Abstract
The bacteriophage resistance plasmid pAH90 (26,490 bp) is a natural cointegrate plasmid formed via homologous recombination between the type I restriction-modification specificity determinants (hsdS) of two smaller lactococcal plasmids, pAH33 (6,159 bp) and pAH82 (20,331 bp), giving rise to a bacteriophage-insensitive mutant following phage challenge (D. O'Sullivan, D. P. Twomey, A. Coffey, C. Hill, G. F. Fitzgerald, and R. P. Ross, Mol. Microbiol. 36:866–876; 2000). In this communication we provide evidence that the recombination event is favored by phage infection. The entire nucleotide sequence of plasmid pAH90 was determined and found to contain 24 open reading frames (ORFs) responsible for phenotypes which include restriction-modification, phage adsorption inhibition, plasmid replication, cadmium resistance, cobalt transport, and conjugative mobilization. The cadmium resistance property, encoded by the cadA gene, which has an associated regulatory gene (cadC), is of particular interest, as it facilitated the selection of pAH90 in other phage-sensitive lactococci after electroporation. In addition, we report the identification of a group II self-splicing intron bounded by two exons which have the capacity to encode a relaxase implicated in conjugation in gram-positive bacteria. The functionality of this intron was evident by demonstrating splicing in vivo. Given that pAH90 encodes potent phage defense systems which act at different stages in the phage lytic cycle, the linkage of these with a food-grade selectable marker on a replicon that can be mobilized among lactococci has significant potential for natural strain improvement for industrial dairy fermentations which are susceptible to phage inhibition.
Lactococcus lactis is widely used as a starter bacterium for the manufacture of Cheddar cheese and other fermented products by the dairy industry (9) and for that reason has attracted intensive research over the last 20 years (1, 4, 27, 70). Lactococci normally contain a rich diversity of plasmids, many of which are responsible for key industrial traits including lactose catabolism, proteinase production, polysaccharide production, bacteriocin production and immunity, and bacteriophage resistance (11, 19, 20, 21, 28, 39). Consequently, plasmid mobilization has formed the backbone of numerous strain enhancement regimens to facilitate the transfer of desirable properties into specific bacterial strains that can be employed in fermentations as starter cultures. The conjugal transfer of phage resistance plasmids to phage-sensitive dairy starters has been the focus of considerable research attention (7, 23, 33, 47, 51, 56). These strategies are aimed at protecting lactococcal strains from the ever-present threat of phage infection during industrial dairy fermentations (16, 33, 44).
Natural phage defense mechanisms such as adsorption inhibition (Ads), injection blocking, restriction/modification (R/M), and abortive infection (Abi) systems are widespread in lactococci and can coexist and complement each other in cheese-making strains, helping prevent the infection and proliferation of phage (13, 47). However, there are limits to the extent to which natural (food-grade) strategies can be used to introduce and artificially stack phage resistance mechanisms within strains. This is principally because of the paucity of readily selectable markers for phage resistance plasmids, leading to difficulties in recognizing those genuine phage-resistant transconjugants which may arise (47). Antibiotic resistance has been rejected as a selectable marker for microorganisms destined for use in food production because of the risk of promoting antibiotic resistance in human intestinal microflora. Hence, the identification of alternative selectable markers (e.g., bacteriocin immunity and heavy metal resistance) that can be considered safe for use in food has received considerable interest recently (7, 18, 23, 36, 47, 50, 51).
Plasmid pAH90 was originally identified in L. lactis subsp. lactis biovar diacetylactis DPC721 (24), a spontaneous bacteriophage-insensitive mutant of L. lactis DPC220, a strain specifically used in the manufacture of cultured butter. The bacteriophage insensitivity phenotype was found to be associated with the loss of two native plasmids (pAH33 and pAH82) in the DPC220 parent and the acquisition of a novel plasmid (pAH90) in the insensitive derivative DPC721. Plasmid pAH90 was demonstrated to be a natural cointegrate plasmid formed by homologous recombination between the two smaller lactococcal plasmids (24). The exact nature of the cointegration event has been determined following sequence analysis of the cointegration site. This site was located within two type I R/M specificity determinants (hsdS), one of which was linked to R/M determinants on pAH82, the other of which was located on pAH33. Homologous recombination between a conserved region of the hsdS determinants on these plasmids facilitated the formation of pAH90 and the evolution of two novel HsdS subunits. The novel HsdS chimeras formed had interchanged the C- and N-terminal variable domains of the parent plasmids, generating two new R/M specificities (49).
This report concerns the detailed genetic and functional characterization of the entire phage resistance plasmid pAH90. Given that the plasmid fortuitously contains the genetic determinants for mobility and cadmium resistance, it can be easily transferred between lactococcal starters in a food-grade manner and is likely to be a valuable asset for the genetic enhancement of starter cultures. Interestingly, pAH90 also contains a group II self-splicing intron of the type found in the chromosomally located sex factor of L. lactis subsp. cremoris 712 (61) and in the conjugative transfer region of the lactococcal plasmid pRS01 (41).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
All bacterial strains and plasmids used in this study are listed in Table 1. Lactococcal strains were routinely propagated at 30°C in M17 medium (Difco Laboratories, Detroit, Mich.) supplemented with 0.5% (wt/vol) glucose or lactose (G/LM17). Solid medium contained 1.0% agar (Oxoid, Basingstokes, Hampshire, United Kingdom). Selective medium contained cadmium chloride (Sigma Chemical Company, Poole, Dorset, United Kingdom) at a final concentration of 0.1 mM. All strains were stocked in G/LM17 containing 40% (vol/vol) glycerol and stored at −80°C. Working cultures were stored at 4°C and transferred periodically. All strains are held in the culture collection at Moorepark.
TABLE 1.
Bacterial strains, plasmids, and bacteriophages
| Strain, phage, or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| L. lactis MG1614 | Plasmid-free derivative of L. lactis subsp. cremoris 712 | 21 |
| L. lactis DPC3333 | MG1614 containing pAH33 | 24 |
| L. lactis DPC3343 | MG1614 containing pAH82 | 24 |
| L. lactis DPC5348 | MG1614 containing pAH82 and pAH33 | 49 |
| L. lactis DPC3290 | MG1614 containing pAH90 | 24 |
| L. lactis DRC3 | Standard size reference strain | 39 |
| Bacteriophages | ||
| D1 | Industrial phage isolate | 24 |
| D2 | Industrial phage isolate | 23 |
| Plasmids | ||
| pAH33 | r/m33 | 24 |
| pAH82 | r/m82, cadR | 24 |
| pAH90 | r/m90, cadR, adsR | 24 |
Growth inhibition assays.
The sensitivity of L. lactis MG1614 and its plasmid-containing derivatives to both CdCl2 and CoCl2 (Sigma) was assessed as follows. Exponentially growing cultures were used to inoculate 10 ml of GM17 broth containing CdCl2 (0 to 0.3 mM) or CoCl2 (0 to 6.0 mM). These were then incubated for 16 h at 30°C, after which time growth was assessed by measuring absorbance at 600 nm.
Generation and characterization of BIMs.
Bacteriophage-insensitive mutants (BIMs) of L. lactis DPC220 were generated using a plaque assay technique (64) with strain DPC220 as the host. This strain was challenged with two bacteriophages isolated from a dairy plant where DPC220 had been used industrially. Each bacteriophage was propagated individually on DPC220, and a cocktail (1:1) of the two bacteriophage (each at a titer of >109 PFU/ml) was used to challenge DPC220. Phage-resistant mutant colonies which appeared on plates having received 105 PFU or greater were propagated individually overnight in 10 ml of GM17. Total DNA was extracted from fully grown cultures as described previously (48). Oligonucleotide primers (5′-CCTGTGTCTACAAATTCATT-3′ and 5′-TGAAATTTTCTACACTGCCT-3′) for PCR were designed to one of the hybrid hsdS determinants (orf17) of the cointegrate plasmid pAH90. These were designed to amplify a 1.03-kb intergenic product, which is only amplified when a cointegrate form is present. Plasmid profile analysis of BIMs was used to confirm that the two smaller plasmids in L. lactis DPC220 had formed a 26-kb cointegrate plasmid. In order to confirm that all BIMs were derived from the parent strain, oligonucleotide primers (5′-CGTCGATGCAGCTAATGCTG-3′ and 5′-ACAGTTAGATGATGAAGTG-3′) for PCR were designed to amplify a 1.06-kb intergenic portion of the cadmium resistance gene present on both pAH82 and pAH90. The frequency of BIM formation was expressed as the number of BIM colonies per parent cell challenged with phage.
DNA preparation.
Lactococcal plasmid DNA was isolated using the lysis method of Anderson and McKay (3). Plasmid preparations were further purified by two rounds of cesium chloride-ethidium bromide density gradient ultracentrifugation followed by dialysis in a 10 mM Tris acetate–10 mM EDTA solution. Restriction enzymes used were purchased from New England Biolabs (Boston, Mass.) and used according to the manufacturer's instructions.
DNA sequencing.
Plasmids were sequenced using the LI-COR 4200L automated DNA sequencer and dye primer chemistry using a cycle sequencing protocol (MWG-Biotech, Milton Keynes, United Kingdom). Oligonucleotide primers were synthesized using standard phosphoramidite chemistry, and oligonucleotides were purified by the purified salt-free procedure or the highly purified salt-free procedure.
Sequence analysis.
Open reading frames (ORFs) were identified initially using DNAStar output (DNAStar, Madison, Wis.), translated, and searched against the nonredundant protein database using the Blast algorithm (2). Sequence alignments were performed using the Clustal method (26) of the Megalign program of the DNAStar software package.
Electroporation and transformation.
L. lactis MG1614 and DPC3333 were made competent and electrotransformed using a Bio-Rad gene pulser apparatus (Bio-Rad Corp., Richmond, Calif.) as described previously (49).
RT-PCR.
For reverse transcriptase-PCR (RT-PCR), total RNA was isolated from lactococci using the method of Keilhauer et al. (30) with several modifications. A 500-ml amount of culture was harvested at mid-log phase (optical density at 600 nm [OD600] = 0.5) by centrifugation. The resultant pellet was resuspended in 5 ml of sterile distilled H2O containing lysozyme (10 mg/ml) and incubated on ice for 30 min. Five milliliters of a 10 mM sodium acetate (pH 5.0)–1% sodium dodecyl sulfate (SDS) solution (65°C) was added, and samples were incubated for 10 min at 65°C. Ten milliliters of water-saturated phenol (pH 5.5) was mixed with the lysate, which was then incubated for a further 10 min at 65°C, after which the aqueous phase was separated by centrifugation (15 min at 14,000 rpm). The phenol extraction was repeated twice, after which cellular RNA was precipitated with absolute ethanol (−20°C), washed with 70% ethanol, dried, and resuspended in 20 μl of diethylpyrocarbonate-treated water containing 80 U of RNasin (Promega Corp., Madison, Wis.). The RNA isolate was then treated with RNase-free DNase I (Gibco-BRL Life Technologies Ltd.) in 1× DNase buffer at room temperature for 15 min. The DNase was inactivated by the addition of 20 mM EDTA, and the mixture was heated at 65°C for 10 min. Reverse transcription was carried out with random hexamers using the Superscript preamplification system (Gibco-BRL Life Technologies Ltd.) both with and without RT. The cDNA produced was used in PCRs which typically employed Taq polymerase with 0.1 μmol of primers specific to the mobDEI and mobDEII exons (intX [5′-CGA CAC GAA CAA TGG CTA TCG CTT-3′] and intY [5′-GTC TTG TGT AAG TCT GTT CGT TCC T -3′] , respectively) and ∼60 ng of DNA template. PCR was performed using a Hybaid PCR express unit (Hybaid Ltd., Middlesex, United Kingdom). Ten microliters of the PCR/RT-PCR was electrophoresed on a 1.0% (wt/vol) agarose gel using 1× TAE buffer (40 mM Tris acetate, 1 mM EDTA [pH 8.0]) containing ethidium bromide (200 ng/ml). Marker X (Roche Diagnostics Ltd., East Sussex, United Kingdom) was employed as a molecular size standard.
RESULTS AND DISCUSSION
Following discovery that the cointegrate plasmid pAH90 encodes two distinct phage resistance mechanisms (24, 47, 49), complete nucleotide sequence analysis revealed that this plasmid combines a number of desirable traits and may form the basis for future strain improvement strategies.
Structural analysis of pAH90.
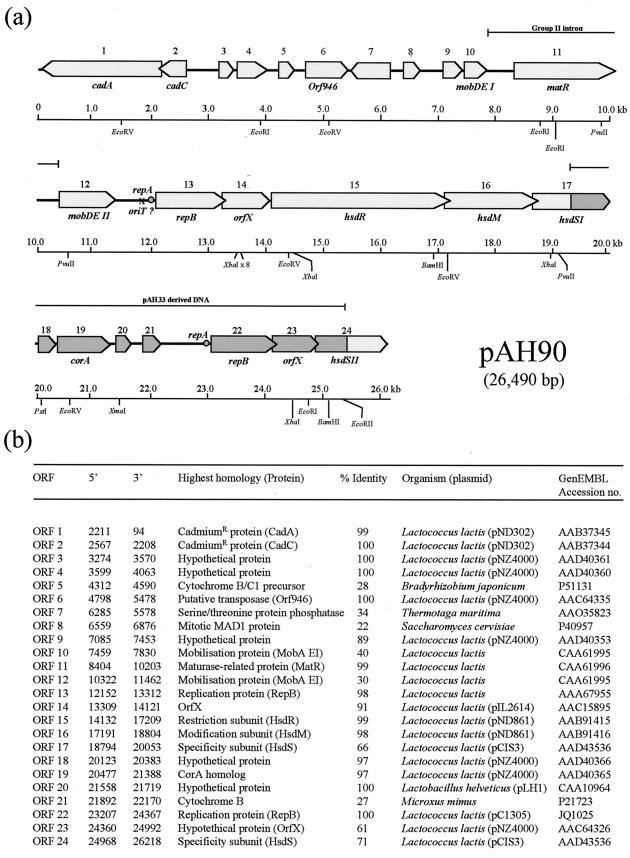
The complete sequence of plasmid pAH82 (GenBank accession no. AF243383) was determined and found to comprise 20,331 bp. Given that the entire nucleotide sequence of pAH33 (6,159 bp; GenBank no. AF207855) and the exact nature of the cointegration event had been previously elucidated (49), the complete sequence of pAH90 (26,490 bp) was then compiled and a detailed map was constructed (Fig. 1). The average G + C content of pAH90 is 34.8%, which is in agreement with the G + C content (35.4%) of the recently presented complete genome sequence of L. lactis IL 1403 (4). Comparisons with DNA databases revealed that certain regions of the plasmid were almost identical to sequences on other lactococcal plasmids. For example, bp 90 to 2700 show 99% identity with lactococcal plasmid pND302 (37), while bp 3240 to 4063 have 99% identity with L. lactis plasmid pNZ4000 (67). Such similarities between plasmid sequences indicates a high level of horizontal gene transfer between lactococci and an overall plasticity in plasmid structures. pAH90 contains 24 ORFs (Fig. 1a), 16 of which are derived from pAH82 and 6 from pAH33, with two hybrid hsdS ORFs formed at the cointegrate junctions which alter R/M specificity. All ORFs identified were analyzed for similarity to known protein sequences, and proteins with the greatest identity are indicated in Fig. 1b. A number of functional regions were identified on pAH90, including phage resistance (49), cadmium resistance (which can be exploited as a selectable marker), conjugative mobilization, and replication functions.
FIG. 1.
(a) ORFs identified in the 26,490-bp cointegrate plasmid pAH90. Shaded region corresponds to DNA derived from pAH33, and nonshaded region corresponds to DNA derived from pAH82. Putative repA and oriT regions are indicated. A selection of relevant restriction sites are also indicated. (b) The columns list ORF number, 5′ end of the ORF, 3′ end of the ORF, identification of similar protein and organism, and percent identity of the pAH90 ORF to the homologous protein.
cadA/C.
Although some metal ions are essential in low concentrations for cellular metabolism (e.g., iron, copper, and zinc), other metals, including cadmium, mercury, and lead, do not play any known physiological role and are in fact toxic for cells (55). Cadmium is extremely toxic even in low concentrations and has been shown to induce DNA breakage (42, 54, 66). Bacteria have evolved strategies to cope with toxic metals in the environment, and bacterial operons that confer resistance to cadmium, mercury, copper, and arsenic have been described (45, 72). Many of these metal resistance operons are encoded on transposons and plasmids (63). It was inferred that pAH90 had the capacity to encode resistance to cadmium, as the deduced proteins for orf1 (705 amino acids) and orf2 (119 amino acids) show high identity to CadA and CadC proteins, respectively, from the L. lactis plasmid pND302 (37) and cadmium resistance systems from several other gram-positive bacteria (10, 29, 35, 46). CadA is a member of the superfamily of cation-translocating P-type ATPases, which allow efflux of cadmium, resulting in reduced accumulation of the toxic cation. In addition, CadA has recently been shown to confer resistance against other cations such as lead and zinc (52). CadC encodes a small protein, belonging to the ArsR family of transcriptional repressors, which acts as a transcriptional regulator (14) and is responsive to the presence of soft metals in the order Pb>Cd>Zn (52).
repB.
The replication regions of pAH33 and pAH82 exhibited 73% identity and are similar to the replication determinants of several theta-replicating lactococcal plasmids, as first identified for plasmid pCI305 (25). In pAH82, a repB gene (orf13) is capable of encoding a protein (386 amino acids) with high identity to RepB proteins of the lactococcal plasmids pND608 (98%; accession number AAA67955), pIL2614 (98% [58]), pWV02 (79% [32]), and pIL103 (79% [58]). The region upstream of repB has features conserved in pCI305-type theta replicons and is also proposed to contain the origin of replication (ori). These features include a 22-bp sequence directly repeated 3.5 times (bp 11967 to 12043), which have a replicon-specific regulatory role in plasmid replication, and two inverted repeat sequences, IR1 and IR2, which overlap the putative −35 region of the repB promoter and serve as a RepB binding site (15). Similarly, the repB gene (orf22) of pAH33 could encode a protein (386 amino acids) identical to the RepB proteins of lactococcal plasmids pCI305 (25) and pCIS3 (60). The cointegrate pAH90 thus possesses two highly homologous functional theta-type replicons. The existence of multiple replicons on lactococcal plasmids is not unique to pAH90. Van Kranenburg and de Vos (68) identified four highly homologous replicons on the lactococcal plasmid pNZ4000, which were demonstrated to be compatible and to function independently. Since pNZ4000 replication requires only one of the four replicons, it was speculated that pNZ4000 could have derived fragments of several plasmids, which might have formed cointegrates during conjugation processes. In this study, it is evident from sequence analysis that the replication regions encoded by cointegrate pAH90 are derived from the progenitor plasmids pAH33 and pAH82.
orfX.
An ORF designated orfX is located immediately downstream of the repB gene on pAH82 and encodes a putative protein, OrfX (270 amino acids), which has high homology to proteins encoded by lactococcal plasmids pIL2614 (91% identity [57]) and pJW565 (67% identity; GenBank no. CAA73267). Similarly, a determinant showing 32% homology to the orfX gene of pAH82 was identified upstream of the hsdS locus on pAH33. The homology lies in two domains localized at the N- and C-terminal parts of these proteins. Such proteins are typically associated with pCI305-type replicons and are transcriptionally coupled to repB (59). However, OrfX does not participate in plasmid replication, and its function remains unknown (17). OrfX does contain a conserved helix-turn-helix motif that suggests a possible involvement in DNA-protein interactions (15). The location of orfX immediately upstream of a type I R/M system was also observed on plasmid pIL2614 (56). Schouler et al. (56) proposed that type I activity or expression may be linked to replication for pIL2614. In the same way, the gene organization of ORFs 13 to 17 and ORFs 22 to 24 of pAH90 suggests that transcription of type I genes is directed by promoters located upstream of orf13 and orf22, respectively.
Group II intron.
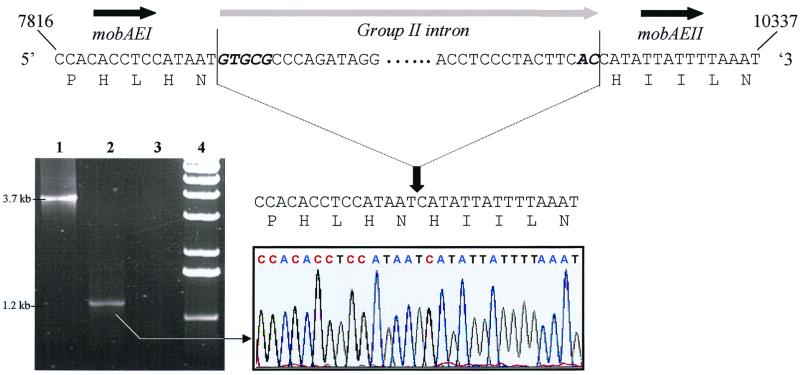
The genetic organization of the region encompassing orf10, orf11, and orf12 (Fig. 1) suggested that this locus was responsible for the conjugative mobilization of pAH82 or pAH90, as it contained a group II self-splicing intron and was bounded by two exons which have the capacity to encode a relaxase implicated in conjugation in gram-positive bacteria. The pAH90 group II intron is 99% identical to an intron which has been identified in the chromosomally located sex factor of L. lactis subsp. cremoris 712 (61) and in the conjugative transfer region of the lactococcal plasmid pRS01 (41). These lactococcal group II introns encode a maturase, designated MatR (61), with homology to both eukaryotic and bacterial intron-encoded maturases (41, 61). As with other group II intron-associated maturases, the pAH90 sequence contains domains I to VII, conserved among reverse transcriptases (43), a C-terminal Zn2+ finger-like region, and domain X, associated with endonuclease activity (22, 62). This sequence architecture is consistent with the mechanism of action of group II introns, which involves a reverse transcriptase that promotes RNA splicing and, in conjunction with the excised intron, forms an endonuclease which mediates intron mobility (34). The splice sites for the pRS01 intron and the one identified in the sex factor of L. lactis 712 were characterized and found to be consistent with the consensus 5′ and 3′ splice site sequences (GUGYG and AY, respectively) observed in eukaryotic group II introns (40, 41, 61). Considering the high similarity observed between this intron and the one located in pAH82/pAH90 and the presence of conserved intron splice sites, the putative pAH90 intron was also indicated to be precisely 2,492 nucleotides in length.
There are only five nucleotide changes between the group II intron already described for L. lactis and the one present in pAH82/pAH90. Four of these changes occur within the matR coding sequence, two of which account for silent mutations. The fifth change occurs within an unpaired bulge loop of domain III. Apart from this change, the lactococcal introns described to date should undergo similar folding and have similar RNA structures. The in vivo splicing of the intron was demonstrated by isolating an RNA sample from L. lactis DPC3290 and subjecting it to RT-PCR with primers specific for the mobDEI and mobDEII exons as described in Materials and Methods. The resulting 1.2-kb product (Fig. 2, lane 2) was approximately 2.5 kb smaller than the product amplified by PCR from pAH90 DNA with the identical primers (Fig. 2, lane 1), and this matched the predicted size of the intron. Direct sequence analysis of the 1.2-kb reverse transcription-PCR fragment revealed an in-frame ligation of the mobDEI and mobDEII reading frames (Fig. 2). Until recently, introns were believed to be relatively rare in bacteria (for a review, see Dunny and McKay [12]), but research over the last 5 years has identified their widespread distribution in the prokaryotic world (38). Where they have been reported in L. lactis, they have been associated with transfer or mobilization functions (41, 61).
FIG. 2.
In vivo splicing of intron RNA. Electrophoretic analysis of the PCR products obtained by employing primers designed specifically to mobDEI and mobDEII exons of plasmid pAH90 with the reverse transcription products from MG1614 containing pAH90 (with and without RT enzyme [lanes 2 and 3, respectively]) and with DNA isolated from the same strain using the same primers (lane 1). Lane 4 contains molecular size standards (Roche). Direct sequence analysis of the 1.2-kb reverse transcription-PCR fragment revealed an in-frame ligation of the mobDEI and mobDEII reading frames (see chromatogram).
Mobilization region.
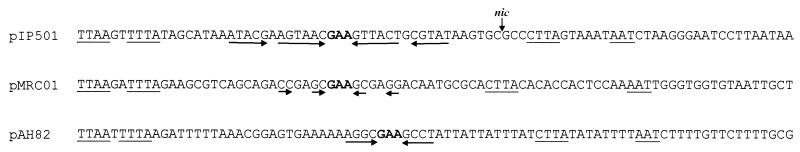
It was previously demonstrated in this laboratory that the conjugative phage resistance plasmid pNP40 was capable of promoting mobilization of pAH82 when it was coresident in the same strain (24). The pAH82/pAH90 intron is bounded by two exons (mobDE1 and mobDE2), which together are capable of encoding a protein of 524 amino acids, which has been designated MobD. Greatest identity (33 and 29%, respectively) was observed with the mobilization and relaxase proteins from L. lactis subsp. cremoris 712 (61) and pRS01 sex factors (41), both of which are associated with group II introns. MobD also showed homology to several gram-positive intron-free mobilization proteins, including 29% identity with the Tn5252-encoded relaxase enzyme in Streptococcus pneumoniae (32) and 22% identity to the RLX protein of Staphylococcus aureus (5). Mobilization and relaxase proteins are known to play an essential role in conjugation and act by introducing a single-stranded nick at the origin of transfer (oriT). The equivalent mobilization proteins in pRS01 and in L. lactis 712 have been shown to play a key role in their respective conjugation processes, and therefore intron splicing is necessary for conjugative transfer. However, deletion of the intron from strains had no effect on conjugation, indicating that RNA splicing is not a rate-limiting step during conjugation (61). While pAH82 is non-self-transmissible, it can be mobilized in the presence of another plasmid (e.g., pNP40 [24]). It is not known whether this mobilization is mediated by a pNP40 relaxase or by the pAH82-encoded MobD. A putative oriT site which could be acted upon by either a cis-acting or trans-acting relaxase, facilitating transfer to a recipient cell, was identified upstream of the pAH82 replication region. This putative oriT (bp 11841 to 11926 in Fig. 3) has some similarities to the oriT of pIP501 and the putative oriT of pMRC01 and contains a 5-nucleotide segment (CGAAG) common to many conjugative plasmids (71).
FIG. 3.
Comparison of the putative oriT of pAH82 with oriT regions of plasmids pIP501 and pMRC01. The pIP501 sequence represents the complement of bp 1229 to 1314, the pMRC01 sequence represents bp 1770 to 1855, and the pAH82 sequence represents bp 11841 to 11926. Inverted repeats are indicated by solid arrows. A consensus GAA sequence central to many of the inverted repeat structures is highlighted in bold. Sequence motifs shared by pIP501, pMRC01, and pAH82 are underlined. Where known, the nic site is indicated by a vertical arrow.
orf946.
A single copy of the insertion sequence element orf946, identical to IS946 of the lactococcal plasmid pNZ4000 (69), was identified on pAH82 (bp 4798 to 5478). The presence of IS946 on lactococcal plasmids is widespread and has previously been reported for pTR2030 (53) and pMRC01 (11). On pAH82, this element (680 bp) is bounded by a 16-bp imperfect inverted repeat at its extremities.
corA.
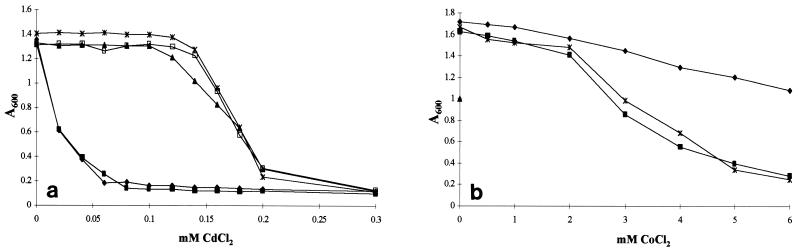
Comparison with databases indicated that orf19 of pAH90 has the capacity to encode a protein (303 amino acids) with high homology to a magnesium transporter with affinity for cobalt. This putative magnesium and cobalt transporter is a member of the CorA family of transport proteins, having homologs across a wide range of prokaryotic micro organisms, including Methanococcus jannaschi (6), Helicobacter pylori (65), and L. lactis (60, 69). The functionality of the corA determinants on pAH33 and pAH90 was demonstrated by evaluating the cobalt tolerance of L. lactis MG1614 and the same strain containing either pAH33 or pAH90 (Fig. 4b). L. lactis MG1614 was able to tolerate CoCl2 concentrations of up to 5 mM. When pAH33 or pAH90 was present in this strain background, its growth was affected by concentrations as low as 0.5 mM CoCl2. Thus, the presence of corA increases the sensitivity of host strains to cobalt.
FIG. 4.
Final OD600 value after 16 h for L. lactis strains MG1614 (⧫), MG1614(pAH33) (■), MG1614(pAH82) (▴), MG1614(pAH33, pAH82) (□), and MG1614(pAH90) (∗) grown on GM17 containing increasing concentrations of cadmium chloride (a) and cobalt chloride (b).
Evolution of pAH90 may be strain or phage specific.
Plasmid pAH90 is a natural cointegrate plasmid formed by homologous recombination between two smaller lactococcal plasmids following phage challenge (24, 49). The recent evolution of pAH90 from the progenitor plasmids pAH33 and pAH82 was explored by determining the frequency of the cointegration event leading to its formation. BIMs of the transformant strain L. lactis DPC5348 (L. lactis MG1614 containing the parent plasmids pAH33 and pAH82 as discrete elements), generated following exposure to phage c2, were assessed for cointegrate formation. Surprisingly, none of 100 phage-insensitive isolates examined was found to possess a 26-kb cointegrate plasmid. Consequently, subsequent investigations were performed with the original strain-phage combination first reported by Harrington and Hill (24).
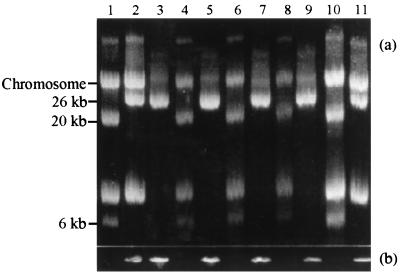
L. lactis DPC220, the original parent strain of plasmids pAH33 and pAH82, was challenged by plaque assay with a blend of two phages. Plaque assays with 1.4 × 108 CFU of strain DPC220 and at least 105 PFU or greater yielded approximately 76 phage-resistant mutant colonies, indicating a frequency of BIM formation of 5.4 × 10−7 per parent cell. When 50 BIM colonies were tested for the presence of a 1.03-kb PCR product specific for the cointegration event, 35 were positive and 15 were negative. Cointegrate formation was confirmed by plasmid analysis of selected BIMs (Fig. 5). Interestingly, the pAH90-containing BIMs thus generated lacked a plasmid band of approximately 8 kb present in L. lactis DPC220 and DPC721. The molecular basis of phage resistance in the 15 negative isolates was unrelated to pAH90 formation. The appearance of a 1.06-kb product for all 50 of the BIMs tested following PCR with primers specific for the cadmium resistance determinant of pAH82 or pAH90 confirmed that all the BIMs were derived from strain DPC220. The frequency of plasmid cointegration was calculated as 3.8 × 10−7 per parent cell. The ability to generate BIMs (due to cointegrate formation) in this manner is significant, but perhaps more intriguing was the inability to demonstrate plasmid cointegration in the MG1614 background. The selective pressure offered by high-titer phage c2 challenge failed to generate cointegrate formation. This suggests that the evolution of pAH90 may be strain or phage specific.
FIG. 5.
(a) Agarose gel electrophoresis of plasmids isolated from DPC220 (lanes 1 and 10), DPC721 (lanes 2 and 11), and seven BIMs of DPC220 origin (lanes 3 to 9) generated by challenge with a cocktail of phages. The positions of plasmids pAH82, pAH33, and pAH90 are indicated. (b) Gel obtained by PCR using primers designed to one of the hybrid hsdS determinants (orf17) of the cointegrate plasmid pAH90. A PCR product of approximately 1.03 kb in length is only possible when a cointegrate form is present. Lanes are the same as in panel a.
Exploiting cadmium resistance as a selectable marker for the transfer of pAH90.
The functionality of the cadmium resistance determinants on pAH82 and pAH90 was demonstrated by evaluating the MIC of cadmium chloride (CdCl2) on plasmid-free L. lactis MG1614 and the same strain containing pAH33, pAH82, or pAH90 (Fig. 4a). Strains MG1614 and MG1614 containing pAH33 were inhibited by 0.05 mM CdCl2, whereas when pAH82 or pAH90 was present, the MG1614 derivative could grow in the presence of CdCl2 at concentrations as high as 0.1 mM. Furthermore, the utility of the cadmium resistance gene as a selectable marker was demonstrated by successfully transforming both pAH90 and pAH82 into MG1614 by electroporation and selecting on GM17 plates containing 0.1 mM CdCl2 (data not shown). Plate assays were also performed to evaluate the level of resistance conferred on L. lactis MG1614 to other heavy metal compounds by pAH82, including zinc and lead. When pAH82 or pAH90 was present in MG1614, it could grow in the presence of ZnCl2 at concentrations as high as 6.0 mM, whereas the parent strain could not tolerate concentrations above 2.0 mM. No enhancement in resistance to lead carbonate was detected, in that both MG1614 and its pAH90-containing derivative tolerated lead carbonate concentrations in excess of 10 mM.
Conclusions.
This report concerns the detailed genetic and functional characterization of the lactococcal plasmid pAH90. The evolution of this cointegrate plasmid provides further evidence for the plasticity in structure of lactococcal plasmids facilitating the formation of stable replicons, which confer highly desirable and dominant traits on the starter organism. Indeed, the utility of the CadA selectable marker was demonstrated in that plasmids pAH82 and pAH90, which are both in excess of 20 kb in size, could be electroporated into a lactococcal host at frequencies of up to 2.0 × 103 transformants/μg of DNA. Two distinct phage resistance mechanisms are encoded by pAH90. Sequence analysis of this plasmid identified the determinants of a type I R/M system with a broad specificity range as a consequence of encoding two hybrid hsdS determinants (49). In addition, pAH90 encodes a potent phage adsorption blocking system (Ads) (24, 47, 49), the nature of which remains to be elucidated. The combination of phage resistance mechanisms, which act at two different points of the phage lytic cycle, provides excellent protection for phage-sensitive industrial lactococcal strains against phage attack (16). Where phage are not countered by the early-acting Ads mechanism (at the cell surface), the R/M system (intracellular) provides a further impediment to their proliferation. The fortuitous association of multiple phage resistance systems with a food-grade selectable marker on a mobilizable plasmid is of tremendous benefit for starter improvement regimens, given the widespread difficulties encountered in delivering many natural phage resistance systems to industrial lactococci (7, 8, 47). Given that many phage resistance plasmids which have been identified worldwide for use in starter improvement programs either lack an easily selectable marker (47) or are nonmobile, pAH90 represents an ideal candidate for use in food-grade starter improvement.
ACKNOWLEDGMENTS
This research has been partly funded by grant aid under the Food Sub-Programme of the Operational Programme for Industrial Development which is administered by the Department of Agriculture, Food and Forestry and supported by national and European Union funds. D.O.S. was supported by a Teagasc Walsh Fellowship.
We thank Derek Butler (University College Cork) for assistance with RNA isolation.
REFERENCES
- 1.Allison G E, Klaenhammer T R. Phage resistance mechanisms in lactic acid bacteria. Int Dairy J. 1998;8:207–226. [Google Scholar]
- 2.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactisIL1403 genome. Antonie van Leeuwenhoek. 1999;76:27–76. [PubMed] [Google Scholar]
- 5.Brennar D G, Shaw W V. The use of synthetic oligonucleotides with universal templates for rapid DNA sequencing: results with staphylococcal replicon pC221. EMBO J. 1985;4:561–568. doi: 10.1002/j.1460-2075.1985.tb03665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C L, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Borodovsky B P, M, Klenk H P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Coakley M, Fitzgerald G F, Ross R P. Application and evaluation of the phage resistance- and bacteriocin-encoding plasmid pMRC01 for the improvement of dairy starter cultures. Appl Environ Microbiol. 1997;63:1434–1440. doi: 10.1128/aem.63.4.1434-1440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffey A, Coakley M, McGarry A, Fitzgerald G F, Ross R P. Increasing phage resistance of cheese starters: a case study using Lactococcus lactisDPC4268. Lett Appl Microbiol. 1998;26:51–55. [Google Scholar]
- 9.Cogan T M, Accolas J-P, editors. Dairy starter cultures. New York, N.Y: VCH Publishers; 1995. [Google Scholar]
- 10.Crupper S S, Worrell V, Stewart G C, Landolo J J. Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J Bacteriol. 1999;181:9193–9199. doi: 10.1128/jb.181.13.4071-4075.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dougherty B A, Hill C, Weldman J F, Richardson D R, Venter J C, Ross R P. Sequence and analysis of the 60kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactisDPC3147. Mol Microbiol. 1998;29:1029–1038. doi: 10.1046/j.1365-2958.1998.00988.x. [DOI] [PubMed] [Google Scholar]
- 12.Dunny G M, McKay L L. Group II introns and expression of conjugative transfer functions in lactic acid bacteria. Antonie van Leeuwenhoek. 1999;76:77–88. [PubMed] [Google Scholar]
- 13.Durmaz E, Klaenhammer T R. A starter culture rotation strategy incorporating paired restriction/modification and abortive infection bacteriophage defences in a single Lactococcus lactisstrain. Appl Environ Microbiol. 1995;61:1266–1273. doi: 10.1128/aem.61.4.1266-1273.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endo G, Silver S. CadC, the transcriptional regulatory protein of the cadmium resistance system in Staphylococcus aureusplasmid p1258. J Bacteriol. 1995;177:4437–4441. doi: 10.1128/jb.177.15.4437-4441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foley S, Bron S, Venema G, Daly C, Fitzgerald G F. Molecular analysis of the replication origin of the Lactococcus lactisplasmid pCI305. Plasmid. 1996;36:125–141. doi: 10.1006/plas.1996.0040. [DOI] [PubMed] [Google Scholar]
- 16.Forde A, Fitzgerald G F. Bacteriophage defence systems in lactic acid bacteria. Antonie van Leeuwenhoek. 1999;76:89–113. [PubMed] [Google Scholar]
- 17.Frere J, Noveland M, Novel G. Molecular analysis of the Lactococcus lactis subspecies lactisCNRZ270 bi-directional theta replicating lactose plasmid pUCL22. Mol Microbiol. 1993;10:1113–1124. doi: 10.1111/j.1365-2958.1993.tb00981.x. [DOI] [PubMed] [Google Scholar]
- 18.Froseth B R, McKay L L. Molecular characterisation of the nisin resistance region of Lactococcus lactis subsp. lactis biovar diacetylactisDRC3. Appl Environ Microbiol. 1991;57:804–811. doi: 10.1128/aem.57.3.804-811.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garvey P, Fitzgerald G F, Hill C. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl Environ Microbiol. 1995;61:4321–4328. doi: 10.1128/aem.61.12.4321-4328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garvey P, van Sinderen D, Twomey D P, Hill C, Fitzgerald G F. Molecular genetics of bacteriophage and natural phage defense systems in the genus Lactococcus. Int Dairy J. 1995;5:905–947. [Google Scholar]
- 21.Gasson M J. Plasmid complements of Streptococcus lactisNCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbalenya A. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 1994;3:1117–1120. doi: 10.1002/pro.5560030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington A, Hill C. Construction of a bacteriophage-resistant derivative of Lactococcus lactis subsp. lactis425A by using the conjugal plasmid pNP40. Appl Environ Microbiol. 1991;57:3405–3409. doi: 10.1128/aem.57.12.3405-3409.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington A, Hill C. Plasmid involvement in the formation of a spontaneous bacteriophage insensitive mutant of Lactococcus lactis. FEMS Microbiol Lett. 1992;96:135–142. doi: 10.1016/0378-1097(92)90393-3. [DOI] [PubMed] [Google Scholar]
- 25.Hayes F, Vos P, Fitzgerald G F, de Vos W M, Daly C. Molecular organization of the minimal replicon of narrow-host-range lactococcal plasmid pCI305. Plasmid. 1991;25:16–26. doi: 10.1016/0147-619x(91)90003-f. [DOI] [PubMed] [Google Scholar]
- 26.Higgins D G, Sharpe P M. Fast and sensitive multiple sequence alignments on a microcomputer. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 27.Hill C, Ross R P. Starter cultures for the dairy industry. In: Roller S, Harlander S, editors. Genetic modification in the food industry. London, United Kingdom: Blackie Academic and Professional; 1998. pp. 174–192. [Google Scholar]
- 28.Hill C, Miller L A, Klaenhammer T R. Nucleotide sequence and distribution of the pTR2030 resistance determinant (hsp) which aborts bacteriophage infection in lactococci. Appl Environ Microbiol. 1990b;56:2255–2258. doi: 10.1128/aem.56.7.2255-2258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivey D M, Guffanti A A, Shen Z, Kudyan N, Krulwich T A. The cadC gene product of alkaliphilic Bacillus firmus OT4 partially restores Na+ resistance to an Eschericia coli strain lacking an Na+/H+antiporter (NhaA) J Bacteriol. 1992;174:4878–4884. doi: 10.1128/jb.174.15.4878-4884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keilhauer C, Eggeling L, Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvCoperon. J Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiewiet R, Bron S, de Jonge K, Venema G, Seegers J F. Theta replication of the lactococcal plamid pWV02. Mol Microbiol. 1993;10:319–327. [PubMed] [Google Scholar]
- 32.Kilic A O, Vijayakumar M N, al-Khaldi S F. Identification and nucleotide sequence analysis of a transfer-related region in the streptococcal conjugative transposon Tn5252. J Bacteriol. 1994;176:5145–5150. doi: 10.1128/jb.176.16.5145-5150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klaenhammer T R. Development of bacteriophage resistant strains of lactic acid bacteria. Biochem Soc Trans. 1991;3:675–681. doi: 10.1042/bst0190675. [DOI] [PubMed] [Google Scholar]
- 34.Lambowitz A M, Belfort M. Introns as mobile genetic elements. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 35.Lebrun M, Audurier A, Cossart P. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are similar to cadA and cadC of Staphylococcus aureusand are induced by cadmium. J Bacteriol. 1994;176:3040–3048. doi: 10.1128/jb.176.10.3040-3048.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C-Q, Leelawatcharamas V, Harvey M L, Dunn N W. Cloning vectors for lactococci based on a plasmid encoding resistance to cadmium. Curr Microbiol. 1996;33:35–39. doi: 10.1007/s002849900070. [DOI] [PubMed] [Google Scholar]
- 37.Liu C-Q, Dunn N W. Genetic analysis of regions involved in replication and cadmium resistance of the plasmid pND302 from Lactococcus lactis. Plasmid. 1997;38:79–90. doi: 10.1006/plas.1997.1301. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Abarca F, Garcia-Rodriguez F M, Toro N. Homing of a bacterial group II intron with an intron-encoded protein lacking a recognisable endonuclease domain. Mol Microbiol. 2000;35:1405–1412. doi: 10.1046/j.1365-2958.2000.01804.x. [DOI] [PubMed] [Google Scholar]
- 39.McKay L L, Baldwin K A. Conjugative 40-megadalton plasmid in Streptococcus lactis subsp. diacetylactisDRC3 is associated with resistance to nisin and bacteriophage. Appl Environ Microbiol. 1984;46:68–74. doi: 10.1128/aem.47.1.68-74.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michel F, Umesomo K, Ozeki H. Comparative and functional anatomy of group II introns—a review. Gene. 1989;82:3–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 41.Mills D A, McKay L L, Dunny G M. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J Bacteriol. 1996;178:3531–3538. doi: 10.1128/jb.178.12.3531-3538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitra R S, Bernstein I A. Single-strand breakage in DNA of Escherichia coli exposed to Cd2+ J Bacteriol. 1978;133:75–80. doi: 10.1128/jb.133.1.75-80.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohr G, Perlman P S, Lambowitz A M. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moineau S. Applications of phage resistance in lactic acid bacteria. Antonie van Leeuwenhoek. 1999;76:377–382. [PubMed] [Google Scholar]
- 45.Nies D H, Silver S. Ion efflux systems involved in bacterial metal resistance. J Ind Microbiol. 1995;14:186–199. doi: 10.1007/BF01569902. [DOI] [PubMed] [Google Scholar]
- 46.Nucifora G, Chu L, Misra T K, Silver S. Cadmium resistance from Staphylococcal aureus p1258 cadAgene results from a cadmium-efflux ATPase. Proc Natl Acad Sci USA. 1989;86:3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Sullivan D, Coffey A, Fitzgerald G F, Hill C, Ross R P. Design of a phage-insensitive lactococcal dairy starter via sequential transfer of naturally occurring conjugative plasmids. Appl Environ Microbiol. 1998;64:4618–4622. doi: 10.1128/aem.64.11.4618-4622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Sullivan D, Ross R P, Fitzgerald G F, Coffey A. Investigation of the relationship between lysogeny and lysis of Lactococcus lactisin cheese using prophage-targeted PCR. Appl Environ Microbiol. 2000;66:2192–2198. doi: 10.1128/aem.66.5.2192-2198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Sullivan D, Twomey D P, Coffey A, Hill C, Fitzgerald G F, Ross R P. Novel type I restriction-modification specificities through domain shuffling of HsdS subunits in Lactococcus lactis. Mol Microbiol. 2000;36:866–876. doi: 10.1046/j.1365-2958.2000.01901.x. [DOI] [PubMed] [Google Scholar]
- 50.Powell I B, Ward C, Hillier A J, Davidson B E. Simultaneous conjugal transfer in Lactococcus of genes involved in bacteriocin production and reduced susceptibility to bacteriophages. FEMS Microbiol Lett. 1990;72:209–214. doi: 10.1016/0378-1097(90)90373-x. [DOI] [PubMed] [Google Scholar]
- 51.Powell I B, Romano G M, Hillier A J, Davidson B E. Genetic enhancement of phage resistance in a commercial cheese starter. Aust J Dairy Technol. 1994;49:30–33. [Google Scholar]
- 52.Rensing C, Sun Y, Mitra B, Rosen B P. Pb(II)-translocating P-type ATPases. J Biol Chem. 1998;273:32614–32617. doi: 10.1074/jbc.273.49.32614. [DOI] [PubMed] [Google Scholar]
- 53.Romero D A, Klaenhammer T R. Characterization of insertion sequence IS946, an iso-IS1element, isolated from the conjugative lactococcal plasmid pTR2030. J Bacteriol. 1990;172:4151–4160. doi: 10.1128/jb.172.8.4151-4160.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ron E Z, Minz D, Finkelstein N P, Rosenberg E. Interactions of bacteria with cadmium. Biodegradation. 1992;3:161–170. [Google Scholar]
- 55.Rossbach S, Kukuk M L, Wilson T L, Feng S F, Pearson M M, Fisher M A. Cadmium-regulated gene fusions in Pseudomonas florescens. Environ Microbiol. 2000;2:373–382. doi: 10.1046/j.1462-2920.2000.00117.x. [DOI] [PubMed] [Google Scholar]
- 56.Sanders M E, Leonerd P J, Sing W D, Klaenhammer T R. Conjugal strategy for the construction of fast-acid producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986;52:1101–1107. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schouler C, Clier F, Lerayer A L, Ehrlich S D, Chopin M C. A type IC restriction-modification system in Lactococcus lactis. J Bacteriol. 1998;180:407–411. doi: 10.1128/jb.180.2.407-411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schouler C, Gautier S, Ehrlich A D, Chopin M C. Combinational variation of restriction-modification specificities in Lactococcus lactis. Mol Microbiol. 1998;28:169–178. doi: 10.1046/j.1365-2958.1998.00787.x. [DOI] [PubMed] [Google Scholar]
- 59.Seegers J F, Bron S, Franke C M, Venema G, Kiewiet R. The majority of lactococcal plasmids carry a highly related replicon. Microbiology. 1994;140:1291–1300. doi: 10.1099/00221287-140-6-1291. [DOI] [PubMed] [Google Scholar]
- 60.Seegers J F M L, van Sinderen D, Fitzgerald G F. Molecular characterization of the lactococcal plasmid pCIS3: natural stacking of specificity subunits of a type I restriction/modification system in a single lactococcal strain. Microbiology. 2000;146:435–443. doi: 10.1099/00221287-146-2-435. [DOI] [PubMed] [Google Scholar]
- 61.Shearman C, Godon J-J, Gasson M J. Splicing of a group II intron in a functional transfer gene of Lactococcus lactis. Mol Microbiol. 1996;21:45–53. doi: 10.1046/j.1365-2958.1996.00610.x. [DOI] [PubMed] [Google Scholar]
- 62.Shub D A, Goodrich-Blair H, Eddy S R. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem Sci. 1994;19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 63.Silver S, Misra T K. Plasmid-mediated heavy metal resistances. Annu Rev Microbiol. 1988;42:717–743. doi: 10.1146/annurev.mi.42.100188.003441. [DOI] [PubMed] [Google Scholar]
- 64.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenny K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1995;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 66.Trevors J T, Stratton G W, Gadd G M. Cadmium transport, resistance, and toxicity in bacteria, algae, and fungi. Can J Microbiol. 1986;32:447–464. doi: 10.1139/m86-085. [DOI] [PubMed] [Google Scholar]
- 67.Van Kranenburg R, Marugg J D, van Swam I I, Willem N J, de Vos W M. Molecular characterisation of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 68.Van Kranenburg R, de Vos W M. Characterisation of multiple regions involved in replication and mobilisation of plasmid pNZ4000 coding for exopolysaccharide production in Lactococcus lactis. J Bacteriol. 1998;20:5285–5290. doi: 10.1128/jb.180.20.5285-5290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Kranenburg R, van Swam I I, Marugg J D, Kleerebezem M, de Vos W M. Exopolysaccharide biosynthesis in Lactococcus lactisNIZO B40: functional analysis of the glycosyltransferase genes involved in synthesis of the polysaccharide backbone. J Bacteriol. 1999;181:338–340. doi: 10.1128/jb.181.1.338-340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Venema G, Kok J, van Sinderen D. From DNA sequence to application: possibilities and complications. Antonie van Leeuwenhoek. 1999;76:3–23. [PubMed] [Google Scholar]
- 71.Wang A, Macrina F L. Streptococcal plasmid pIP501 has a functional oriTsite. J Bacteriol. 1995;177:4199–4206. doi: 10.1128/jb.177.15.4199-4206.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu C, Zhou T, Kuroda M, Rosen B P. Metalloid resistance mechanisms in prokaryotes. J Biochem. 1998;123:16–23. doi: 10.1093/oxfordjournals.jbchem.a021904. [DOI] [PubMed] [Google Scholar]