Abstract
The electrocardiogram (ECG) empowered clinician scientists to measure the electrical activity of the heart noninvasively to identify arrhythmias and heart disease. Shortly after the standardization of the 12-lead ECG for the diagnosis of heart disease, several families with autosomal recessive (Jervell and Lange-Nielsen Syndrome) and dominant (Romano–Ward Syndrome) forms of long QT syndrome (LQTS) were identified. An abnormally long heart rate-corrected QT-interval was established as a biomarker for the risk of sudden cardiac death. Since then, the International LQTS Registry was established; a phenotypic scoring system to identify LQTS patients was developed; the major genes that associate with typical forms of LQTS were identified; and guidelines for the successful management of patients advanced. In this review, we discuss the molecular and cellular mechanisms for LQTS associated with missense variants in KCNQ1 (LQT1) and KCNH2 (LQT2). We move beyond the “benign” to a “pathogenic” binary classification scheme for different KCNQ1 and KCNH2 missense variants and discuss gene- and mutation-specific differences in K+ channel dysfunction, which can predispose people to distinct clinical phenotypes (e.g., concealed, pleiotropic, severe, etc.). We conclude by discussing the emerging computational structural modeling strategies that will distinguish between dysfunctional subtypes of KCNQ1 and KCNH2 variants, with the goal of realizing a layered precision medicine approach focused on individuals.
Keywords: long QT syndrome, KCNQ1, KCNH2, K+ channel, heart, arrhythmia, electrocardiogram, molecular dynamics
1. Introduction
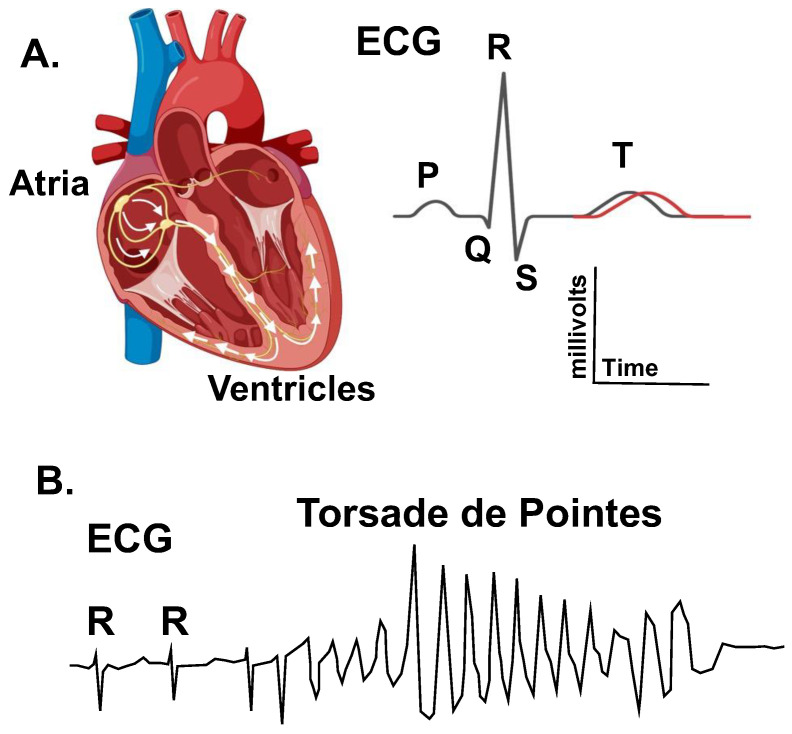
People with long QT syndrome (LQTS) are at increased risk for syncope and ventricular arrhythmias (e.g., Torsade de pointes) that may result in sudden cardiac death (SCD) (Figure 1) [1,2]. Congenital LQTS is most commonly caused by autosomal dominant mutations in the cardiac K+ channel genes, KCNQ1 (LQTS type 1 or LQT1) and KCNH2 (LQTS type 2 or LQT2) [3]. KCNQ1 and KCNH2 each encode a different pore-forming α-subunit of voltage-gated channel proteins, Kv7.1 and Kv11.1, respectively. The Kv7.1 and Kv11.1 channels conduct the delayed-rectifier K+ currents important for normal ventricular repolarization.
Figure 1.
Long-QT syndrome (LQTS) is an arrhythmia disorder that sometimes causes a prolongation in the QT interval as measured on an electrocardiogram (ECG). (A). The left diagram shows a cross section of the heart illustrating the cardiac conduction pathway in the atrial and ventricles. The electrical activation of the heart (arrows) is measured using electrocardiography. The right image shows an ECG trace for a single cardiac cycle. The depolarization of the atria generates the P-wave, the depolarization of the ventricle generates the QRS complex, and the repolarization of the ventricle generates the T-wave. The QT-interval changes as a function of the heart rate (as measured by the RR-interval). (A) Prolongation in the heart rate-corrected QT-interval is a biomarker for an increased risk of polymorphic ventricular tachycardia Torsade de pointes. (B). Diagram of an ECG trace showing a typical Torsade de pointes arrhythmia. Torsade results in a loss in cardiac output and can cause syncope, seizures, and sudden cardiac death.
Most people diagnosed with LQT1 and LQT2 can be successfully managed with medications that block β-adrenergic receptors (β-blockers). Examples of β-blockers used to manage LQTS include the non-selective β-blockers nadolol and propranolol [4]. Clinical management also includes an avoidance of hypokalemia and medications that are known to prolong the QT interval [5]. People who suffered cardiac arrest or have higher-risk clinical phenotypes can be managed using implantable cardioverter defibrillators and cardiac sympathetic denervation [6,7,8]. Although people with LQT1 and LQT2 are managed similarly, they tend to show gene-specific differences in symptoms. For example, the timing of arrhythmogenic events in people with LQT1 tend to occur during the daytime, and these events are commonly triggered by activity and physical exertion [9]. In contrast, people who have LQT2 tend to suffer events in the morning, and these events associate with sudden arousal (e.g., standing, telephone ringing, alarm clock, etc.) [10]. The expressivity and penetrance of the LQT1- and LQT2-phenotypes are also influenced by the type of mutation; the location of the mutation; the impact that the mutation has on the channel function; and several additional biological, environmental, and genetic factors [1,11,12,13,14,15]. This review provides a brief background on the initial phenotypic identification of congenital LQTS and discusses the different mechanisms by which KCNQ1 (Kv7.1) or KCNH2 (Kv11.1) mutations can impact K+ currents and channel function to cause LQT1 and LQT2, respectively. Specifically, we use simplified computational models and simulations to illustrate how different LQT1- and LQT2-linked mutations can impact the human ventricular action potential (AP) duration (a cellular surrogate to the QT-interval). The AP simulations are also useful for understanding how gene-specific differences in the loss of delayed-rectifier K+ currents may generate distinct cellular and clinical phenotypes. We conclude with a discussion summarizing the computational approaches and strategies that are being developed to navigate the increasingly complex genotype-phenotype landscape and understand variant-specific differences in channel structure and dysfunction. The goal of this review is to compliment several existing reviews that focus on the genetics or ventricular repolarization and use of novel in vitro strategies (e.g., human inducible pluripotent stem-cell-derived cardiomyocytes) to understand inherited arrhythmia syndromes [16,17,18].
2. The QT-Interval: From Biomarker to Clinical Genetics
Surface electrodes placed on the skin can record the electrical activity of excitable tissues in the body including the heart. The electrocardiogram (ECG) was introduced over 120 years ago, and it was adapted to provide clinician scientists with a valuable instrument to measure the structure and function of the heart noninvasively [19,20]. Initially the ECG was used to identify cardiac arrhythmias (e.g., atrial fibrillation) [21], but it was also used to recognize ECG patterns indicative of heart disease (e.g., ischemic heart disease) [22]. In 1954, the American Heart Association published their recommendation for the standardization of the 12-lead ECG, and the 12-lead ECG remains the gold-standard for the noninvasive diagnosis of arrhythmias, conduction abnormalities, and several types of heart disease [23,24].
In 1957, Drs. Jervell and Lange-Nielsen first described a family with a “peculiar” heart disease that caused syncope and sudden death in several children [25]. The clinicians noted that ECG testing in one of the children showed an abnormally long QT-interval that worsened with exercise. Exercise seemed to trigger arrhythmias because the children suffered syncope or sudden death while running, swimming, or playing. There was no evidence that the children’s hearts were structurally or functionally abnormal, but the afflicted children were deaf. This was the first known documented case of the very rare Jervell Lange-Neilson syndrome (JLNS), the autosomal recessive form of LQTS that sometimes associates with deafness.
In the years that followed, families who had the much more common autosomal dominant form of LQTS without deafness (Romano–Ward Syndrome or RWS) were identified. In 1963 and 1964, Dr. Romano and colleagues [26], as well as Dr. Ward [27], each described families with children who suffered abnormally long QT-intervals, syncope, and SCD. In his published case report, Dr. Ward noted [27]:
“The QT-interval in the present cases is, however, so very prolonged that it must be regarded as pathological by any standard. The E.C.G. changes suggest a metabolic disorder in the myocardium which slows repolarization of the muscle after systole… Undue sensitivity of the myocardium to sympathetic stimulation is postulated. Excessive sympathetic activity may prolong QT-interval and may also contribute to ventricular fibrillation. The clinical observation that the children have improved on a drug which blocks the effect of the sympathetic at the beta receptors in the myocardium might confirm this hypothesis…”.
Clinician-scientists soon began documenting more cases of congenital LQTS and an international LQTS registry was established in 1979 [28,29]. The identification and registry of families with JLNS or RWS eventually led to an evolving diagnostic scoring scheme (the Schwartz score) that is used to identify and treat people who most likely have LQTS [7,30,31,32]. These studies also facilitated the identification of the major genes that cause JLNS and RWS [11,33,34,35,36,37]. Families with members who suffer from JLNS have loss-of-function mutations in the K+ channel genes that underlie the slowly activating delayed-rectifier K+ current (IKs) in the heart (KCNQ1 and/or KCNE1). Most cases of RWS are linked to loss-of-function mutations in either KCNQ1 (LQT1) or KCNH2 (LQT2), or gain-of-function mutations in the Na+ channel gene SCN5A (LQT3). KCNH2 encodes the channel proteins that conduct the rapidly activating delayed-rectifier K+ current (IKr). Phenotypical manifestations of different SCN5A mutations are complex and are not discussed in this review, but they have been reviewed in detail elsewhere [38,39,40].
3. LQT1 and LQT2 Clinical Phenotypes
Despite all the progress made over the past 50 years, accurately identifying people with LQTS is challenging [41]. The reason is mainly because clinicians use several different approaches to measure the QT-interval. Manually defining the QT-interval on an ECG is subjective and highly individualized, and computerized approaches to measure the ECG can produce widely variable results [42,43,44,45,46]. The advent of inexpensive genetic screens held the promise for improving an accurate LQTS diagnosis and patient management [47]. However, a large number of rare missense variants in LQTS-susceptibility genes exist in the general population, and as such, it is difficult to determine whether a genotype-positive test for a rare missense variant in an LQTS-susceptibility gene is causative, a genetic modifier, or neutral [48].
Additional ECG testing methods have been developed to improve the identification of people with abnormalities in ventricular repolarization, such as having the person stand from a supine position quickly [49,50]. Subsequent to standing, abnormalities in the heart rate-corrected QT-interval (QTc-interval) between the peak heart rate (QT stretching) and return to baseline (QT stunning) could be an indicator of LQTS. Another method is to measure the QTc-interval after an exercise stress test [51]. Measuring the QTc-interval immediately after an exercise stress test is particularly useful for identifying people with “concealed LQTS”. Concealed LQTS is characterized by a normal to borderline QTc-interval at rest but prolongation with provocation [52]. People with concealed LQTS tend to show abnormal QTc-intervals four minutes after stopping maximal exercise [32,51].
The exercise stress test is particularly sensitive for identifying people with LQT1 [53,54,55,56]. This is likely because the relative contribution of IKs for driving ventricular repolarization is small in basal conditions but increases during adrenergic stimulation. People with LQT2 tend to show an increase in QT-interval hysteresis with exercise [57,58]. QT-interval hysteresis is characterized by longer QT-intervals at a given RR-interval while heart rates increase during exercise, and shorter QT-intervals at the same RR-interval while heart rates decrease during recovery from exercise. QT-interval hysteresis in humans likely depends in part on the dynamic change in the relative contribution of IKs to IKr to drive ventricular repolarization before, during, and immediately after exercise.
Artificial intelligence and machine learning approaches are now being developed for the identification of people with LQTS, including concealed LQTS [59,60]. Early studies suggest that artificial-intelligence-enhanced ECGs can reliably distinguish people with concealed LQTS or the three main genotypic subgroups (LQT1, LQT2, or LQT3) with ≈80% accuracy prior to a genetic test. However, as with any emerging strategies, there are limitations to generalizing this approach, and its validation across different clinics will need to be continually assessed.
In isolation, neither standing, exercise, nor artificial enhanced ECG tests are sufficient to diagnose LQTS. However, ECG screening methods that show QTc-interval adaptation and the use of artificial intelligence will help to improve the identification of people with LQTS [32]. In the next section, we use simplified ventricular AP simulations to better illustrate how a loss in IKs or IKr can predict differences in the LQT1 and LQT2-related cellular phenotypes in basal conditions or with β-adrenergic stimulation.
4. Importance of IKs and IKr in Cardiac Action Potential Repolarization
The normal function of IKs and IKr is to drive cardiac repolarization and contribute to repolarization reserve. Repolarization reserve is the concept that there exists repolarization redundancy (i.e., multiple repolarizing K+ currents) in the normal ventricle and conduction system to aid in preventing re-entry and early-after depolarizations [61,62]. IKs and IKr can serve both primary and secondary roles in repolarization because there is a large drop in total membrane conductance that occurs during the repolarization phase of the ventricular AP.
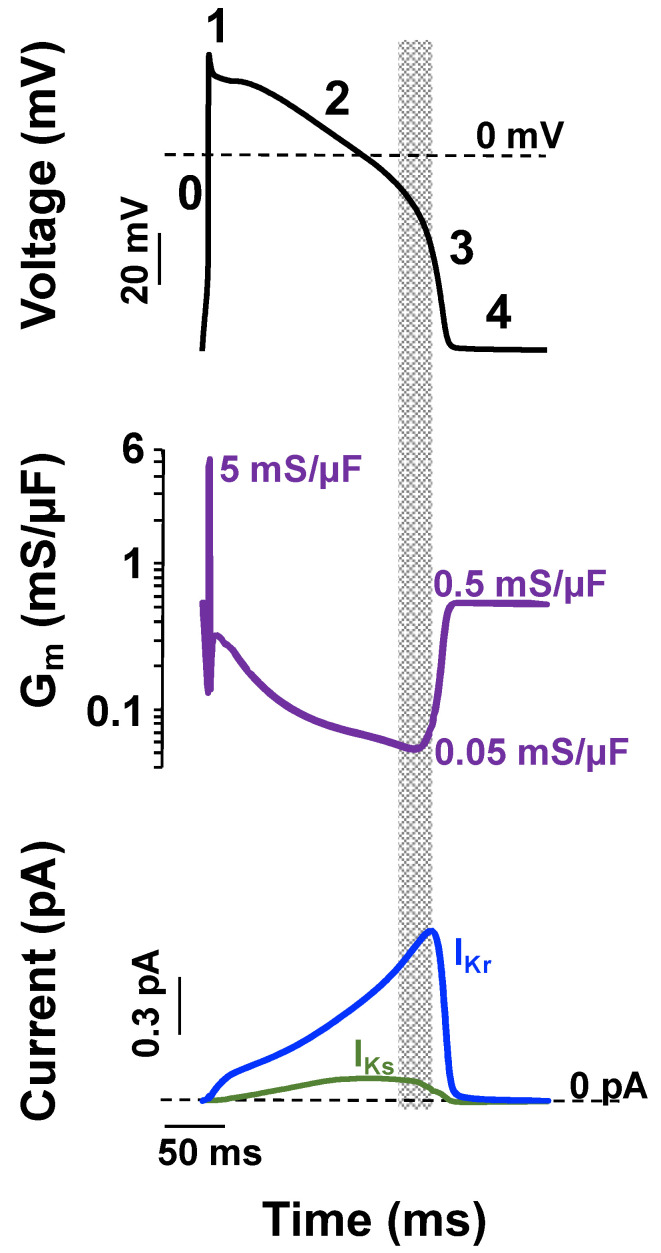
The ventricular AP waveform is defined as having five phases: 0, 1, 2, 3, and 4. Phase 0 describes the rapid membrane depolarization; phase 1 describes a brief but rapid repolarization that creates a notch; phase 2 refers to the plateau phase; phase 3 describes the rapid repolarization phase; and phase 4 corresponds to the stable resting membrane potential (Figure 2). IKs and IKr peak at the end of phase 2 and the start of phase 3. Notice that the absolute magnitude of IKs and IKr is very small. At first glance this seems counterintuitive to their functional role to drive membrane repolarization. However, recall that the voltage response of the membrane to changes in ionic currents depends on the absolute conductance of the cell. The small IKs and IKr have a large role in driving membrane repolarization because the absolute conductance of the cell membrane drops substantially between phases 2 and 3 of the ventricular AP (Figure 2).
Figure 2.
Small IKs and IKr can drive ventricular action potential repolarization and contribute to repolarization reserve because of low membrane conductance. Shown is a computational simulation for the change in membrane potential using the Soltis–Saucerman ventricular myocyte AP model [63] for normal conditions (37 °C and 5.4 mM extracellular [K+], black traces) driven at 1 Hz (upper panel). The phases of the AP are numbered (0–4). The middle panel shows the corresponding changes in membrane conductance (Gm), and the bottom panel shows the corresponding IKs (green line) and IKr (blue line). The dashed lines represent 0 mV or 0 pA levels in Vm and macroscopic current recordings, respectively. The start of the rapid repolarization phase occurs when Gm is at its lowest value and IKs and IKr are near their maximal values (shaded region).
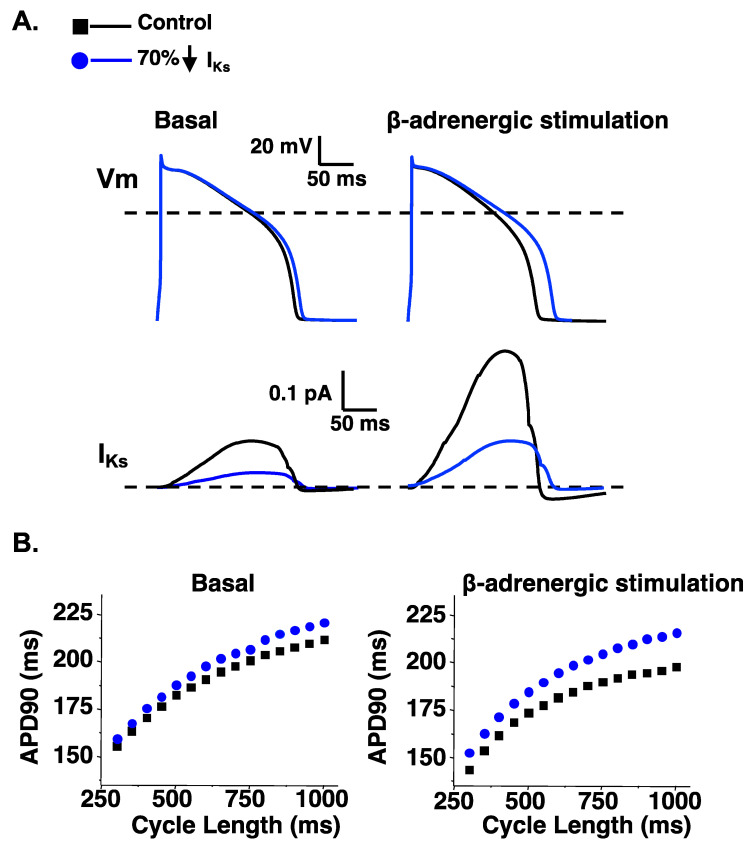
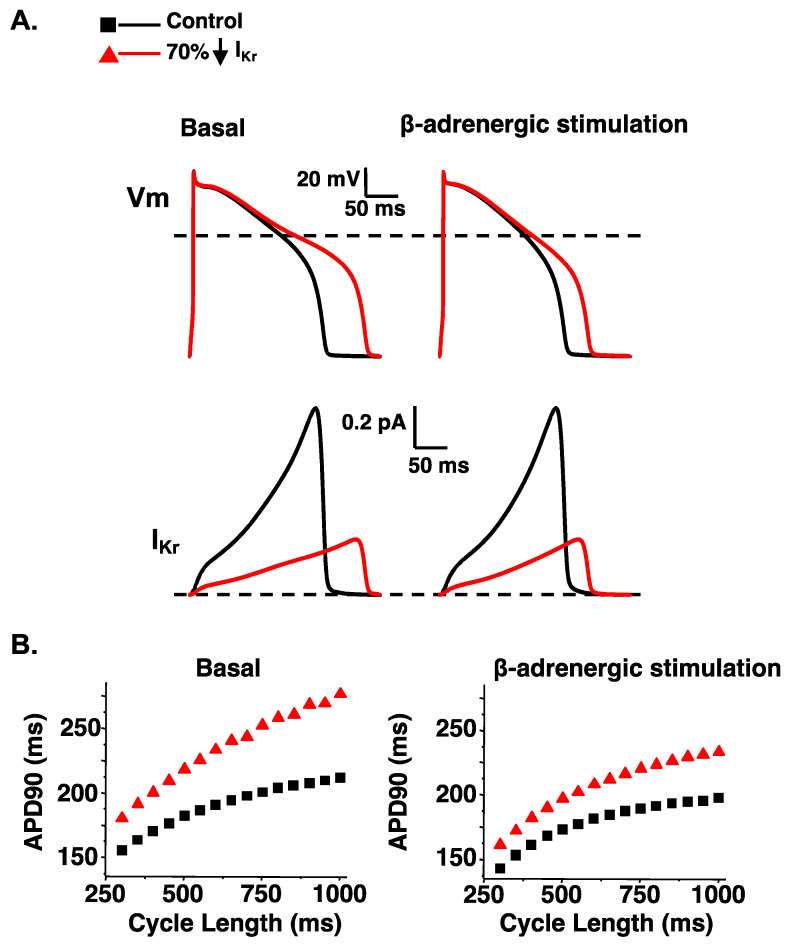
Computational simulations of the ventricular AP are useful for illustrating how a loss in either IKs or IKr can impact the AP duration at many different cycle lengths in basal conditions or conditions that simulate β-adrenergic stimulation. A reduction in IKs or IKr by 70% (to mimic LQT1 or LQT2, respectively) both predict a prolongation in the ventricular AP duration in basal conditions (as measured by the time to 90% repolarization or APD90) (Figure 3 and Figure 4). However, a 70% reduction in IKs predicts a smaller prolongation in the APD90 compared to a 70% reduction in IKr. The importance of IKs in regulating ventricular repolarization increases following β-adrenergic stimulation [63,64,65]. β-adrenergic stimulation increases inward calcium current (ICa), and the increase in inward ICa is normally offset by an increase in outward IKs to maintain or shorten the APD90 (Figure 3). Therefore, a 70% reduction in IKs predicts a longer prolongation in the APD90 during β-adrenergic stimulation compared to basal conditions. In contrast, the increase in IKs during β-adrenergic stimulation mitigates the prolongation in the APD90 in simulations of reduced IKr (Figure 4). The change in IKs with β-adrenergic stimulation helps to explain why some people with LQT1 show exaggerated QT-interval prolongation when β-adrenergic stimulation is high (e.g., during or immediately after exercise). The increase in IKs with β-adrenergic stimulation also helps to explain why some people with LQT2 have longer QT-intervals when β-adrenergic stimulation is low compared to when β-adrenergic stimulation is high (e.g., QT-interval hysteresis immediately before and after intense exercise).
Figure 3.
A reduction of IKs predicts a larger prolongation in the APD90 following β-adrenergic stimulation. (A). Representative AP waveforms and the corresponding IKs for control simulations (black line) and simulations in which the IKs component was reduced by 70% (blue line) for basal conditions (left) and β-adrenergic stimulation (right). (B). The steady state duration to 90% AP repolarization (APD90) was plotted as a function of the cycle length for basal conditions (left) or with β-adrenergic stimulation (right). Shown are the corresponding steady-state APD90 calculated for simulations at cycle lengths between 300 and 1000 ms for control simulations (black squares) and simulations in which the IKs component was reduced by 70% (blue circles) in basal conditions (left) or conditions that mimic β-adrenergic stimulation (right). The methodology and some of the data for producing these data is adapted from [66].
Figure 4.
A reduction in IKr predicts a disproportionate prolongation in the AP duration in the absence of β-adrenergic stimulation. (A). Representative AP waveforms and the corresponding IKr for control simulations (black line); simulations in which the IKr component was reduced by 70% (red line). On the left shows basal conditions and on the right show conditions that simulate β-adrenergic stimulation. (B). The duration APD90 was plotted as a function of the cycle length for basal conditions (left) or with β-adrenergic stimulation (right). Shown are the corresponding steady-state APD90 calculated for simulations at cycle lengths between 300 and 1000 ms for control simulations (black squares) and simulations in which the IKr component was reduced by 70% (red triangles) in basal conditions (left) or conditions that mimic β-adrenergic stimulation (right). Some data was adapted from [67].
5. Mechanistic Classification of LQT1- and LQT2-Linked Mutations
In addition to phenotypic differences in people with LQT1 and LQT2, LQT1, or LQT2, mutations can cause a loss of function in Kv7.1 and Kv11.1 by several different mechanisms. There is no one dominant disease-causing mutation for LQT1 or LQT2, and LQT1 and LQT2 each associate with hundreds of different rare mutations that span the length of the entire Kv7.1 or Kv11.1 channel protein [14,15,68,69]. It is increasingly clear that the type and location of different LQT1- or LQT2-linked mutations impact the severity of the molecular and clinical phenotype [14,70]. Kv7.1 and Kv11.1 channels are formed by the tetramerization channel proteins. Missense mutations that have dominant negative effects and interfere with the function of the wild-type (WT) protein tend to associate with worse clinical phenotypes than those that are haploinsufficient and primarily disrupt the function of the mutant protein [14,15,68,71].
To better illustrate how different types of LQT1- or LQT2-linked mutations reduce IKs or IKr, it is useful to break down the biophysical components that define the amplitude of the IKs or IKr [72]. Macroscopic current (I) is equal to the number of ion channels expressed in the cell surface membrane (n), the open probability of the channels (Po), and the amplitude of ionic current through an individual channel (i), such that I = n*(Po)*(i) [73]. A mutation that reduces IKs or IKr disrupts one or more of these factors. LQT1- and LQT2-linked mutations can be categorized into four distinct classes based on the dominant mechanism that causes a loss of function. Class 1 LQT1- or LQT2-linked mutations decrease n by disrupting the synthesis of the respective Kv7.1 or Kv11.1 channel proteins; Class 2 LQT1- or LQT2-linked mutations decrease n by decreasing the insertion of Kv7.1 or Kv11.1 channel proteins in the cell surface membrane (i.e., disrupt vesicular transport/secretory pathways); Class 3 LQT1- or LQT2-linked mutations disrupt normal Po by altering normal Kv7.1 or Kv11.1 channel gating (i.e., decreasing Po); and Class 4 LQT1- or LQT2-linked mutations decrease i (i.e., disrupt ionic selectivity or K+ permeation through the pore) [68,70,71,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89].
Many LQT1- and LQT2-linked mutations are radical mutations (i.e., mutations that shift the codon reading frame, disrupt mRNA transcript splicing, result in premature termination of translation, or large nucleotide deletions or insertions) and are expected to cause haploinsufficiency via a Class 1 mechanism [47,72]. Studies suggest a common mechanism by which many LQT1- and most LQT2-linked missense mutations cause a loss of channel function is to disrupt channel protein folding/trafficking/turnover and decrease the number of channel proteins expressed in the cell surface membrane (a Class 2 mechanism) [68,89]. Less common are LQT1- and LQT2-linked mutations that decrease the current by disrupting normal channel gating or single channel current levels, a Class 3 or 4 mechanism, respectively. In the next two sections we discuss how different Classes of LQT1 and LQT2 mutations associate with distinct clinical phenotypes.
6. LQT1-Linked Mutation Dysfunctional Phenotypes: PKA, CaM, PIP2, and Pleiotropy
IKs is upregulated by β-adrenergic stimulation through the activation of protein kinase A (PKA) [64]. Kv7.1 channel proteins are phosphorylated by protein kinase A (PKA) in the amino-terminus at Ser27 (S27) and S92 [90,91,92]. Phosphorylation of Kv7.1 channel proteins increases IKs by increasing the open probability of Kv7.1 channels and the amplitude of single channel currents [90]. Kv7.1 channel proteins are part of a macromolecular signaling complex that minimally includes Kv7.1, the auxiliary β-subunit, KCNE1, and the A-kinase anchoring protein, AKAP9 (Yotiao) [64,93,94]. There are also multiple interactions with other ion channel subunit proteins, but KCNE1 alters the biophysical properties of Kv7.1 channels to generate native-like IKs currents. Studies show that both KCNE1 and AKAP9 may be required for normal PKA regulation of Kv7.1 channels [64,95,96].
Genetic testing suggests 80% of suspected LQT1 mutations are missense (i.e., point mutation that leads to an amino acid substitution in frame) [47], and although in vitro studies show that different mutations reduce IKs by different absolute amounts, the reduction in IKs does not always correlate with clinical phenotypes or the risk of life-threatening events [97,98,99]. The lack of a clear association between the reduction IKs and clinical severity of LQT1 suggests additional factors are important for determining how individual LQT1 mutations impact IKs to cause life-threatening symptoms. Hiejman and colleagues (2012) demonstrated that the LQT1 missense mutation A341V-Kv7.1, which causes a severe LQT1 clinical phenotype, generates Kv7.1 channels that are insensitive to PKA. These data suggest that mutations which disrupt the PKA regulation of Kv7.1 channels can contribute to the more severe clinical phenotype in some people [100]. Barsheshet and colleagues (2012) identified several other LQT1-linked mutations that generate Kv7.1 channels which disrupt the PKA regulations of Kv7.1 channels [101]. Many of the PKA insensitive LQT1-linked mutations were found to localize to cytoplasmic loops and, similar to A341V-Kv7.1, these PKA insensitive mutations appear to confer an increased risk for life-threatening events in people.
Several KCNQ1 mutations can also cause a “concealed LQT1” phenotype [52,66,102]. KCNQ1 variants linked to concealed LQT1 generate Kv7.1 channels that do not cause large reductions in IKs under basal conditions, but rather, prevent the upregulation of IKs following the activation of PKA [66,102]. Studies focused on KCNQ1 mutations that cause a concealed LQT1 phenotype used engineered phosphomimetic variants at S27 to demonstrate that these LQT1 mutations may prevent the conformational changes in Kv7.1 channels that increase IKs subsequent to PKA phosphorylation [66,102]. Unfortunately, the PKA sensitivity for only a small fraction of LQT1-linked mutations (<3%) of KCNQ1 variants has been studied [103]. A deeper knowledge in which the mechanisms that mutant Kv7.1 channels impair Kv7.1 channel regulation by PKA is needed to better understand the potential clinical impact of different LQT1-linked mutations.
Kv7.1 channels are also modified by calmodulin (CaM). Earlier studies show that CaM is a component of Kv7.1 channels, and CaM regulates Kv7.1 channel assembly and gating [104,105]. Several Kv7.1 mutations linked to LQT1 can disrupt the CaM interaction to prevent the assembly of Kv7.1 channels. Kv7.1 channel gating is also influenced by the membrane phospholipid phosphoinositol bisphosphate (PIP2) [106,107]. Basic amino acid residues in the intracellular regions the Kv7.1 channels are thought to interact with PIP2. Several Kv7.1 missense mutations that alter these basic amino acid residues can decrease the sensitivity of Kv7.1 channels to PIP2 [108]. Interestingly, subsequent studies investigating protein kinase C (PKC) show PKC activation of IKs can regulate the Kv7.1 channel response to PIP2 [109]. After PKC activation, Kv7.1 channels are less sensitive to changes in the membrane levels of PIP2. Moreover, PKC partially normalizes the response of LQTS-linked Kv7.1 mutant channels to PIP2 by improving channel interactions with PIP2. Thus, the regulation of Kv7.1 channel assembly, gating, and conductance is regulated by phosphorylation, protein–protein interactions, and interactions between the channel and the phospholipids. LQTS-linked mutations in Kv7.1 channels that differentially impact the regulation of Kv7.1 channels by these mechanisms can generate distinct clinical phenotypes.
Another surprising mutation-specific phenotype associated with several LQT1-linked missense mutations is a pleiotropic effect on atrial excitability. Several LQT1-linked missense mutations can associate with familial Atrial Fibrillation (fAF) [69,110,111,112,113,114,115,116,117,118]. This is unexpected because AF is commonly attributed to reduction in the atrial refractoriness. Most of the LQT1/fAF-linked mutations generate macroscopic currents with two distinct biophysical IKs components, a voltage-dependent component, and a smaller voltage-independent component. Computational simulations of atrial APs that incorporate the functional changes of LQT1/fAF-linked mutations predict that the voltage-independent component leads to a shortening of atrial action potential duration [112,113,119]. However, computational simulations that incorporate the functional changes of LQT1/fAF-linked mutations in the ventricular action potential show they can prolong the ventricular action potential by reducing the larger voltage-dependent IKs and rendering the larger component insensitive to modulation by PKA. Although rare, LQT1/fAF-linked mutations highlight an important functional difference for IKs in the atria and ventricle, and they underscore how different KCNQ1 mutations generate an umbrella of different LQT1-linked molecular and clinical phenotypes.
7. LQT2-Linked Mutation Dysfunctional Phenotypes: Trafficking in Heterotetramers
IKr is conducted by the KCNH2-encoded Kv11.1a and Kv11.1b channel proteins and is regulated by the KCNE2 encoded MiRP1 auxiliary subunit [67,120,121]. Most functional expression analyses that study KCNH2 mutations focus on studying the mutations in the Kv11.1a α-subunit [68,70]. Roughly half of LQT2-linked mutations disrupt the synthesis of Kv11.1 channel proteins and many these mutations are predicted to cause haploinsufficiency (Class 1 mechanism) [74,76,83]. The remaining LQT2-linked mutations are mostly missense, and of the >150 missense mutations in the Kv11.1a channel that have been studied, ≈90% disrupt the trafficking of Kv11.1a channels to the cell surface membrane (Class 2 mechanism) [72,75,76,77,78,79,80,81,82].
Class 2 LQT2-linked missense mutations localize to four major structural domains in the full length Kv11.1a α-subunit: the N-terminal Per-Arnt-Sim domain (PASD), the voltage-sensor domain, the pore domain, or the C-terminal cyclic nucleotide-binding domain (CNBD). Class 2 LQT2-linked mutations are postulated to decrease the folding efficiency of Kv11.1 channel proteins and increase their retention and associated degradation by cellular quality control mechanisms in the Endoplasmic Reticulum (ER). Although Class 2 LQT2-linked mutations disrupt Kv11.1 channel protein folding, studies show domain-specific differences in molecular and clinical properties [67,68,70,122,123].
LQT2 mutations in the pore domain confer the highest risk of arrhythmic events [15]. Most Class 2 mutations in the pore domain decrease the trafficking of WT-Kv11.1 channel proteins, causing dominant negative effects [68]. In contrast, Class 2 LQT2 mutations in the CNBD appear to disrupt the co-assembly of mutant LQT2 channel proteins [122,124,125]. Class 2 LQT2-linked mutations in the CNBD that disrupt Kv11.1 channel protein co-assembly are expected to be haploinsufficient [122,124,126]. Consistent with CNBD LQT2-linked mutations causing a haploinsufficiency, clinical data show some CNBD domain mutations confer a moderate to mild clinical phenotype that is unmasked in the presence of genetic modifiers [126]. Not surprisingly, immunocytochemistry studies show that cells expressing Class 2 LQT2-linked pore or CNBD mutations show dramatically different immunostaining patterns in mutant Kv11.1a channel proteins, suggesting they are regulated by distinct quality control mechanisms [125,127].
Unlike Kv7.1 channels, Kv11.1 channels do not appear to be part of a larger macromolecular complex whose proper assembly is necessary to assess function and second messenger regulation. Through this lens, functional assessment LQT2-linked mutations using the full-length Kv11.1a channel protein appears justified. However, layered on this are several channel specific aspects that confound functional pathogenic assignment using in vitro systems. In cardiomyocytes, the Kv11.1b channel protein forms tetrameric channels with Kv11.1a to conduct IKr [128,129]. Kv11.1b channel proteins are generated from an alternate start site and have a unique 5′ exon amino terminus lacking PASD [130]. Interestingly, some work has shown that Kv11.1a/Kv11.1b heterotetrametric channels have distinct biophysical current characteristics compared to Kv11.1a homotetrameric channels. In a recent study, the functional assessment of patient-specific induced pluripotent stem-cell-derived cardiomyocytes of a KCNH2 variant located in the PASD revealed a relative increase in the transcription of Kv11.1b protein that altered IKr biophysical properties [131]. Such nuance of variant effect is not readily appreciated without the incorporation of both Kv11.1a and Kv11.1b channel isoforms, as well as their relative transcriptional regulation.
These and other features of KCNH2 regulation add to the complexity of associating variant dysfunction with LQT2 using in vitro systems including patient-specific induced pluripotent stem-cell-derived cardiomyocytes. For example, when determining risk of arrhythmia for people with LQT2, one factor to consider is dependent on the presence or absence of sex hormones. Postpubescent males in general have lower event risk while the opposite is true for females. Moreover, sex-dependent differences may depend on mutation location, as only certain regions of the Kv11.1a channel protein appear to associate with estrogen increasing arrhythmogenic risk if the mutation stabilizes estrogen binding in the channel [132]. It has yet to be definitively determined whether these specific estrogen binding regions in Kv11.1 channel proteins are the reason for an increased risk of arrhythmic events for some LQT2 women postpartum [133]. Similar to different LQT1 mutations, the mutation-specific functional impacts that different KCNH2 mutations have on Kv11.1 channel protein functions also generate an umbrella of different LQT2-linked phenotypes—some being more severe than others.
8. Visualizing Mutation-Specific Differences in Kv7.1 and Kv11.1 Channel Structures
Given the complexity and cumbersome nature of the functional assessment of individual LQT1- and LQT2-linked mutations using in vitro systems, alternative molecular-based structure–function simulations are being developed to predict the impact that individual missense variants have on Kv7.1 and Kv11.1 channel structures [134,135,136,137]. LQT1 and LQT2-linked missense mutations perturb the structure and the physiochemical properties of the channel, and these structural perturbations are expected to correlate with different mechanisms of dysfunction (e.g., Class 2, Class 3, and Class 4 mechanisms). The mutant-induced perturbations in Kv7.1 or Kv11.1 channel physicochemical properties could be orthosteric (local to the mutation position) and allosteric (far removed from the site). For mutations primarily causing orthosteric perturbations, linking structure to function changes is challenging, and mutations that cause allosteric effects are unlikely to be inferred from visualizing the Kv7.1 or Kv11.1 channel protein structures [138].
Advances in molecular simulations will be important for accurately determining orthosteric and allosteric effects that mutations have on Kv7.1 or Kv11.1 channel structure and function. Molecular simulations are widely used for predicting protein function by animating proteins based on their three-dimensional structure and molecular interactions. Recent molecular dynamic simulations of Kv7.1 and Kv11.1 channel protein structures have provided insights into how individual missense mutations perturb the channel structure in Kv7.1 and Kv11.1 [134,135,138,139,140]. As such, computational tools are beginning to serve as useful tools for mapping how mutations in Kv7.1 and Kv11.1 channel proteins may impact channel structure to cause dysfunction.
Molecular simulations rely on high (subnanometer) resolution three-dimensional structures of proteins that depict the location of individual atoms and the conformations of the amino acids. Simulations of channel structures can be used to predict changes in protein structure that underlie channel function [135], such as the shifting of the voltage-sensor in response to changes in membrane potential. Molecular dynamics and Monte Carlo simulations are among the most used for predicting conformational changes in proteins, including ion channels [141]. Molecular simulations commonly use physics-based equations and parameters to describe Å-level interactions between sets of atoms including bonds, angles, and torsions, as well as long-range potentials that encompass van der Waals forces and electrostatic interactions. Molecular simulations can predict the energies of distinct conformational states. Molecular dynamics can specifically predict how proteins dynamically ebb and flow in response to long- and short- range physical interactions by solving equations of motion. Structural simulations of LQT1- and LQT2-linked mutations are expected to identify the dynamic conformations in the Kv7.1 and Kv11.1 protein structures that associate with mutation-specific mechanisms for channel dysfunction [71,134,135,140,142,143,144].
There are presently two main challenges in using molecular dynamic simulations to accurately predict mutation-specific impacts on channel function: (1) the quality and abundance of input structural data and (2) limitations in molecular simulation. Improved resolution of protein transmembrane regions, as well as the conformations associated with gating (open, closed, inactive), will be essential for predicting channel conformations caused by missense mutations. The highest resolution structure of the transmembrane region in Kv11.1, as an example, is just under 4 Å. Average resolutions in the 3–4 Å range are generally sufficient for the correct placement of helices composing the transmembrane bundle, but will be insufficient in resolving many amino acid side chain orientations and hydrogen atoms [145]. Ambiguities in side-chain placements will almost certainly influence structure–function predictions from molecular simulations.
A key limitation to molecular simulation approaches is the mismatch between typical simulation lengths of microseconds versus the timescales of biological processes that unfold at millisecond and longer timescales [145]. Currently, bridging these disparate timescales necessitates model simplifications [146]. These approximations entail reducing the complexity of fully atomistic physics-based models by representing groups of atoms or amino acids as individual units (e.g., coarse-graining) [147], reducing the energetic detail by neglecting terms like bond vibrations (such as ‘SHAKE’), or even long-range interactions (e.g., Go models) [146,148], or by utilizing statistical and machine learning techniques in lieu of time-dependent equations of motion [149]. The balance between the aforementioned approximations and the physical detail needed to relate channel structure to phenotype has yet to be determined and will likely differ on a case-by-case basis. However, continued advances in algorithms, software, and hardware will be integral to fully realizing the potential for identifying mutation-specific differences associated with LQT1 or LQT2.
In recent years, machine learning has exploded in popularity, including its applications for predicting impacts of genetic variants on protein structures [150,151]. Applications of machine learning applied to Kv11.1 channels thus far have largely focused on predicting responses to medications that increase the risk for arrhythmias. Examples include using decision trees to determine which medications that block IKr associate with torsade de pointes or the disruption of K+ permeation by using neural networks [152,153]. More recent applications used ensemble methods and techniques, including naive Bayes and logistic regression, for predicting drug cardiotoxicity and variant pathogenicity [154,155,156]. Applications of machine learning to map genotype to electrophysiological cellular phenotype have been fewer in number. Along these lines, a recent study classified many different KCNH2 variants by their potential to influence trafficking, in part by categorizing how an amino acid substitution might impact the site’s occupied volume, polarity, and steric interactions [157]. The classification of the variants included structural information that leveraged by the Kv11.1 channel structure resolved by cryo-electron microscopy [145].
While the application of machine learning approaches to voltage-gated channel function is rather recent, there is a rich history of machine learning techniques applied to protein structure predictions in general. For over a decade, machine learning techniques including Rosetta [158] and MODELLER [159] have been developed to predict protein structure from primary sequence or the three dimensional structures of homologous proteins, as has been extensively reviewed in [160]. The popularity of machine learning algorithms in the prediction of protein structure from primary sequence is reflected in the growing participation in competitions such as the Critical Assessment of Methods of Protein Structure Prediction (CASP). CASP in particular has culminated in the development of computational tools like Alpha fold [161] and RosettaTTa fold [162] for modeling protein structure [163,164], which together have been used to predict the structures of thousands of proteins they have not yet been determined by crystallography or related approaches. Additional machine learning approaches have been used for the design of proteins with user-defined tertiary structures [165]. Similarly, several approaches for predicting effects of missense mutations on protein structure have been reported [166,167,168]. Modeling mutation-specific differences in LQT1- and LQT2-linked mutations will likely benefit from recent advances in machine-learning-guided protein structure prediction.
In addition to predicting or refining protein structure, machine learning approaches have also been used to extract meaningful data from molecular dynamic simulations to predict diverse phenomena including protein–protein interactions and binding thermodynamics [169,170,171,172]. Machine learning techniques such as logistic regression, decision trees, and multilayer perceptron were recently used to predict interactions between the SARS-CoV-2 Spike Protein and Angiotensin converting enzyme 2 [173]. Additional approaches have been used to highlight physiochemical features in amyloid beta that promote their aggregation [174]. Machine learning-based refinement of computational protein structure prediction from molecular dynamic simulations has also been reported [175]. Extensions to missense variants include approaches for predicting the stability and oligomerization of epistatic enzyme [176], and impacts on protein–protein interactions [177]. Progress toward using these approaches with K+ channels is evident in a recent characterization of renal outer medullary K+ channel using in silico and experimental techniques [178]. Therefore, the wide-ranging applications of machine learning methods to molecular dynamic simulations will undoubtedly advance the forefront of understanding the structural and functional impacts of different LQT1- and LQT2-linked mutations on channel function.
9. Summary
Early case studies identified abnormal QT-interval prolongation as a biomarker for increased risk of SCD. Clinician-scientists were able to identify families who have autosomal recessive and dominant form of LQTS, begin the International LQTS Registry, and develop an evolving phenotypic score to identify people at risk for LQTS. The major genetic causes for LQTS were identified and most patients were found to have mutations in K+ channel genes important for ventricular repolarization (LQT1 and LQT2). Subsequent clinical studies identified genotype-specific differences in phenotypes and symptoms, and functional studies demonstrated that these differences are in part due to changes in IKs and IKr during β-adrenergic activation. Additional studies found subsets of LQT1- and LQT2-linked mutations showed mutation-specific differences in clinical phenotypes. Advances in molecular simulations, including molecular dynamics and machine learning, hold the promise of identifying mutation-specific differences in LQT1- and LQT2-linked mutations without the need for comprehensive in vitro testing. We expect that advances in understanding clinical phenotypes, in combination with computational approaches, will improve the clinical management of people with mutation-specific subtypes of LQT1 and LQT2.
Acknowledgments
Biorender was used to make some of the Figures in the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This project was supported in part by the Gary and Marie Weiner Professorship in Cardiovascular Medicine Research (LLE), NHLBI R01 HL139738-01 (LLE), NHLBI R01HL141342-01 (BD PI; LLE and CTJ co-I), and the American Heart Association Grant 20IPA35320141 (PKH).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Moss A.J. Long QT Syndrome. JAMA. 2003;289:2041–2044. doi: 10.1001/jama.289.16.2041. [DOI] [PubMed] [Google Scholar]
- 2.Priori S.G., Wilde A.A., Horie M., Cho Y., Behr E.R., Berul C., Blom N., Brugada J., Chiang C.E., Huikuri H., et al. HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: Document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm. 2013;10:1932–1963. doi: 10.1016/j.hrthm.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Adler A., Novelli V., Amin A.S., Abiusi E., Care M., Nannenberg E.A., Feilotter H., Amenta S., Mazza D., Bikker H., et al. An International, Multicentered, Evidence-Based Reappraisal of Genes Reported to Cause Congenital Long QT Syndrome. Circulation. 2020;141:418–428. doi: 10.1161/CIRCULATIONAHA.119.043132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han L., Liu F., Li Q., Qing T., Zhai Z., Xia Z., Li J. The Efficacy of Beta-Blockers in Patients with Long QT Syndrome 1-3 According to Individuals’ Gender, Age, and QTc Intervals: A Network Meta-analysis. Front. Pharm. 2020;11:579525. doi: 10.3389/fphar.2020.579525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilde A.A.M., Amin A.S., Postema P.G. Diagnosis, management and therapeutic strategies for congenital long QT syndrome. Heart. 2022;108:332–338. doi: 10.1136/heartjnl-2020-318259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Priori S.G., Wilde A.A., Horie M., Cho Y., Behr E.R., Berul C., Blom N., Brugada J., Chiang C.E., Huikuri H., et al. Executive Summary: HRS/EHRA/APHRS Expert Consensus Statement on the Diagnosis and Management of Patients with Inherited Primary Arrhythmia Syndromes. Heart Rhythm Off. J. Heart Rhythm Soc. 2013;15:1389–1406. doi: 10.1016/j.hrthm.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz P.J., Crotti L., Insolia R. Long-QT syndrome: From genetics to management. Circ. Arrhythm Electrophysiol. 2012;5:868–877. doi: 10.1161/CIRCEP.111.962019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q., Chen Q., Towbin J.A. Genetics, molecular mechanisms and management of long QT syndrome. Ann. Med. 1998;30:58–65. doi: 10.3109/07853899808999385. [DOI] [PubMed] [Google Scholar]
- 9.Takigawa M., Kawamura M., Noda T., Yamada Y., Miyamoto K., Okamura H., Satomi K., Aiba T., Kamakura S., Sakaguchi T., et al. Seasonal and circadian distributions of cardiac events in genotyped patients with congenital long QT syndrome. Circ. J. 2012;76:2112–2118. doi: 10.1253/circj.CJ-12-0213. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz P.J., Priori S.G., Spazzolini C., Moss A.J., Vincent G.M., Napolitano C., Denjoy I., Guicheney P., Breithardt G., Keating M.T., et al. Genotype-phenotype correlation in the long-QT syndrome: Gene-specific triggers for life-threatening arrhythmias. Circulation. 2001;103:89–95. doi: 10.1161/01.CIR.103.1.89. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu W., Moss A.J., Wilde A.A., Towbin J.A., Ackerman M.J., January C.T., Tester D.J., Zareba W., Robinson J.L., Qi M., et al. Genotype-phenotype aspects of type 2 long QT syndrome. J. Am. Coll. Cardiol. 2009;54:2052–2062. doi: 10.1016/j.jacc.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa J., Lopes C.M., Barsheshet A., Moss A.J., Migdalovich D., Ouellet G., McNitt S., Polonsky S., Robinson J.L., Zareba W., et al. Combined assessment of sex- and mutation-specific information for risk stratification in type 1 long QT syndrome. Heart Rhythm. 2012;9:892–898. doi: 10.1016/j.hrthm.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilde A.A., Moss A.J., Kaufman E.S., Shimizu W., Peterson D.R., Benhorin J., Lopes C., Towbin J.A., Spazzolini C., Crotti L., et al. Clinical Aspects of Type 3 Long-QT Syndrome: An International Multicenter Study. Circulation. 2016;134:872–882. doi: 10.1161/CIRCULATIONAHA.116.021823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moss A.J., Shimizu W., Wilde A.A., Towbin J.A., Zareba W., Robinson J.L., Qi M., Vincent G.M., Ackerman M.J., Kaufman E.S., et al. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation. 2007;115:2481–2489. doi: 10.1161/CIRCULATIONAHA.106.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss A.J., Zareba W., Kaufman E.S., Gartman E., Peterson D.R., Benhorin J., Towbin J.A., Keating M.T., Priori S.G., Schwartz P.J., et al. Increased risk of arrhythmic events in long-QT syndrome with mutations in the pore region of the human ether-a-go-go-related gene potassium channel. Circulation. 2002;105:794–799. doi: 10.1161/hc0702.105124. [DOI] [PubMed] [Google Scholar]
- 16.Ponce-Balbuena D., Deschenes I. Long QT syndrome—Bench to bedside. Heart Rhythm O2. 2021;2:89–106. doi: 10.1016/j.hroo.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krahn A.D., Laksman Z., Sy R.W., Postema P.G., Ackerman M.J., Wilde A.A.M., Han H.C. Congenital Long QT Syndrome. JACC Clin. Electrophysiol. 2022;8:687–706. doi: 10.1016/j.jacep.2022.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Pandit M., Finn C., Tahir U., Frishman W.H. Congenital Long QT Syndrome: A Review of Genetic and Pathophysiologic Etiologies, Phenotypic Subtypes and Clinical Management. Cardiol. Rev. 2022 doi: 10.1097/CRD.0000000000000459. [DOI] [PubMed] [Google Scholar]
- 19.AlGhatrif M., Lindsay J. A brief review: History to understand fundamentals of electrocardiography. J. Commun. Hosp. Intern. Med. Perspect. 2012;2:14383. doi: 10.3402/jchimp.v2i1.14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisch C. Evolution of the clinical electrocardiogram. J. Am. Coll. Cardiol. 1989;14:1127–1138. doi: 10.1016/0735-1097(89)90407-5. [DOI] [PubMed] [Google Scholar]
- 21.Lewis T. Report Cxix. Auricular Fibrillation: A Common Clinical Condition. Br. Med. J. 1909;2:1528. doi: 10.1136/bmj.2.2552.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine S.A. Coronary thrombosis: Its various clinical features. Medicine. 1929;8:245–418. doi: 10.1097/00005792-192909000-00001. [DOI] [Google Scholar]
- 23.RECOMMENDATIONS for standardization of electrocardiographic and vectorcardiographic leads. Circulation. 1954;10:564–573. doi: 10.1161/01.CIR.10.4.564. [DOI] [PubMed] [Google Scholar]
- 24.Kadish A.H., Buxton A.E., Kennedy H.L., Knight B.P., Mason J.W., Schuger C.D., Tracy C.M., Winters W.L., Jr., Boone A.W., Elnicki M., et al. ACC/AHA clinical competence statement on electrocardiography and ambulatory electrocardiography: A report of the ACC/AHA/ACP-ASIM task force on clinical competence (ACC/AHA Committee to develop a clinical competence statement on electrocardiography and ambulatory electrocardiography) endorsed by the International Society for Holter and noninvasive electrocardiology. Circulation. 2001;104:3169–3178. [PubMed] [Google Scholar]
- 25.Jervell A., Lange-Nielsen F. Congenital deaf-mutism, functional heart disease with prolongation of the Q-T interval and sudden death. Am. Heart J. 1957;54:59–68. doi: 10.1016/0002-8703(57)90079-0. [DOI] [PubMed] [Google Scholar]
- 26.Romano C., Gemme G., Pongiglione R. Rare Cardiac Arrythmias of the Pediatric Age. Ii. Syncopal Attacks Due to Paroxysmal Ventricular Fibrillation (Presentation of 1st Case in Italian Pediatric Literature) Clin. Pediatr. 1963;45:656–683. [PubMed] [Google Scholar]
- 27.Ward O.C. A New Familial Cardiac Syndrome in Children. J. Ir. Med. Assoc. 1964;54:103–106. [PubMed] [Google Scholar]
- 28.Schwartz P.J. 1970-2020: 50 years of research on the long QT syndrome-from almost zero knowledge to precision medicine. Eur. Heart J. 2021;42:1063–1072. doi: 10.1093/eurheartj/ehaa769. [DOI] [PubMed] [Google Scholar]
- 29.Moss A.J., Schwartz P.J. 25th anniversary of the International Long-QT Syndrome Registry: An ongoing quest to uncover the secrets of long-QT syndrome. Circulation. 2005;111:1199–1201. doi: 10.1161/01.CIR.0000157069.91834.DA. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz P.J. Idiopathic long QT syndrome: Progress and questions. Am. Heart J. 1985;109:399–411. doi: 10.1016/0002-8703(85)90626-X. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz P.J., Moss A.J., Vincent G.M., Crampton R.S. Diagnostic criteria for the long QT syndrome. An update. Circulation. 1993;88:782–784. doi: 10.1161/01.CIR.88.2.782. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz P.J., Crotti L. QTc behavior during exercise and genetic testing for the long-QT syndrome. Circulation. 2011;124:2181–2184. doi: 10.1161/CIRCULATIONAHA.111.062182. [DOI] [PubMed] [Google Scholar]
- 33.Curran M.E., Splawski I., Timothy K.W., Vincent G.M., Green E.D., Keating M.T. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 34.Sanguinetti M.C., Jiang C., Curran M.E., Keating M.T. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 35.Trudeau M.C., Warmke J.W., Ganetzky B., Robertson G.A. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 1995;269:92–95. doi: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q., Shen J., Splawski I., Atkinson D., Li Z., Robinson J.L., Moss A.J., Towbin J.A., Keating M.T. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q., Curran M.E., Splawski I., Burn T.C., Millholland J.M., VanRaay T.J., Shen J., Timothy K.W., Vincent G.M., de Jager T., et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat. Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 38.Wilde A.A., Brugada R. Phenotypical manifestations of mutations in the genes encoding subunits of the cardiac sodium channel. Circ. Res. 2011;108:884–897. doi: 10.1161/CIRCRESAHA.110.238469. [DOI] [PubMed] [Google Scholar]
- 39.Abriel H. Cardiac sodium channel Na(v)1.5 and interacting proteins: Physiology and pathophysiology. J. Mol. Cell Cardiol. 2010;48:2–11. doi: 10.1016/j.yjmcc.2009.08.025. [DOI] [PubMed] [Google Scholar]
- 40.Remme C.A., Wilde A.A., Bezzina C.R. Cardiac sodium channel overlap syndromes: Different faces of SCN5A mutations. Trends Cardiovasc. Med. 2008;18:78–87. doi: 10.1016/j.tcm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Viskin S., Rosovski U., Sands A.J., Chen E., Kistler P.M., Kalman J.M., Rodriguez Chavez L., Iturralde Torres P., Cruz F.F., Centurion O.A., et al. Inaccurate electrocardiographic interpretation of long QT: The majority of physicians cannot recognize a long QT when they see one. Heart Rhythm Off. J. Heart Rhythm Soc. 2005;2:569–574. doi: 10.1016/j.hrthm.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 42.Garg A., Lehmann M.H. Prolonged QT interval diagnosis suppression by a widely used computerized ECG analysis system. Circ. Arrhythm Electrophysiol. 2013;6:76–83. doi: 10.1161/CIRCEP.112.976803. [DOI] [PubMed] [Google Scholar]
- 43.Garson A., Jr. How to measure the QT interval—What is normal? Am. J. Cardiol. 1993;72:14B–16B. doi: 10.1016/0002-9149(93)90034-A. [DOI] [PubMed] [Google Scholar]
- 44.Lepeschkin E., Surawicz B. The measurement of the Q-T interval of the electrocardiogram. Circulation. 1952;6:378–388. doi: 10.1161/01.CIR.6.3.378. [DOI] [PubMed] [Google Scholar]
- 45.Postema P.G., Wilde A.A. The measurement of the QT interval. Curr. Cardiol. Rev. 2014;10:287–294. doi: 10.2174/1573403X10666140514103612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taggart N.W., Haglund C.M., Tester D.J., Ackerman M.J. Diagnostic miscues in congenital long-QT syndrome. Circulation. 2007;115:2613–2620. doi: 10.1161/CIRCULATIONAHA.106.661082. [DOI] [PubMed] [Google Scholar]
- 47.Kapa S., Tester D.J., Salisbury B.A., Harris-Kerr C., Pungliya M.S., Alders M., Wilde A.A., Ackerman M.J. Genetic testing for long-QT syndrome: Distinguishing pathogenic mutations from benign variants. Circulation. 2009;120:1752–1760. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giudicessi J.R., Kapplinger J.D., Tester D.J., Alders M., Salisbury B.A., Wilde A.A., Ackerman M.J. Phylogenetic and physicochemical analyses enhance the classification of rare nonsynonymous single nucleotide variants in type 1 and 2 long-QT syndrome. Circ. Cardiovasc. Genet. 2012;5:519–528. doi: 10.1161/CIRCGENETICS.112.963785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viskin S., Postema P.G., Bhuiyan Z.A., Rosso R., Kalman J.M., Vohra J.K., Guevara-Valdivia M.E., Marquez M.F., Kogan E., Belhassen B., et al. The response of the QT interval to the brief tachycardia provoked by standing: A bedside test for diagnosing long QT syndrome. J. Am. Coll Cardiol. 2010;55:1955–1961. doi: 10.1016/j.jacc.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adler A., van der Werf C., Postema P.G., Rosso R., Bhuiyan Z.A., Kalman J.M., Vohra J.K., Guevara-Valdivia M.E., Marquez M.F., Halkin A., et al. The phenomenon of “QT stunning”: The abnormal QT prolongation provoked by standing persists even as the heart rate returns to normal in patients with long QT syndrome. Heart Rhythm. 2012;9:901–908. doi: 10.1016/j.hrthm.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sy R.W., van der Werf C., Chattha I.S., Chockalingam P., Adler A., Healey J.S., Perrin M., Gollob M.H., Skanes A.C., Yee R., et al. Derivation and validation of a simple exercise-based algorithm for prediction of genetic testing in relatives of LQTS probands. Circulation. 2011;124:2187–2194. doi: 10.1161/CIRCULATIONAHA.111.028258. [DOI] [PubMed] [Google Scholar]
- 52.Ackerman M.J., Khositseth A., Tester D.J., Hejlik J.B., Shen W.K., Porter C.B. Epinephrine-induced QT interval prolongation: A gene-specific paradoxical response in congenital long QT syndrome. Mayo Clin. Proc. 2002;77:413–421. doi: 10.1016/S0025-6196(11)62209-X. [DOI] [PubMed] [Google Scholar]
- 53.Horner J.M., Horner M.M., Ackerman M.J. The diagnostic utility of recovery phase QTc during treadmill exercise stress testing in the evaluation of long QT syndrome. Heart Rhythm. 2011;8:1698–1704. doi: 10.1016/j.hrthm.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 54.Shimizu W., Noda T., Takaki H., Nagaya N., Satomi K., Kurita T., Suyama K., Aihara N., Sunagawa K., Echigo S., et al. Diagnostic value of epinephrine test for genotyping LQT1, LQT2, and LQT3 forms of congenital long QT syndrome. Heart Rhythm Off. J. Heart Rhythm Soc. 2004;1:276–283. doi: 10.1016/j.hrthm.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 55.Vyas H., Ackerman M.J. Epinephrine QT stress testing in congenital long QT syndrome. J. Electrocardiol. 2006;39:S107–S113. doi: 10.1016/j.jelectrocard.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 56.Peroz D., Rodriguez N., Choveau F., Baro I., Merot J., Loussouarn G. Kv7.1 (KCNQ1) properties and channelopathies. J. Physiol. 2008;586:1785–1789. doi: 10.1113/jphysiol.2007.148254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong J.A., Gula L.J., Klein G.J., Yee R., Skanes A.C., Krahn A.D. Utility of treadmill testing in identification and genotype prediction in long-QT syndrome. Circ. Arrhythm Electrophysiol. 2010;3:120–125. doi: 10.1161/CIRCEP.109.907865. [DOI] [PubMed] [Google Scholar]
- 58.Krahn A.D., Klein G.J., Yee R. Hysteresis of the RT interval with exercise: A new marker for the long-QT syndrome? Circulation. 1997;96:1551–1556. doi: 10.1161/01.CIR.96.5.1551. [DOI] [PubMed] [Google Scholar]
- 59.Bos J.M., Attia Z.I., Albert D.E., Noseworthy P.A., Friedman P.A., Ackerman M.J. Use of Artificial Intelligence and Deep Neural Networks in Evaluation of Patients With Electrocardiographically Concealed Long QT Syndrome From the Surface 12-Lead Electrocardiogram. JAMA Cardiol. 2021;6:532–538. doi: 10.1001/jamacardio.2020.7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aufiero S., Bleijendaal H., Robyns T., Vandenberk B., Krijger C., Bezzina C., Zwinderman A.H., Wilde A.A.M., Pinto Y.M. A deep learning approach identifies new ECG features in congenital long QT syndrome. BMC Med. 2022;20:162. doi: 10.1186/s12916-022-02350-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roden D.M. Taking the “idio” out of “idiosyncratic”: Predicting torsades de pointes. Pacing Clin. Electrophysiol. 1998;21:1029–1034. doi: 10.1111/j.1540-8159.1998.tb00148.x. [DOI] [PubMed] [Google Scholar]
- 62.Roden D.M. Repolarization reserve: A moving target. Circulation. 2008;118:981–982. doi: 10.1161/CIRCULATIONAHA.108.798918. [DOI] [PubMed] [Google Scholar]
- 63.Soltis A.R., Saucerman J.J. Synergy between CaMKII substrates and beta-adrenergic signaling in regulation of cardiac myocyte Ca(2+) handling. Biophys. J. 2010;99:2038–2047. doi: 10.1016/j.bpj.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marx S.O., Kurokawa J., Reiken S., Motoike H., D’Armiento J., Marks A.R., Kass R.S. Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]
- 65.Zipes D.P., Jalife J., Stevenson W.G. Cardiac Electrophysiology: From Cell to Bedside. Elsevier Health Science; Philadelphia, PA, USA: 2018. [Google Scholar]
- 66.Bartos D.C., Giudicessi J.R., Tester D.J., Ackerman M.J., Ohno S., Horie M., Gollob M.H., Burgess D.E., Delisle B.P. A KCNQ1 mutation contributes to the concealed type 1 long QT phenotype by limiting the Kv7.1 channel conformational changes associated with protein kinase A phosphorylation. Heart Rhythm. 2014;11:459–468. doi: 10.1016/j.hrthm.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith J.L., Anderson C.L., Burgess D.E., Elayi C.S., January C.T., Delisle B.P. Molecular pathogenesis of long QT syndrome type 2. J. Arrhythm. 2016;32:373–380. doi: 10.1016/j.joa.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson C.L., Kuzmicki C.E., Childs R.R., Hintz C.J., Delisle B.P., January C.T. Large-scale mutational analysis of Kv11.1 reveals molecular insights into type 2 long QT syndrome. Nat. Commun. 2014;5:5535. doi: 10.1038/ncomms6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Napolitano C., Priori S.G., Schwartz P.J., Bloise R., Ronchetti E., Nastoli J., Bottelli G., Cerrone M., Leonardi S. Genetic testing in the long QT syndrome: Development and validation of an efficient approach to genotyping in clinical practice. JAMA. 2005;294:2975–2980. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- 70.Anderson C.L., Delisle B.P., Anson B.D., Kilby J.A., Will M.L., Tester D.J., Gong Q., Zhou Z., Ackerman M.J., January C.T. Most LQT2 mutations reduce Kv11.1 (hERG) current by a class 2 (trafficking-deficient) mechanism. Circulation. 2006;113:365–373. doi: 10.1161/CIRCULATIONAHA.105.570200. [DOI] [PubMed] [Google Scholar]
- 71.Burgess D.E., Bartos D.C., Reloj A.R., Campbell K.S., Johnson J.N., Tester D.J., Ackerman M.J., Fressart V., Denjoy I., Guicheney P., et al. High-risk long QT syndrome mutations in the Kv7.1 (KCNQ1) pore disrupt the molecular basis for rapid K(+) permeation. Biochemistry. 2012;51:9076–9085. doi: 10.1021/bi3009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delisle B.P., Anson B.D., Rajamani S., January C.T. Biology of cardiac arrhythmias: Ion channel protein trafficking. Circ. Res. 2004;94:1418–1428. doi: 10.1161/01.RES.0000128561.28701.ea. [DOI] [PubMed] [Google Scholar]
- 73.Hille B. Ion Channels of Excitable Membranes. 3rd ed. Sinauer Associates, Inc.; Sunderland, MA, USA: 2001. p. 814. [Google Scholar]
- 74.Gong Q., Zhang L., Vincent G.M., Horne B.D., Zhou Z. Nonsense mutations in hERG cause a decrease in mutant mRNA transcripts by nonsense-mediated mRNA decay in human long-QT syndrome. Circulation. 2007;116:17–24. doi: 10.1161/CIRCULATIONAHA.107.708818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dausse E., Berthet M., Denjoy I., Andre-Fouet X., Cruaud C., Bennaceur M., Faure S., Coumel P., Schwartz K., Guicheney P. A mutation in HERG associated with notched T waves in long QT syndrome. J. Mol. Cell Cardiol. 1996;28:1609–1615. doi: 10.1006/jmcc.1996.0151. [DOI] [PubMed] [Google Scholar]
- 76.Sanguinetti M.C., Curran M.E., Spector P.S., Keating M.T. Spectrum of HERG K+-channel dysfunction in an inherited cardiac arrhythmia. Proc. Natl. Acad. Sci. USA. 1996;93:2208–2212. doi: 10.1073/pnas.93.5.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Satler C.A., Walsh E.P., Vesely M.R., Plummer M.H., Ginsburg G.S., Jacob H.J. Novel missense mutation in the cyclic nucleotide-binding domain of HERG causes long QT syndrome. Am. J. Med. Genet. 1996;65:27–35. doi: 10.1002/(SICI)1096-8628(19961002)65:1<27::AID-AJMG4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 78.Li X., Xu J., Li M. The human delta1261 mutation of the HERG potassium channel results in a truncated protein that contains a subunit interaction domain and decreases the channel expression. J. Biol. Chem. 1997;272:705–708. doi: 10.1074/jbc.272.2.705. [DOI] [PubMed] [Google Scholar]
- 79.Nakajima T., Furukawa T., Tanaka T., Katayama Y., Nagai R., Nakamura Y., Hiraoka M. Novel mechanism of HERG current suppression in LQT2: Shift in voltage dependence of HERG inactivation. Circ. Res. 1998;83:415–422. doi: 10.1161/01.RES.83.4.415. [DOI] [PubMed] [Google Scholar]
- 80.Zhou Z., Gong Q., Epstein M.L., January C.T. HERG channel dysfunction in human long QT syndrome. Intracellular transport and functional defects. J. Biol. Chem. 1998;273:21061–21066. doi: 10.1074/jbc.273.33.21061. [DOI] [PubMed] [Google Scholar]
- 81.Chen J., Zou A., Splawski I., Keating M.T., Sanguinetti M.C. Long QT syndrome-associated mutations in the Per-Arnt-Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J. Biol. Chem. 1999;274:10113–10118. doi: 10.1074/jbc.274.15.10113. [DOI] [PubMed] [Google Scholar]
- 82.Yoshida H., Horie M., Otani H., Takano M., Tsuji K., Kubota T., Fukunami M., Sasayama S. Characterization of a novel missense mutation in the pore of HERG in a patient with long QT syndrome. J. Cardiovasc. Electrophysiol. 1999;10:1262–1270. doi: 10.1111/j.1540-8167.1999.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 83.Stump M.R., Gong Q., Zhou Z. LQT2 nonsense mutations generate trafficking defective NH2-terminally truncated channels by the reinitiation of translation. Am. J. Physiol. Heart Circ. Physiol. 2013;305:H1397–H1404. doi: 10.1152/ajpheart.00304.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bianchi L., Priori S.G., Napolitano C., Surewicz K.A., Dennis A.T., Memmi M., Schwartz P.J., Brown A.M. Mechanisms of I(Ks) suppression in LQT1 mutants. Am. J. Physiol. Heart Circ. Physiol. 2000;279:H3003–H3011. doi: 10.1152/ajpheart.2000.279.6.H3003. [DOI] [PubMed] [Google Scholar]
- 85.Chouabe C., Neyroud N., Guicheney P., Lazdunski M., Romey G., Barhanin J. Properties of KvLQT1 K+ channel mutations in Romano-Ward and Jervell and Lange-Nielsen inherited cardiac arrhythmias. EMBO J. 1997;16:5472–5479. doi: 10.1093/emboj/16.17.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shalaby F.Y., Levesque P.C., Yang W.P., Little W.A., Conder M.L., Jenkins-West T., Blanar M.A. Dominant-negative KvLQT1 mutations underlie the LQT1 form of long QT syndrome. Circulation. 1997;96:1733–1736. doi: 10.1161/01.CIR.96.6.1733. [DOI] [PubMed] [Google Scholar]
- 87.Murray A., Donger C., Fenske C., Spillman I., Richard P., Dong Y.B., Neyroud N., Chevalier P., Denjoy I., Carter N., et al. Splicing mutations in KCNQ1: A mutation hot spot at codon 344 that produces in frame transcripts. Circulation. 1999;100:1077–1084. doi: 10.1161/01.CIR.100.10.1077. [DOI] [PubMed] [Google Scholar]
- 88.Franqueza L., Lin M., Shen J., Splawski I., Keating M.T., Sanguinetti M.C. Long QT syndrome-associated mutations in the S4-S5 linker of KvLQT1 potassium channels modify gating and interaction with minK subunits. J. Biol. Chem. 1999;274:21063–21070. doi: 10.1074/jbc.274.30.21063. [DOI] [PubMed] [Google Scholar]
- 89.Huang H., Kuenze G., Smith J.A., Taylor K.C., Duran A.M., Hadziselimovic A., Meiler J., Vanoye C.G., George A.L., Jr., Sanders C.R. Mechanisms of KCNQ1 channel dysfunction in long QT syndrome involving voltage sensor domain mutations. Sci. Adv. 2018;4:eaar2631. doi: 10.1126/sciadv.aar2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thompson E., Eldstrom J., Westhoff M., McAfee D., Balse E., Fedida D. cAMP-dependent regulation of IKs single-channel kinetics. J. Gen. Physiol. 2017;149:781–798. doi: 10.1085/jgp.201611734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lopes C.M., Remon J.I., Matavel A., Sui J.L., Keselman I., Medei E., Shen Y., Rosenhouse-Dantsker A., Rohacs T., Logothetis D.E. Protein kinase A modulates PLC-dependent regulation and PIP2-sensitivity of K+ channels. Channels. 2007;1:124–134. doi: 10.4161/chan.4322. [DOI] [PubMed] [Google Scholar]
- 92.Lundby A., Andersen M.N., Steffensen A.B., Horn H., Kelstrup C.D., Francavilla C., Jensen L.J., Schmitt N., Thomsen M.B., Olsen J.V. In vivo phosphoproteomics analysis reveals the cardiac targets of beta-adrenergic receptor signaling. Sci. Signal. 2013;6:rs11. doi: 10.1126/scisignal.2003506. [DOI] [PubMed] [Google Scholar]
- 93.Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 94.Sanguinetti M.C., Curran M.E., Zou A., Shen J., Spector P.S., Atkinson D.L., Keating M.T. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 95.Kurokawa J., Motoike H.K., Rao J., Kass R.S. Regulatory actions of the A-kinase anchoring protein Yotiao on a heart potassium channel downstream of PKA phosphorylation. Proc. Natl. Acad. Sci. USA. 2004;101:16374–16378. doi: 10.1073/pnas.0405583101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen L., Marquardt M.L., Tester D.J., Sampson K.J., Ackerman M.J., Kass R.S. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc. Natl. Acad. Sci. USA. 2007;104:20990–20995. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schwartz P.J., Vanoli E., Crotti L., Spazzolini C., Ferrandi C., Goosen A., Hedley P., Heradien M., Bacchini S., Turco A., et al. Neural control of heart rate is an arrhythmia risk modifier in long QT syndrome. J. Am. Coll. Cardiol. 2008;51:920–929. doi: 10.1016/j.jacc.2007.09.069. [DOI] [PubMed] [Google Scholar]
- 98.Thomas D., Khalil M., Alter M., Schweizer P.A., Karle C.A., Wimmer A.B., Licka M., Katus H.A., Koenen M., Ulmer H.E., et al. Biophysical characterization of KCNQ1 P320 mutations linked to long QT syndrome 1. J. Mol. Cell. Cardiol. 2010;48:230–237. doi: 10.1016/j.yjmcc.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 99.Brink P.A., Crotti L., Corfield V., Goosen A., Durrheim G., Hedley P., Heradien M., Geldenhuys G., Vanoli E., Bacchini S., et al. Phenotypic variability and unusual clinical severity of congenital long-QT syndrome in a founder population. Circulation. 2005;112:2602–2610. doi: 10.1161/CIRCULATIONAHA.105.572453. [DOI] [PubMed] [Google Scholar]
- 100.Heijman J., Spatjens R.L., Seyen S.R., Lentink V., Kuijpers H.J., Boulet I.R., de Windt L.J., David M., Volders P.G. Dominant-negative control of cAMP-dependent IKs upregulation in human long-QT syndrome type 1. Circ. Res. 2012;110:211–219. doi: 10.1161/CIRCRESAHA.111.249482. [DOI] [PubMed] [Google Scholar]
- 101.Barsheshet A., Goldenberg I., O-Uchi J., Moss A.J., Jons C., Shimizu W., Wilde A.A., McNitt S., Peterson D.R., Zareba W., et al. Mutations in cytoplasmic loops of the KCNQ1 channel and the risk of life-threatening events: Implications for mutation-specific response to beta-blocker therapy in type 1 long-QT syndrome. Circulation. 2012;125:1988–1996. doi: 10.1161/CIRCULATIONAHA.111.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu J., Naiki N., Ding W.G., Ohno S., Kato K., Zang W.J., Delisle B.P., Matsuura H., Horie M. A molecular mechanism for adrenergic-induced long QT syndrome. J. Am. Coll Cardiol. 2014;63:819–827. doi: 10.1016/j.jacc.2013.08.1648. [DOI] [PubMed] [Google Scholar]
- 103.Policarova M., Novotny T., Bebarova M. Impaired Adrenergic/Protein Kinase A Response of Slow Delayed Rectifier Potassium Channels as a Long QT Syndrome Motif: Importance and Unknowns. Can. J. Cardiol. 2019;35:511–522. doi: 10.1016/j.cjca.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 104.Ghosh S., Nunziato D.A., Pitt G.S. KCNQ1 assembly and function is blocked by long-QT syndrome mutations that disrupt interaction with calmodulin. Circ. Res. 2006;98:1048–1054. doi: 10.1161/01.RES.0000218863.44140.f2. [DOI] [PubMed] [Google Scholar]
- 105.Shamgar L., Ma L., Schmitt N., Haitin Y., Peretz A., Wiener R., Hirsch J., Pongs O., Attali B. Calmodulin is essential for cardiac IKS channel gating and assembly: Impaired function in long-QT mutations. Circ. Res. 2006;98:1055–1063. doi: 10.1161/01.RES.0000218979.40770.69. [DOI] [PubMed] [Google Scholar]
- 106.Loussouarn G., Park K.H., Bellocq C., Baro I., Charpentier F., Escande D. Phosphatidylinositol-4,5-bisphosphate, PIP2, controls KCNQ1/KCNE1 voltage-gated potassium channels: A functional homology between voltage-gated and inward rectifier K+ channels. EMBO J. 2003;22:5412–5421. doi: 10.1093/emboj/cdg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zaydman M.A., Silva J.R., Delaloye K., Li Y., Liang H., Larsson H.P., Shi J., Cui J. Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc. Natl. Acad. Sci. USA. 2013;110:13180–13185. doi: 10.1073/pnas.1305167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Park K.H., Piron J., Dahimene S., Merot J., Baro I., Escande D., Loussouarn G. Impaired KCNQ1-KCNE1 and phosphatidylinositol-4,5-bisphosphate interaction underlies the long QT syndrome. Circ. Res. 2005;96:730–739. doi: 10.1161/01.RES.0000161451.04649.a8. [DOI] [PubMed] [Google Scholar]
- 109.Matavel A., Medei E., Lopes C.M. PKA and PKC partially rescue long QT type 1 phenotype by restoring channel-PIP2 interactions. Channels. 2010;4:3–11. doi: 10.4161/chan.4.1.10227. [DOI] [PubMed] [Google Scholar]
- 110.Chen Y.H., Xu S.J., Bendahhou S., Wang X.L., Wang Y., Xu W.Y., Jin H.W., Sun H., Su X.Y., Zhuang Q.N., et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 111.Hong K., Piper D.R., Diaz-Valdecantos A., Brugada J., Oliva A., Burashnikov E., Santos-de-Soto J., Grueso-Montero J., Diaz-Enfante E., Brugada P., et al. De novo KCNQ1 mutation responsible for atrial fibrillation and short QT syndrome in utero. Cardiovasc. Res. 2005;68:433–440. doi: 10.1016/j.cardiores.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 112.Bartos D.C., Duchatelet S., Burgess D.E., Klug D., Denjoy I., Peat R., Lupoglazoff J.M., Fressart V., Berthet M., Ackerman M.J., et al. R231C mutation in KCNQ1 causes long QT syndrome type 1 and familial atrial fibrillation. Heart Rhythm Off. J. Heart Rhythm Soc. 2011;8:48–55. doi: 10.1016/j.hrthm.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bartos D.C., Anderson J.B., Bastiaenen R., Johnson J.N., Gollob M.H., Tester D.J., Burgess D.E., Homfray T., Behr E.R., Ackerman M.J., et al. A KCNQ1 Mutation Causes a High Penetrance for Familial Atrial Fibrillation. J. Cardiovasc. Electrophysiol. 2013;24:562–569. doi: 10.1111/jce.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Das S., Makino S., Melman Y.F., Shea M.A., Goyal S.B., Rosenzweig A., Macrae C.A., Ellinor P.T. Mutation in the S3 segment of KCNQ1 results in familial lone atrial fibrillation. Heart Rhythm Off. J. Heart Rhythm Soc. 2009;6:1146–1153. doi: 10.1016/j.hrthm.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hasegawa K., Ohno S., Ashihara T., Itoh H., Ding W.G., Toyoda F., Makiyama T., Aoki H., Nakamura Y., Delisle B.P., et al. A novel KCNQ1 missense mutation identified in a patient with juvenile-onset atrial fibrillation causes constitutively open IKs channels. Heart Rhythm. 2014;11:67–75. doi: 10.1016/j.hrthm.2013.09.073. [DOI] [PubMed] [Google Scholar]
- 116.Henrion U., Zumhagen S., Steinke K., Strutz-Seebohm N., Stallmeyer B., Lang F., Schulze-Bahr E., Seebohm G. Overlapping cardiac phenotype associated with a familial mutation in the voltage sensor of the KCNQ1 channel. Cell Physiol. Biochem. 2012;29:809–818. doi: 10.1159/000178470. [DOI] [PubMed] [Google Scholar]
- 117.Knoche J.W., Orland K.M., January C.T., Maginot K.R. Atrial Fibrillation and Long QT Syndrome Presenting in a 12-Year-Old Girl. Case Rep. Pediatr. 2012;2012:124838. doi: 10.1155/2012/124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tamargo M., Espinosa M.Á., Gómez-Carrillo V., Juárez M., Fernández-Avilés F., Yotti R. Sudden Death in a Young Patient with Atrial Fibrillation. Cardiogenetics. 2017;7:18–21. doi: 10.4081/cardiogenetics.2017.6304. [DOI] [Google Scholar]
- 119.Kharche S., Adeniran I., Stott J., Law P., Boyett M.R., Hancox J.C., Zhang H. Pro-arrhythmogenic effects of the S140G KCNQ1 mutation in human atrial fibrillation—Insights from modelling. J. Physiol. 2012;590:4501–4514. doi: 10.1113/jphysiol.2012.229146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jonsson M.K., van der Heyden M.A., van Veen T.A. Deciphering hERG channels: Molecular basis of the rapid component of the delayed rectifier potassium current. J. Mol. Cell Cardiol. 2012;53:369–374. doi: 10.1016/j.yjmcc.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 121.Abbott G.W., Sesti F., Splawski I., Buck M.E., Lehmann M.H., Timothy K.W., Keating M.T., Goldstein S.A. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97:175–187. doi: 10.1016/S0092-8674(00)80728-X. [DOI] [PubMed] [Google Scholar]
- 122.Ficker E., Thomas D., Viswanathan P.C., Dennis A.T., Priori S.G., Napolitano C., Memmi M., Wible B.A., Kaufman E.S., Iyengar S., et al. Novel characteristics of a misprocessed mutant HERG channel linked to hereditary long QT syndrome. Am. J. Physiol. Heart Circ. Physiol. 2000;279:H1748–H1756. doi: 10.1152/ajpheart.2000.279.4.H1748. [DOI] [PubMed] [Google Scholar]
- 123.Ono M., Burgess D.E., Schroder E.A., Elayi C.S., Anderson C.L., January C.T., Sun B., Immadisetty K., Kekenes-Huskey P.M., Delisle B.P. Long QT Syndrome Type 2: Emerging Strategies for Correcting Class 2 KCNH2 (hERG) Mutations and Identifying New Patients. Biomolecules. 2020;10:1144. doi: 10.3390/biom10081144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ficker E., Obejero-Paz C.A., Zhao S., Brown A.M. The binding site for channel blockers that rescue misprocessed human long QT syndrome type 2 ether-a-gogo-related gene (HERG) mutations. J. Biol. Chem. 2002;277:4989–4998. doi: 10.1074/jbc.M107345200. [DOI] [PubMed] [Google Scholar]
- 125.Ficker E., Dennis A.T., Wang L., Brown A.M. Role of the cytosolic chaperones Hsp70 and Hsp90 in maturation of the cardiac potassium channel HERG. Circ. Res. 2003;92:e87–e100. doi: 10.1161/01.RES.0000079028.31393.15. [DOI] [PubMed] [Google Scholar]
- 126.Chai S., Wan X., Ramirez-Navarro A., Tesar P.J., Kaufman E.S., Ficker E., George A.L., Jr., Deschenes I. Physiological genomics identifies genetic modifiers of long QT syndrome type 2 severity. J. Clin. Investig. 2018;128:1043–1056. doi: 10.1172/JCI94996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hall A.R., Anderson C.L., Smith J.L., Mirshahi T., Elayi C.S., January C.T., Delisle B.P. Visualizing Mutation-Specific Differences in the Trafficking-Deficient Phenotype of Kv11.1 Proteins Linked to Long QT Syndrome Type 2. Front. Physiol. 2018;9:584. doi: 10.3389/fphys.2018.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jones E.M., Roti Roti E.C., Wang J., Delfosse S.A., Robertson G.A. Cardiac IKr channels minimally comprise hERG 1a and 1b subunits. J. Biol. Chem. 2004;279:44690–44694. doi: 10.1074/jbc.M408344200. [DOI] [PubMed] [Google Scholar]
- 129.McNally B.A., Pendon Z.D., Trudeau M.C. hERG1a and hERG1b potassium channel subunits directly interact and preferentially form heteromeric channels. J. Biol. Chem. 2017;292:21548–21557. doi: 10.1074/jbc.M117.816488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.London B., Trudeau M.C., Newton K.P., Beyer A.K., Copeland N.G., Gilbert D.J., Jenkins N.A., Satler C.A., Robertson G.A. Two isoforms of the mouse ether-a-go-go-related gene coassemble to form channels with properties similar to the rapidly activating component of the cardiac delayed rectifier K+ current. Circ. Res. 1997;81:870–878. doi: 10.1161/01.RES.81.5.870. [DOI] [PubMed] [Google Scholar]
- 131.Feng L., Zhang J., Lee C., Kim G., Liu F., Petersen A.J., Lim E., Anderson C.L., Orland K.M., Robertson G.A., et al. Long QT Syndrome KCNH2 Variant Induces hERG1a/1b Subunit Imbalance in Patient-Specific Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ. Arrhythm Electrophysiol. 2021;14:e009343. doi: 10.1161/CIRCEP.120.009343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang P.C., Perissinotti L.L., Lopez-Redondo F., Wang Y., DeMarco K.R., Jeng M.T., Vorobyov I., Harvey R.D., Kurokawa J., Noskov S.Y., et al. A multiscale computational modelling approach predicts mechanisms of female sex risk in the setting of arousal-induced arrhythmias. J. Physiol. 2017;595:4695–4723. doi: 10.1113/JP273142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meregalli P.G., Westendorp I.C., Tan H.L., Elsman P., Kok W.E., Wilde A.A. Pregnancy and the risk of torsades de pointes in congenital long-QT syndrome. Neth. Heart J. 2008;16:422–425. doi: 10.1007/BF03086191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang M., Gao M., Fang S., Zheng R., Peng D., Luo Q., Yu B. L51P, a novel mutation in the PAS domain of hERG channel, confers long QT syndrome by impairing channel activation. Am. J. Transl. Res. 2020;12:8040–8049. [PMC free article] [PubMed] [Google Scholar]
- 135.Miranda W.E., DeMarco K.R., Guo J., Duff H.J., Vorobyov I., Clancy C.E., Noskov S.Y. Selectivity filter modalities and rapid inactivation of the hERG1 channel. Proc. Natl. Acad. Sci. USA. 2020;117:2795–2804. doi: 10.1073/pnas.1909196117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kroncke B.M., Yang T., Kannankeril P., Shoemaker M.B., Roden D.M. Exploiting ion channel structure to assess rare variant pathogenicity. Heart Rhythm. 2018;15:890–894. doi: 10.1016/j.hrthm.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brewer K.R., Kuenze G., Vanoye C.G., George A.L., Jr., Meiler J., Sanders C.R. Structures Illuminate Cardiac Ion Channel Functions in Health and in Long QT Syndrome. Front. Pharm. 2020;11:550. doi: 10.3389/fphar.2020.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.DeMarco K.R., Bekker S., Vorobyov I. Challenges and advances in atomistic simulations of potassium and sodium ion channel gating and permeation. J. Physiol. 2019;597:679–698. doi: 10.1113/JP277088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Paquin A., Ye D., Tester D.J., Kapplinger J.D., Zimmermann M.T., Ackerman M.J. Even pore-localizing missense variants at highly conserved sites in KCNQ1-encoded Kv7.1 channels may have wild-type function and not cause type 1 long QT syndrome: Do not rely solely on the genetic test company’s interpretation. Heart Rhythm Case Rep. 2018;4:37–44. doi: 10.1016/j.hrcr.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Agudelo W.A., Gil-Quinones S.R., Fonseca A., Arenas A., Castro L., Sierra-Diaz D.C., Patarroyo M.A., Laissue P., Suarez C.F., Cabrera R. Structural Modelling of KCNQ1 and KCNH2 Double Mutant Proteins, Identified in Two Severe Long QT Syndrome Cases, Reveals New Insights into Cardiac Channelopathies. Int. J. Mol. Sci. 2021;22:12861. doi: 10.3390/ijms222312861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Paquet E., Viktor H.L. Molecular dynamics, monte carlo simulations, and langevin dynamics: A computational review. Biomed. Res. Int. 2015;2015:183918. doi: 10.1155/2015/183918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Costa F., Guardiani C., Giacomello A. Molecular dynamics simulations suggest possible activation and deactivation pathways in the hERG channel. Commun. Biol. 2022;5:165. doi: 10.1038/s42003-022-03074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kuenze G., Duran A.M., Woods H., Brewer K.R., McDonald E.F., Vanoye C.G., George A.L., Jr., Sanders C.R., Meiler J. Upgraded molecular models of the human KCNQ1 potassium channel. PLoS ONE. 2019;14:e0220415. doi: 10.1371/journal.pone.0220415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tobelaim W.S., Dvir M., Lebel G., Cui M., Buki T., Peretz A., Marom M., Haitin Y., Logothetis D.E., Hirsch J.A., et al. Competition of calcified calmodulin N lobe and PIP2 to an LQT mutation site in Kv7.1 channel. Proc. Natl. Acad. Sci. USA. 2017;114:E869–E878. doi: 10.1073/pnas.1612622114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang W., MacKinnon R. Cryo-EM Structure of the Open Human Ether-a-go-go-Related K(+) Channel hERG. Cell. 2017;169:422–430.e410. doi: 10.1016/j.cell.2017.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.van Gunsteren W.F., Daura X., Fuchs P.F.J., Hansen N., Horta B.A.C., Hunenberger P.H., Mark A.E., Pechlaner M., Riniker S., Oostenbrink C. On the Effect of the Various Assumptions and Approximations used in Molecular Simulations on the Properties of Bio-Molecular Systems: Overview and Perspective on Issues. Chemphyschem. 2021;22:264–282. doi: 10.1002/cphc.202000968. [DOI] [PubMed] [Google Scholar]
- 147.Meier K., Choutko A., Dolenc J., Eichenberger A.P., Riniker S., van Gunsteren W.F. Multi-resolution simulation of biomolecular systems: A review of methodological issues. Angew. Chem. Int. Ed. Engl. 2013;52:2820–2834. doi: 10.1002/anie.201205408. [DOI] [PubMed] [Google Scholar]
- 148.Takada S. Go model revisited. Biophys. Phys. 2019;16:248–255. doi: 10.2142/biophysico.16.0_248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Torrisi M., Pollastri G., Le Q. Deep learning methods in protein structure prediction. Comput. Struct. Biotechnol. J. 2020;18:1301–1310. doi: 10.1016/j.csbj.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pandurangan A.P., Blundell T.L. Prediction of impacts of mutations on protein structure and interactions: SDM, a statistical approach, and mCSM, using machine learning. Protein Sci. 2020;29:247–257. doi: 10.1002/pro.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li B., Mendenhall J.L., Kroncke B.M., Taylor K.C., Huang H., Smith D.K., Vanoye C.G., Blume J.D., George A.L., Jr., Sanders C.R., et al. Predicting the Functional Impact of KCNQ1 Variants of Unknown Significance. Circ. Cardiovasc. Genet. 2017;10:e001754. doi: 10.1161/CIRCGENETICS.117.001754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gepp M.M., Hutter M.C. Determination of hERG channel blockers using a decision tree. Bioorg. Med. Chem. 2006;14:5325–5332. doi: 10.1016/j.bmc.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 153.Zhang Y., Zhao J., Wang Y., Fan Y., Zhu L., Yang Y., Chen X., Lu T., Chen Y., Liu H. Prediction of hERG K+ channel blockage using deep neural networks. Chem. Biol. Drug Des. 2019;94:1973–1985. doi: 10.1111/cbdd.13600. [DOI] [PubMed] [Google Scholar]
- 154.Lee H.M., Yu M.S., Kazmi S.R., Oh S.Y., Rhee K.H., Bae M.A., Lee B.H., Shin D.S., Oh K.S., Ceong H., et al. Computational determination of hERG-related cardiotoxicity of drug candidates. BMC Bioinform. 2019;20:250. doi: 10.1186/s12859-019-2814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu M., Zhang L., Li S., Yang T., Liu L., Zhao J., Liu H. Prediction of hERG potassium channel blockage using ensemble learning methods and molecular fingerprints. Toxicol. Lett. 2020;332:88–96. doi: 10.1016/j.toxlet.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 156.Draelos R.L., Ezekian J.E., Zhuang F., Moya-Mendez M.E., Zhang Z., Rosamilia M.B., Manivannan P.K.R., Henao R., Landstrom A.P. GENESIS: Gene-Specific Machine Learning Models for Variants of Uncertain Significance Found in Catecholaminergic Polymorphic Ventricular Tachycardia and Long QT Syndrome-Associated Genes. Circ. Arrhythm Electrophysiol. 2022;15:e010326. doi: 10.1161/CIRCEP.121.010326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Oliveira-Mendes B., Feliciangeli S., Menard M., Chatelain F., Alameh M., Montnach J., Nicolas S., Ollivier B., Barc J., Baro I., et al. A standardised hERG phenotyping pipeline to evaluate KCNH2 genetic variant pathogenicity. Clin. Transl. Med. 2021;11:e609. doi: 10.1002/ctm2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W., et al. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzym. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Webb B., Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016;54:5–6. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jisna V.A., Jayaraj P.B. Protein Structure Prediction: Conventional and Deep Learning Perspectives. Protein. J. 2021;40:522–544. doi: 10.1007/s10930-021-10003-y. [DOI] [PubMed] [Google Scholar]
- 161.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., Wang J., Cong Q., Kinch L.N., Schaeffer R.D., et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.AlQuraishi M. ProteinNet: A standardized data set for machine learning of protein structure. BMC Bioinform. 2019;20:311. doi: 10.1186/s12859-019-2932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kryshtafovych A., Schwede T., Topf M., Fidelis K., Moult J. Critical assessment of methods of protein structure prediction (CASP)-Round XIII. Proteins. 2019;87:1011–1020. doi: 10.1002/prot.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Meinen B.A., Bahl C.D. Breakthroughs in computational design methods open up new frontiers for de novo protein engineering. Protein Eng. Des. Sel. 2021;34:gzab007. doi: 10.1093/protein/gzab007. [DOI] [PubMed] [Google Scholar]
- 166.Sruthi C.K., Balaram H., Prakash M.K. Toward Developing Intuitive Rules for Protein Variant Effect Prediction Using Deep Mutational Scanning Data. ACS Omega. 2020;5:29667–29677. doi: 10.1021/acsomega.0c02402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Thusberg J., Vihinen M. Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum. Mutat. 2009;30:703–714. doi: 10.1002/humu.20938. [DOI] [PubMed] [Google Scholar]
- 168.Coskuner-Weber O., Uversky V.N. Insights into the Molecular Mechanisms of Alzheimer’s and Parkinson’s Diseases with Molecular Simulations: Understanding the Roles of Artificial and Pathological Missense Mutations in Intrinsically Disordered Proteins Related to Pathology. Int. J. Mol. Sci. 2018;19:336. doi: 10.3390/ijms19020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sagar A., Xue B. Recent Advances in Machine Learning Based Prediction of RNA-protein Interactions. Protein Pept. Lett. 2019;26:601–619. doi: 10.2174/0929866526666190619103853. [DOI] [PubMed] [Google Scholar]
- 170.Terayama K., Shinobu A., Tsuda K., Takemura K., Kitao A. evERdock BAI: Machine-learning-guided selection of protein-protein complex structure. J. Chem. Phys. 2019;151:215104. doi: 10.1063/1.5129551. [DOI] [PubMed] [Google Scholar]
- 171.Bitencourt-Ferreira G., de Azevedo W.F., Jr. Machine Learning to Predict Binding Affinity. Methods Mol. Biol. 2019;2053:251–273. doi: 10.1007/978-1-4939-9752-7_16. [DOI] [PubMed] [Google Scholar]
- 172.Wang D.D., Ou-Yang L., Xie H., Zhu M., Yan H. Predicting the impacts of mutations on protein-ligand binding affinity based on molecular dynamics simulations and machine learning methods. Comput. Struct. Biotechnol. J. 2020;18:439–454. doi: 10.1016/j.csbj.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pavlova A., Zhang Z., Acharya A., Lynch D.L., Pang Y.T., Mou Z., Parks J.M., Chipot C., Gumbart J.C. Machine Learning Reveals the Critical Interactions for SARS-CoV-2 Spike Protein Binding to ACE2. J. Phys. Chem. Lett. 2021;12:5494–5502. doi: 10.1021/acs.jpclett.1c01494. [DOI] [PubMed] [Google Scholar]
- 174.Gurunathan V., Hamre J., 3rd, Klimov D.K., Jafri M.S. Data Mining of Molecular Simulations Suggest Key Amino Acid Residues for Aggregation, Signaling and Drug Action. Biomolecules. 2021;11:1541. doi: 10.3390/biom11101541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Heo L., Feig M. High-accuracy protein structures by combining machine-learning with physics-based refinement. Proteins. 2020;88:637–642. doi: 10.1002/prot.25847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Li G., Qin Y., Fontaine N.T., Ng Fuk Chong M., Maria-Solano M.A., Feixas F., Cadet X.F., Pandjaitan R., Garcia-Borras M., Cadet F., et al. Machine Learning Enables Selection of Epistatic Enzyme Mutants for Stability Against Unfolding and Detrimental Aggregation. Chembiochem. 2021;22:904–914. doi: 10.1002/cbic.202000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Babbitt G.A., Fokoue E.P., Evans J.R., Diller K.I., Adams L.E. DROIDS 3.0-Detecting Genetic and Drug Class Variant Impact on Conserved Protein Binding Dynamics. Biophys. J. 2020;118:541–551. doi: 10.1016/j.bpj.2019.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ponzoni L., Nguyen N.H., Bahar I., Brodsky J.L. Complementary computational and experimental evaluation of missense variants in the ROMK potassium channel. PLoS Comput. Biol. 2020;16:e1007749. doi: 10.1371/journal.pcbi.1007749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.