Abstract
Due to their sessile state, plants are inevitably affected by and respond to the external environment. So far, plants have developed multiple adaptation and regulation strategies to abiotic stresses. One such system is epigenetic regulation, among which DNA methylation is one of the earliest and most studied regulatory mechanisms, which can regulate genome functioning and induce plant resistance and adaption to abiotic stresses. In this review, we outline the most recent findings on plant DNA methylation responses to drought, high temperature, cold, salt, and heavy metal stresses. In addition, we discuss stress memory regulated by DNA methylation, both in a transient way and the long-term memory that could pass to next generations. To sum up, the present review furnishes an updated account of DNA methylation in plant responses and adaptations to abiotic stresses.
Keywords: plant epigenetics, DNA methylation, abiotic stresses, stress memory
1. Introduction
Most plants are in a sessile state during the entire growth cycle, and they cannot avoid the stress of the natural environment as sensitively as animals. In order to better adapt to the environment and improve the survival probability, plants have formed a series of complex mechanisms. Among them, the regulation of plant responses to external stresses by epigenetic mechanisms is one of the important mechanisms discovered in recent years. Abiotic stress could induce epigenetic changes at many different levels, transmit stress signals, and regulate plants stress responses [1]. Epigenetic changes involve three main mechanisms: DNA methylation, histone modifications, and RNA-mediated gene silencing [2]. DNA methylation is one of the earliest discovered and most studied regulatory mechanisms in epigenetics, and is considered to be a relatively stable, heritable, transgenerational mark, involving a series of biological processes such as temporal and spatial gene expression, transposable element activity, and genomic imprinting [3].
DNA methylation generally refers to the transfer of a methyl group onto the C5 position of the cytosine to form 5 methylcytosine (5mC) [4]. In mammals, the main type of DNA methylation is CG methylation, while non-CG methylation is limited in specific tissues such as in embryonic stem cells [5,6]. In contrast, both CG and non-CG methylation, such as DNA methylation in CHG (symmetric) and CHH (asymmetric) environments (H = A, C, or T), are ubiquitously detected in higher plants [3]. DNA methylation is not limited to the promoter regions of genes, but also their coding regions. There are three processes of DNA methylation involved in plants: DNA methylation maintenance (the methylation of hemimethylated symmetrical sequences), de novo DNA methylation (methylate at a previously unmethylated C), and DNA demethylation (the methylation state can be reversed). The methylome in plants is primarily maintained during DNA replication and cell division by DNA methyltransferases, including maintenance methylases and de novo methylases [7]. The maintenance of DNA methylation of symmetrical sites, such as CG and CHG sequences, is completed through the methyltransferase 1 (MET1) [8], and chromomethylase 3 (CMT3) [9], respectively. The RNA-directed DNA methylation (RdDM) pathway and chromomethylase 2 (CMT2) are necessary for maintaining CHH methylation. The methylation of asymmetric site CHH is achieved by de novo methylases, via domains rearranged methyltransferase 1 (DRM1) and domains rearranged methyltransferase 2 (DRM2) by small RNAs through RdDM [10]. The RdDM pathway and the action of RNA interference (RNAi) machinery, DRMs, CMT3, and RNA polymerases are required in de novo methylation processes of all sequence contexts. DNA demethylation can be divided into active and passive processes: passive DNA demethylation is caused by the reduction or inactivation of enzymes important in DNA methylation during DNA replication, while the mechanism of active DNA demethylation is a complex process mediated by the base excision repair (BER) pathway. Active DNA demethylation in plants involves 5mC DNA glycosylase, and studies in Arabidopsis have shown that repressor of silencing 1 (AtROS1), DNA glycosylases demeter (AtDME), and demeter-like proteins 2 and 3 (DML2 and DML3) take roles in DNA demethylation [11,12,13].
Several methods could be used to detect DNA methylation, including methods based on antibodies or specific proteins with an affinity for methylcytosine, such as methylated DNA immunoprecipitation (MEDIP) and the capture of methylated DNA by methyl-CpG binding domain-based (MBD) proteins (MBDCap) [14], methods using methyl-sensitive restriction enzymes, such as methyl-sensitive amplification polymorphism (MSAP) [15,16], and methods involving the treatment of DNA with sodium bisulfite [17], such as whole-genome bisulfite sequencing (WGBS). According to their resolution and cost, affinity enrichment-based methods are suitable for rapid, large scale, and low-resolution studies, restriction enzymes-based methods are suitable for site-specific/targeted studies, and bisulfite conversion-based methods are suitable for high resolution studies.
Abiotic stresses include high temperature stress, cold stress, drought stress, salt stress, and heavy metal stress, which threaten plant growth and result in reduced crop yields and species diversity. Existing studies have shown that DNA methylation is a mechanism by which plants adapt to abiotic stresses [18,19]. Different stresses can trigger specific and dynamic DNA methylation changes in plants, regulating gene expression levels of stress-responsive genes, and controlling the activity of transposable elements (TEs), thereby inducing and improving the response and adaption of plants to stresses [20]. Most abiotic stress-induced DNA methylation modifications are transient and return to initial levels upon stress elimination; however, studies have also shown that some short-term or long-term “memory effect” could be induced in plants [21]. Due to transient changes in morphological and biochemical metabolites, short-term stress memory allows plants to maintain stress resistance for a short period of time or throughout their life, whereas long-term stress memory may be transferred to offspring. DNA methylation causes heritable epigenetic modifications in the absence of sequence changes, and the methylation changes in the offspring of stressed plants related to the parental environment [21].
This review summarizes the different roles of DNA methylation in plant responses to abiotic stresses and proposes the future development direction of DNA methylation research, which facilitates the development of stress-resistant plants that can deal with abiotic stresses such as drought, high temperature, cold, salt, and heavy metals.
2. DNA Methylation and Drought Stress
Drought is the most damaging environmental stress on plant yield and growth rate in the past few decades, due to climate change [22,23]. Plants have evolved complex mechanisms in response to drought stress, and DNA methylation regulation plays a pivotal role in regulating gene expression. By studying the methylation status of a single cytosine in the whole genome under drought stress, the overall methylation level of plants under drought stress are higher than that of control plants, such as 8.64% higher in mulberry (Morus alba) and 2.29% higher in Populus trichocarpa [24,25].
Correlation analysis has shown that DNA methylation has multiple effects on gene expression under drought stress, indicating that it directly or indirectly affects gene expression through multiple regulatory pathways. Drought-tolerant plants were found to have a more stable methylome under drought conditions, with differentially methylated regions (DMRs)-related genes mainly related to the stress response, programmed cell death, and other pathways in rice (Oryza sativa L.) [26], mulberry (Morus alba L.) [27], mungbean (Vigna radiata L.) [28], and maize (Zea mays L.) [29]. For example, an in-silico genome-wide DNA methylation (5mC) analysis was performed in rice (Oryza sativa cv. Zhonghua11) where 14 unique genes of the eukaryotic gene superfamily cytochrome P450 with different methylation levels were identified in the rice genome under drought stress [30], cytosine methylation at a single-base resolution and methylation patterns associated with water scarcity in representative drought-sensitive and drought-tolerant varieties in the genome of commercial apple (Malus x domestica) [31], and so on.
Global methylation and transcription analysis revealed that promoter unmethylated genes were expressed at higher levels than promoter methylated genes. In maize, DNA methylation in the ZmNAC111 promoter represses ZmNAC111 expression, resulting in an increased drought sensitivity [32]. Drought stress is also associated with changes in the methylation of the gene body of many genes, including those encoding transcription factors (TFs). A negative correlation between gene body methylation and gene expression was found in maize roots under water stress [29]. Changes of DNA methylation occurs in TEs as well. In poplar (Populus trichocarpa), transcription factors affecting gene expression after drought treatment were affected by methylated transposons. Methylated transposons involved in drought signal transduction pathways were found in C2C2, WRKY, MYB, and other families [25].
Cytosine-5-methyltransferases and demethylases are two important enzymes that play an important role in dynamically maintaining the DNA methylation status of plant genomes under drought stress. Drought stress induced the up-regulation of 5mC methyltransferase and demethylase in plants, such as apple (Malus x domestica) [31], tomato (Solanum lycopersicum) [33], and eggplant (Solanum melongena L.) [34].
The degree, level, and polymorphism of plant DNA methylation under water-deficient conditions were found to exhibit tissue-specific and genotype-specific characteristics. Higher levels of DNA methylation and demethylation, and higher methylation polymorphisms were found in wheat (Triticum aestivum L.) drought tolerant genotype AK58 compared with those of the common wheat genotype MinMai 13 [35]. In addition, methylation polymorphisms in the root were higher than that in the leaf under a water deficit, especially in AK58, which might be one of possible explanations that AK58 responds more quickly to water deprivation through changes in DNA methylation [35].
2.1. DNA Methylation and High Temperature Stress
High temperature stress is a serious threat to crop growth and development worldwide, which causes a series of morphological, physiological, and biochemical changes in plants. As a result, nearly all organisms have evolved signaling pathways to sense changes in ambient temperature and adjust their metabolism and cellular functions to prevent heat-related damage [36]. Recently, great progress has been made in the epigenetic regulation of thermal responses, including DNA methylation [37,38].
Results have shown that the overall methylation level of plants under heat are lower than that of control plants in most cases, such as in Populus simonii and Brassica napus [39,40]. Cytosine methylation changes in a large number of different genes were affected by heat stress. Under heat treatment, the degree of methylation of heat-sensitive genotypes was higher than that of heat-resistant genotypes, and more DNA demethylation events occurred in heat-resistant genotypes, while more DNA methylation occurred in heat-sensitive genotypes [41]. Studies have shown that heat stress induces DNA demethylation in genes in Arabidopsis thaliana, rather than in the intergenic regions [42]. There is also strong evidence that TEs, which have been implicated in the up-regulation of gene expressions under heat stress [43], can be induced or activated in response to heat stress [44,45]. For example, the LTR-copia-type retrotransposon ONSEN in Arabidopsis is activated by heat stress [46]. DMRs were identified in both gene sequences and promoter regions under both mild heat stress and severe heat stress, as well as in mitochondrial DNA in Arabidopsis [47].
DNA methylases and demethylase genes play critical roles in dynamically maintaining the DNA methylation status of plant genomes under heat stress. Twenty-two DNA methylase genes and six DNA demethylase genes were identified in the rapeseed (Brassica napus L.) genome [48]. Expression analysis by RNA-seq and qRT-PCR indicated that these DNA methylation/demethylation-related genes may be involved in the heat/salt stress response of rapeseed [48]. Xia Shen et al., studied two Arabidopsis germplasms from Eurasia and discovered 16 novel loci, including an association between CMT2 and temperature seasonality. cmt2 deletion mutants showed a higher tolerance to heat stress, strongly suggesting a role for the genetic regulation of epigenetic modifications in natural adaptation to temperature [49]. Notably, DNA (de)methylation may be a key regulatory process to ensure the proper germination of seeds produced under heat stress. In Arabidopsis, gene expression is strongly altered under severe heat stress during seed development, promoting heat stress response mechanisms. It was observed that DNA demethylation caused by the ROS1 gene could impair seed germination by affecting the expression of germination-related genes. On the other hand, under severe heat stress, most of the DMRs are located in the promoters and gene sequences of germination-related genes [47].
2.2. DNA Methylation and Cold Stress
Freezing or extremely low temperatures are key factors affecting plant growth, development, and crop yield. In response to cold stress, plants develop several mechanisms to minimize the potential damage caused by low temperature [50]. DNA methylation changes are an important way for plants to regulate gene expression in response to cold stress [51,52].
Cold exposure resulted in significantly lower DNA methylation levels in sugar beet [53]. Total methylation was decreased under high chill conditions, while no significant decrease was found in low chill conditions in apple (Malus x domestica Borkh.) [54]. Under cold stress, the tolerant genotype prevented the accumulation of H2O2, resulting in lower damage indices, such as malondialdehyde and electrolyte leakage indices, compared with the sensitive genotype [55]. For prolonged cold stress, the changes of demethylation bands in tolerant genotypes were higher than those in sensitive genotypes, indicating a higher activation potential of cold-stress-responsive genes in tolerant genotypes in chickpea [55].
As a direct and/or indirect product of gene expression regulated by different factors such as DNA methylation, cold stress signals are translated into physiological changes. Genes involved in cellular metabolism, the stress response, the antioxidant system, the lysine metabolic pathway, and transcriptional regulation showed a correlation between methylation and their expression in chickpeas and tartary buckwheat [55,56]. The decrease in DNA methylation was accompanied by the transcriptional down-regulation of the CMT2 gene and strong up-regulation of several genes mediating active DNA demethylation such as HbICE1, HbCBF2, and HbMET [53].
Both methylation and demethylation occur during cold adaptation. In Brassica, totally 1562 differentially methylated genes were identified during cold acclimation, including BrammDH1, BraKAT2, BraSHM4, and Bra4CL2, whose promoters were demethylated and resulted in an increase in their transcriptional activity [57]. It was found that in the rice cold-tolerant variety P427, 51 genes showed both methylation and expression level changes under cold stress, involved in the ICE–CBF–COR (CBF expression inducer—C-repeat binding factor—cold regulation) pathway and plays a crucial role in cold tolerance [58]. It was found that cold stress may lead to decreased DNA methylation in the promoter of the homologous gene of the open stomatal 1 in rice (Os03g0610900), which could interact with and phosphorylates ICE1, and increases its gene expression [58]. The correlation between gene body methylation and gene expression during chilling dormancy in apple was analyzed by bisulfite sequencing and qRT-PCR. Low temperature was associated with the hypermethylation of gene bodies, which may lead to the repression of their expression [54]. It was also found that TE families may be associated with the triggering of the cold stress-responsive expression of nearby genes, with responses highly variable between genotypes [44].
DNA demethylation take roles in cold responses as well. Cold treatment increased the transcriptional activity of cold-related genes and cold-responsive genes, such as HbICE1, HbCBF2, accompanied with induced expression of DNA methylation related genes, and also induces DNA demethylation of their promoters in rubber tree (Hevea brasiliensis) [59], Brassica rapa [57], tomato [60], and sweet orange (Citrus sinensis L.) [61]. Cold stress also resulted in a reduction in DNA methylation levels in the CHH context, accompanied by transcriptional down-regulation of CMT2, and the strong up-regulation of several genes mediating DNA demethylation [53]. Reduced genomic DNA methylation in the apex tissue of poplar (Populus L.) was found to be correlated with the induction of chilling-dependent DEMETER-LIKE DNA demethylase 10, which was involved in bud break [62].
2.3. DNA Methylation and Salt Stress
Soil salinization has become a serious environmental problem, threatening sustainable agriculture and future food security [63]. High salinity negatively affects osmotic and ionic balance, protein synthesis, photosynthesis, energy, and lipid metabolism [64]. Accumulating evidence suggests that DNA methylation plays an important role in regulating the gene expression response to salinity [65].
Salinity induces genome-wide changes in DNA methylation status, and different effects on DNA methylation in diverse plant species or specific genes were induced by salt stress [66]. Salinity stress increased the methylome content of alfalfa (Medicago spp.) plants, and the treatment with 5-AzaC (a DNA methylation inhibitor) on alfalfa seedlings resulted in a significant decrease in salt tolerance [67]. On the other hand, CG methylation levels were significantly reduced in the genomic regions analyzed within the epidermis under NaCl stresses, and the reduction was more robust in severely stressed Arabidopsis plants [68]. An increase in 5mC has been detected in CHG and CHH in the shoot under a salt-sensitive wheat genotype, while reduced 5mC levels were found in a salinity-tolerant wheat cultivar SR3 [69]. In rice, hypermethylation was found in tolerant genotypes, whereas sensitive genotypes displayed demethylation [70]. Different levels of DNA methylation were found in the root and shoot system during salt stress in rice and wheat [71,72]. In olive (Olea europaea subsp. europaea var. europaea) plants, DNA methylation levels increased when plants were subjected to salt stress. These changes were more pronounced in salt-tolerant cultivars, with higher DNA methylation events in royal cultivars than in Koroneiki [73]. These findings further indicated the possible regulatory roles of DNA methylation in conditioning the tolerance to high salinity depending on different species and tissues.
Under salt stress, DNA methylation regulates the expression of genes, including membrane transporter genes, heavy metal transporter genes, and organic acid secretion genes, thereby controlling stress signals and causing stress responses in plants. A genotype- and tissue-specific increase in cytosine methylation on the high-affinity potassium transporters TaHKT2;1 and TaHKT2;3 under NaCl treatment, were found down-regulated in wheat genotype Kharchia-65, contributing to the improved salt-tolerance ability [74]. Methylation in salinity responsive genes were found could induce the salinity tolerance of plants, such as the flavonol synthase genes TaFLS1 and TaWRS15 in wheat and barley (Hordeum vulgare) [75].
DNA demethylation plays crucial roles in salt stress as well. The exposure of plants to salt stress induced the expression of genes encoding enzymes of the flavonoid biosynthesis pathway (CHS, CHI, F3H, FLS, DFR, ANS) and the antioxidant pathway (GST, APx, GPx, GR), which correlated with their methylated status and AtROS1 demethylase activity [76]. Salt stress reduced the CG methylation level of the Glabra-2 (GL2, a master gene associated with root epidermal cell differentiation) in its gene body region, which related to its lower expression levels [68]. Compared with IR64 in rice, the expression of OsBZ8 (Abscisic acid Responsive Element -binding factor) was highly induced in the salt-tolerant variety Nonabokra under salinity stress, along with the loss of DNA methylation was observed at OsBZ8 locus [77].
The majority of the gene promoters exhibiting changes in methylation were hypermethylated under salt stress, and gene bodies in the progeny of stressed plants as well, accompanied with most of the hypermethylated genes having a lower gene expression in Arabidopsis [78]. Furthermore, cytosine alterations found in the UTRs and exons of rice under salinity stress indicated a significant role of gene body methylation in regulating gene expression [79].
Salt stress can affect the expression of CMT, DNMTs, DRMs, DMEs, and DMLs and induce methylation variation in plant DNA, providing plasticity for plants to adapt to salt stress. The expression levels of some members of the CMT and MET family were significantly down-regulated in response to salt stress, while DNMT2 showed up-regulation in rapeseed (Brassica napus L.) [48]. Genes in the DRMs family were down-regulated in response to salt stress, while BnaDRMa, BnaDRMg, and BnaDRMh were significantly up-regulated after salt treatment, and most demethylase genes such as DMEs, DML3s, and ROS1 were mildly up-regulated in rapeseed [48]. Furthermore, it was reported that in P. betulaefolia, the expression of DME responds to salt in a tissue-specific pattern, with down-regulation in leaves and up-regulation in roots [80]. Overexpressing the Arabidopsis demethylase gene AtROS1 in tabacco increases the demethylation levels of both promoters and gene bodies of genes in flavonoid biosynthetic and antioxidant pathways under salt stress, which showed a higher gene expression and a higher tolerance to salt stress [78].
It is worth noting that various single salt treatments and their mixed salts may have different effects on plants. Research found that in the halophyte Chloris Virgata, the effects of salt on DNA methylation was ranked as Na2CO3 > NaHCO3 > Na2SO4 > NaCl, and the mixed salts showed tissue-specific effects. Furthermore, they concluded that mixed salts are not a simple combination of a single salt [81].
2.4. DNA Methylation and Heavy Metal Stress
Industrial pollution has led to changes in the balance of some heavy metals, and excessive accumulation of heavy metals in soil is toxic to most plants [82]. Plant roots absorb excess heavy metal ions from the environment and transfer them to their shoots, which affects their metabolism and hinders their growth [83]. Under heavy metal stress, there is a close relationship between physiological responses, gene expression levels, and DNA methylation patterns [84]. Much attention has been paid to decipher the mechanisms by which plants resist heavy metal stress.
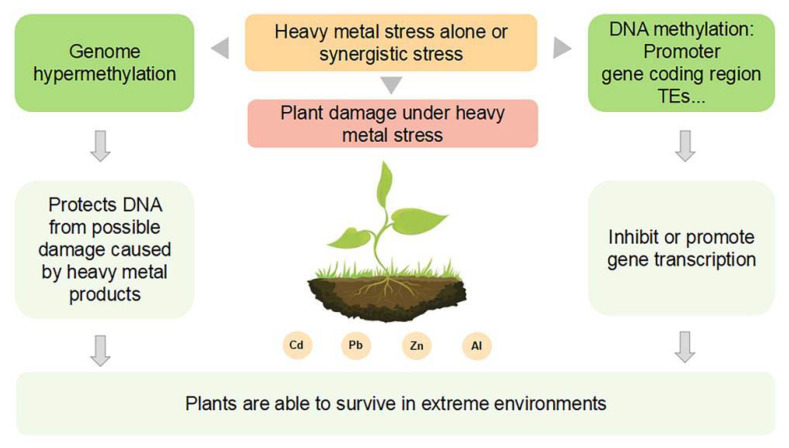
Hypermethylation has been viewed as one of the defense strategies for plants to protect themselves from possible damage by heavy metal products, allowing them to survive in extreme environments (Figure 1). DNA methylation changes induced by cadmium (Cd) stresses were analyzed in rice plants (Oryza sativa ssp japonica cv. Nipponbare), and more hypermethylated genes were found than hypomethylated ones [85]. The level of total methylation in radish and soybean (Glycine max) increased after Cd exposure with a significant dose-dependent relationship [86,87]. Methylation levels in the roots of heavy metal tolerant plants is significantly higher than that of sensitive plants, comparing clover (Trifolium repens L., sensitive species) with hemp (Cannabis sativa L., partial tolerant) [88], as well as Ni-tolerant Noccaea caerulescens with Ni-susceptible A. thaliana [89]. However, DNA hypomethylation was observed in the wheat resistant variety Pirsabak 2004 in response to lead (Pb), Cd, and zinc (Zn), in promoter regions of metal detoxification transporters [90]. For heavy metal such as Aluminum (Al), the exposition induced demethylation in both tolerant and non-tolerant plants, while Al stress triggered DNA hypermethylation as a protective response in Zea mays [91]. Furthermore, it was reported that in triticale lines, DNA methylation increased in Al tolerant lines and reduced in non-tolerant lines [92]. The complicated responsive patterns between them reveal that there might be different mechanisms for plants to protect themselves depending on whether the heavy metal element is essential for plant growth or not, or depending on the plant species and their developmental stages.
Figure 1.
DNA methylation and heavy metal stresses. Under heavy metal stress alone or in concert, DNA methylation may play a role in regulating plant responses to heavy metals through at least two mechanisms. The first mechanism is hypermethylation, a defense strategy of plants that can prevent genomic instability and protect DNA from possible damage by heavy metal products, allowing plants to survive in extreme environments. The second mechanism involves gene expression regulation, which is not limited to the promoter regions of genes, but also their coding regions.
Gene expression controlled by DNA methylation is another epigenetic response to heavy metal stress. In rice after Cd treatment, most of the DNA methylation modified genes show altered transcription levels, and methylation patterns were closely associated with transcriptional differences in stress-responsive genes involved in metal transport, metabolic processes, and transcriptional regulation [85]. An analysis of maize roots under normal Pb treatment revealed that 140 genes exhibited an altered DNA methylation status, including some well-known stress-responsive transcription factors and proteins such as MYB, AP2/ERF, bZIP, serine-threonine/tyrosine-protein, pentapeptide repeat protein, RING zinc finger protein, F-box protein, leucine-rich repeat protein, and tetrapeptide repeat protein [84]. Of the 30 differentially methylated DNA fragments of characterized soybean after Cd stress, fifteen were found associated with plant stress responses [87].
Methylation on gene promoters usually inhibits gene transcription, but in some cases it can also promote it under heavy metal stress [93]. In barley, 97.8% of whole cytosine of the promoter of HvAACT1, a major gene responsible for exogenous detoxification of Al, was unmethylated in the Al-tolerant cultivar FM404, whereas the Al-sensitive cultivar SV239 showed a much higher rate of methylation in the promoter [94]. The regulation by DNA methylation under heavy metal stresses is not limited to the promoter regions of genes, but also their coding regions and TEs. DNA methylation and DMRs were found in upstream, gene body, and downstream regions in rice after Cd treatment [85]. In maize root under Pb stress, results of whole-genome bisulfite sequencing showed that the average methylation density of the introns was higher than the UTRs and exons [74]. Transposable elements are one of the most heavily methylated DNA regions and were shown to play a role in Al stress responses in tolerant accessions of barley [94]. Furthermore, it was also found that a high density of TEs were strongly methylated in radish under Pb treatment [86].
DNA methylation and demethylation were found in response to heavy metals. The upregulation of CMT1 was found related with induced DNA hypermethylation in Posidonia oceanica after Cd treatment [95]. Mutation of MET1 and DRM2 resulted in significantly reduced transcript levels of the genes such as OsIRO2 and OsPR1b in rice seedlings under Cd stress as well, suggesting that DNA methylation participated in the plant response to Cd [85]. Moreover, heavy metals uptake and translocation have an interplay and coordination effect and recent findings have shown that Zn, Pb, and Cd could differentially regulate the expression level of DNA methyltransferases and the DNA methylation levels in maize, individually and in combinations [96].
3. DNA Methylation and Stress Memory
Plants are continuously challenged by different environment stresses in their lifetime, and interestingly, some plants could become more tolerant during subsequent stresses to the similar stress after a first mild exposure to better survive. In this way, it is widely accepted that plants have “stress memory”. Therefore, processes such as priming are proposed as a promising approach and have been used for plant adaption to future exposure through the acquisition of memory in plants, such as in Zea mays [97], Arabidopsis [98], and Brachypodium distachyon [99].
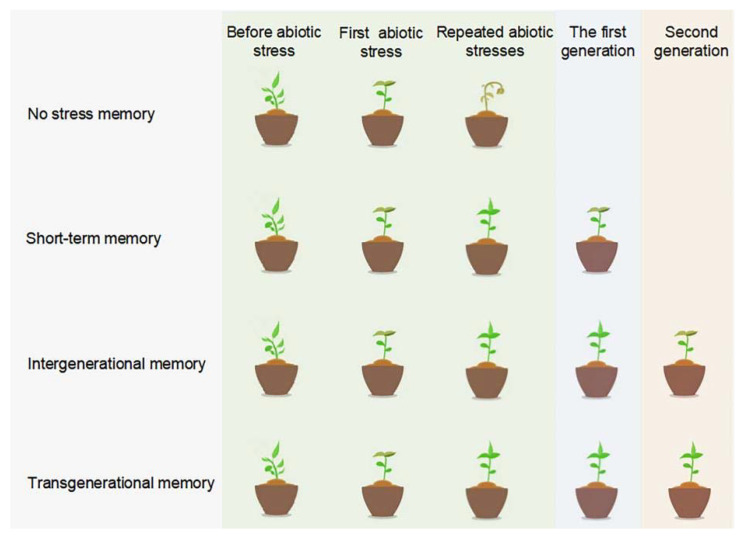
Sometimes, transgenerational priming effects could be observed between generations [100], passing down the memories to offspring and are inheritable. Therefore, the stress memory induced in plants could be classified as short-term stress memory and long-term stress memory (Figure 2). Short-term stress memory allows plants maintain resistance to stress in a short time or throughout the lifespan, due to temporary changes in morphology and biochemical metabolites [101], while long-term stress memory could be transferred to offspring. Among long-term stress memory, memory only passed on to immediate offspring is called intergenerational stress memory, while some stress responses could be remembered for at least two subsequent stress-free generations, called transgenerational stress memory [102].
Figure 2.
Plant memory in abiotic stresses. Plants that have not obtained stress memory in the first stress could not survive after repeated abiotic stresses. Plants that gain a short-term memory could enhance their resistance to the second stress, but only last for a short time or during the lifespan, not possible to pass on to the progeny. Plants that gain intergenerational memory exhibit stress memory only in the first stress-free offspring generation. The stress memory is heritable and observable for at least two generations in plants that achieve transgenerational memory.
In recent years, epigenetic modulations such as DNA methylation have been recognized as important components in stress memory, enabling plants to respond efficiently to recurring stress and prepare their offspring for potential stresses. It has been observed that the DNA methylation level changes during exposure to stress conditions, and the effects could be repeatedly established. During the recovery stages after stress conditions, some methylation changes were gradually restored while some others remained for a better and quicker response against stresses. After cycles of mild drought and re-watering treatment in rice, short-term drought stress memory was established, and genes that participated in the rice drought short-term memory response were identified and grouped into 16 distinct memory patterns [103]. Transcriptome analysis and whole genome bisulfite sequencing results showed a linkage between DNA methylation and drought memory transcripts, which provide evidence that DNA methylation participated in plant stress short-term memory formation. Furthermore, methylation status was found highly dynamic throughout stress duration and recovery. Memory DMRs could directly regulate rice drought memory genes and were associated with drought stress short-term memory [104].
DNA methylation could also be stably inherited through generations, and play important roles in plant long-term stress memory, including intergenerational and transgenerational memory (Figure 2). An analysis of the DNA methylation patterns in leaf tissues of rice (Oryza sativa) plants treated with heavy metals showed that CHG demethylation status could be inherited via the maternal and paternal germline and was often accompanied by further hypomethylation [105]. Intergenerational stress memory was also found associated with DNA methylation in species other than rice, such as in Arabidopsis [106], oilseed rape (Brassica napus) [107], soybean [108], and maize under various abiotic stresses [109]. A high proportion of drought induced DNA methylation status that was maintained in advanced generations, resulted in transgenerational stress memory, and improved the drought adaptability of rice offspring in upland fields [110]. Differential methylation between maize inbred lines is heritable, and DMRs could shift from one epiallele to others stably inherited in recombinant inbred lines [111]. Weixuan Cong et al., investigated a Tos17 retrotransposon for its methylation state for three generations and found that the DNA methylation in response to heavy metal stress was transgenerational inherited [100]. The heritability of DNA methylation was also found in the annual plant Polygonum persicaria. Their offspring of drought-stressed parents developed longer root systems and gained greater biomass than the offspring of well-watered parents of the same genetic line [112].
However, the inheritance of long-term memory varies in how many generations it lasts. The effect of salt stress on DNA methylation of Thlaspi arvense was investigated at the population level under the stimulation of salinity stress, and results showed that their stress memory could pass to at least two generations of offspring in a non-stress environment [113]. Xiaoguo Zheng et al., evaluated the DNA methylation of the original generation and the sixth generation of two rice varieties with different drought resistance levels under drought stress. The results showed that DNA methylation induced by drought stress could be preserved in the subsequent six generations and the drought stress had a cumulative effect on the DNA methylation patterns of both varieties [114]. Multigenerational exposure was performed to understand epigenetic variations in offspring. Ten generations of cultivation under salt stress could increase 45% spontaneous epigenetic changes in Arabidopsis [115]. Over 25 consecutive generations repeatedly exposed to heat stress, Arabidopsis line F25H was compared with its parental and parallel control progenies, results showed significantly lower global methylation levels of CHH and CHG in F25H than in control plants and more pronounced methylation changes in the gene body than TEs in F25H stressed progeny [116]. Hence, DNA methylation induced by heat stress in the progeny could result in a phenotypic resilience to adverse environments under long-term stress conditions.
4. Concluding Remarks
Continuous environmental changes pose serious abiotic threats to plant survival, and certain environmental stresses are repetitive, such as heat, drought, salinity, etc. Being sessile in nature, plants have evolved a complex response to these unfavorable and recurrent environmental conditions that involve transcription control, epigenetic regulation, and physiological and metabolic reprogramming for instance. DNA methylation is one of reversible epigenetic modifications that responds to abiotic stresses and regulates the spatial and temporal expression of genes in the short-term and long-term. Understanding the mechanisms in depth could aid in the development of genetic tools to improve plant stress resistance and benefit us in molecular plant breeding. With the advance of current molecular technologies, our knowledge of plant epigenic responses to various stresses is growing rapidly. In this review, we discuss the recent advances in plant responses and adaptions to abiotic stresses, focusing on the DNA methylation response to stresses and its roles in plant stress memory.
The important physiological significance of cytosine DNA methylation in plant responses and adaptions to various abiotic stresses has been indicated in many studies. Changes in the DNA methylation state in specific regions or the global state regulate the expression of genes in response to stimuli requirements and improve their defense systems. However, more work on a wider range of plant species will be a benefit for us to gain a more complete understanding of the mechanisms of DNA methylation in plant evolution and adaption. With the advantages of the explosion data of genome-wide bisulfite DNA methylome data by next generation sequencing, a large-scale meta-analysis across plant species will provide us with more information about DNA methylation and plant abiotic stresses.
Plant stress memory and their capacity to influence plant tolerance to a changing environment and crop productivity have been considered to play an important role in the adaptation and evolution of plants [117]. However, there is still a knowledge gap in understanding the molecular mechanisms underlying the establishment and maintenance of plant stress memory, since such memories are not established in many cases, and how the ability maintained is not fully illustrated as well. Further studies on stress memory mechanisms will largely improve plant tolerance to rapidly changing environmental conditions.
The development of methodologies of epigenome editing tools that specifically target genomic loci to alter epigenetic modifications, such as cytosine methylation and demethylation, could enable the precise generation of artificial epigenomic diversity and site-specific epigenetic changes. Methods such as chemical treatment, target-specific epigenetic engineering, including exogenous RNAi mediated by virus-induced gene silencing and the CRISPR-Cas9 system, will also facilitate the efforts for epibreeding in the future [118,119,120].
Author Contributions
Conceptualization, L.L. and L.D.; writing—original draft preparation, M.S.; writing—review and editing, L.L., L.D., Z.Y. and M.S.; funding acquisition, L.L., and L.D. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by Science Foundation of Hubei University (Grant No. 210803000013).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gutzat R., Scheid O.M. Epigenetic responses to stress: Triple defense? Curr. Opin. Plant Biol. 2012;15:568–573. doi: 10.1016/j.pbi.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pikaard C.S., Scheid O.M. Epigenetic Regulation in Plants. Cold Spring Harb. Perspect. Biol. 2014;6:a019315. doi: 10.1101/cshperspect.a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartels A., Han Q., Nair P., Stacey L., Gaynier H., Mosley M., Huang Q.Q., Pearson J.K., Hsieh T.F., An Y.Q.C., et al. Dynamic DNA Methylation in Plant Growth and Development. Int. J. Mol. Sci. 2018;19:2144. doi: 10.3390/ijms19072144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lister R., O’Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Law J.A., Jacobsen S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Mendoza A., Lister R., Bogdanovic O. Evolution of DNA Methylome Diversity in Eukaryotes. J. Mol. Biol. 2019;11:1687–1705. doi: 10.1016/j.jmb.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Huang J.J., Wang H.H., Xie X.J., Zhang D., Liu Y., Guo G.Q. Roles of DNA methyltransferases in Arabidopsis development. Afr. J. Biotechnol. 2010;9:8506–8514. [Google Scholar]
- 8.Zubko E., Gentry M., Kunova A., Meyer P. De novo DNA methylation activity of METHYLTRANSFERASE 1 (MET1) partially restores body methylation in Arabidopsis thaliana. Plant J. 2012;71:1029–1037. doi: 10.1111/j.1365-313X.2012.05051.x. [DOI] [PubMed] [Google Scholar]
- 9.Wendte J.M., Zhang Y.W., Ji L.X., Shi X.L., Hazarika R.R., Shahryary Y., Johannes F., Schmitz R.J. Epimutations are associated with CHROMOMETHYLASE 3-induced de novo DNA methylation. eLife. 2019;8:e47891. doi: 10.7554/eLife.47891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong X.H., Du J.M., Hale C.J., Gallego-Bartolome J., Feng S.H., Vashisht A.A., Chory J., Wohlschlegel J.A., Patel D.J., Jacobsen S.E. Molecular Mechanism of Action of Plant DRM De novo DNA Methyltransferases. Cell. 2014;157:1050–1060. doi: 10.1016/j.cell.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parrilla-Doblas J.T., Roldan-Arjona T., Ariza R.R., Cordoba-Canero D. Active DNA Demethylation in Plants. Int. J. Mol. Sci. 2019;20:4683. doi: 10.3390/ijms20194683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L.P., Lang C.J., Wu Y.J., Meng D.W., Yang T.B., Li D.Q., Jin T.C., Zhou X.F. ROS1-mediated decrease in DNA methylation and increase in expression of defense genes and stress response genes in Arabidopsis thaliana due to abiotic stresses. BMC Plant Biol. 2022;22:104. doi: 10.1186/s12870-022-03473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C.Q., Hung Y.H., Rim H.J., Zhang D.P., Frost J.M., Shin H., Jang H.S., Liu F., Xiao W.Y., Iyer L.M., et al. The catalytic core of DEMETER guides active DNA demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2019;116:17563–17571. doi: 10.1073/pnas.1907290116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu H.K., Weng Y.I., Hsu P.Y., Huang T.H., Huang Y.W. Detection of DNA methylation by MeDIP and MBDCap assays: An overview of techniques. Methods Mol. Biol. 2014;1105:61–70. doi: 10.1007/978-1-62703-739-6_5. [DOI] [PubMed] [Google Scholar]
- 15.Yaish M.W., Peng M., Rothstein S.J. Global DNA methylation analysis using methyl-sensitive amplification polymorphism (MSAP) Methods Mol. Biol. 2014;1062:285–298. doi: 10.1007/978-1-62703-580-4_16. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Benito M.E., Ibanez M.A., Pirredda M., Mira S., Martin C. Application of the MSAP Technique to Evaluate Epigenetic Changes in Plant Conservation. Int. J. Mol. Sci. 2020;21:7459. doi: 10.3390/ijms21207459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouil Q., Keniry A. Latest techniques to study DNA methylation. Essays Biochem. 2019;63:639–648. doi: 10.1042/EBC20190027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashapkin V.V., Kutueva L.I., Aleksandrushkina N.I., Vanyushin B.F. Epigenetic Mechanisms of Plant Adaptation to Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2020;21:7457. doi: 10.3390/ijms21207457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhter Z., Bi Z., Ali K., Sun C., Fiaz S., Haider F.U., Bai J. In Response to Abiotic Stress, DNA Methylation Confers EpiGenetic Changes in Plants. Plants. 2021;10:1096. doi: 10.3390/plants10061096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S., Mohapatra T. Dynamics of DNA Methylation and Its Functions in Plant Growth and Development. Front. Plant Sci. 2021;12:596236. doi: 10.3389/fpls.2021.596236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhoeven K.J.F., Jansen J.J., van Dijk P.J., Biere A. Stress-induced DNA methylation changes and their heritability in Asexual dandelions. New Phytol. 2010;185:1108–1118. doi: 10.1111/j.1469-8137.2009.03121.x. [DOI] [PubMed] [Google Scholar]
- 22.Lobell D.B., Gourdji S.M. The Influence of Climate Change on Global Crop Productivity. Plant Physiol. 2012;160:1686–1697. doi: 10.1104/pp.112.208298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhry S., Sidhu G.P.S. Climate change regulated abiotic stress mechanisms in plants: A comprehensive review. Plant Cell Rep. 2022;41:1–31. doi: 10.1007/s00299-021-02759-5. [DOI] [PubMed] [Google Scholar]
- 24.Li R.X., Hu F., Li B., Zhang Y.P., Chen M., Fan T., Wang T.C. Whole genome bisulfite sequencing methylome analysis of mulberry (Morus alba) reveals epigenome modifications in response to drought stress. Sci. Rep. 2020;10:8013. doi: 10.1038/s41598-020-64975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang D., Zhang Z.J., Wu H.L., Huang C.Y., Shuai P., Ye C.Y., Tang S., Wang Y.J., Yang L., Wang J., et al. Single-base-resolution methylomes of Populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genet. 2014;15:S9. doi: 10.1186/1471-2156-15-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W.S., Qin Q., Sun F., Wang Y.X., Xu D.D., Li Z.K., Fu B.Y. Genome-Wide Differences in DNA Methylation Changes in Two Contrasting Rice Genotypes in Response to Drought Conditions. Front. Plant Sci. 2016;7:1675. doi: 10.3389/fpls.2016.01675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ackah M., Guo L., Li S., Jin X., Asakiya C., Aboagye E.T., Yuan F., Wu M., Essoh L.G., Adjibolosoo D., et al. DNA Methylation Changes and Its Associated Genes in Mulberry (Morus alba L.) Yu-711 Response to Drought Stress Using MethylRAD Sequencing. Plants. 2022;11:190. doi: 10.3390/plants11020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao P.L., Ma B., Cai C.M., Xu J.H. Transcriptome and methylome changes in two contrasting mungbean genotypes in response to drought stress. BMC Genom. 2022;23:80. doi: 10.1186/s12864-022-08315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q., Xu J., Pu X.M., Lv H.Z., Liu Y.J., Ma H.L., Wu F.K., Wang Q.J., Feng X.J., Liu T.H., et al. Maize DNA Methylation in Response to Drought Stress Is Involved in Target Gene Expression and Alternative Splicing. Int. J. Mol. Sci. 2021;22:8285. doi: 10.3390/ijms22158285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waseem M., Huang F.Y., Wang Q.Y., Aslam M.M., Abbas F., Ahmad F., Ashraf U., Hassan W., Fiaz S., Ye X.W., et al. Identification, methylation profiling, and expression analysis of stress-responsive cytochrome P450 genes in rice under abiotic and phytohormones stresses. GM Crops. Food. 2021;12:551–563. doi: 10.1080/21645698.2021.1908813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J.D., Zhou S.S., Gong X.Q., Song Y., van Nocker S., Ma F.W., Guan Q.M. Single-base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnol. J. 2018;16:672–687. doi: 10.1111/pbi.12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao H.D., Wang H.W., Liu S.X., Li Z., Yang X.H., Yan J.B., Li J.S., Tran L.S.P., Qin F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015;6:8326. doi: 10.1038/ncomms9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar R., Chauhan P.K., Khurana A. Identification and expression profiling of DNA methyltransferases during development and stress conditions in Solanaceae. Funct. Integr. Genomic. 2016;16:513–528. doi: 10.1007/s10142-016-0502-3. [DOI] [PubMed] [Google Scholar]
- 34.Moglia A., Gianoglio S., Acquadro A., Valentino D., Milani A.M., Lanteri S., Comino C. Identification of DNA methyltransferases and demethylases in Solanum melongena L., and their transcription dynamics during fruit development and after salt and drought stresses. PLoS ONE. 2019;14:e0223581. doi: 10.1371/journal.pone.0223581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan H.Y., Li J.Y., Zhu Y.Q., Jia W.J., Wang H.H., Jiang L.N., Zhou Y.Q. Responsive changes of DNA methylation in wheat (Triticum aestivum) under water deficit. Sci. Rep. 2020;10:7938. doi: 10.1038/s41598-020-64660-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mittler R., Finka A., Goloubinoff P. How do plants feel the heat? Trends Biochem. Sci. 2012;37:118–125. doi: 10.1016/j.tibs.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Liu J.Z., Feng L.L., Li J.M., He Z.H. Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 2015;6:267. doi: 10.3389/fpls.2015.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J.G., Lu Z.G., Wang L., Jin B. Plant Responses to Heat Stress: Physiology, Transcription, Noncoding RNAs, and Epigenetics. Int. J. Mol. Sci. 2021;22:117. doi: 10.3390/ijms22010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ci D., Song Y.P., Tian M., Zhang D.Q. Methylation of miRNA genes in the response to temperature stress in Populus simonii. Front. Plant Sci. 2015;6:921. doi: 10.3389/fpls.2015.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Huang Q., Sun M.X., Zhang T.Y., Li H., Chen B.Y., Xu K., Gao G.Z., Li F., Yan G.X., et al. Global DNA methylation variations after short-term heat shock treatment in cultured microspores of Brassica napus cv. Topas. Sci. Rep. 2016;6:38401. doi: 10.1038/srep38401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao G.Z., Li J., Li H., Li F., Xu K., Yan G.X., Chen B.Y., Qiao J.W., Wu X.M. Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breed Sci. 2014;64:125–133. doi: 10.1270/jsbbs.64.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korotko U., Chwialkowska K., Sanko-Sawczenko I., Kwasniewski M. DNA Demethylation in Response to Heat Stress in Arabidopsis thaliana. Int. J. Mol. Sci. 2021;22:1555. doi: 10.3390/ijms22041555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makarevitch I., Waters A.J., West P.T., Stitzer M., Hirsch C.N., Ross-Ibarra J., Springer N.M. Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress. PLoS Genet. 2015;11:e1004915. doi: 10.1371/journal.pgen.1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang Z.K., Anderson S.N., Noshay J.M., Crisp P.A., Enders T.A., Springer N.M. Genetic and epigenetic variation in transposable element expression responses to abiotic stress in maize. Plant Physiol. 2021;186:420–433. doi: 10.1093/plphys/kiab073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hossain M.S., Kawakatsu T., Do Kim K., Zhang N., Nguyen C.T., Khan S.M., Batek J.M., Joshi T., Schmutz J., Grimwood J., et al. Divergent cytosine DNA methylation patterns in single-cell, soybean root hairs. New Phytol. 2017;214:808–819. doi: 10.1111/nph.14421. [DOI] [PubMed] [Google Scholar]
- 46.Cavrak V.V., Lettner N., Jamge S., Kosarewicz A., Bayer L.M., Scheid O.M. How a Retrotransposon Exploits the Plant’s Heat Stress Response for Its Activation. PLoS Genet. 2014;10:e1004115. doi: 10.1371/journal.pgen.1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malabarba J., Windels D., Xu W.J., Verdier J. Regulation of DNA (de)Methylation Positively Impacts Seed Germination during Seed Development under Heat Stress. Genes. 2021;12:457. doi: 10.3390/genes12030457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan S.H., Liu H.F., Liu J., Hua W., Xu S.M., Li J. Systematic Analysis of the DNA Methylase and Demethylase Gene Families in Rapeseed (Brassica napus L.) and Their Expression Variations After Salt and Heat stresses. Int. J. Mol. Sci. 2020;21:953. doi: 10.3390/ijms21030953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen X., De Jonge J., Forsberg S.K.G., Pettersson M.E., Sheng Z.Y., Hennig L., Carlborg O. Natural CMT2 Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality. PLoS Genet. 2014;10:e1004842. doi: 10.1371/journal.pgen.1004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miura K., Furumoto T. Cold Signaling and Cold Response in Plants. Int. J. Mol. Sci. 2013;14:5312–5337. doi: 10.3390/ijms14035312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan H.H., Wei J., Li T.C., Li Z.P., Guo N., Cai Y.P., Lin Y. DNA methylation alterations of upland cotton (Gossypium hirsutum) in response to cold stress. Acta Physiol. Plant. 2013;35:2445–2453. doi: 10.1007/s11738-013-1278-x. [DOI] [Google Scholar]
- 52.Thiebaut F., Hemerly A.S., Ferreira P.C.G. A Role for Epigenetic Regulation in the Adaptation and Stress Responses of Non-model Plants. Front. Plant Sci. 2019;10:246. doi: 10.3389/fpls.2019.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gutschker S., Corral J.M., Schmiedl A., Ludewig F., Koch W., Fiedler-Wiechers K., Czarnecki O., Harms K., Keller I., Rodrigues C.M., et al. Multi-omics data integration reveals link between epigenetic modifications and gene expression in sugar beet (Beta vulgaris subsp. vulgaris) in response to cold. BMC Genom. 2022;23:1–25. doi: 10.1186/s12864-022-08312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar G., Rattan U.K., Singh A.K. Chilling-Mediated DNA Methylation Changes during Dormancy and Its Release Reveal the Importance of Epigenetic Regulation during Winter Dormancy in Apple (Malus x domestica Borkh.) PLoS ONE. 2016;11:e0149934. doi: 10.1371/journal.pone.0149934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rakei A., Maali-Amiri R., Zeinali H., Ranjbar M. DNA methylation and physio-biochemical analysis of chickpea in response to cold stress. Protoplasma. 2016;253:61–76. doi: 10.1007/s00709-015-0788-3. [DOI] [PubMed] [Google Scholar]
- 56.Song Y., Jia Z.F., Hou Y.K., Ma X., Li L.Z., Jin X., An L.Z. Roles of DNA Methylation in Cold Priming in Tartary Buckwheat. Front. Plant Sci. 2020;11:608540. doi: 10.3389/fpls.2020.608540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu T.K., Li Y., Duan W.K., Huang F.Y., Hou X.L. Cold acclimation alters DNA methylation patterns and confers tolerance to heat and increases growth rate in Brassica rapa. J. Exp. Bot. 2017;68:1213–1224. doi: 10.1093/jxb/erw496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo H., Wu T.K., Li S.X., He Q., Yang Z.L., Zhang W.H., Gan Y., Sun P.Y., Xiang G.L., Zhang H.Y., et al. The Methylation Patterns and Transcriptional Responses to Chilling Stress at the Seedling Stage in Rice. Int. J. Mol. Sci. 2019;20:5089. doi: 10.3390/ijms20205089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang X., Wang Q.C., Yuan H.M., Huang X. Chilling-induced DNA Demethylation is associated with the cold tolerance of Hevea brasiliensis. BMC Plant Biol. 2018;18:70. doi: 10.1186/s12870-018-1276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang B., Tieman D.M., Jiao C., Xu Y.M., Chen K.S., Fei Z.J., Giovannoni J.J., Klee H.J. Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc. Natl. Acad. Sci. USA. 2016;113:12580–12585. doi: 10.1073/pnas.1613910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sicilia A., Scialo E., Puglisi I., Lo Piero A.R. Anthocyanin Biosynthesis and DNA Methylation Dynamics in Sweet Orange Fruit [Citrus sinensis L. (Osbeck)] under Cold Stress. J. Agric. Food Chem. 2020;68:7024–7031. doi: 10.1021/acs.jafc.0c02360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conde D., Le Gac A.L., Perales M., Dervinis C., Kirst M., Maury S., Gonzalez-Melendi P., Allona I. Chilling-responsive DEMETER-LIKE DNA demethylase mediates in poplar bud break. Plant. Cell Environ. 2017;40:2236–2249. doi: 10.1111/pce.13019. [DOI] [PubMed] [Google Scholar]
- 63.Rahman M.M., Mostofa M.G., Keya S.S., Siddiqui M.N., Ansary M.M.U., Das A.K., Rahman M.A., Tran L.S.P. Adaptive Mechanisms of Halophytes and Their Potential in Improving Salinity Tolerance in Plants. Int. J. Mol. Sci. 2021;22:10733. doi: 10.3390/ijms221910733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muchate N.S., Nikalje G.C., Rajurkar N.S., Suprasanna P., Nikam T.D. Plant Salt Stress: Adaptive Responses, Tolerance Mechanism and Bioengineering for Salt Tolerance. Bot. Rev. 2016;82:371–406. doi: 10.1007/s12229-016-9173-y. [DOI] [Google Scholar]
- 65.Al-Harrasi I., Al-Yahyai R., Yaish M.W. Differential DNA methylation and transcription profiles in date palm roots exposed to salinity. PLoS ONE. 2018;13:e0191492. doi: 10.1371/journal.pone.0191492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mastan S.G., Rathore M.S., Bhatt V.D., Yadav P., Chikara J. Assessment of changes in DNA methylation by methylation-sensitive amplification polymorphism in Jatropha curcas L. subjected to salinity stress. Genes. 2012;508:125–129. doi: 10.1016/j.gene.2012.07.063. [DOI] [PubMed] [Google Scholar]
- 67.Al-Lawati A., Al-Bahry S., Victor R., Al-Lawati A.H., Yaish M.W. Salt stress alters DNA methylation levels in alfalfa (Medicago spp) Genet. Mol. Res. 2016;15:15018299. doi: 10.4238/gmr.15018299. [DOI] [PubMed] [Google Scholar]
- 68.Beyrne C.C., Iusem N.D., Gonzalez R.M. Effect of Salt Stress on Cytosine Methylation within GL2, An Arabidopsis thaliana Gene Involved in Root Epidermal Cell Differentiation. Absence of Inheritance in the Unstressed Progeny. Int. J. Mol. Sci. 2019;20:4446. doi: 10.3390/ijms20184446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miryeganeh M. Plants’ Epigenetic Mechanisms and Abiotic Stress. Genes. 2021;12:1106. doi: 10.3390/genes12081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feng Q., Yang C., Lin X., Wang J., Liu B. Salt and alkaline stress induced transgenerational alteration in DNA methylation of rice (Oryza sativa) Aust. J. Crop. Sci. 2012;6:877–883. [Google Scholar]
- 71.Ferreira L.J., Azevedo V., Maroco J., Oliveira M.M., Santos A.P. Salt Tolerant and Sensitive Rice Varieties Display Differential Methylome Flexibility under Salt Stress. PLoS ONE. 2015;10:e0124060. doi: 10.1371/journal.pone.0124060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang Y.N., Zhu C., Jiang J., Zhang H., Zhu J.K., Duan C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020;62:563–580. doi: 10.1111/jipb.12901. [DOI] [PubMed] [Google Scholar]
- 73.Mousavi S., Regni L., Bocchini M., Mariotti R., Cultrera N.G.M., Mancuso S., Googlani J., Chakerolhosseini M.R., Guerrero C., Albertini E., et al. Physiological, epigenetic and genetic regulation in some olive cultivars under salt stress. Sci. Rep. 2019;9:1093. doi: 10.1038/s41598-018-37496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar S., Beena A.S., Awana M., Singh A. Salt-Induced Tissue-Specific Cytosine Methylation Downregulates Expression of HKT Genes in Contrasting Wheat (Triticum aestivum L.) Genotypes. DNA Cell Biol. 2017;36:283–294. doi: 10.1089/dna.2016.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kong L.Y., Liu Y.N., Wang X.Y., Chang C. Insight into the Role of Epigenetic Processes in Abiotic and Biotic Stress Response in Wheat and Barley. Int. J. Mol. Sci. 2020;21:1480. doi: 10.3390/ijms21041480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bharti P., Mahajan M., Vishwakarma A.K., Bhardwaj J., Yadav S.K. AtROS1 overexpression provides evidence for epigenetic regulation of genes encoding enzymes of flavonoid biosynthesis and antioxidant pathways during salt stress in transgenic tobacco. J. Exp. Bot. 2015;66:5959–5969. doi: 10.1093/jxb/erv304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paul A., Dasgupta P., Roy D., Chaudhuri S. Comparative analysis of Histone modifications and DNA methylation at OsBZ8 locus under salinity stress in IR64 and Nonabokra rice varieties. Plant Mol. Biol. 2017;95:63–88. doi: 10.1007/s11103-017-0636-2. [DOI] [PubMed] [Google Scholar]
- 78.Bilichak A., Ilnystkyy Y., Hollunder J., Kovalchuk I. The Progeny of Arabidopsis thaliana Plants Exposed to Salt Exhibit Changes in DNA Methylation, Histone Modifications and Gene Expression. PLoS ONE. 2012;7:e30515. doi: 10.1371/journal.pone.0030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karan R., DeLeon T., Biradar H., Subudhi P.K. Salt Stress Induced Variation in DNA Methylation Pattern and Its Influence on Gene Expression in Contrasting Rice Genotypes. PLoS ONE. 2012;7:e40203. doi: 10.1371/journal.pone.0040203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu C.X., Li H., Lin J., Wang Y., Xu X.Y., Cheng Z.M., Chang Y.H. Genome-Wide Characterization of DNA Demethylase Genes and Their Association with Salt Response in Pyrus. Genes. 2018;9:398. doi: 10.3390/genes9080398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao X., Cao D.H., Liu J., Wang X.P., Geng S.J., Liu B., Shi D.C. Tissue-Specific and Cation/Anion-Specific DNA Methylation Variations Occurred in C. virgata in Response to Salinity Stress. PLoS ONE. 2013;8:e78426. doi: 10.1371/journal.pone.0078426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kushwaha A., Rani R., Kumar S., Gautam A. Heavy metal detoxification and tolerance mechanisms in plants: Implications for phytoremediation. Environ. Rev. 2016;24:39–51. doi: 10.1139/er-2015-0010. [DOI] [Google Scholar]
- 83.Rizwan M., Ali S., Qayyum M.F., Ok Y.S., Adrees M., Ibrahim M., Zia-ur-Rehmand M., Farid M., Abbas F. Effect of metal and metal oxide nanoparticles on growth and physiology of globally important food crops: A critical review. J. Hazard. Mater. 2017;322:2–16. doi: 10.1016/j.jhazmat.2016.05.061. [DOI] [PubMed] [Google Scholar]
- 84.Ding H.P., Gao J., Qin C., Ma H.X., Huang H., Song P., Luo X.R., Lin H.J., Shen Y.O., Pan G.T., et al. The Dynamics of DNA Methylation in Maize Roots under Pb Stress. Int. J. Mol. Sci. 2014;15:23537–23554. doi: 10.3390/ijms151223537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Feng S.J., Liu X.S., Tao H., Tan S.K., Chu S.S., Oono Y., Zhang X.D., Chen J., Yang Z.M. Variation of DNA methylation patterns associated with gene expression in rice (Oryza sativa) exposed to cadmium. Plant Cell Environ. 2016;39:2629–2649. doi: 10.1111/pce.12793. [DOI] [PubMed] [Google Scholar]
- 86.Tang M.J., Xu L.A., Wang Y., Dong J.H., Zhang X.L., Wang K., Ying J.L., Li C., Liu L.W. Melatonin-induced DNA demethylation of metal transporters and antioxidant genes alleviates lead stress in radish plants. Hortic. Res. 2021;8:124. doi: 10.1038/s41438-021-00561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun D.D., Sun J.W., Huang L.Y., Chen N., Wang Q.W. Effects of cadmium stress on DNA methylation in soybean. Biotechnol. Biotec. Eq. 2021;35:1696–1705. doi: 10.1080/13102818.2021.1980107. [DOI] [Google Scholar]
- 88.Aina R., Sgorbati S., Santagostino A., Labra M., Ghiani A., Citterio S. Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol. Plant. 2004;121:472–480. doi: 10.1111/j.1399-3054.2004.00343.x. [DOI] [Google Scholar]
- 89.Gulli M., Marchi L., Fragni R., Buschini A., Visioli G. Epigenetic modifications preserve the hyperaccumulator Noccaea caerulescens from Ni geno-toxicity. Environ. Mol. Mutagen. 2018;59:464–475. doi: 10.1002/em.22191. [DOI] [PubMed] [Google Scholar]
- 90.Shafiq S., Zeb Q., Ali A., Sajjad Y., Nazir R., Widemann E., Liu L.Y. Lead, Cadmium and Zinc Phytotoxicity Alter DNA Methylation Levels to Confer Heavy Metal Tolerance in Wheat. Int. J. Mol. Sci. 2019;20:4676. doi: 10.3390/ijms20194676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taspinar M.S., Aydin M., Sigmaz B., Yagci S., Arslan E., Agar G. Aluminum-Induced Changes on DNA Damage, DNA Methylation and LTR Retrotransposon Polymorphism in Maize. Arab. J. Sci. Eng. 2018;43:123–131. doi: 10.1007/s13369-017-2697-6. [DOI] [Google Scholar]
- 92.Agnieszka N. The influence of Al3+ on DNA methylation and sequence changes in the triticale (x Triticosecale Wittmack) genome. J. Appl. Genet. 2018;59:405–417. doi: 10.1007/s13353-018-0459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gallo-Franco J.J., Sosa C.C., Ghneim-Herrera T., Quimbaya M. Epigenetic Control of Plant Response to Heavy Metal Stress: A New View on Aluminum Tolerance. Front. Plant Sci. 2020;11:602625. doi: 10.3389/fpls.2020.602625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kashino-Fujii M., Yokosho K., Yamaji N., Yamane M., Saisho D., Sato K., Ma J.F. Retrotransposon Insertion and DNA Methylation Regulate Aluminum Tolerance in European Barley Accessions. Plant Physiol. 2018;178:716–727. doi: 10.1104/pp.18.00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Greco M., Chiappetta A., Bruno L., Bitonti M.B. In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 2012;63:695–709. doi: 10.1093/jxb/err313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shafiq S., Ali A., Sajjad Y., Zeb Q., Shahzad M., Khan A.R., Nazir R., Widemann E. The Interplay between Toxic and Essential Metals for Their Uptake and Translocation Is Likely Governed by DNA Methylation and Histone Deacetylation in Maize. Int. J. Mol. Sci. 2020;21:6959. doi: 10.3390/ijms21186959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ding Y., Virlouvet L., Liu N., Riethoven J.J., Fromm M., Avramova Z. Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol. 2014;14:141. doi: 10.1186/1471-2229-14-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zuther E., Schaarschmidt S., Fischer A., Erban A., Pagter M., Mubeen U., Giavalisco P., Kopka J., Sprenger H., Hincha D.K. Molecular signatures associated with increased freezing tolerance due to low temperature memory in Arabidopsis. Plant Cell Environ. 2019;42:854–873. doi: 10.1111/pce.13502. [DOI] [PubMed] [Google Scholar]
- 99.Mayer B.F., Charron J.B. Transcriptional memories mediate the plasticity of cold stress responses to enable morphological acclimation in Brachypodium distachyon. New Phytol. 2021;229:1615–1634. doi: 10.1111/nph.16945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cong W.X., Miao Y.L., Xu L., Zhang Y.H., Yuan C.L., Wang J.M., Zhuang T.T., Lin X.Y., Jiang L.L., Wang N.N., et al. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.) BMC Plant Biol. 2019;19:282. doi: 10.1186/s12870-019-1887-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Crisp P.A., Ganguly D., Eichten S.R., Borevitz J.O., Pogson B.J. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2016;2:e1501340. doi: 10.1126/sciadv.1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lamke J., Baurle I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017;18:124. doi: 10.1186/s13059-017-1263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li P., Yang H., Wang L., Liu H.J., Huo H.Q., Zhang C.J., Liu A.Z., Zhu A.D., Hu J.Y., Lin Y.J., et al. Physiological and Transcriptome Analyses Reveal Short-Term Responses and Formation of Memory Under Drought Stress in Rice. Front. Genet. 2019;10:55. doi: 10.3389/fgene.2019.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kou S.Y., Gu Q.Y., Duan L., Liu G.J., Yuan P.R., Li H.H., Wu Z.G., Liu W.H., Huang P., Liu L. Genome-Wide Bisulphite Sequencing Uncovered the Contribution of DNA Methylation to Rice Short-Term Drought Memory Formation. J. Plant Growth Regul. 2021:1–15. doi: 10.1007/s00344-021-10483-3. [DOI] [Google Scholar]
- 105.Ou X.F., Zhang Y.H., Xu C.M., Lin X.Y., Zang Q., Zhuang T.T., Jiang L.L., von Wettstein D., Liu B. Transgenerational Inheritance of Modified DNA Methylation Patterns and Enhanced Tolerance Induced by Heavy Metal Stress in Rice (Oryza sativa L.) PLoS ONE. 2012;7:e41143. doi: 10.1371/journal.pone.0041143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wibowo A., Becker C., Marconi G., Durr J., Price J., Hagmann J., Papareddy R., Putra H., Kageyama J., Becker J., et al. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife. 2016;5:e13546. doi: 10.7554/eLife.13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hatzig S.V., Nuppenau J.N., Snowdon R.J., Schiessl S.V. Drought stress has transgenerational effects on seeds and seedlings in winter oilseed rape (Brassica napus L.) BMC Plant Biol. 2018;18:297. doi: 10.1186/s12870-018-1531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen R., Li M., Zhang H.Y., Duan L.J., Sun X.J., Jiang Q.Y., Zhang H., Hu Z. Continuous salt stress-induced long non-coding RNAs and DNA methylation patterns in soybean roots. BMC Genom. 2019;20:730. doi: 10.1186/s12864-019-6101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tan M.P. Analysis of DNA methylation of maize in response to osmotic and salt stress based on methylation-sensitive amplified polymorphism. Plant Physiol. Bioch. 2010;48:21–26. doi: 10.1016/j.plaphy.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 110.Zheng X.G., Chen L., Xia H., Wei H.B., Lou Q.J., Li M.S., Li T.M., Luo L.J. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 2017;7:39843. doi: 10.1038/srep39843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Regulski M., Lu Z.Y., Kendall J., Donoghue M.T.A., Reinders J., Llaca V., Deschamps S., Smith A., Levy D., McCombie W.R., et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 2013;23:1651–1662. doi: 10.1101/gr.153510.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Herman J.J., Sultan S.E. DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Proc. Biol. Sci. 2016;283:20160988. doi: 10.1098/rspb.2016.0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Geng Y.P., Chang N., Zhao Y.W., Qin X.Y., Lu S.G., Crabbe M.J.C., Guan Y.B., Zhang T.C. Increased epigenetic diversity and transient epigenetic memory in response to salinity stress in Thlaspi arvense. Ecol. Evol. 2020;10:11622–11630. doi: 10.1002/ece3.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zheng X.G., Chen L., Li M.S., Lou Q.J., Xia H., Wang P., Li T.M., Liu H.Y., Luo L.J. Transgenerational Variations in DNA Methylation Induced by Drought Stress in Two Rice Varieties with Distinguished Difference to Drought Resistance. PLoS ONE. 2013;8:e80253. doi: 10.1371/journal.pone.0080253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Boyko A., Blevins T., Yao Y.L., Golubov A., Bilichak A., Ilnytskyy Y., Hollander J., Meins F., Kovalchuk I. Transgenerational Adaptation of Arabidopsis to Stress Requires DNA Methylation and the Function of Dicer-Like Proteins. PLoS ONE. 2010;5:e9514. doi: 10.1371/annotation/726f31b5-99c4-44e9-9cd6-b8d66b3f6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Singh N., Titov V., Ayemere I., Byeon B., Kovalchuk I. Multigenerational exposure to heat stress induces phenotypic resilience, and genetic and epigenetic variations in Arabidopsis thaliana offspring. Front. Plant Sci. 2020;13:728167. doi: 10.3389/fpls.2022.728167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sun C., Ali K., Yan K., Fiaz S., Dormatey R., Bi Z.Z., Bai J.P. Exploration of Epigenetics for Improvement of Drought and Other Stress Resistance in Crops: A Review. Plants. 2021;10:1226. doi: 10.3390/plants10061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hou Q.C., Wan X.Y. Epigenome and Epitranscriptome: Potential Resources for Crop Improvement. Int. J. Mol. Sci. 2021;22:12912. doi: 10.3390/ijms222312912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim E.Y., Kim K.D., Cho J. Harnessing epigenetic variability for crop improvement: Current status and future prospects. Genes Genom. 2022;44:259–266. doi: 10.1007/s13258-021-01189-7. [DOI] [PubMed] [Google Scholar]
- 120.Gallego-Bartolome J. DNA methylation in plants: Mechanisms and tools for targeted manipulation. New Phytol. 2020;227:38–44. doi: 10.1111/nph.16529. [DOI] [PubMed] [Google Scholar]