Abstract
The PST-01 protease is secreted by the organic solvent-tolerant microorganism Pseudomonas aeruginosa PST-01 and is stable in the presence of various organic solvents. Therefore, the PST-01 strain and the PST-01 protease are very useful for fermentation and reactions in the presence of organic solvents, respectively. The organic solvent-stable PST-01 protease has two disulfide bonds (between Cys-30 and Cys-58 and between Cys-270 and Cys-297) in its molecule. Mutant PST-01 proteases in which one or both of the disulfide bonds were deleted were constructed by site-directed mutagenesis, and the effect of the disulfide bonds on the activity and the various stabilities was investigated. The disulfide bond between Cys-270 and Cys-297 in the PST-01 protease was found to be essential for its activity. The disulfide bond between Cys-30 and Cys-58 played an important role in the organic solvent stability of the PST-01 protease.
The PST-01 protease produced by the organic solvent tolerant microorganism Pseudomonas aeruginosa PST-01 is very stable in the presence of organic solvents (16). It is a metalloendopeptidase and is stabler than subtilisin Carlsberg, thermolysin, and α-chymotrypsin, especially in the presence of water-soluble organic solvents (14). The PST-01 protease catalyzes peptide synthesis with higher yields and higher reaction rates in the presence of organic solvents such as dimethyl sulfoxide, N,N-dimethylformamide, and methanol than in the absence of organic solvents (17, 18). In our previous study, the PST-01 protease gene was cloned and sequenced (15). The amino acid sequence of the PST-01 protease was found to be the same as that of pseudolysin, an elastase from P. aeruginosa PAO1, whose three-dimensional structure has been analyzed (22). The amino acid sequence and overall three-dimensional structure of the PST-01 protease were very similar to those of thermolysin (1, 3, 6, 9, 11, 13, 15, 22, 23). One difference is the presence of disulfide bonds in the PST-01 protease.
The presence of a disulfide bond at the appropriate location is very important for enzyme stability in the presence of temperature stress or attack by proteases, as demonstrated by the introduction of disulfide bonds into enzyme molecules (2, 4, 7, 8, 10, 19, 21, 24–26). The PST-01 protease has two disulfide bonds in its molecule; however its thermostability is not higher than that of thermolysin, which has no disulfide bond. On the other hand, the stability of the PST-01 protease in the presence of organic solvents is higher than that of thermolysin (14). Therefore, the disulfide bonds in the PST-01 protease molecule may play an important role in the organic solvent stability of the enzyme. In this study, the effect of the disulfide bonds on the activity, pH stability, heat stability, and organic solvent stability was investigated by substitution of cysteine.
MATERIALS AND METHODS
Organism.
Escherichia coli JM109 (recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi λ− Δ(lac-proAB) F′ [traD36, proAB+, lacIqZΔM15]) (27) was used as the host for the recombinant plasmids and production of the wild-type and mutant PST-01 proteases. The transformed E. coli JM109 cells were cultivated on a Luria-Bertani (LB) agar medium (20) containing 50 mg of ampicillin sodium salt per liter, 1.0% (wt/vol) skim milk powder (Wako Pure Chemical Industries, Ltd., Osaka, Japan), and 1.5% (wt/vol) agar.
Plasmids.
pPC1, which was constructed in our previous work (15) by cloning the PST-01 protease gene, including its promoter region in the BamHI site of pUC19, was used as a template for constructing other plasmids. It was also used for producing the wild-type PST-01 protease. All the plasmids used are summarized in Table 1.
TABLE 1.
Plasmids, proteins, and activities of transformants
| Plasmida | Proteaseb | Activityc |
|---|---|---|
| pPC1 | PST-01 | + |
| pPC1-T763G | PST-01-C58G | − |
| pPC1-T1399G | PST-01-C270G | − |
| pPC1-T763G-T1399G | PST-01-C58G-C270G | − |
| pPC1-C1482A | PST-01-C297Stop | − |
| pPC1-T1399G-C1482A | PST-01-C270G-C297Stop | − |
pPC1 was constructed in our previous study (15). All the other plasmids were constructed in the present work.
Proteases produced from the transformants harboring each plasmid are shown.
The proteolytic activities of the transformants harboring each plasmid were tested on LB agar medium containing skim milk.
Replacement of Cys-58 by Gly.
The replacement of cysteine at amino acid 58 (Cys-58) of the PST-01 protease by glycine (C58G) was performed by replacing thymine at nucleotide 756 position (T-763) of the PST-01 protease gene with guanine (T763G) using the technique of splicing by overlap extension by PCR (5). The first PCR was performed using pPC1/HindIII as a template, 5′-TGC CGA CGA ACT GAA AGC GAT C-3′ and 5′-TGT AGG TGT TGG TCG GGC C*GG CGA AGC GGA-3′ (the asterisk indicates a mismatch) as primers, and Z-Taq (Takara Shuzo Co., Ltd., Kyoto, Japan). The second PCR was performed using pPC1/HindIII as a template, 5′- TCC GCT TCG CCG∗ GCC CGA CCA ACA CCT ACA-3′ and 5′-AAC AGC AGA CTC ATG GCA GGA C-3′ as primers, and Z-Taq. The first and second PCR products, which were purified by agarose gel electrophoresis using a QIAquick gel extraction kit (Qiagen GmbH, Hilden, Germany), were used as templates of the third PCR with 5′-TGC CGA CGA ACT GAA AGC GAT C-3′ and 5′-AAC AGC AGA CTC ATG GCA GGA C-3′ as primers. Repeated cycles of thermal denaturation, annealing, and extension/termination were performed using a GeneAmp PCR system 2400 apparatus (Perkin-Elmer Co., Norwalk, Conn.). The purified third PCR product was cleaved with BbsI and NotI and ligated with the 4.2-kbp fragment of pPC1 which was cleaved with the same restriction endonucleases.
Replacement of Cys-270 by Gly and of Cys-297 by a stop codon.
The replacement of cysteine at amino acid 270 (Cys-270) of the PST-01 protease by glycine (C270G) was performed by replacing thymine at nucleotide 1399 (T-1399) of the PST-01 protease gene by guanine (T1399G) using a QuikChange site-directed mutagenesis kit (Stratagene Cloning Systems, La Jolla, Calif.) with two primers, 5′-ACA ACA GCG GCG CCG∗ GCG GGG TGA TTC GCT CGG CGC-3′ and 5′-GCG CCG AGC GAA TCA CCC CGC C∗GG CGC CGC TGT TGT-3′. The replacement of cysteine at amino acid 297 (Cys-297) of the PST-01 protease by the stop codon (C297Stop) was performed by replacing cytosine at nucleotide 1482 (C-1482) of the PST-01 protease gene by adenine (C1399A) using the same kit with two primers, 5′-CGG CGT GAC CTG A∗CC GAG CGC GTT GT-3′ and 5′-ACA ACG CGC TCG GT∗C AGG TCA CGC CG-3′.
Identification of the mutated gene fragment.
Clones containing the mutated gene fragment were identified by restriction enzyme analysis using NaeI and the nucleotide sequence of the full inset fragment from the genomic DNA of P. aeruginosa PST-01. The nucleotide sequence was determined by cycle sequencing (12) using a Thermo Sequenase fluorescence-labeled primer cycle-sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech Ltd., Uppsala, Sweden) with the following fluorescein-labeled primers: 5′-fluorescein-CGC CAG GGT TTT CCC AGT CAC GAC-3′ (universal M13 forward primer), 5′-fluorescein-GAG CGG ATA ACA ATT TCA CAC AGG-3′ (universal M13 reverse primer), 5′-fluorescein-AGC AGA CTC ATG GCA GGA-3′, 5′-fluorescein-CCA AGC GAC ATA AAG CAG CCG CCC-3′, 5′-fluorescein-GCG AAT TGG CCA ACA GGT-3′, 5′-fluorescein-GCC GCG CAT ATA GAA CTC GGC AGC-3′, 5′-fluorescein-CCC GTA GTG CAC CTT CAT-3′, 5′-fluorescein-CTT CCC ACT GAT CGA GCA-3′, 5′-fluorescein-CGT ACG CCG TTG TGG AAT-3′, 5′-fluorescein-AGA AGG AAC TGC ACT CCC-3′, 5′-fluorescein-AGG GGA GTG CAG TTC CTT-3′, 5′-fluorescein-CGC TAC GAG CAA TTC CAC-3′, 5′-fluorescein-AAC GTC TCC TAC CTG ATT CCC GGC-3′, 5′-fluorescein-GCA ACC AGA AGA TCG GCA-3′, 5′-fluorescein-GTT CTA TCC GCT GGT GTC GCT GGA-3′, 5′-fluorescein-CGT TCT ACC TGT TGG CCA-3′, and 5′-fluorescein-GAG TCC TGC CAT GAG TCT-3′. Denaturing polyacrylamide gel electrophoresis, detection of fluorescein, and signal analysis were performed with an automated fluorescent DNA sequencer (DSQ-2000L; Shimadzu Co., Kyoto, Japan).
Expression of PST-01 protease and its mutants in E. coli JM109 and their purification.
E. coli JM109 cells transformed with pPC1 (E. coli JM109/pPC1) or pPC1-T763G (E. coli JM109/pPC1-T763G) were cultured in LB liquid medium containing 50 mg of ampicillin sodium salt per liter. The medium was adjusted to pH 7.2 using 1 M NaOH. A 500-ml baffled Erlenmeyer flask containing 200 ml of the medium was inoculated with the transformed cells and incubated at 37°C with rotary shaking (150 rpm, 7-cm-diameter shaking). The culture supernatants were prepared by removing the cells in the culture by centrifuging 2,000 ml of the culture at 10,000 × g at 4°C for 5 min. The precipitated cells were resuspended in 200 ml of a Tris-HCl buffer (pH 7.5) and disrupted by ultrasonic disintegration using an ultrasonic disruptor (UD-200; Tomy Seiko Co., Ltd., Tokyo, Japan) at 97 W for 5 min intermittently in an ice bath. Cell extract was obtained by centrifugation at 10,000 × g at 4°C for 5 min. After incubation at 30°C for 2 h, solid ammonium sulfate was added to 40% saturation. The precipitate formed was removed by centrifugation at 10,000 × g at 4°C for 5 min. After further addition of solid ammonium sulfate to the supernatant to 75% saturation, the resulting precipitate was collected by centrifugation at 10,000 × g at 4°C for 5 min. The collected precipitate was dissolved in a 10 mM borax-HCl buffer (pH 8.5) containing 1 M ammonium sulfate. The PST-01 protease and its mutant contained in these crude cell extracts were purified by hydrophobic interaction chromatography with a TSKgel Butyl-Toyopearl 650M (Tosoh, Tokyo, Japan) as described in our previous paper (15).
Measurement of proteolytic activity.
The protease activity was routinely determined by the casein hydrolysis method (16). A reaction mixture of 5 ml of a 50 mM borax-HCl buffer (pH 8.5) containing 0.6% (wt/vol) Hammarsten casein (E. Merck, Darmstadt, Germany) and 0.1 ml of an enzyme solution was incubated at 30°C for 10 min. The reaction was stopped by the addition of 1 ml of a trichloroacetic acid solution consisting of 5.44% (wt/vol) trichloroacetic acid, 6% (wt/vol) acetic acid, and 5.46% (wt/vol) sodium acetate. The mixture was further incubated at 4°C for 30 min and then filtered using a no. 5C filter paper (Toyo Roshi Kaisha Ltd., Tokyo, Japan). The concentration of digested casein in the filtrate was determined by measuring the absorbance at 280 nm using tyrosine as a standard. One unit of the proteolytic activity was defined as the amount of enzyme which produces the casein digest equivalent to 1 μmol of tyrosine in the filtrate per min at 30°C.
Measurement of the activity of peptide synthesis.
As the acid and amine components of the substrates for the peptide synthesis, N-α-carbobenzoxy-l-arginine (Cbz-Arg) (Nacalai Tesque, Inc. Kyoto, Japan) and l-leucineamide (Leu-NH2) were used, respectively. Leu-NH2 was prepared by the following method. l-Leucineamide hydrochloride (30 mmol; Nacalai Tesque, Inc.) and 0.033 mol of NaOH were dissolved in 50 ml of distilled water. Leu-NH2 was extracted with 200 ml of dichloromethane and recovered by evaporating dichloromethane with a rotary evaporator at room temperature. About 0.027 mol of Leu-NH2 was recovered, and it was identified using a 200-MHz nuclear magnetic resonance spectrometer (Gemini-2000; Varian, Palo Alto, Calif.).
The reaction mixtures (pH 6.8) containing 130 kU of protease per liter, 10 mM Cbz-Arg as a carboxyl component, 500 mM Leu-NH2 as an amine component, and 50% (vol/vol) dimethyl sulfoxide were incubated at 30°C.
The amount of the produced Cbz-Arg-Leu-NH2 was analyzed by the following procedure. Aliquots of sample solutions were taken from the reaction mixture after 0, 30, 60, 90, 120, and 180 min of incubation and diluted with an eluent (acetonitrile–50 mM sodium phosphate buffer [pH 3.0], 80:20 [vol/vol]) to 1:20 for high-performance liquid chromatography. After the enzymatic reaction was quenched by adding the eluent and the mixture was stored at −20°C, an aliquot of the mixture was analyzed by reversed-phase high-performance liquid chromatography using a Shimadzu CL-10A chromatograph system equipped with a DGU-12A on-line degasser, an LC-10ADvp solvent delivery unit, a SIL-10ADvp autoinjector, a CTO-10Avp column oven, an SPD-10Avp UV-VIS detector, and a C-R6A Chromatopac integrator (Shimadzu Corp.). The column used was an ODS column packed with Cosmosil 5C18-AR-II (4.6 by 150 mm) (Nacalai Tesque, Inc.). The flow rate of the eluent was 1.0 ml/min. The oven temperature was 35°C. The eluted reactants and product were detected at a wavelength of 257 nm. The retention times of Cbz-Arg and Cbz-Arg-Leu-NH2 were 4 and 14 min, respectively. The product yield was calculated based on the amount of the limiting substrate, the acid component. The initial rates were calculated from the slopes of the curves of amount of product versus reaction time.
Measurement of the organic solvent stability of the enzyme.
The 10 mM borax-HCl buffer (pH 8.5) containing about 5 U of the purified PST-01 or PST-01-C58G proteases per ml was filtered with a cellulose acetate membrane filter (pore size, 0.2 μm). A 1-ml volume of an organic solvent was added to 3 ml of the filtrate in a test tube (16.5 mm in diameter) with a screw cap and was incubated at 30°C with shaking at 160 strokes per min. The time courses of the remaining proteolytic activity were determined by the casein hydrolysis method.
RESULTS AND DISCUSSION
Construction of the mutated PST-01 protease and its activity.
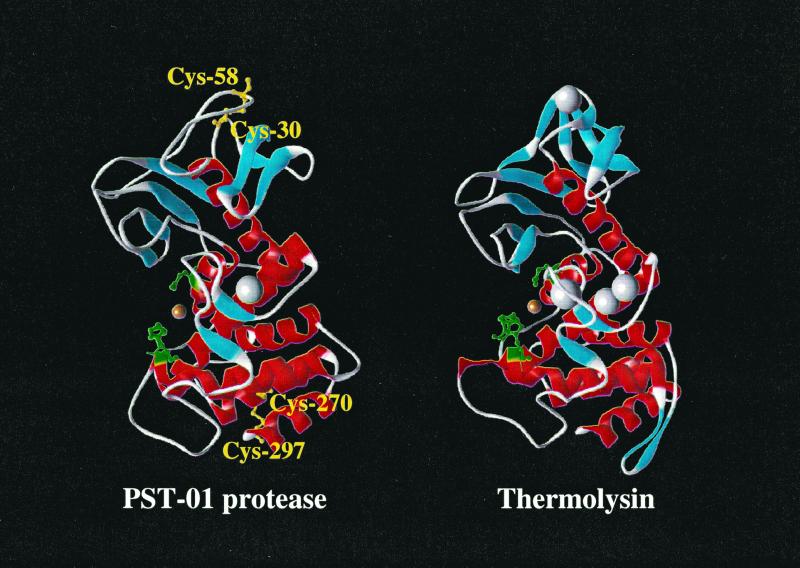
The amino acid sequence of the PST-01 protease was very similar to that of thermolysin. The homology of the primary structure was 33%, and the overall three-dimensional structure of the PST-01 protease was very similar to that of thermolysin (1, 3, 6, 9, 11, 13, 15, 22, 23), as shown in Fig. 1. One difference was the presence of the disulfide bonds in the PST-01 protease. The PST-01 protease has two disulfide bonds between Cys-30 and Cys-58 and between Cys-270 and Cys-297; however, thermolysin has no disulfide bond in its molecule. Therefore, mutant PST-01 proteases from which the disulfide bonds were deleted were constructed by replacement of amino acids in the PST-01 protease by using site-directed mutagenesis.
FIG. 1.
Comparison of the three-dimensional structure of the PST-01 protease with that of thermolysin. Solid ribbon diagrams of the PST-01 protease and thermolysin are shown. α-Helices, β-strands, active centers, and disulfide bonds are colored red, blue, green, and yellow, respectively. The magnesium and calcium ions are shown as gray and brown spheres, respectively. The diagrams of the PST-01 protease and thermolysin were created using MSI WebLab ViewerLite with the structure data of the elastase from P. aeruginosa (PDB code 1EZM) and thermolysin (PDB code 1LNF), respectively.
Five mutant plasmids, pPC1-T763G, pPC1-T1399G, pPC1-T763G-T1399G, pPC1-C1482A, and pPC1-T1399G-C1482A, were constructed (Table 1). The PST-01-C58G protease, in which Cys-58 of the PST-01 protease was replaced by glycine, was produced with pPC1-T763G, in which T-763 of the PST-01 protease gene was replaced by guanine. Similarly, the PST-01-C270G protease, in which Cys-270 was replaced by glycine, was produced with pPC1-T1399G, in which T-1399 was replaced by guanine. The PST-01-C58G-C270G protease, in which Cys-58 and Cys-270 were both replaced by glycine, was produced with pPC1-T763G-T1399G, in which T-763 and T-1399 were both replaced by guanine. The PST-01-C297Stop protease, in which 5 amino acids at positions 297 (cysteine) to 301 (C terminus) were deleted, was produced with pPC1-C1482A, in which C-1482 was replaced by adenine. The PST-01-C270G-C297Stop protease, in which Cys-270 was replaced by glycine and 5 amino acids at positions 297 (cysteine) to 301 (C terminus) were deleted was produced with pPC1-C1399G-C1482A, in which C-1399 and C-1482 was replaced by guanine and adenine, respectively. The wild-type and mutant PST-01 protease genes were expressed in E. coli. E. coli containing pPC1 and pPC1-T763G formed clear zones on LB agar media containing skim milk. However, E. coli containing pPC1-T1399G, pPC1-T763G-T1399G, pPC1-C1482A, and pPC1-C1399G-C1482A did not form clear zones on LB agar medium containing skim milk (Table 1). These results indicated that the disulfide bond between Cys-270 and Cys-297 was necessary for exhibiting the proteolytic activity of the PST-01 protease. Therefore, E. coli containing pPC1 and pPC1-T763G were cultured in a liquid medium and the expressed proteases (PST-01 protease and PST-01-C58G protease, respectively) were purified by hydrophobic interaction chromatography.
The specific activities of the proteolytic reaction using casein as a substrate, the initial rates of the peptide synthesis catalyzed by the PST-01 protease and the PST-01-C58G protease, and the equilibrium yields are summarized in Table 2. Both the specific activity of the proteolytic reaction and the initial rate of the peptide synthesis catalyzed by the PST-01-C58G protease were one-fourth of those of the reaction and synthesis catalyzed by the PST-01 protease. These results showed that replacing Cys-58 with Gly resulted in a decrease in the specific activities of the proteolytic reaction and the peptide synthesis. However, the equilibrium yields were almost the same irrespective of the protease.
TABLE 2.
Activities of the proteolytic reaction and the peptide synthesis catalyzed by the PST-01 and PST-01-C58G proteasesa
| Protease | Activity of proteolytic reaction (kU/g) | Peptide synthesis
|
|
|---|---|---|---|
| Initial rate (μmol/min/g of enzyme) | Equilibrium yield (%) | ||
| PST-01 | 35.3 ± 3.0 | 330 ± 2 | 74.9 ± 0.3 |
| PST-01-C58G | 9.27 ± 1.0 | 82.1 ± 1.5 | 73.7 ± 0.6 |
The wild-type (PST-01) and the mutant (PST-01-C58G) proteases were both purified by ammonium sulfate fractionation and successive hydrophobic interaction chromatography steps using a Butyl-Toyopearl gel.
Effect of pH on the activity.
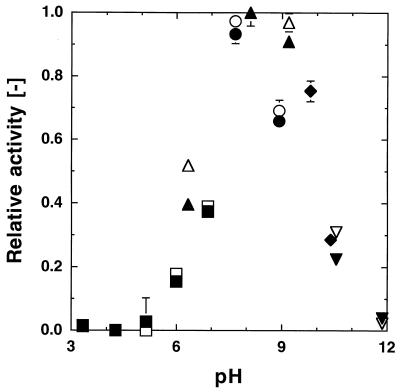
The effect of pH on the proteolytic activity of the purified PST-01 and PST-01-C58G proteases was investigated using 50 mM McIlvain buffer (pH 3.3 to 6.9), 50 mM Tris-HCl buffer (pH 6.3 to 9.2), 50 mM borax-HCl buffer (pH 7.7 and 8.9), 50 mM borax-NaOH buffer (pH 9.8 and 10.4), and 50 mM sodium phosphate buffer (pH 10.6 and 11.9). The relative activities at various pH values are shown in Fig. 2, in which the activity measured using the 50 mM Tris-HCl buffer (pH 8.1) is taken as 1. No difference in the pH dependency of the activity was observed between the PST-01 and PST-01-C58G proteases.
FIG. 2.
Effect of pH on the proteolytic activities of the PST-01 and PST-01-C58G proteases. The wild-type (PST-01) and mutant (PST-01-C58G) proteases were both purified by ammonium sulfate fractionation and successive hydrophobic interaction chromatography steps using a Butyl-Toyopearl gel. About 5 kU of these purified proteases per liter was used. The activities of the PST-01 (open symbols) and PST-01-C58G (solid symbols) proteases at 30°C were measured using 50 mM McIlvain buffer (□, ■), 50 mM Tris-HCl buffer (▵, ▴), 50 mM borax-HCl buffer (○, ●), 50 mM borax-NaOH buffer (◊, ⧫), and sodium phosphate buffer (▿, ▾) with casein as the substrate. The activities at various pHs relative to that measured using 50 mM Tris-HCl buffer (pH 8.1) are shown. The error bars on the symbols indicate deviation. The absence of a bar indicates that the deviation was smaller than the symbol.
Effect of temperature on the activity.
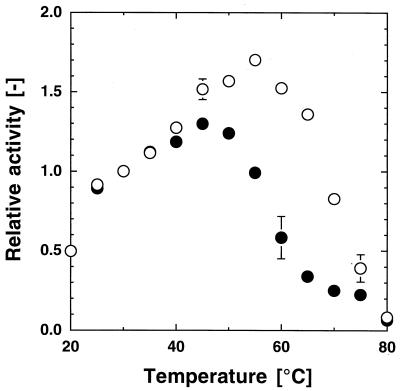
The activities of the PST-01 and PST-01-C58G proteases measured at pH 8.5 and various temperatures are shown in Fig. 3, taking the activities at 30°C to be 1. Although the maximum activity of the PST-01 protease was observed at approximately 55°C, that of the PST-01-C58G protease was observed at approximately 45°C. The optimum temperature at which the maximum activity was exhibited decreased due to deletion of the disulfide bond between Cys-30 and Cys-58 of the PST-01 protease.
FIG. 3.
Effect of temperature on the proteolytic activities of the PST-01 and PST-01-C58G proteases. The wild-type and mutant proteases were both purified by ammonium sulfate fractionation and successive hydrophobic interaction chromatography steps using a Butyl-Toyopearl gel. The activities of the PST-01 (○) and PST-01-C58G (●) proteases (about 5 kU/liter) at various temperatures were determined using casein as the substrate at pH 8.5. The activities at various temperatures relative to those at 30°C are shown. The error bars on the symbols indicate deviation. The absence of a bar indicates that the deviation was smaller than the symbol.
Effect of temperature on the stability.
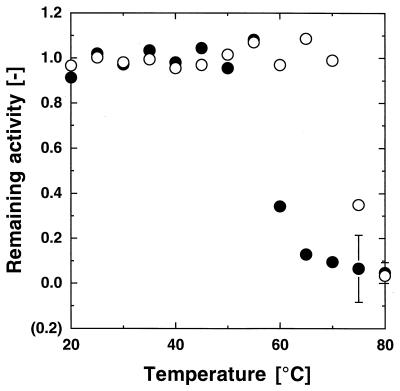
The heat stability of the enzymes was studied by assaying the residual activities after 10 min of incubation at various temperatures at pH 8.5 and is shown in Fig. 4. Although the PST-01 protease was stable up to 70°C, the PST-01-C58G protease was stable only below 55°C. The temperature at which the enzyme is stable decreased due to the deletion of the disulfide bond between Cys-30 and Cys-58 of the PST-01 protease.
FIG. 4.
Heat stabilities of the PST-01 and PST-01-C58G proteases. The wild-type (PST-01) and mutant (PST-01-C58G) proteases were both purified by ammonium sulfate fractionation and successive hydrophobic interaction chromatography steps using a Butyl-Toyopearl gel. Solutions containing the PST-01 (○) and PST-01-C58G (●) proteases (about 5 kU/liter [pH 8.5]) were incubated at various temperatures for 10 min. The remaining activities were measured at 30°C and are shown as the values relative to those before incubation. The error bars on the symbols indicate deviation. The absence of a bar indicates that the deviation was smaller than the symbol.
Organic solvent stability.
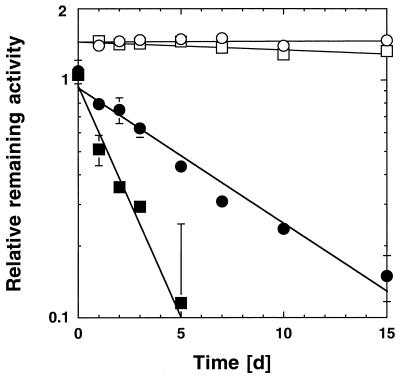
The effect of various organic solvents on the stability of the PST-01 and PST-01-C58G proteases was studied. Figure 5 shows the typical time courses of the remaining activity of the PST-01 and PST-01-C58G proteases in the presence of 1,5-pentanediol and acetone. Deactivation of both proteases in the presence of organic solvents obeyed the first-order kinetics. The half-lives of the activities of both proteases are summarized in Table 3. The PST-01 protease was stable in the presence of water-soluble organic solvents or alcohols such as 1,4-butanediol, 1,5-pentanediol, ethanol, methanol, dimethyl sulfoxide, 2-propanol, and N,N-dimethylformamide. Inactivation of the PST-01 protease in the presence of these organic solvents was not observed at all during the 15-day experiment. These results were in good agreement with those of our previous study (14). However, the half-lives of the PST-01-C58G protease in the presence of organic solvents were much shorter than those of the PST-01 protease in the same organic solvents. Therefore, the disulfide bond between Cys-30 and Cys-58 of the PST-01 protease plays an important role in exhibiting the organic solvent stability of the PST-01 protease.
FIG. 5.
Time courses of relative remaining activity of the PST-01 and PST-01-C58G proteases in the presence of 1, 5-pentanediol and acetone. The wild-type (PST-01)and mutant (PST-01-C58G) proteases were both purified by ammonium sulfate fractionation and successive hydrophobic interaction chromatography steps using a Butyl-Toyopearl gel. Solutions containing the PST-01 (open symbols) and PST-01-C58G (solid symbols) proteases (about 5 kU/liter [pH 8.5]) were incubated at 30°C with shaking in the presence of 1,5-pentanediol (○, ●) or acetone (□, ■). The relative remaining activities based on the activity before addition of organic solvents are shown. The error bars on the symbols indicate deviation. The absence of a bar indicates that the deviation was smaller than the symbol.
TABLE 3.
Half-lives of the wild-type and mutant proteases in the presence of organic solventsa
| Organic solvent | Half-life (days)b of:
|
|
|---|---|---|
| PST-01 protease | PST-01-C58G protease | |
| Dimethyl sulfoxide | >100 | 33.3 |
| Ethylene glycol | >100 | 20.3 |
| 1,4-Butanediol | >100 | 14.5 |
| Methanol | >100 | 5.8 |
| 1,5-Pentanediol | >100 | 5.3 |
| Ethanol | >100 | 5.0 |
| N,N-Dimethylformamide | >100 | 3.5 |
| 2-Propanol | >100 | 2.4 |
| Acetone | 85.6 | 1.6 |
| Acetonitrile | 2.4 | <1 |
| Cyclohexane | 1.7 | <1 |
The wild-type PST-01 protease and the mutant protease (PST-01-C58G protease) were both purified by ammonium sulfate fractionation and successive hydrophobic interaction chromatography steps using a Butyl-Toyopearl gel. The filtered solutions (3 ml) containing the PST-01 protease or the PST-01-C58G protease (about 5 kU/liter) were incubated for 15 days at pH 8.5 at 30°C with shaking at 160 strokes per min in the presence of 1 ml of an organic solvent. The remaining activities were measured after 0, 1, 2, 3, 5, 7, 10, and 15 days.
The half-lives were calculated from the exponential regression curve.
In this study, the effects of disulfide bonds, which are present in the PST-01 protease, on the heat and organic solvent stabilities of the protease were studied. The disulfide bond between Cys-270 and Cys-297 was necessary for exhibiting the proteolytic activity of the PST-01 protease. However, the disulfide bond between Cys-30 and Cys-58 played an important role in not only the heat stability but also the organic solvent stability. To our knowledge, this is the first experimental demonstration that the disulfide bond has played an important role in the organic solvent stability of an enzyme. Although further extensive research on the organic solvent stability of the mutant enzymes as well as of the wild-type enzyme is needed, the present work should lead us to a fuller understanding of the organic solvent stability of enzymes and to the development of new organic solvent-stable enzymes.
ACKNOWLEDGMENTS
A part of this work was supported by the Proposal-Based Immediate-Effect R&D Promotion Program from the New Energy and Industrial Technology Development Organization (NEDO, project ID 98Z36-013-1) of Japan and a Grant-in-Aid for Scientific Research (B) (11555209) from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Bever R A, Iglewski B H. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J Bacteriol. 1988;170:4309–4314. doi: 10.1128/jb.170.9.4309-4314.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho Y, Gu W, Watkins S, Lee S P, Kim T R, Brady J W, Batt C A. Thermostable variants of bovine beta-lactoglobulin. Protein Eng. 1994;7:263–270. doi: 10.1093/protein/7.2.263. [DOI] [PubMed] [Google Scholar]
- 3.Fukushima J, Yamamoto S, Morihara K, Atsumi Y, Takeuchi H, Kawamoto S, Okuda K. Structural gene and complete amino acid sequence of Pseudomonas aeruginisa IFO 3455 elastase. J Bacteriol. 1989;171:1698–1704. doi: 10.1128/jb.171.3.1698-1704.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gokhale R S, Agarwalla S, Francis V S, Santi D V, Balaram P. Thermal stabilization of thymidylate synthase by engineering two disulfide bridges across the dimer interface. J Mol Biol. 1994;235:89–94. doi: 10.1016/s0022-2836(05)80018-x. [DOI] [PubMed] [Google Scholar]
- 5.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 6.Holland D R, Tronrud D E, Pley H W, Flaherty K M, Stark W, Jansonius J N, McKay D B, Matthews B W. Structural comparison suggests that thermolysin and related neutral proteases undergo hinge-bending motion during catalysis. Biochemistry. 1992;31:11310–11316. doi: 10.1021/bi00161a008. [DOI] [PubMed] [Google Scholar]
- 7.Ikegaya K, Ishida Y, Murakami K, Masaki A, Sugio N, Takechi K, Murakami S, Tatsumi H, Ogawa Y, Nakano E, Motai H, Kawabe H. Enhancement of the thermostability of the alkaline protease from Aspergillus oryzae by introduction of a disulfide bond. Biosci Biotechnol Biochem. 1992;56:326–327. doi: 10.1271/bbb.56.326. [DOI] [PubMed] [Google Scholar]
- 8.Kanaya S, Katsuda C, Kimura S, Nakai T, Kitakuni E, Nakamura H, Katayanagi K, Morikawa K, Ikehara M. Stabilization of Escherichia coli ribonuclease H by introduction of an artificial disulfide bond. J Biol Chem. 1991;266:6038–6044. [PubMed] [Google Scholar]
- 9.Kawamoto S, Shibano Y, Fukushima J, Ishii N, Morihara K, Okuda K. Site-directed mutagenesis of Glu-141 and His-223 in Pseudomonas aeruginosa elastase: catalytic activity, processing, and protective activity of the elastase against Pseudomonas infection. Infect Immun. 1993;61:1400–1405. doi: 10.1128/iai.61.4.1400-1405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko J H, Jang W H, Kim E K, Lee H B, Park K D, Chung J H, Yoo O J. Enhancement of thermostability and catalytic efficiency of AprP, an alkaline protease from Pseudomonas sp., by the introduction of a disulfide bond. Biochem Biophys Res Commun. 1996;221:631–635. doi: 10.1006/bbrc.1996.0647. [DOI] [PubMed] [Google Scholar]
- 11.Kubo M, Imanaka T. Cloning and nucleotide sequence of the highly thermostable neutral protease gene from Bacillus stearothermophilus. J Gen Microbiol. 1988;134:1883–1892. doi: 10.1099/00221287-134-7-1883. [DOI] [PubMed] [Google Scholar]
- 12.Murray V. Improved double-stranded DNA sequencing using the linear polymerase chain reaction. Nucleic Acids Res. 1989;17:8889. doi: 10.1093/nar/17.21.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishiya Y, Imanaka T. Cloning and nucleotide sequences of the Bacillus stearothermophilus neutral protease gene and its transcriptional activator gene. J Bacteriol. 1990;172:4861–4869. doi: 10.1128/jb.172.9.4861-4869.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogino H, Watanabe F, Yamada M, Nakagawa S, Hirose T, Noguchi A, Yasuda M, Ishikawa H. Purification and characterization of organic solvent-stable protease from organic solvent-tolerant Pseudomonas aeruginosa PST-01. J Biosci Bioeng. 1999;87:61–68. doi: 10.1016/s1389-1723(99)80009-7. [DOI] [PubMed] [Google Scholar]
- 15.Ogino H, Yokoo J, Watanabe F, Ishikawa H. Cloning and sequencing of a gene of organic solvent-stable protease secreted from Pseudomonas aeruginosa PST-01 and its expression in Escherichia coli. Biochem Eng J. 2000;5:191–200. doi: 10.1016/s1369-703x(00)00060-7. [DOI] [PubMed] [Google Scholar]
- 16.Ogino H, Yasui K, Shiotani T, Ishihara T, Ishikawa H. Organic solvent-tolerant bacterium which secretes an organic solvent-stable proteolytic enzyme. Appl Environ Microbiol. 1995;61:4258–4262. doi: 10.1128/aem.61.12.4258-4262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogino H, Yamada M, Watanabe F, Ichinose H, Yasuda M, Ishikawa H. Peptide synthesis catalyzed by organic solvent-stable protease from Pseudomonas aeruginosa PST-01 in monophasic aqueous-organic solvent systems. J Biosci Bioeng. 1999;88:513–518. doi: 10.1016/s1389-1723(00)87668-9. [DOI] [PubMed] [Google Scholar]
- 18.Ogino H, Gemba Y, Yamada M, Shizuka M, Yasuda M, Ishikawa H. The rates of peptide synthesis catalyzed by organic solvent-stable protease from Pseudomonas aeruginosa PST-01 in the presence of water-soluble organic solvents. Biochem Eng J. 2000;5:219–223. doi: 10.1016/s1369-703x(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 19.Pantoliano M W, Ladner R C, Bryan P N, Rollence M L, Wood J F, Poulos T L. Protein engineering of subtilisin BPN': enhanced stabilization through the introduction of two cysteines to form a disulfide bond. Biochemistry. 1987;26:2077–2082. doi: 10.1021/bi00382a002. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Takagi H, Takahashi T, Momose H, Inouye M, Maeda Y, Matsuzawa H, Ohta T. Enhancement of the thermostability of subtilisin E by introduction of a disulfide bond engineered on the basis of structural comparison with a thermophilic serine protease. J Biol Chem. 1990;265:6874–6878. [PubMed] [Google Scholar]
- 22.Thayer M M, Flaherty K M, McKay D B. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5 A resolution. J Biol Chem. 1991;266:2864–2871. doi: 10.2210/pdb1ezm/pdb. [DOI] [PubMed] [Google Scholar]
- 23.Titani K, Hermodson M A, Ericsson L H, Walsh K A, Neurath H. Amino-acid sequence of thermolysin. Nat New Biol. 1972;238:35–37. doi: 10.1038/newbio238035a0. [DOI] [PubMed] [Google Scholar]
- 24.van den Akker F, Feil I K, Roach C, Platas A A, Merritt E A, Hol W G. Crystal structure of heat-labile enterotoxin from Escherichia coli with increased thermostability introduced by an engineered disulfide bond in the A subunit. Protein Sci. 1997;6:2644–2649. doi: 10.1002/pro.5560061219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Burg B, Dijkstra B W, van der Vinne B, Stulp B K, Eijsink V G, Venema G. Introduction of disulfide bonds into Bacillus subtilis neutral protease. Protein Eng. 1993;6:521–527. doi: 10.1093/protein/6.5.521. [DOI] [PubMed] [Google Scholar]
- 26.Wakarchuk W W, Sung W L, Campbell R L, Cunningham A, Watson D C, Yaguchi M. Thermostabilization of the Bacillus circulans xylanase by the introduction of disulfide bonds. Protein Eng. 1994;7:1379–1386. doi: 10.1093/protein/7.11.1379. [DOI] [PubMed] [Google Scholar]
- 27.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]