Abstract
We have developed a three-component system for microbial identification that consists of (i) a universal syringe-operated silica minicolumn for successive DNA and RNA isolation, fractionation, fragmentation, fluorescent labeling, and removal of excess free label and short oligonucleotides; (ii) microarrays of immobilized oligonucleotide probes for 16S rRNA identification; and (iii) a portable battery-powered device for imaging the hybridization of fluorescently labeled RNA fragments with the arrays. The minicolumn combines a guanidine thiocyanate method of nucleic acid isolation with a newly developed hydroxyl radical-based technique for DNA and RNA labeling and fragmentation. DNA and RNA can also be fractionated through differential binding of double- and single-stranded forms of nucleic acids to the silica. The procedure involves sequential washing of the column with different solutions. No vacuum filtration steps, phenol extraction, or centrifugation is required. After hybridization, the overall fluorescence pattern is captured as a digital image or as a Polaroid photo. This three-component system was used to discriminate Escherichia coli, Bacillus subtilis, Bacillus thuringiensis, and human HL60 cells. The procedure is rapid: beginning with whole cells, it takes approximately 25 min to obtain labeled DNA and RNA samples and an additional 25 min to hybridize and acquire the microarray image using a stationary image analysis system or the portable imager.
Traditional methods of bacterial identification are usually based on morphological and/or physiological features of a microorganism or on analysis of 16S rRNA gene sequences (59). These methods can require considerable amounts of time. Recently, PCR and other amplification technologies were introduced for bacterial identification (33). Immunological methods (16) and mass spectrometry (18) have also been adapted for this purpose but are expensive or cumbersome. DNA microchip technology (37) advantageously combines a rapid, high-throughput platform for nucleic acid hybridization with low cost and the potential for automation, although sample preparation procedures, including DNA and RNA isolation, fragmentation, and labeling, are still limiting steps (32, 44). Another limitation of microarray technology is the lack of portable and inexpensive devices for the acquisition of hybridization patterns (5). We have addressed these shortcomings through the development of a rapid and simple system for sample preparation and microarray analysis.
MATERIALS AND METHODS
Preparation of silica syringe-operated columns.
A silica suspension (50 μl) was prepared as described previously (4) and loaded into a 25-mm-long sterile centrifuge device containing a polysulfone filter with a diameter of 6.5 mm and a pore size of 0.2 μm (Whatman, Fairfield, N.J.). The column was sealed against the end of a 10-ml syringe without any glue, using the O-ring from a 1.5-ml screw-cap microcentrifuge tube introduced between the syringe and the top of the column, and washed once with 500 μl of diethylpyrocarbonate-treated water.
Isolation of total nucleic acids.
Bacterial strains Bacillus subtilis B-459, B. thuringiensis 4Q281, and Escherichia coli BL21, as well as human HL60 cells, were used as the starting material. Gram-positive cells were pretreated by incubation with 25 μl of a lysozyme solution (100 mg/ml) at 37°C for 5 min before lysis. A cell pellet obtained from 1 ml of log-phase bacterial cells (2 × 107 to 2 × 108 cells/ml) grown in standard Luria-Bertani medium (45) or human HL60 cell cultures (6 × 106 cells/ml) grown as described previously (49) was lysed by adding 550 μl of mixture (9:4) of lysis (L) and binding (B) buffers. L buffer was composed of 4.5 M guanidine thyocianate (GuSCN) and 100 mM EDTA (pH 8); B buffer contained 4 M GuSCN, 135 mM Tris-HCl (pH 6.4), 3.5% (wt/vol) Triton X-100, 17.5 mM EDTA, and 215 mM MgCl2. The lysate was applied to a silica minicolumn, which was washed by using a syringe with 0.5 ml of the applied L-B buffer mixture (9:4) (twice), 0.5 ml of 70% (vol/vol) ethanol (twice), and 0.5 ml of 100% ethanol (once). The column was dried by forcing 5 ml of air through it with a syringe. The bound nucleic acids were either eluted from the column with 1 mM HEPES (pH 7.5) or directly subjected to labeling and fragmentation.
RNA and DNA isolation and fractionation.
A cell pellet obtained from 1 ml of log-phase culture was lysed by the addition of 450 μl of L buffer (gram-positive cells were pretreated with lysozyme as described above). DNA was isolated by passing the lysate over a syringe-operated column, allowing DNA to bind to the silica. B buffer (200 μl) was added to the flowthrough RNA fraction, which was then applied to the analogous fresh column. The first column, containing bound DNA, was washed five times with 0.5 ml of L buffer, twice with 0.5 ml of 70% (vol/vol) ethanol, and once with 0.5 ml of 100% (vol/vol) ethanol. The second column, containing bound RNA, was washed twice with 0.5 ml of the L-B buffer mixture (9:4) and then with ethanol as described for isolation of total nucleic acids (see above). Fractionated DNA or RNA was either eluted as described above or directly subjected to labeling and fragmentation on the column.
Labeling, fragmentation, and hybridization.
The silica column containing bound RNA, DNA, or both was sealed at the bottom with a cap from a microcentrifuge tube and preheated in a sand bath at 95°C for 2 min. Freshly prepared labeling cocktail (150 μl) containing 5 mM 1,10-phenanthroline, 500 μM CuSO4, 1 mM lissamine-rhodamine B ethylenediamine (Molecular Probes, Inc., Eugene, Oreg.), 2 mM H2O2, 20 mM sodium phosphate (pH 7.0), and 20 mM NaCNBH3 was applied to the minicolumn (the H2O2 was added immediately before application of the cocktail to the column), which was then sealed to prevent evaporation. After incubation of the mixture for 10 min at 95°C, the reaction was stopped by adding 9 μl of 500 mM EDTA (pH 8.0). Nucleic acids were precipitated on the column by adding 15 μl of 5 M ammonium acetate and 450 μl of 100% (vol/vol) ethanol followed by a 5-min incubation at room temperature. Excess fluorescent label was removed by washing the column twice with 1.5 ml of 100% (vol/vol) ethanol. The column was then dried with forced air. The labeled product was eluted twice with 45 to 60 μl of 1 mM HEPES (pH 7.5). The eluant was adjusted to contain 5 mM EDTA, 1 M GuSCN, and 50 mM HEPES (pH 7.5) and filtered through a 0.45-μm-pore-size Millex-HV syringe filter (Millipore, Bedford, Mass.). The resulting solution (30 μl), containing 5 to 15 μg of nucleic acids, including 1 to 3.5 μg of 16S rRNA, was applied to the oligonucleotide microarray covered with a 0.5-mm-deep, 13-mm-diameter CoverWell gasketed incubation chamber (Grace Bio-Labs, Inc., Bend, Oreg.) and incubated for 20 min at room temperature.
Optional removal of small fragments and traces of free label.
A polypropylene, 4.5-mm-diameter, Wizard syringe Minicolumn (Promega, Inc., Madison, Wis.) containing 70 μl of Q Sepharose (Pharmacia Biotech, Uppsala, Sweden) was conditioned by being washed twice with 0.5 ml of diethylpyrocarbonate-treated H2O, once with 0.5 ml of 2 M LiClO4, and then twice again with 0.5 ml of H2O. After the 10-min labeling and fragmentation step (see above), the contents of a silica minicolumn were expelled into a microcentrifuge tube containing 9 μl of 500 mM EDTA (pH 8.0). The same tube was used to collect material rinsed from the silica column with 1 ml of hot (95°C) 1 mM sodium phosphate (pH 7.0). This solution of labeled nucleic acids and free label was then applied to the Q Sepharose column. The Q Sepharose column was washed with 1 ml of 100 mM LiClO4 to remove unincorporated label and small nucleic acid fragments (shorter than 20 bases). Nucleic acids were eluted with 100 μl of 0.5 M GuSCN. The eluant was adjusted to contain 5 mM EDTA, 1 M GuSCN, and 50 mM HEPES (pH 7.5), and 30 μl of the resulting solution was applied to the microarray as described above.
Oligonucleotide synthesis and oligonucleotide array fabrication.
Oligonucleotide microarrays were constructed with 10 oligonucleotide probes, each approximately 20 bases in length, with the following sequences (5′→3′): EU1, ACCGCTTGTGCGGGCCC; EU2, TGCCTCCCGTAGGAGTCT; U1, GA/TATTACCGCGGCT/GGCTG; U2, ACGGGCGGTGTGTA/GCAA; BSG1, ATTCCAGCTTCACGCAGTC; BSG2, ACAGATTTGTGGGATTGGCT; BS1, AAGCCACCTTTTATGTTTGA; BS2, CGGTTCAAACAACCATCCGG; BCG1, CGGTCTTGCAGCTCTTTGTA; and BCG2, CAACTAGCACTTGTTCTTCC (Probe targets are described in the legend to Fig. 3). The sequences of probes U1, U2, EU1 and EU2 were chosen following the recommendations of Amann et al. (1). All other probe sequences were selected using original software developed in our laboratory (Y. Lysov, unpublished data), where each potential probe was tested against all available 16S rRNA sequences (from GenBank and RDP) by a function that estimates the relative duplex stability according to the number and position of mismatches. If the 16S rRNA of any microorganism that did not belong to the genus of interest formed stable duplexes with any oligonucleotide being considered as a potential probe for the microchip, this oligonucleotide was excluded from the list of probes. Oligonucleotides were synthesized with a 394 DNA/RNA Synthesizer (Perkin-Elmer/Applied BioSystems, Foster City, Calif.) using standard phosphoramidite chemistry. 5′-Amino-Modifier C6 (Glen Research, Sterling, Va.) was linked to the 5′ ends of oligonucleotides. The microarray matrix, containing 100- by 100- by 20-μm polyacrylamide gel pads fixed on a glass slide and placed 200 μm from each other, was manufactured using photopolymerization (25) and activated as described previously (41). Individual 1 mM amino-oligonucleotide solutions (0.006 μl) were applied to each gel pad containing aldehyde groups (53) which were designed and implemented by the Argonne National Laboratory state-of-the-art computer-based robot-arrayer Quadrate II (61). Schiff bases coupling the oligonucleotides with aldehyde groups within the gel pads were stabilized by reduction with NaCNBH3 (41).
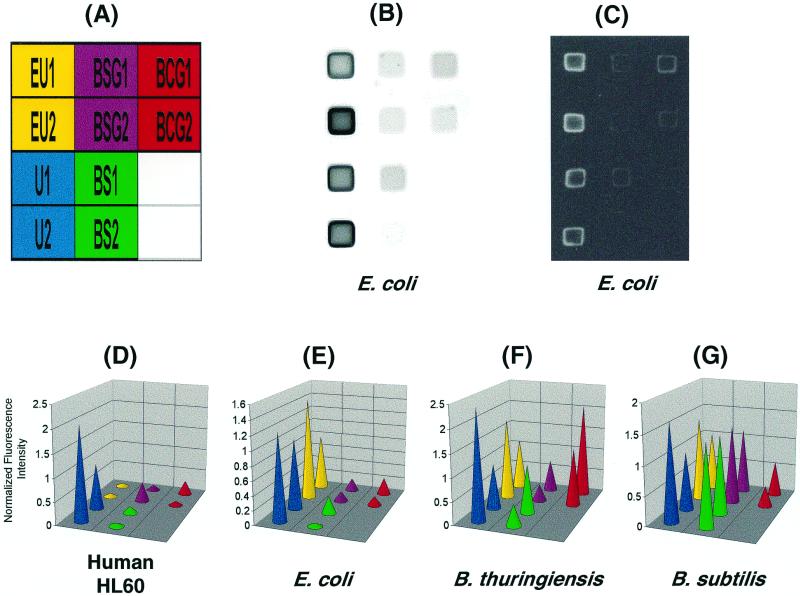
FIG. 3.
Hybridization of total nucleic acids with an oligonucleotide microarray. Total nucleic acids were isolated and labeled using the silica minicolumn. (A) The arrangement of probes (see Materials and Methods for a list of sequences) immobilized on the microarray for identification of U1 and U2 (“all life”), EU1 and EU2 (all eubacteria), BSG1 and BSG2 (B. subtilis group bacteria), BS1 and BS2 (B. subtilis spp), and BCG1 and BCG2 (B. cereus group bacteria). (B and C) Analysis of E. coli with a stationary microscope (B) and the portable imager (C). (D to G) Normalized fluorescent signal intensities for labeled total nucleic acids from human HL60 cells (D), E. coli (E), B. thuringiensis (F), and B. subtilis (G). Hybridization results were obtained with the stationary fluorescent microscope (B and D to G) or with the portable imager (C). Fluorescence intensities were quantified using Image, a custom LabVIEW program (National Instruments, Austin, Tex.).
Microchip analysis.
Hybridization signals were acquired with a stationary wide-field fluorescence microscope (2, 61) or with the portable imager. Before analysis, hybridization solution was removed from the microarrays, which were then washed twice at room temperature with 150 μl of washing buffer (60 mM sodium phosphate [pH 7.4], 900 mM NaCl, 6mM EDTA, 1% [wt/vol] Tween 20) for 15 s. The microarrays were imaged wet (covered with a thin film of washing buffer) when used with the fluorescence microscope or dry when used with the portable analyzer (2 to 30 s of exposure).
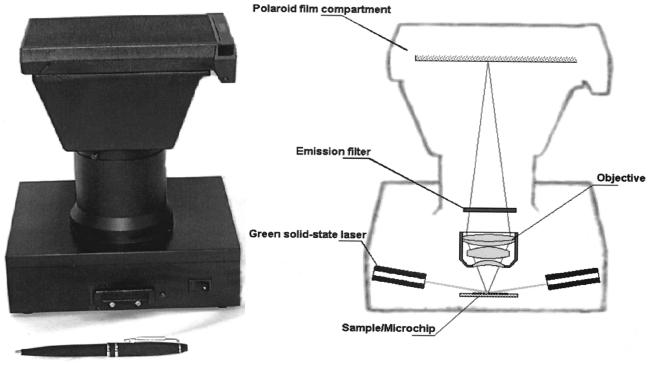
The portable imager was designed and manufactured in collaboration with the State Optical Institute (GOI, St. Petersburg, Russia). The portable battery-powered imager utilizes a wide-field, high-aperture, long-working-distance lens objective with the following parameters: field of view, 4.2 mm in diameter; numeric aperture, 0.5; working distance, 2.0 mm; magnification, ×20; spatial resolution, 1.5 μm. Microarrays are fixed at the focal point of the objective and illuminated by two 3-mW green (532-nm) diode lasers (DeHarpporte Trading Co., Eden Prairie, Minn.). The lasers are situated near the body of the objective such that the excitation light strikes the sample at an angle of 82° to the objective axis. Cylindrical lenses are positioned at the ends of the lasers to provide uniform illumination of the objective field of view. An XF3024 (590DF35) emission filter (Omega Optical, Brattleboro, Vt.) with a transmission maximum at 590 nm is used to cut off excitation light. In contrast to the use of expensive scanners that measure fluorescence intensity in one moment for one spot only and summarize signals from the photomultiplier, in our analyzer the images of 180 (12 × 15) individual gel elements are simultaneously projected onto the surface of ISO-3000 Polaroid film (3.25 by 4.25 in.).
RESULTS
System overview.
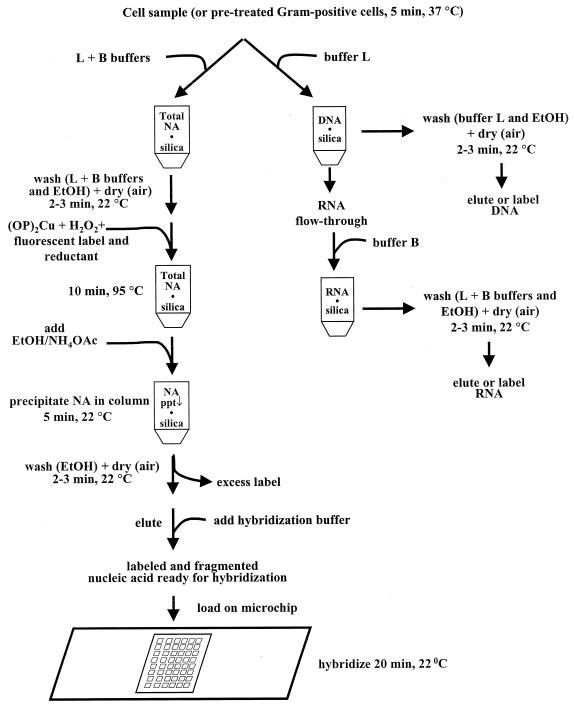
We combined a silica minicolumn, oligonucleotide microarrays, and a portable imager to produce a simple and inexpensive system for bacterial identification. A procedure was developed for nucleic acid isolation, labeling, and fragmentation within a single syringe-operated silica minicolumn. The process requires no vacuum filtration step, phenol-chloroform extraction, CsCl fractionation, or centrifugation. A flowchart of the protocol is shown in Fig. 1. There are three main steps in the procedure: (i) cell lysis and nucleic acid isolation (this may also include DNA and RNA fractionation), (ii) fluorescent labeling and fragmentation of nucleic acids (DNA or RNA can be labeled using the same protocol), and (iii) removal of short oligonucleotides and unbound dye.
FIG. 1.
Flowchart of the isolation, fractionation, fragmentation, and labeling of nucleic acids with subsequent removal of excess free label, using a silica minicolumn.
Nucleic acid purification and fractionation.
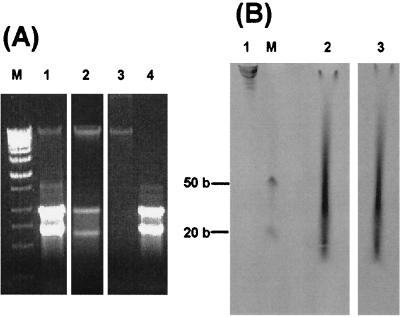
Using the silica minicolumn, one can isolate total nucleic acids or fractionated DNA and RNA from gram-negative bacteria within several minutes; the procedure requires only an additional 5 min of lysozyme pretreatment for gram-positive microorganisms (Fig. 1). Electrophoretic analysis of total nucleic acids and fractionated DNA and RNA isolated from B. subtilis using the minicolumn is shown in Fig. 2A. The yields of isolated total nucleic acids, pure RNA, and pure DNA were 91, 77, and 34%, respectively (Table 1). The recovery of fractionated DNA could be increased considerably, up to an 86% yield, by reducing the number of L buffer washes applied to the DNA-silica column, but this resulted in increased RNA admixture.
FIG. 2.
Nucleic acids isolated, fractionated, labeled with lissamine-rhodamine B, and fragmented on a silica syringe-operated column. (A) Isolated total nucleic acids (lane 1), partially fractionated DNA (lane 2), purified DNA (lane 3), and purified RNA (lane 4) from B. subtilis were analyzed by electrophoresis in 1% agarose. M, λ-HindIII DNA marker. (B) Total nucleic acids from B. thuringiensis fractionated in a denaturing 7.5% polyacrylamide gel (46) before (lane 1) and after (lanes 2 and 3) labeling and fragmentation; fluorescence of labeled and fragmented product before (lane 3) and after (lane 2) ethidium bromide staining. M, single-stranded 20- and 50-base size markers.
TABLE 1.
Isolation of nucleic acids from B. subtilis using the silica minicolumn
| Nucleic acid fraction | Cell contenta (μg) | Amt isolated (μg) | Yield (%) |
|---|---|---|---|
| Total nucleic acid | 35 | 32 | 91 |
| RNA | 30 | 23 | 77 |
| Partially fractionated DNAb | 5 | 4.3 | 86 |
| DNA | 5 | 1.7 | 34 |
Theoretical content of 1 ml of log-phase bacterial culture (3 × 108 cells) calculated from data reported for E. coli or Salmonella enterica servar Typhimurium (Qiagen Product Guide, Qiagen Inc., Valencia, Calif., 1999).
Samples of partially fractionated DNA were obtained after washing the DNA-silica column (Fig. 1) with only two portions of 500 μl of L buffer instead of five portions of 500 μl (see Materials and Methods).
Nucleic acid fragmentation and fluorescent labeling.
The newly developed labeling and fragmentation procedure that was performed with the syringe-operated column devised in this study requires only 10 to 12 min to complete (Fig. 1). The extent of fluorescent-dye incorporation and the length of the nucleic acid fragments may be varied over a wide range through manipulation of bis(1,10-phenanthroline)copper(I) [(OP)2Cu] and H2O2 concentrations, reaction temperature, and duration of the reaction. To avoid the influence of secondary structure on nucleic acid fragmentation and to increase the rate of the reaction, we performed the reaction at 95°C (for 10 min). This resulted in the production of labeled fragments 20 to 100 bases in length (Fig. 2B) with the same efficiency for both RNA and DNA (data not shown). The intrinsic fluorescence of lissamine-rhodamine-labeled nucleic acids was apparent when this material was subjected to denaturing polyacrylamide gel electrophoresis and viewed with a transilluminator (Fig. 2B, lane 3). The same gel stained with ethidium bromide (lanes 1, 2, and M) revealed the total population of nucleic acid fragments. The coincidence of the patterns appearing as smears without any visible bands suggests this hydroxyl radical-based method provides sequence-independent labeling and fragmentation.
Removal of short nucleic acid fragments and unbound label.
On completion of the labeling reaction, nucleic acids were precipitated by the addition of ethanol to the minicolumn and free dye was eliminated by washing the column with 100% ethanol. This procedure removed most of the free dye and oligonucleotides shorter than 5 bases (45). The resulting samples were hybridized on microarrays containing 20-mer oligonucleotide probes and, after a standard washing procedure, were visualized (Fig. 3) with both a stationary fluorescence microscope and the portable device (Fig. 4). When signals are to be measured during hybridization (e.g., in kinetics experiments), the trace amounts of unbound dye and labeled fragments shorter than 15 to 20 bases may be removed to further minimize background by using a Sepharose Q minicolumn instead of ethanol precipitation and washing (see Materials and Methods). This step requires only 5 min.
FIG. 4.
Photograph and schematic of the portable imager.
Hybridization and visualization of hybridization results.
Labeled nucleic acids were eluted from the silica minicolumn with low-ionic-strength buffer. The fragmented and labeled 16S rRNA (1 to 3.5 μg in 30 μl of hybridization buffer) was applied to a microarray of immobilized 20-mer oligonucleotide probes for recognition of “life” in general, all eubacteria, and microorganisms that belong to the B. subtilis group, the B. cereus group, and B. subtilis spp. (Fig. 3A).
To provide for hybridization of labeled nucleic acids at room temperature, we developed a GuSCN-based hybridization buffer. GuSCN destabilizes nucleic acid duplexes and increases hybridization rates (52, 55). In our hands, unambiguous diagnostic hybridization patterns on the microarray could be detected within 20 min of hybridization (Fig. 3).
After hybridization, the microchip was washed and then analyzed using either a stationary wide-field fluorescence microscope coupled with a cooled charge-coupled device (CCD) camera (2, 61) or the portable microchip imager. Exposure times of 2 to 30 s produced clear images on the Polaroid film (Fig. 3C). The sample patterns and intensities obtained with the portable device (Fig. 3C) were very similar to the images obtained with the stationary fluorescence microscope (Fig. 3B). Labeled nucleic acids from Escherichia coli, B. subtilis, B. thuringiensis, and human HL60 leukemia cells produced hybridization patterns characteristic for each organism (Fig. 3D to 3G). We carried out our experiments with two to four repeats, and the most common data are shown in Fig. 3. Hybridization experiments were performed several times both on the same and on the different microchips, and similar results were obtained in both cases (data not shown).
The portable Polaroid microchip analyzer allows qualitative determination of microorganisms in collected samples. For fast and simple detection of targeted microorganisms and approximate estimation of their amounts in the sample, the portable microchip analyzer should be provided with standard images obtained from a chip after hybridization with nucleic acids obtained from a known number of analyzed bacterial cells and photographed with a fixed exposure time.
The design of the analyzer allows a lens adapter to be attached and coupled with 35-mm film or a CCD camera. Polaroid or 35-mm negative films can be scanned to obtain 8-bit digital images; a CCD camera allows images to be obtained with a larger dynamic range and provides a quantitative estimation of obtained images. The analyzer with a CCD camera tested successfully for identification of drug-resistant strains of Mycobacterium tuberculosis (Y. Barsky et. al., unpublished data).
DISCUSSION
The main goal of this work was to develop a rapid and inexpensive procedure for analysis of different microorganisms using biological microchip technology. One of the bottlenecks in the use of biological microchips for nucleic acid analyses is sample preparation time (32, 44). A number of standard biochemical procedures such as cell fractionation and lysis (9), chromatography (43), electrophoresis (6, 28, 43), sample concentration (28), PCR (29), DNA ligation and phosphorylation (15), thermodynamic analysis of hybridization (17), immunoassay (34, 35), and single-base extension analysis (15) are already performed routinely on microchips. Moreover, some current microchips combine microarrays and biological microlaboratories in the same device (6, 9, 28, 29, 35, 43, 44, 46, 50). Therefore, we have sought to develop our procedures with future miniaturization and automation in mind.
rRNA is a “universal chronometric cellular molecule” (59). Up to 80% of bacterial RNA is rRNA. One cell of E. coli can contain about 20,000 copies of rRNA. Therefore, rRNA analysis is a common, rather sensitive, and relatively simple method of bacterial identification (59). The use of microarrays in microbial identification has been demonstrated (21, 24). We recently utilized oligonucleotide probes to rRNA to develop a microarray that is able to differentiate very closely related microorganisms within the B. cereus group, i.e., organisms whose 16S rRNAs differ from each other in only one nucleotide (unpublished data). In the present study we demonstrate the potential of our new multicomponent system by using a simple 16S rRNA microarray containing 20-mer probes (Fig. 3A). This limited microarray should not be considered a final device for identification of bacterial groups or species but only a tool for demonstration of perfect work by the three-component system for bacteria identification as a whole.
GuSCN is known to be powerful lysing agent for many types of cell and also an inactivator of various nucleases (4, 10, 11, 38, 45). Nucleic acids bind to silica in the presence of high concentrations of salt (3, 4). To create a syringe-operated minicolumn for nucleic acid purification and fractionation, we modified the previously developed batch protocols (3, 4) by simplifying the procedure and making it more rapid. To eliminate all centrifugation steps, we used a syringe-operated column format. As a result, our protocol requires only two buffers, and it is possible to isolate total nucleic acids or fractionate DNA and RNA from gram-negative bacteria in 3 to 5 min (Fig. 1) instead of the previously described 40- to 60-min procedure requiring four buffers (3, 4).
Free radical oxidants are well-known tools for the modification of DNA and RNA (7). Redox-active coordination complexes such as (OP)2Cu and Fe · EDTA, are commonly used as “chemical nucleases” to introduce single-strand breaks in nucleic acids (36, 39, 51). Treatment of DNA or RNA with (OP)2Cu results in abstraction of a hydrogen atom from the sugar moiety, producing a carbon-based radical that can rearrange to an abasic site as a result of deglycosylation followed by fragmentation of the nucleic acid (39). Aldehydes and lactones formed at the site of scission may be used for conjugation of amino derivatives with the nucleic acid fragments (19, 42). We recently used this idea to create a new method for sequence-independent fragmentation and fluorescent labeling of nucleic acids with (OP)2Cu and Fe · EDTA complexes (unpublished data). We utilize (OP)2Cu chemistry for sample preparation on the silica minicolumn. Here we demonstrated labeling and fragmentation of total nucleic acids for cell identification (Fig. 3). The (OP)2Cu silica minicolumn method may also be used for labeling and fragmentation of pure RNA and DNA (data not shown). We successfully recruited this method for on-microchip identification of whole (about 1,550 bases in length) B. subtilis 16S rRNA, utilizing the same experimental conditions. However, it was necessary to change the concentration of hydrogen peroxide or (OP)2Cu complex in the labeling cocktail considerably (see Materials and Methods) for identification of 300-base RNA fragments and PCR-amplified double-stranded DNA of 16S rRNA genes of B. cereus group bacteria.
The most popular methods for nucleic acid labeling are currently based on time-consuming enzymatic procedures such as those involving reverse transcriptases (13, 48, 56, 57, 60), terminal transferases (23, 58), kinases (58), random priming (22, 27), or PCR (8, 12, 20, 21, 26, 30, 31, 47, 54). Most of these protocols also demand careful nucleic acid purification, separate sample fragmentation procedures (which considerably improve the specificity of hybridization), and a final precipitation or gel filtration step to eliminate excess label. As a result, sample isolation and fractionation steps (generally 1 h or more) usually precede separate labeling-fragmentation-purification routines, which adds 2 to 3 h. Recently developed chemical labeling methods also require a considerable time to perform (more than 3 h) (14, 40). Our entire minicolumn procedure, from cell lysis to removal of excess fluorescent label, can be executed within 20 to 30 min.
Instrumentation required for the detection and identification of fluorescent hybridization signals represents one of the most expensive aspects of microarray technology. Our stationary laboratory fluorescent microscope was assembled at a price of about $60,000, while the market cost of a laser scanner is generally $40,000-$110,000 (5). In contrast, the projected cost of our laser diode/Polaroid film-based portable imager is considerably lower (about $2,000). The wide-field, high-aperture, long-working-distance objective provides the ability to analyze 180 individual probes simultaneously, which is enough to permit the design of arrays specific for many different microorganisms. Coupling of our portable analyzer with a CCD camera and PC provides the possibility of quantifying image analysis while not substantially increasing its price; however, this converts the system to a stationary device.
We think that our portable multicomponent system can be successfully used under laboratory or field conditions for rapid microbial (or eukaryote) identification in medical, agricultural, or environmental applications.
ACKNOWLEDGMENTS
This work was supported by the Defense Advanced Research Project Agency under Interagency Agreement AO-E428 and by the Russian Human Genome Program grant 5/2000.
We express our gratitude to John Kelly, Isaac Barsky, and Lev Agroskin for many helpful consultations. We are also grateful to Yuri Lysov for selection of probe sequences and to Gennadiy Yershov and Anne Gemmell for chip manufacture.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barsky Y, Grammatin A, Ivanov A, Kreindlin E, Kotova E, Barskii V, Mirzabekov A. Wide-field luminescence microscopes for analyzing biological microchips. J Opt Technol. 1998;65:938–941. [Google Scholar]
- 3.Beld M, Sol C, Goudsmit J, Boom R. Fractionation of nucleic acids into single-stranded and double-stranded forms. Nucleic Acids Res. 1996;24:2618–2619. doi: 10.1093/nar/24.13.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowtell D D L. Options available—from start to finish—for obtaining expression data by microarray. Nat Genet. 1999;21:25–32. doi: 10.1038/4455. [DOI] [PubMed] [Google Scholar]
- 6.Burns M A, Johnson B N, Brahmasandra S N, Handique K, Webster J R, Krishnan M, Sammarco T S, Man P M, Jones D, Heldsinger D, Mastrangelo C H, Burke D T. An integrated nanoliter DNA analysis device. Science. 1998;282:484–487. doi: 10.1126/science.282.5388.484. [DOI] [PubMed] [Google Scholar]
- 7.Burrows C J, Muller J G. Oxidative nucleobase modifications leading to strand scission. Chem Rev. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 8.Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P A. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 9.Cheng J, Sheldon E L, Wu L, Uribe A, Gerrue L O, Carrino J, Heller M J, O'Connell J P. Preparation and hybridization analysis of DNA/RNA from E. coli on microfabricated bioelectronic chips. Nat Biotechnol. 1998;16:541–546. doi: 10.1038/nbt0698-541. [DOI] [PubMed] [Google Scholar]
- 10.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;24:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 11.Ciulla T A, Sklar R M, Hauser S L. A simple method for DNA purification from peripheral blood. Anal Biochem. 1988;174:485–488. doi: 10.1016/0003-2697(88)90047-4. [DOI] [PubMed] [Google Scholar]
- 12.Cronin M T, Fucini R V, Kim S M, Masino R S, Wespi R M, Miyada C G. Cystic fibrosis mutation detection by hybridization to light-generated DNA probe arrays. Hum Mutat. 1996;7:244–255. doi: 10.1002/(SICI)1098-1004(1996)7:3<244::AID-HUMU9>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 13.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 14.de Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 15.Dubilei S, Kirillov E, Lysov Y, Mirzabekov A. Fractionation, phosphorylation and ligation on oligonucleotide microchip to enhance sequencing by hybridization. Nucleic Acids Res. 1997;25:2259–2265. doi: 10.1093/nar/25.12.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezzell J W, Jr, Abshire T G, Little S F, Lidgerding B C, Brown C. Identification of Bacillus anthracis by using monoclonal antibody to cell wall galactose-N-acetylglucosamine polysaccharide. J Clin Microbiol. 1990;28:223–231. doi: 10.1128/jcm.28.2.223-231.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fotin A V, Drobyshev A L, Proudnikov D Y, Perov A N, Mirzabekov A D. Parallel thermodynamic analysis of duplex on oligodeoxyribonucleotide microchips. Nucleic Acids Res. 1998;26:1515–1521. doi: 10.1093/nar/26.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox A J, Gilbart J, Morgan S. Analytical microbiology: a perspective. In: Fox A, Larsson L, Morgan S, Odham G, editors. Analytical microbiology methods: chromatography and mass spectrometry. New York, N.Y: Plenum Press; 1990. pp. 1–17. [Google Scholar]
- 19.Gavin I M, Melnik S M, Yurina N P, Khabarova M I, Bavykin S G. Zero-length protein-nucleic acid crosslinking by radical-generating coordination complexes as a probe for analysis of protein-DNA interactions in vitro and in vivo. Anal Biochem. 1998;263:26–30. doi: 10.1006/abio.1998.2827. [DOI] [PubMed] [Google Scholar]
- 20.Gilles P N, Wu D J, Foster C B, Dillon P J, Chanock S J. Single nucleotide polymorphic discrimination by an electronic dot blot assay on semiconductor microchips. Nat Biotechnol. 1999;17:365–370. doi: 10.1038/7921. [DOI] [PubMed] [Google Scholar]
- 21.Gingeras T R, Ghandour G, Wang E, Berno A, Small P M, Drobniewski F, Alland D, Desmond E, Holodniy M, Drenkow J. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 22.Guiliano D, Ganatra M, Ware J, Parrot J, Daub J, Moran L, Brennecke H, Foster J M, Supali T, Blaxter M, Scott A L, Williams S A, Slatko B E. Chemiluminescent detection of sequential DNA hybridizations to high-density, filter-arrayed cDNA libraries: a subtraction method for novel gene discovery. BioTechniques. 1999;27:146–152. doi: 10.2144/99271rr03. [DOI] [PubMed] [Google Scholar]
- 23.Gunderson K L, Huang X C, Morris M S, Lipshutz R J, Lockhart D J, Chee M. Mutation detection by ligation to complete n-mer DNA arrays. Genome Res. 1998;8:1142–1153. doi: 10.1101/gr.8.11.1142. [DOI] [PubMed] [Google Scholar]
- 24.Guschin D Y, Mobarry B K, Proudnikov D, Stahl D A, Rittmann B E, Mirzabekov A D. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl Environ Microbiol. 1997;63:2397–2402. doi: 10.1128/aem.63.6.2397-2402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guschin D, Yershov G, Zaslavsky A, Gemmell A, Shick V, Proudnikov D, Arenkov P, Mirzabekov A. Manual manufacturing of oligonucleotide, DNA, and protein microchips. Anal Biochem. 1997;250:203–211. doi: 10.1006/abio.1997.2209. [DOI] [PubMed] [Google Scholar]
- 26.Hacia J G, Lawrence C B, Chee M S, Fodor S P A, Collins F S. Detection of heterozygous mutations in BRCAI using high density oligonucleotide arrays and two-color fluorescence analysis. Nat Genet. 1996;14:441–447. doi: 10.1038/ng1296-441. [DOI] [PubMed] [Google Scholar]
- 27.Hermanson G T. Bioconjugate techniques. San Diego, Calif: Academic Press, Inc.; 1996. [Google Scholar]
- 28.Khandurian J, Jacobson S C, Waters L C, Foote R S, Ramsey J M. Microfabricated porous membrane structure for sample concentration and electrophoretic analysis. Anal Chem. 1999;71:1815–1819. doi: 10.1021/ac981161c. [DOI] [PubMed] [Google Scholar]
- 29.Kopp M U, de Mello A J, Manz A. Chemical amplification: continuous-flow PCR on a chip. Science. 1998;280:1046–1048. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- 30.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 31.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 32.Marshall A, Hodgson J. DNA chips: an array of possibilities. Nat Biotechnol. 1998;16:27–31. doi: 10.1038/nbt0198-27. [DOI] [PubMed] [Google Scholar]
- 33.Martin G P, Timmers E. PCR and its modifications for the detection of infectious diseases. In: Lee H, Morse S, Olsvik O, editors. Nucleic acid amplification technologies: application to disease diagnosis. Natick, Mass: Eaton Publishing; 1997. pp. 79–99. [Google Scholar]
- 34.Mendoza L G, McQuary P, Mongan A, Gangadharan R, Brignac S, Eggers M. High-throughput microarray-based enzyme-linked immunosorbent assay (ELISA) BioTechniques. 1999;27:778–788. doi: 10.2144/99274rr01. [DOI] [PubMed] [Google Scholar]
- 35.Ouellette J. Biosensors: microelectronics marries biology. Ind Physicist. 1998;4:11–14. [Google Scholar]
- 36.Papavassiliou A G. Chemical nucleses as probes for studying DNA-protein interactions. Biochem J. 1995;305:345–357. doi: 10.1042/bj3050345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phimister B, editor. The chipping forecast. Nat Genet. 1999;21(Suppl.):1–60. [Google Scholar]
- 38.Pitcher D G, Saunders N A, Owens R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 39.Pogozelski W K, Tullius T D. Oxidative strand scission of nucleic acids: routes initiated by hydrogen abstraction from the sugar moiety. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 40.Proudnikov D, Mirzabekov A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 1996;24:4535–4532. doi: 10.1093/nar/24.22.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Proudnikov D, Timofeev E, Mirzabekov A. Immobilization of DNA in polyacrylamide gel for the manufacture of DNA and DNA-oligonucleotide microchips. Anal Biochem. 1998;259:34–41. doi: 10.1006/abio.1998.2620. [DOI] [PubMed] [Google Scholar]
- 42.Pruss D, Gavin I M, Melnik S, Bavykin S. DNA-protein cross-linking applications for chromatin studies in vitro and in vivo. Methods Enzymol. 1999;304:516–533. doi: 10.1016/s0076-6879(99)04030-6. [DOI] [PubMed] [Google Scholar]
- 43.Regnier F E, He B, Lin S, Busse J. Chromatography and electrophoresis on chips: critical elements of future integrated, microfluidic analytical systems for life science. Trends Biotechnol. 1999;17:101–106. doi: 10.1016/s0167-7799(98)01294-3. [DOI] [PubMed] [Google Scholar]
- 44.Robertson D. Chips advance but cost constraints remain. Nat Biotechnol. 1998;16:509. [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Santini J T, Jr, Cima M J, Langer R. A controlled-release microchip. Nature. 1999;397:335–338. doi: 10.1038/16898. [DOI] [PubMed] [Google Scholar]
- 47.Sapolsky R J, Lipshutz R J. Mapping genomic library clones using oligonucleotide arrays. Genomics. 1996;33:445–456. doi: 10.1006/geno.1996.0219. [DOI] [PubMed] [Google Scholar]
- 48.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 49.Semizarov D, Glesne D, Laouar A, Schiebel K, Huberman E. A lineage-specific protein kinase crucial for myeloid maturation. Proc Natl Acad Sci USA. 1998;95:15412–15417. doi: 10.1073/pnas.95.26.15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Service R F. Microchip arrays put DNA on the spot. Science. 1998;282:396–399. doi: 10.1126/science.282.5388.396. [DOI] [PubMed] [Google Scholar]
- 51.Sigman D S. Chemical nucleases. Biochemistry. 1990;29:9097–9105. doi: 10.1021/bi00491a001. [DOI] [PubMed] [Google Scholar]
- 52.Thompson J, Gillespie D. Molecular hybridization with RNA probes in concentrated solutions of guanidine thiocyanate. Anal Biochem. 1987;163:281–291. doi: 10.1016/0003-2697(87)90225-9. [DOI] [PubMed] [Google Scholar]
- 53.Timofeev E, Kochetkova S, Mirzabekov A, Florentiev V. Regioselective immobilization of short oligonucleotides to acrylic copolymer gels. Nucleic Acids Res. 1996;24:3142–3148. doi: 10.1093/nar/24.16.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyagi S, Bratu D P, Kramer F R. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 55.Van Ness J, Chen L. The use of oligodeoxynucleotide probes in chaotrope-based hybridization solutions. Nucleic Acids Res. 1991;19:5143–5151. doi: 10.1093/nar/19.19.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang K, Gan L, Jeffery E, Gayle M, Gown A M, Skelly M, Nelson P S, Ng W V, Schummer M, Hood L, Mulligan J. Monitoring gene expression profile changes in ovarian carcinomas using cDNA microarray. Gene. 1999;229:101–108. doi: 10.1016/s0378-1119(99)00035-9. [DOI] [PubMed] [Google Scholar]
- 57.Wilson M, DeRisi J, Kristensen H, Imboden P, Rane S, Brown P O, Schoolnik G K. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wodicka L, Dong H, Mittmann M, Ho M, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 59.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang G P, Ross D R, Kuang W W, Brown P O, Weigel R J. Combining SSH and cDNA microarrays for rapid identification of differentially expressed genes. Nucleic Acids Res. 1999;27:1517–1523. doi: 10.1093/nar/27.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yershov G, Barsky V, Belgovskiy A, Kirillov E, Kreindlin E, Ivanov I, Parinov S, Guschin D, Drobishev A, Dubiley S, Mirzabekov A. DNA analysis and diagnostics on oligonucleotide microchips. Proc Natl Acad Sci USA. 1996;93:4915–4918. doi: 10.1073/pnas.93.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]