Abstract
A new fluorescence in situ hybridization method using peptide nucleic acid (PNA) probes for identification of Brettanomyces is described. The test is based on fluorescein-labeled PNA probes targeting a species-specific sequence of the rRNA of Dekkera bruxellensis. The PNA probes were applied to smears of colonies, and results were interpreted by fluorescence microscopy. The results obtained from testing 127 different yeast strains, including 78 Brettanomyces isolates from wine, show that the spoilage organism Brettanomyces belongs to the species D. bruxellensis and that the new method is able to identify Brettanomyces (D. bruxellensis) with 100% sensitivity and 100% specificity.
Brettanomyces is a well-recognized wine spoilage yeast that causes an undesirable flavor. The sensory character of this “Bretty” flavor is often described as mousiness, barnyard, horse sweat, or Band-Aid (5, 9). Current methods for identification and enumeration of Brettanomyces contamination take 1 to 2 weeks and rely on growth on a semiselective culture medium, followed by final identification by biochemical and physiological analysis and morphology as determined by microscopic examination (3). Morphological characterization of Brettanomyces is somewhat subjective, and there have been various morphological descriptions, such as bud scars, bullet shape, and Mickey Mouse-like. Newer techniques for rapid detection and identification of Brettanomyces, such as an enzyme-linked immunosorbent assay (7) and, more recently, PCR (6), have also been described.
The nomenclature of Brettanomyces used in the wine industry differs from that of the recently revised taxonomy of yeasts (11, 12). Enologists refer to the spoilage organism as Brettanomyces or “Brett” or, in some publications, by the species names Dekkera intermedia and Brettanomyces intermedius (3), Brettanomyces lambicus (3), Brettanomyces custersii, or Dekkera bruxellensis (6). Today, only D. bruxellensis is an accepted species name, and the other names are considered synonyms.
Peptide nucleic acid (PNA) molecules are pseudopeptides which are able to hybridize to complementary nucleic acid targets (RNA and DNA) obeying Watson-Crick base pairing rules (2, 10). Due to their uncharged, neutral backbone, PNA probes exhibit favorable hybridization characteristics, such as high specificity, strong affinity, and rapid kinetics resulting in improved hybridization to highly structured targets, such as rRNA (13). In addition, the relatively hydrophobic character of PNAs compared to DNA oligonucleotides makes PNA probes capable of penetrating the hydrophobic cell wall following mild fixation conditions that do not lead to disruption of cell morphology (14). These unique characteristics of PNA have opened new possibilities for molecular diagnostic assays.
The D1-D2 region of 26S ribosomal DNA (rDNA) of eucaryotic organisms shows a high degree of species variation and has been used for identification and taxonomy of yeast species (1, 8). In this study, 26S rDNA sequence information was used to design species-specific probes targeting the rRNA of D. bruxellensis. These probes were used to develop a new fluorescence in situ hybridization (FISH) method for identification of Brettanomyces.
MATERIALS AND METHODS
Yeast strains.
Five type strains representing the five Dekkera and Brettanomyces species, 10 reference strains representing synonyms of D. bruxellensis, and 26 yeast species potentially found in wine were obtained from the Agricultural Research Service Culture Collection (Peoria, Ill.) and the American Type Culture Collection (Manassas, Va.). Seventy-eight wine isolates of Brettanomyces were kindly provided by E&J Gallo (Modesto, Calif.), California State University at Fresno (Fresno, Calif.), Sutter Home (St. Helena, Calif.), Robert Mondavi Winery (Oakville, Calif.), and Boston Probes, Inc. (Bedford, Mass.). Eight wine isolates of cycloheximide-resistant spheroidal yeasts were kindly provided by Beringer (St. Helena, Calif.), Vinquiry, Inc. (Windsor, Calif.), Columbia Winery (Woodinville, Wash.), and Robert Mondavi Winery. The spheroid yeasts were included because they grow relatively slowly on cycloheximide containing media, like Brettanomyces, and may therefore be misidentified as Brettanomyces.
Wine samples.
Three wine samples confirmed to be positive for Brettanomyces by microscopy were kindly provided by Vinquiry, Inc.
Culture media and growth conditions.
A nonselective yeast and mold medium (YM) (Difco Laboratories, Detroit, Mich.) and a Brettanomyces-selective medium (BSM) (Millipore Corp., Bedford, Mass.) were used. BSM contains cycloheximide as well as antibiotics that inhibit bacterial growth. Yeast strains were propagated in YM at 25°C.
For FISH analysis, strains were spread onto YM agar and incubated at 30°C, whereas wine samples were filtered through 47-mm-diameter, 0.45-μm-pore-size HVLP filter membranes (Millipore) and then incubated at 30°C on a pad soaked with 2 ml of BSM in a small petri dish.
Preparation of smears.
For each smear, 1 drop of phosphate-buffered saline was placed in the well of a Teflon-coated microscope slide (Erie Scientific, Portsmouth, N.H.). A small portion of a colony was picked with a clean, sterile toothpick and suspended in the phosphate-buffered saline by gentle mixing in the microscope well. The slide was then placed on a 50°C slide warmer for 30 min, after which the smears were dry.
Selection of probe sequence.
Sequence processing was performed by using computer software from DNASTAR (Madison, Wis.). Alignment of closely related yeast D1-D2 26S rDNA sequences (1, 8) was performed by using the Megalign (version 4.03) program. From the alignments, species-specific sequences of D. bruxellensis were identified and subsequently checked for significant sequence similarity with the whole GenBank database by using the GeneMan (version 3.30) software and an Advanced BLAST search of the GenBank nr-database (www.ncbi.nlm.nih.govlast). Complementary 15-mer probe sequences were checked for significant levels of secondary structure by using the PrimerSelect program (version 4.03).
Synthesis of fluorescein-labeled PNA probes.
PNAs were synthesized by using an Expedite 8909 nucleic acid synthesis system with the PNA option and reagents from PE Biosystems, Foster City, Calif. The aqueous solubility of the PNAs was enhanced by flanking the nucleobase sequence with solubility enhancers (4). The N terminus of each PNA was extended by using an 8-amino-3,6-dioxaoctanoic acid spacer (PE Biosystems). Following removal of the terminal Fmoc protecting group, the N terminus of the resin-bound PNA was labeled with 5(6)-carboxyfluorescein. Specifically, the resin was treated with 250 μl of a solution containing 0.5 M 5(6)-carboxyfluorescein (Aldrich, Milwaukee, Wis.), 0.5 M N,N′-diisopropylcarbodiimide (Aldrich), and 0.5 M 1-hydroxy-7-azabenzotriazole (PE Biosystems) in dimethylformamide (Burdick & Jackson, Muskegon, Mich.) (15). The synthesis support was then washed and dried under a high vacuum. After removal from the synthesis cartridge, the resin was transferred to an Ultrafree spin cartridge (Millipore Corp.) for cleavage and deprotection (User's Guide. PNA Chemistry for the Expedite Nucleic Acid Synthesis System, Perspective Biosystems, Inc., Framingham, Mass.). The product was analyzed by high-performance liquid chromatography and matrix-assisted laser desorption ionization-time of flight mass spectrometry to confirm its purity and identity. The fluorescein-labeled PNA probe was finally purified by using standard reversed-phase C18 chromatographic methods.
FISH.
Smears were covered with approximately 20 μl of a hybridization solution containing 10% (wt/vol) dextran sulfate (Sigma Chemical Co., St. Louis, Mo.), 10 mM NaCl (J. T. Baker), 30% (vol/vol) formamide (Sigma), 0.1% (wt/vol) sodium pyrophosphate (Sigma), 0.2% (wt/vol) polyvinylpyrrolidone (Sigma), 0.2% (wt/vol) Ficoll (Sigma), 5 mM Na2EDTA (Sigma), 0.1% (vol/vol) Triton X-100 (Aldrich), 50 mM Tris-HCl (pH 7.5), and 100 nM fluorescein-labeled PNA probe. Coverslips were put on the smears to ensure even coverage with hybridization solution, and the slides were subsequently placed on a slide warmer with a humidity chamber (Slidemoat, Boeckel, Germany) and incubated for 30 min at 50°C. Following hybridization, the coverslips were removed by submerging the slides in approximately 20 ml of prewarmed 5 mM Tris–15 mM NaCl–0.1% (vol/vol) Triton X-100 (pH 10) per slide in a water bath at 50°C and washed for 30 min. The slides were then cooled to room temperature by brief immersion in H2O and air dried following brief immersion in ethanol. Each smear was finally mounted by using 1 drop of IMAGEN mounting fluid (DAKO, Ely, United Kingdom) and covered with a coverslip. Microscopic examination was conducted with a fluorescence microscope (Optiphot; Nikon Corporation, Tokyo, Japan) equipped with a 60×/1.4 oil objective (Nikon), an HBO 100-W mercury lamp, and a fluorescein isothiocyanate-Texas Red dual-band filter set (Chroma Technology Corp., Brattleboro, Vt.). Images were obtained by using a color charge-coupled device camera (Diagnostic Instruments, Inc., Sterling Heights, Mich.) connected to a computer system.
RESULTS
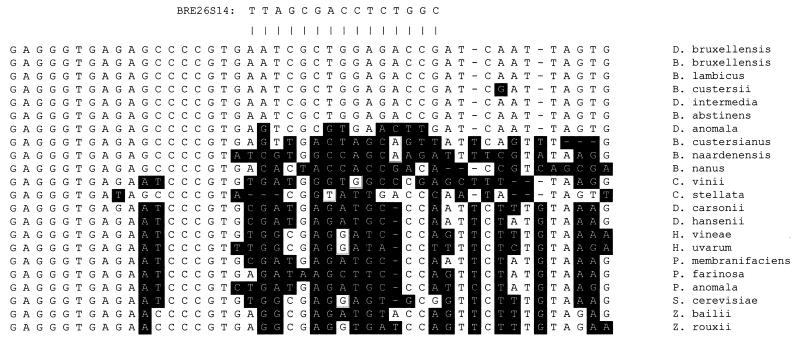
Sequences of D1-D2 26S rDNA from yeast species potentially found in wine were aligned in order to identify species-specific target regions of D. bruxellensis rRNA. The optimal target sequence was found in all synonyms of D. bruxellensis and differed by at least four bases from the sequences of other yeast species (Fig. 1). In addition, a BLAST search did not reveal other eucaryotic or bacterial rDNA sequences with the exact same target sequence.
FIG. 1.
Alignment of partial yeast D1-D2 26S rDNA sequences for probe selection. The anti-parallel hybridization sequence of the BRE26S14 PNA probe is shown above the alignment. Base differences between the target sequences and other sequences are highlighted.
Initially, the specificity of BRE26S14 labeled with fluorescein (BRE26S14/Flu) was tested by FISH by using the type strains of the five species of Dekkera and Brettanomyces (Table 1), as well as 10 reference strains representing different synonyms of D. bruxellensis (Table 2). Twenty-six other yeast species potentially found in wine were also examined for reactivity with the probe (Table 3). As predicted from the alignment of sequences in the target area, BRE26S14/Flu hybridized only to the type strain of D. bruxellensis and synonyms thereof, whereas it did not detect any of the other 26 yeast species. In addition, BRE26S14/Flu did not react with any of eight isolates of spheroid yeasts capable of growing on BSM. These unidentified spheroid yeasts grow relatively slowly on cycloheximide-containing media, like Brettanomyces, and are therefore among the species most likely to be misidentified as Brettanomyces by persons without experience with identification of Brettanomyces.
TABLE 1.
Detection of type strains of Dekkera and Brettanomyces accepted species with BRE26S14/Flu
| Organism | Straina | Result |
|---|---|---|
| Dekkera anomala | NRRL Y-17522 | − |
| Dekkera bruxellensis | NRRL Y-12961 | + |
| Brettanomyces naardenensis | NRRL Y-17526 | − |
| Brettanomyces custersianus | NRRL Y-6653 | − |
| Brettanomyces nanus | NRRL Y-17527 | − |
NRRL, Agricultural Research Service Culture Collection, Peoria, Ill.
TABLE 2.
Detection of D. bruxellensis reference strains (synonyms) with BRE26S14/Flu
| Organism | Straina | Result |
|---|---|---|
| Brettanomyces bruxellensis | NRRL Y-1411 | + |
| Brettanomyces lambicus | NRRL Y-1413 | + |
| Mycotorula intermedia | NRRL Y-17534 | + |
| Brettanomyces bruxellensis | NRRL Y-1412 | + |
| Brettanomyces schanderlii | NRRL Y-17523 | + |
| Brettanomyces abstinens | NRRL Y-17525 | + |
| Dekkera intermedia | ATCC 52904b | + |
| Dekkera intermedia | ATCC 56869 | + |
| Dekkera intermedia | ATCC 64276 | + |
| Dekkera lambica | ATCC 10563c | + |
NRRL, Agricultural Research Service Culture Collection, Peoria, Ill.; ATCC, American Type Culture Collection, Manassas, Va.
Equivalent to strain NRRL Y-17523.
Equivalent to strain NRRL Y-1413.
TABLE 3.
Reactions of other yeast species potentially found in wine with BRE26S14/FLU
| Organism | Straina | Result |
|---|---|---|
| Hanseniaspora uvarum | NRRL Y-1614 | − |
| Hanseniaspora guilliermondii | NRRL Y-1625 | − |
| Hanseniaspora occidentalis | NRRL Y-7946 | − |
| Hanseniaspora osmophila | NRRL Y-1613 | − |
| Hanseniaspora valbyensis | NRRL Y-1626 | − |
| Hanseniaspora vineae | NRRL Y-17529 | − |
| Kloeckera lindneri | NRRL Y-17531 | − |
| Torulaspora delbrueckii | NRRL Y-866 | − |
| Debaryomyces hansenii | NRRL Y-7426 | − |
| Debarymyces carsonii | NRRL YB-4275 | − |
| Candida stellata | NRRL Y-1446 | − |
| Metschnikowia pulcherrima | NRRL Y-7111 | − |
| Rhodotorula fujisanensis | NRRL YB-4824 | − |
| Rhodotorula glutinis | NRRL Y-2502 | − |
| Rhodotorula graminis | NRRL Y-2474 | − |
| Schizosaccharomyces pombe | NRRL Y-12796 | − |
| Pichia anomala | NRRL Y-366 | − |
| Pichia membranifaciens | NRRL Y-2026 | − |
| Pichia farinosa | NRRL Y-7553 | − |
| Saccharomyces cerevisiae | ATCC 4098 | − |
| Saccharomyces kluyveri | NRRL Y-12651 | − |
| Saccharomycodes ludwigii | NRRL Y-12793 | − |
| Zygosaccharomyces bailii | ATCC 66825 | − |
| Zygosaccharomyces bisporus | NRRL Y-12626 | − |
| Zygosaccharomyces rouxii | NRRL Y-229 | − |
| Zygosaccharomyces florentinus | NRRL Y-1560 | − |
NRRL, Agricultural Research Service Culture Collection, Peoria, Ill.; ATCC, American Type Culture Collection, Manassas, Va.
The sensitivity of BRE26S14/Flu for detection of the actual spoilage organism, Brettanomyces, was then assessed by analyzing 78 wine isolates of Brettanomyces. All isolates were identified by the probe; thus, there was 100% correlation with the results of methods used by wine makers to identify Brettanomyces isolated from wine. This result provided further proof that the spoilage organism named Brettanomyces belongs to the species D. bruxellensis.
Finally, the routine applicability of the method for identification of colonies of Brettanomyces obtained directly from wine samples was also evaluated with three Brettanomyces-contaminated wines. Colonies from all three wine samples were identified by BRE26S14/Flu.
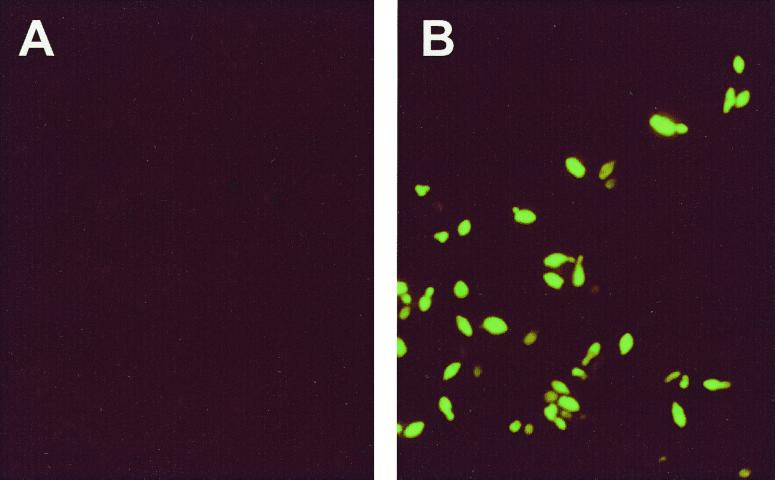
Figure 2 shows images obtained by the FISH method with smears of colonies grown for 1 to 2 weeks on BSM following membrane filtration. Individual cells of Brettanomyces were identified by their bright green fluorescence, whereas undetected cells were reddish brown. Often mixtures of cells exhibiting high, medium, low, and no green fluorescence were observed in smears of cells from a Brettanomyces colony. This was not due to a mixed population as all cells originated from the same colony. Instead, it was most likely a result of variable amounts of target rRNA in the individual cells due to different metabolic stages of the cells in a colony, so that some cells were growing and multiplying while others may have been resting or even dead. Alternatively, the variability in intensity may have been due to variable permeability of the cell wall. The images also demonstrate that the morphology of the cells was not affected by the FISH procedure. However, some of the morphological characteristics were not as pronounced when this method was used as they were when bright-field microscopy was used because the cell membrane was not fluorescent since the rRNA molecules were located in the cell cytoplasm.
FIG. 2.
Microscope images of cells from colonies of a negative control (spheroid yeast #1; Beringer) (A) and a Brettanomyces-positive wine sample (Vinquiry, Inc.) (B).
DISCUSSION
We showed that using fluorescently labeled PNA oligomers is a powerful method for identifying colonies of the spoilage organism Brettanomyces (D. bruxellensis). The FISH method described here provides a combination of the high specificity offered by molecular techniques with the simplicity of microscopy. In contrast to the previous subjective method of identification based on morphology, this new method provides 100% definitive identification of Brettanomyces irrespective of the experience and skill of the wine technologist.
This study also shows that Brettanomyces, the spoilage organism in wine, belongs to the species D. bruxellensis. Probes designed by using sequence data from taxonomic studies have been shown to detect all 78 confirmed isolates of Brettanomyces. To our knowledge, this is the first study that provides a link between the recently revised taxonomy of yeasts and the spoilage organism Brettanomyces. The various descriptions of the flavors caused by Brettanomyces, as well as the many somewhat dubious morphological descriptions and the many synonyms, can all be ascribed to D. bruxellensis. Although D. anomala, the other species of the genus Dekkera, may spoil wine, it is not associated with the wine spoilage organism Brettanomyces.
In summary, our new method for identification of Brettanomyces is easily adapted to microscopic techniques currently used in wine laboratories, except that a fluorescence microscope is required. Furthermore, the uncertainty and subjectivity associated with the currently used methods are eliminated by the specificity of the PNA probe, which provides definitive identification of the spoilage organism.
ACKNOWLEDGMENTS
We thank Rich Morenzoni (E&J Gallo), Kenneth Fugelsang (California State University at Fresno), Glenn Andrade (Sutter Home), Judy Miles (Beringer), Pat Paris (Robert Mondavi Winery), Neil Brown (Vinquiry, Inc.), and Bruce Watson (Columbia Winery) for providing many yeast isolates. S. Casey, J. MacNeill, and S. Voetsch are acknowledged for synthesis of the PNA probes.
REFERENCES
- 1.Boekhout T, Kurtzman C P, O'Donnell K, Smith M T. Phylogeny of the yeast genera Hanseniaspora (anamorph Kloeckera), Dekkera (anamorph Brettanomyces), and Eeniella as inferred from partial 26S ribosomal DNA nucleotide sequences. Int J Syst Bacteriol. 1994;44:781–786. doi: 10.1099/00207713-44-4-781. [DOI] [PubMed] [Google Scholar]
- 2.Egholm M, Buchard O, Christensen L, Behrens C, Freier S M, Driver D A, Berg R H, Kim S K, Norden B, Nielsen P E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen bonding rules. Nature. 1993;365:556–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 3.Fugelsang K C. Yeasts and molds. In: Fugelsang K C, editor. Wine microbiology. New York, N.Y: Chapman & Hall; 1997. pp. 68–116. [Google Scholar]
- 4.Gildea B D, Casey S, MacNeill J, Perry-O'Keefe H, Sørensen D, Coull J M. PNA solubility enhancers. Tetrahedron Lett. 1998;39:7255–7258. [Google Scholar]
- 5.Hereszeztyn T. Formation of substituted tetrahydropyridines by species of Brettanomyces and Lactobacillus isolated from mousy wines. Am J Enol Vitic. 1986;37:127–132. [Google Scholar]
- 6.Ibeas J I, Lozano I, Perdigones F, Jimenez J. Detection of Dekkera-Brettanomyces strains in sherry by a nested PCR method. Appl Environ Microbiol. 1996;62:998–1003. doi: 10.1128/aem.62.3.998-1003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuniyuki A H, Rous C, Sanderson J L. Enzyme-linked immunosorbent assay (ELISA) detection of Brettanomyces contaminants in wine production. Am J Enol Vitic. 1984;35:143–145. [Google Scholar]
- 8.Kurtzman C P, Robnett C J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- 9.Licker J L, Acree T E, Henick-Kling T. What is ‘Brett’ (Brettanomyces) flavor?: a preliminary investigation. Am Chem Soc Symp Ser. 1999;714:96–115. [Google Scholar]
- 10.Nielsen P E, Egholm M, Buchard O. Peptide nucleic acids (PNA). A DNA mimic with a peptide backbone. Bioconj Chem. 1994;5:3–7. doi: 10.1021/bc00025a001. [DOI] [PubMed] [Google Scholar]
- 11.Smith M T. Dekkera van der Walt. In: Kurtzman C P, Fell J W, editors. The yeasts. A taxonomic study. Amsterdam, The Netherlands: Elsevier Science B.V.; 1998. pp. 174–177. [Google Scholar]
- 12.Smith M T. Brettanomyces Kufferath & van Laer. In: Kurtzman C P, Fell J W, editors. The yeasts. A taxonomic study. Amsterdam, The Netherlands: Elsevier Science B.V.; 1998. pp. 450–453. [Google Scholar]
- 13.Stefano K, Hyldig-Nielsen J J. Diagnostic applications of PNA oligomers. In: Minden S A, Savage L M, editors. Diagnostic gene detection & quantification technologies. Southborough, Mass: IBC Library Series; 1997. pp. 19–37. [Google Scholar]
- 14.Stender H, Mollerup T A, Lund K, Petersen K H, Hongmanee P, Godtfredsen S E. Direct detection and identification of Mycobacterium tuberculosis in smear-positive sputum samples by fluorescence in situ hybridization (FISH) using peptide nucleic acid (PNA) probes. Int J Tuberc Lung Dis. 1999;3:830–837. [PubMed] [Google Scholar]
- 15.Weber P J A, Bader J E, Folkes G, Beck-Sickinger A G. A fast and inexpensive method for N-terminal fluorescein-labeling of peptides. Bioorg Med Chem Lett. 1998;8:597–600. doi: 10.1016/s0960-894x(98)00084-5. [DOI] [PubMed] [Google Scholar]