Olsvik R, editors. Vibrio cholerae and cholera—molecular to global perspective. Washington, D.C.: American Society for Microbiology; 1994. pp. 69–76. [Google Scholar]
- 58.Roy S, Mukerjee S, Tanamal S J W. Hemolytic and non-hemolytic El Tor vibrios. Ann Biochem Exp Med. 1963;23:553–558. [PubMed] [Google Scholar]
- 59.Saha P K, Koley H, Nair G B. Purification and characterization of an extracellular secretogenic non-membrane-damaging cytotoxin produced by clinical strains of Vibrio cholerae non-O1. Infect Immun. 1996;64:3101–3108. doi: 10.1128/iai.64.8.3101-3108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saha P K, Koley H, Mukhopadhyay A K, Bhattacharya S K, Nair G B, Ramakrishnan B S, Krishnan S, Takeda T, Takeda Y. Nontoxigenic Vibrio cholerae O1 serotype Inaba biotype E1 Tor associated with cluster of cases in southern India. J Clin Microbiol. 1996;34:1114–1117. doi: 10.1128/jcm.34.5.1114-1117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanyal S C. Vibrio toxin. In: Dorner F, Drews J, editors. Pharmacology of bacterial toxins. International encyclopedia of pharmacology and therapeutics. Vol. 119. Oxford, United Kingdom: Pergamon Press; 1986. pp. 207–225. [Google Scholar]
- 62.Sanyal S C, Alam K, Neogi P K B, Huq M I, Al-Mahmud K A. A new cholera toxin. Lancet. 1983;i:1337. doi: 10.1016/s0140-6736(83)92449-2. [DOI] [PubMed] [Google Scholar]
- 63.Sanyal S C, Neogi P K B, Alam K, Huq M I, Al-Mahmud K A. A new enterotoxin produced by V. cholerae O1. J Diarrh Dis Res. 1984;2:3–12. [PubMed] [Google Scholar]
- 64.Sharma C, Thungapathra M, Ghosh A, Mukhopadhyay A K, Basu A, Mitra R, Basu I, Bhattacharya S K, Takeda T, Yamasaki S, Takeda Y, Nair G B. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J Clin Microbiol. 1998;36:756–763. doi: 10.1128/jcm.36.3.756-763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi L, Miyoshi S, Hiura M, Tomochika K, Shimada T, Shinoda S. Detection of genes encoding cholera toxin (CT), zonula occludens toxin (ZOT), accessory cholera enterotoxin (ACE) and heat stable enterotoxin (ST) in Vibrio mimicus clinical strains. Microbiol Immunol. 1998;42:823–828. doi: 10.1111/j.1348-0421.1998.tb02357.x. [DOI] [PubMed] [Google Scholar]
- 66.Shukla B N, Singh D V, Sanyal S C. Attachment of non-culturable toxigenic Vibrio cholerae O1, and non-O1, and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the River Ganga, Varanasi. FEMS Immunol Med Microbiol. 1995;12:113–120. doi: 10.1111/j.1574-695X.1995.tb00182.x. [DOI] [PubMed] [Google Scholar]
- 67.Singh D V, Tikoo A, Sanyal S C. Production of the new cholera toxin by environmental isolates of Vibrio cholerae non-O1. J Med Microbiol. 1996;45:31–34. doi: 10.1099/00222615-45-1-31. [DOI] [PubMed] [Google Scholar]
- 68.Singh D V, Shukla B N, Sanyal S C. Haemolysin produced by Vibrio cholerae non-O1 is not enterotoxic. J Med Microbiol. 1996;45:35–39. doi: 10.1099/00222615-45-1-35. [DOI] [PubMed] [Google Scholar]
- 69.Smith C L, Carpentor C R. Purification, specific fragmentation, and separation of large DNA molecules. Methods Enzymol. 1987;155:449–467. doi: 10.1016/0076-6879(87)55030-3. [DOI] [PubMed] [Google Scholar]
- 70.Sochard M R, Wilson D F, Austin B, Colwell R R. Bacteria associated with the surface and the gut of marine copepods. Appl Environ Microbiol. 1979;37:750–759. doi: 10.1128/aem.37.4.750-759.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tikoo A, Singh D V, Sanyal S C. Influence of animal passage on haemolysin and enterotoxin production in Vibrio cholerae O1 biotype E1 Tor strains. J Med Microbiol. 1994;40:246–251. doi: 10.1099/00222615-40-4-246. [DOI] [PubMed] [Google Scholar]
- 72.Venkateswaran K, Takai T, Navarro I M, Nakano H, Hashimoto H, Sibling R I. Ecology of Vibrio cholerae non-O1 and Salmonella spp. and the role of zooplankton in their seasonal distribution in Fukuyama coastal water, Japan. Appl Environ Microbiol. 1989;35:1591–1598. doi: 10.1128/aem.55.6.1591-1598.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application of fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vicente A C P, Coelho A M, Salles C A. Detection of Vibrio cholerae and V. mimicus heat stable toxin gene sequence by PCR. J Med Microbiol. 1997;46:398–402. doi: 10.1099/00222615-46-5-398. [DOI] [PubMed] [Google Scholar]
- 75.Vos P, Hogers R, Bleeker M, Reijans M, Lee T, Jornes M, Frijters A, Pot J, Peelman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 77.World Health Organization. Manual for laboratory investigation of acute enteric infections. Publication WHO CDD/83.3. Geneva, Switzerland: Program for Control of Diarrhoeal Diseases, World Health Organization; 1987. [Google Scholar]
- 78.Yamamoto K, Al-Omani M, Honda T, Takeda Y, Miwatani T. Non-O1 Vibrio cholerae hemolysin: purification, partial characterization, and immunological relatedness to E1 Tor hemolysin. Infect Immun. 1984;45:192–196. doi: 10.1128/iai.45.1.192-196.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoh M, Honda T, Miwatani T. Production of non-O1 Vibrio cholerae hemolysin related to thermostable direct hemolysin of Vibrio parahaemolyticus. FEMS Microbiol Lett. 1985;29:197–200. [Google Scholar]