Abstract
The enhanced green fluorescent protein (GFP) gene (egfp) was used as a reporter of gene expression driven by the glyceraldehyde-p-dehydrogenase (gpd) gene promoter and the manganese peroxidase isozyme 1 (mnp1) gene promoter in Phanerochaete chrysosporium. Four different constructs were prepared. pUGGM3′ and pUGiGM3′ contain the P. chrysosporium gpd promoter fused upstream of the egfp coding region, and pUMGM3′ and pUMiGM3′ contain the P. chrysosporium mnp1 promoter fused upstream of the egfp gene. In all constructs, the egfp gene was followed by the mnp1 gene 3′ untranslated region. In pUGGM3′ and pUMGM3′, the promoters were fused directly with egfp, whereas in pUGiGM3′ and pUMiGM3′, following the promoters, the first exon (6 bp), the first intron (55 bp), and part of the second exon (9 bp) of the gpd gene were inserted at the 5′ end of the egfp gene. All constructs were ligated into a plasmid containing the ura1 gene of Schizophyllum commune as a selectable marker and were used to transform a Ural1 auxotrophic strain of P. chrysosporium to prototrophy. Crude cell extracts were examined for GFP fluorescence, and where appropriate, the extracellular fluid was examined for MnP activity. The transformants containing a construct with an intron 5′ of the egfp gene (pUGiGM3′ and pUMiGM3′) exhibited maximal fluorescence under the appropriate conditions. The transformants containing constructs with no introns exhibited minimal or no fluorescence. Northern (RNA) blots indicated that the insertion of a 5′ intron resulted in more egfp RNA than was found in transformants carrying an intronless egfp. These results suggest that the presence of a 5′ intron affects the expression of the egfp gene in P. chrysosporium. The expression of GFP in the transformants carrying pUMiGM3′ paralled the expression of endogenous mnp with respect to nitrogen and Mn levels, suggesting that this construct will be useful in studying cis-acting elements in the mnp1 gene promoter.
The white rot basidiomycete Phanerochaete chrysosporium has been the focus of numerous studies of the degradation of the plant cell wall polymer lignin (14, 19, 30) and various aromatic pollutants (10, 22, 42, 43). Under ligninolytic conditions, this fungus secretes two families of peroxidases, lignin peroxidase (LiP) and manganese peroxidase (MnP), along with an H2O2-generating system, which are the major components of its extracellular lignin-degrading system (16, 19, 21, 28, 30, 51). MnP is an H2O2-dependent, heme-containing glycoprotein with an Mr of ∼46,000 which oxidizes Mn(II) to Mn(III) (6, 21, 38, 40, 47, 48, 50); the latter acts as a diffusible mediator in the oxidation of lignin model compounds (6, 16, 50, 51).
The production of the idiophasic proteins MnP and LiP is activated by the depletion of nutrient nitrogen in culture (13, 19, 33, 40, 49). In addition, production of the MnP protein is dependent on, and transcription of the mnp gene is activated by, the presence of Mn(II) in the culture medium (7, 8, 15, 19). Production of MnP is also activated by oxidative stress (32) and heat shock (9). The promoter regions of the P. chrysosporium mnp genes contain several putative cis-acting elements which may be responsible for regulation in response to these environmental factors (2, 13, 18, 19). To study the regulation of genes encoding lignin-degrading enzymes, a suitable gene reporter system is required. Previously, we developed a reporter system based on the orotidylate decarboxylase gene from Schizophyllum commune (17). While this reporter system is sufficient for reporting the regulation of mnp by Mn(II) and nutrient nitrogen levels, the assay is complicated and uses a radioactive substrate.
Green fluorescent protein (GFP) from Aequorea victoria has several advantages as a gene reporter system, and the enhanced gfp gene (egfp) contains a G + C content of 59%, which is the same as that of the P. chrysosporium genome (11, 12, 41). Furthermore, GFP can be used for monitoring both gene transcription and protein localization (11). In this work, we report that the GFP system faithfully reports gene expression when it is driven by the glyceraldehyde-p-dehydrogenase (gpd) or the mnp gene promoter from P. chrysosporium; however, efficient expression is observed only when an intron is inserted within the egfp gene.
MATERIALS AND METHODS
Organisms.
P. chrysosporium OGC101 and the auxotrophic Ura11 strain were maintained as described previously (1, 4). Escherichia coli strain DH5α was used for subcloning plasmids.
Construction of transformation vectors.
The P. chrysosporium Ura transformation vector (pUB) contained the full coding region of the S. commune ura1 orotidylate decarboxylase gene, including 200 bp of the promoter region, and was constructed as described previously (45).
Construction of pUGGM3′.
The pUGGM3′ construct (no intron) contains the P. chrysosporium gpd promoter (36), the egfp gene coding sequence (CLONTECH) (39), and 250 bp of the mnp1 3′ untranslated region (UTR) immediately following the stop codon. The gpd promoter was modified to contain a KpnI site 5 bp upstream of the translation start site. With a forward primer 5′ upstream of the native BamHI site and a reverse primer located 5 bp upstream of the translation start site containing the introduced KpnI site, a BamHI-KpnI PCR fragment was prepared. This fragment replaced a BamHI-KpnI fragment of the pGPD stu1.8 sequence, which had been constructed previously (36). The entire 1.14-kb gpd promoter, containing an XbaI-KpnI fragment, was subcloned into pUC18 for further use.
The coding region of the egfp gene and the 3′ end of the mnp1 gene (18) were fused at the stop codon using the megaprimer method in a two-step reaction. A 250-bp megaprimer was prepared in an initial PCR using the mnp1 gene as a template. The forward primer contained 18 nucleotides (nt) encoding the 6 C-terminal amino acids of GFP and a stop codon (TAA) followed by 13 nt encoding the proximal end of the mnp1 3′ UTR. The 18-nt reverse primer (ncMR1) annealed 250 bp downstream of the mnp1 gene stop codon and included a new EcoRI site at its 5′ end. The megaprimer was purified by agarose gel electrophoresis and extracted using a gel extraction kit (Qiagen). A second PCR mixture contained pEGFPC-3 (CLONTECH) as the template. The forward primer contained the 5′ end of the gfp coding region with a new 5′ KpnI site 4 nt upstream of the translation start codon. The second PCR mixture also contained the megaprimer described above and the reverse primer used in the first PCR step (ncMR1). The final PCR product was a 1.0-kb fragment containing the gfp coding sequence fused to the mnp1 3′ UTR. This PCR fragment was extracted with CHCl3, precipitated with ethanol, digested with both KpnI and EcoRI, and purified by gel electrophoresis. The KpnI-EcoRI (gfp-mnp) fragment and the XbaI-KpnI gpd promoter fragment described above were then ligated into the XbaI- and EcoRI-digested pUB expression vector in a three-way ligation to yield pUGGM3′ (Fig. 1).
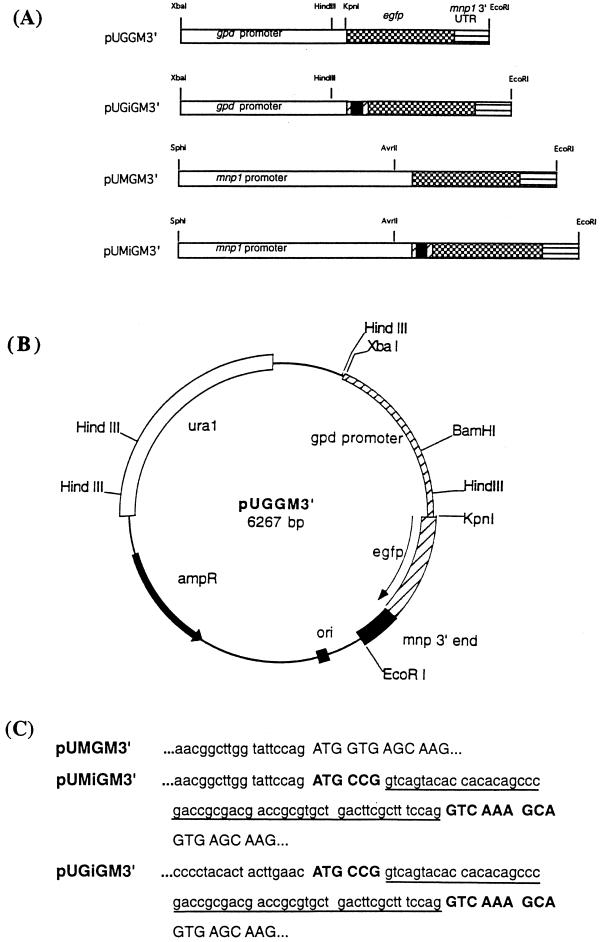
FIG. 1.
(A) Maps of egfp constructs used for transformations. The black boxes represent the first intron from the gpd gene (55 bp). The hatched boxes 3′ and 5′ to the intron represent the coding sequence of the gpd gene. The positions of the gpd promoter (white), the mnp1 promoter (white), the egfp gene (checkered), and the mnp1 3′ UTR (horizontally striped) are indicated. (B) Restriction map of pUGGM3′, containing the gpd promoter, egfp coding region, and mnp 3′ UTR in pUB. The positions of these gene fragments with respect to the ura1 and the ampR genes are indicated and were the same in the other constructs. (C) Sequences surrounding and including the 5′ inserted intron in pUGiGM3′ and pUMGiGM3′. pUMGM3′ is used as a control. The 5′ mnp and the 5′ gpd UTR sequences are in lowercase. The intron sequences are in lowercase and are underlined. The two short gpd coding sequences are in bold uppercase, and the gfp sequences are in uppercase.
pUGiGM3′.
The pUGiGM3′ construct (5′ intron) contained the gpd promoter sequence followed by the fragment iGM3′, which includes, from 5′ to 3′, gpd's exon 1 (6 bp), intron 1 (55 bp), the 5′ end of exon 2 (9 bp), the egfp coding sequence (717 bp), and the mnp1 3′ UTR ending at the EcoRI site. In the first PCR for constructing the megaprimer, the template was pGPD stu1.8, constructed from the gpd gene of P. chrysosporium as described previously (36). The 18-nt forward primer contained a native HindIII restriction site located 170 bp upstream of the translation start codon. The 36-nt reverse primer included an 18-nt nonhybridizing tail encoding the 6 N-terminal amino acids of GFP, excluding the start codon, followed by 9 nt encoding 3 amino acids at the beginning of the second exon of gpd, followed by 9 nt encoding the 3′ end of intron 1 of gpd (23, 36). The 260-bp megaprimer PCR product was amplified and gel purified. The second PCR used pGM3′ as a template. pGM3′ was constructed by subcloning the KpnI-EcoRI fragment from pUGGM3′ into KpnI- and EcoRI-digested pUC18. The reaction also contained the same forward primer used in the first PCR, the ncMR1 reverse primer, and the megaprimer. The 1.3-kb final PCR product was amplified and gel purified as a HindIII-EcoRI fragment. pGPD stu1.8 was doubly digested with XbaI and HindIII. The XbaI-HindIII gpd promoter fragment and the HindIII-EcoRI PCR fragment were ligated to XbaI- and EcoRI-digested pUB in a three-way ligation.
pUMGM3′.
The pUMGM3′ construct is similar to pUGGM3′, except that the gpd promoter is replaced by the mnp1 promoter. The mnp1 promoter and egfp were fused at the start codon using the megaprimer method in a two-step PCR. In the first PCR, we constructed a 158-bp megaprimer containing 140 bp of the 3′ end of the mnp1 promoter followed by 18 bp of the 5′ end of egfp. This megaprimer was used in the second PCR with pUGGM3′ as the template. A 1.1-kb fragment which contains 140 bp of the 3′ end of the mnp1 promoter fused with the entire egfp and mnp1 3′ UTR ending at an EcoRI site was produced. This fragment was cut at a native AvrII site located at the 3′ end of the mnp1 promoter and the EcoRI site. The AvrII-EcoRI fragment was exchanged with the AvrII-EcoRI fragment in pMO (17), yielding pMGM3′. Finally, the whole construct MGM3′, containing 1.5 kb of the mnp1 promoter, the egfp coding region, and the mnp1 3′ UTR, was isolated as an SphI-EcoRI fragment and inserted into pUB to yield pUMGM3′.
pUMiGM3′.
The construct pUGiGM3′ was modified to produce pUMiGM3′ by replacing the gpd promoter with the mnp1 promoter, using the megaprimer method in a two-step PCR. In the first step, a 158-bp megaprimer which includes 140 bp of the 3′ end of the mnp1 promoter followed by 18 bp of the 5′ end of iGM3′ was produced as described above. This megaprimer was used in the second PCR with pUGiGM3′ as the template. The final product was a 1.15-kb fragment containing 140 bp of the 3′ end of the mnp1 promoter fused with the entire iGM3′ sequence (see above). An AvrII-EcoRI digest was performed, and the isolated AvrII-EcoRI fragment was exchanged with the AvrII-EcoRI fragment in pMO (17), yielding pMiGM3′. Finally, the MiGM3′ fragment, containing 1.5 kb of the mnp1 promoter followed by iGM3′, was isolated as an SphI-EcoRI fragment and inserted into pUB to yield pUMiGM3′ (Fig. 1).
Fungal transformations.
Protoplasts (2 × 106) of the P. chrysosporium Ura11 strain (1) and ∼2 μg of each plasmid construct linearized at the unique EcoRI site were used for each transformation as described previously (1, 5, 19), and prototrophic colonies growing in the absence of uracil were selected on plates. Approximately 50 Ura+ transformant colonies obtained for each vector were reexamined for their ability to grow on minimal medium in the absence of uracil and were subsequently purified by fruiting and basidiospore plating as described previously (3, 5).
Culture conditions.
Transformants carrying pUGGM3′ and pUGiGM3′ were grown at 37°C in stationary culture from a conidial inoculum in 250-ml flasks, containing 20 ml of medium containing Kirk's salts, 2% glucose, and 24 mM ammonium tartrate (high carbon, high nitrogen [HCHN]) in 20 mM sodium succinate (pH 4.5) (31, 36). The mycelia from 40-h-old liquid cultures were filtered through Miracloth (Calbiochem), dried between layers of paper towel, frozen in liquid nitrogen, and stored at −80°C. For each transformant, the mycelia from each of three flasks were frozen and processed separately.
Transformants carrying pUMGM3′ and pUMiGM3′ were grown at 37°C from conidial inocula in 20-ml stationary cultures of high-carbon, low-nitrogen (HCLN) (2% glucose, 1.2 mM ammonium tartrate, and Kirk's salt) (31) medium in the presence or absence of 180 μM MnSO4 in 250-ml Erlenmeyer flasks. Cultures were incubated under air for 4 days and then purged with 100% O2 at 48-h intervals. Normally, 7-day-old cultures were harvested as described above, and the extracellular MnP activity and intracellular fluorescence were assayed. For the time course experiment, the cells were harvested on days 4, 5, 6, 7, and 8. For the Mn induction experiment, cultures were grown for 4 days under air in HCLN or HCHN medium without Mn at 37°C. On day 5, 180 μM MnSO4 was added, cultures were purged with O2, and the cells were harvested after additional 12-, 24-, 36-, or 48-h periods.
Intracellular GFP determination.
Mycelia were homogenized with 1 g of acid-washed glass beads in a minibead beater (Biospec) in a buffer containing 10 mM Tris-Cl (pH 8.0), 10 mM EDTA, and 0.002% NaN3. The mixture was centrifuged at 10,000 × g at 4°C. The supernatant was decanted and assayed for protein concentration by the bicinchoninic acid method (Sigma) (44). The crude supernatant was diluted to 1 mg of protein per ml, and a fluorescence spectrum (500 to 600 nm) was determined using a 488-nm excitation wavelength with an SLM Aminco 8000C spectrofluorometer. Maximum fluorescence occurred at ∼509 nm.
RNA extraction and Northern blotting.
Filtered and dried P. chrysosporium mycelia (100 mg) were homogenized in Tri-reagent (Sigma) with 1 g of glass beads as described previously (9). RNA was extracted as described in the protocol supplied with the reagent. The RNA pellet was resuspended in 4 M LiCl and kept at 4°C for 4 h. After microcentrifugation at 10,000 × g, the pellet, containing high-molecular-weight RNA, was washed with ethanol, dissolved in 0.5% sodium dodecyl sulfate, and stored at −80°C. After spectrophotometric quantitation at 260 nm, the RNA (20 μg per lane) was denatured in the presence of 2.2 M formaldehyde for 15 min at 68°C and electrophoresed in a denaturing (0.6 M formaldehyde, 1% agarose) gel. Northern blotting was performed as described previously (9, 33). The coding region of the egfp gene was used as a template for randomly primed synthesis of [α-32P]dCTP probes using a Multiprime DNA labeling kit (Amersham). Probed RNA blots were washed and exposed to XAR-5 X-ray film.
Southern blotting.
DNAs from two transformants for each construct and two control transformants containing only the pUB 1.7 vector were extracted as described previously (3), digested with BamHI, gel electrophoresed, and transferred to nylon membranes. The NheI-HindIII fragment containing the egfp gene was labeled with [α-32P]-dCTP. Southern hybridization and autoradiography were performed as described previously (3).
Fluorescence microscopy.
P. chrysosporium mycelia were grown on microscope coverslips and observed in a Leica model TCS SP microscopic system with appropriate fluorescein isothiocyanate filters (Leica Microsystems, Heidelberg, Germany). Normal phase-contrast images of each sample were used as controls. The digital image was further processed using Photoshop 5.0 (Adobe).
RESULTS
Expression plasmids.
As shown in Fig. 1A, four different expression constructs were made: pUGGM3′, pUGiGM3′, pUMGM3′, and pUMiGM3′. pUGGM3′ contains the gpd gene promoter, followed by the egfp gene and then the mnp1 3′ UTR. pUGiGM3′ contains the gpd promoter, followed by the iGM3′ sequence containing the first intron of the gpd gene just 5′ of egfp, as described above. pUMM3′ is similar to pUGGM3′ except that the mnp gene promoter replaces the gpd gene promoter. pUMiGM3′ contains the mnp1 gene promoter, followed by the iGM3′ sequence as described above. All four constructs were placed in pUB (45), which contains the S. commune ura1 gene as a selectable marker, as illustrated for pUGGM3′ in Fig. 1B. All four constructs were sequenced to confirm that no sequencing errors were introduced during the PCRs. The sequences spanning the intron junction are shown in Fig. 1C.
Transformation and characterization of transformants.
Transformation of the Ura11 mutant strain, with the series of gfp expression vectors containing the constructs shown in Fig. 1, resulted in the isolation of ∼50 prototrophic transformants per 2 μg of DNA of each expression vector. The transformants, containing pUGGM3′ and pUGiGM3′, were grown in stationary culture at 37°C, in 20 ml of HCHN medium (31, 36). After 40 h, the cells were filtered and broken and the fluorescence intensity of the cytosolic fraction was measured. The transformants containing pUMGM3′ and pUMiGM3′ were grown at 37°C in stationary cultures containing 20 ml of HCLN medium in the presence and absence of 180 μM MnSO4 for 7 days. The cells were filtered, broken, and examined for fluorescence intensity in the cytosolic fraction. In addition, the extracellular MnP activity was measured as described previously (51). Under both experimental conditions, the background fluorescence shown in transformants carrying only pUB, the expression vector, was also determined. Only the transformants exhibiting more than three times-the background fluorescence were considered to express gfp. None of the transformants carrying the pUGGM3′ or the pUMGM3′ construct exhibited significant fluorescence under the experimental conditions. In contrast, approximately 50% of the transformants containing constructs with introns (pUGiGM3′ and pUMiGM3′) produced GFP. The other 50% probably contained inserts in a nonexpressible location. Among the transformants carrying pUMGM3′ and pUMiGM3′, ∼80% exhibited extracellular MnP activity.
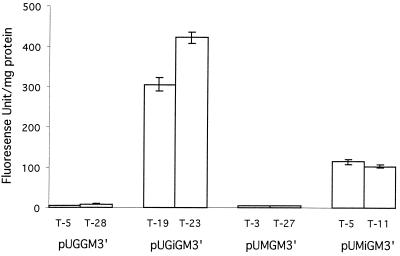
The fluorescence spectra of each transformant exhibited a maximum at 509 nm and a weak shoulder at 545 nm when excited at 488 nm, which is identical to the spectrum of the standard recombinant EGFP protein (data not shown). The two transformants exhibiting the highest fluorescence at 509 nm for each expression vector were used for further analysis. The bar graph (Fig. 2) shows the fluorescence peak at 509 nm for the most fluorescent transformants carrying expression plasmids. The transformants carrying the pUGiGM3′ construct, which contains an intron 5′ of egfp (T-19 and T-23), exhibited high levels of fluorescence, whereas the transformants containing pUGGM3′ (T-5 and T-28) exhibited fluorescence at only background levels. The transformants carrying the construct containing the mnp promoter and the 5′ intron (pUMiGM′3) (T-5 and T-11) also expressed GFP at high levels, whereas the transformants containing pUMGM3′ (T-3 and T-27) expressed GFP at only background levels. This result suggests that the presence of the introduced intron is important for maximal production of the GFP protein.
FIG. 2.
Bar graph showing the fluorescence emission intensity at 509 nm of transformants carrying the various constructs shown in Fig. 1. Transformants exhibiting the highest fluorescence intensity for each construct were grown at 37°C for 40 h as described in the text. Crude extracts from triplicate cultures of the various transformants were prepared, and fluorescence was measured as described in the text. The means and deviations of triplicate values are shown.
Northern blots of transformants of pUGGM3′ and pUGiGM3′.
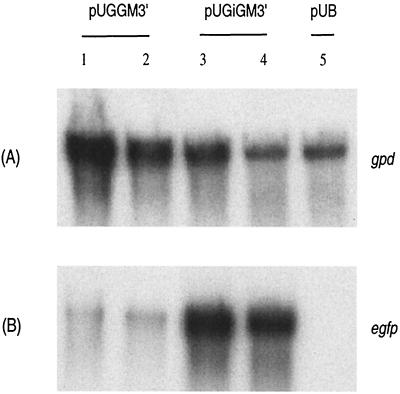
A Northern blot of total RNA from the cultures of the two most fluorescent transformants carrying pUGGM3′ and pUGiGM3′ was probed with the egfp gene (Fig. 3). The weak band from transformants containing pUGGM3′ suggests very weak transcription and/or very low stability of the egfp mRNA. In contrast, a high level of egfp mRNA is seen in the transformants carrying pUGiGM3′, which contains an intron (Fig. 3). The same RNA was also probed with the radiolabeled DNA of the gpd gene, which is constitutively expressed, to determine the quality of the total RNA sample.
FIG. 3.
Northern blot analysis of transformants carrying various constructs. The two transformants exhibiting the highest fluorescence intensity for each construct were grown as described in the text. The preparation of the Northern blot was as described in the text. (A) The blot was probed with the 32P-labeled gpd gene. (B) The blot was probed with the labeled coding sequence of the egfp gene. Lanes 1 and 2, T-5 and T-28 carrying pUGGM3′ with no intron; lanes 3 and 4, T-5 and T-11 carrying pUGiGM3′ with the 5′ intron; lane 5, pUB control vector with no egfp insert.
Southern blotting results confirmed that all of the transformants examined as described above contained the egfp gene, and there were no obvious differences in the egfp gene copy numbers among these transformants containing different egfp constructs (data not shown).
Fluorescent mycelia.
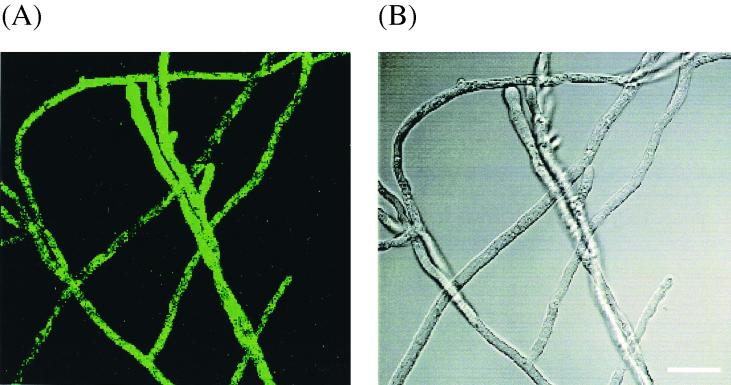
Figure 4A shows a fluorescent micrograph of transformed mycelia carrying the 5′ intron construct pUGiGM3′ (T-23). It is apparent that the green fluorescence of GFP is located within the mycelium. Figure 4B shows a phase-contrast image of the same mycelium. Transformants containing pUGGM3′ did not exhibit green fluorescence under the same conditions (data not shown). Transformants containing pUMiGM3′ also fluoresced, while transformants containing pUMGM3′ did not (data not shown).
FIG. 4.
(A) Fluorescence micrograph of mycelium containing the plasmid pUGiGM3′ (5′ intron). The green color in the mycelium is due to GFP. Control transformants such as pUGGM3′ did not fluoresce significantly (data not shown). (B) Phase-contrast image of the same mycelium. The bar indicates 20 μm.
Effect of Mn on expression of gfp and mnp in culture.
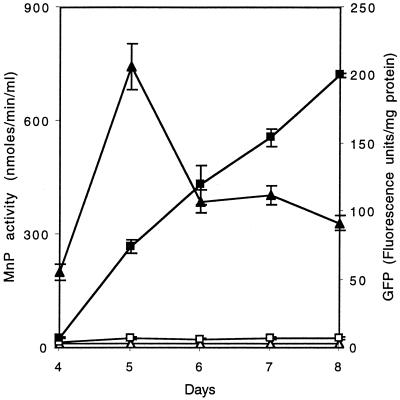
Time courses for the appearance of intracellular GFP and extracellular MnP activity are shown in Fig. 5. Cultures of transformant T-5 carrying pUMiGM3′ grown with no exogenous Mn had no detectable MnP activity through day 8 and had negligible expression of GFP during the same period. In contrast, cultures grown in the presence of 180 μM Mn exhibited both MnP activity and GFP fluorescence, which first appeared on day 4 and were present through day 8. MnP activity peaked on day 5, while GFP fluorescence continued to increase through day 8. Apparently, the GFP mRNA or protein is more stable than the MnP mRNA or protein under these conditions. Cells of transformants carrying the pUMGM3′ construct expressed no detectable fluorescence under any of these conditions (data not shown). These experiments were performed in triplicate, and the deviations are shown in Fig. 5.
FIG. 5.
Effect of Mn supplementation on the production of extracellular MnP and intracellular GFP. Nitrogen-limited cultures of transformant T-5 were grown in the presence of 180 μm Mn or no additional Mn from a conidial inoculum as described in the text. MnP activity from duplicate cultures in the presence of 180 μM (▴) or <1 μM (▵) Mn and GFP fluorescence from duplicate cultures grown in the presence of 180 μM (■) or <1 μM (□) Mn were assayed as described in the text. Experiments were run in triplicate, and the deviations are shown.
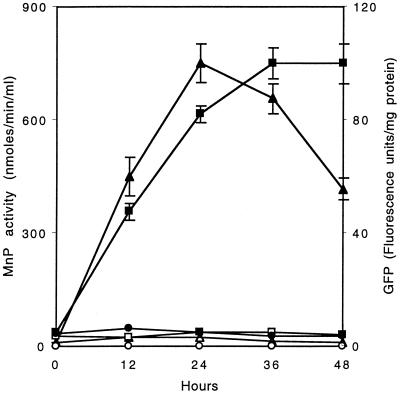
The Mn induction of MnP activity and GFP fluorescence is shown in Fig. 6. In this experiment, the pUMiGM3′ transformant (T-5) was grown for 5 days in HCLN medium in the absence of Mn, after which Mn was added to the experimental cultures. Neither significant MnP activity nor GFP fluorescence was detected in cultures without Mn. After the addition of Mn on day 5, MnP activity was observed and reached a maximum after ∼24 h, slowly declining thereafter. This is similar to our previous results with the wild-type strain (7–9). In parallel with the appearance of MnP activity, GFP fluorescence steadily increased for 36 h after the addition of Mn and leveled off thereafter. Apparently, the GFP protein or mRNA is somewhat more stable than MnP protein or mRNA under these conditions. Transformant T-5 carrying pUMiGM3′ was also grown in HCHN medium and exhibited no MnP activity or GFP fluorescence even after the addition of Mn. All experiments were performed in triplicate, and the deviations are shown in Fig. 6.
FIG. 6.
Induction of extracellular MnP activity and intracellular GFP by Mn. Nitrogen-limited or nitrogen-sufficient, Mn-deficient cultures of transformant T-5 were grown for 5 days, after which 180 μM MnSO4 was added to the experimental flasks. GFP fluorescence in nitrogen-limited cultures induced with Mn (■) or not induced (□) and in nitrogen-sufficient cultures induced with Mn (●) was assayed as described in the text. MnP activity in nitrogen-limited cultures induced with Mn (▴) or not induced (▵) and in nitrogen-sufficient cultures induced with Mn (○) was assayed as described in the text. Experiments were run in triplicate, and the deviations are shown.
Effect of Mn on the expression of GFP and MnP mRNA in cells transformed with pUMiGM3′.
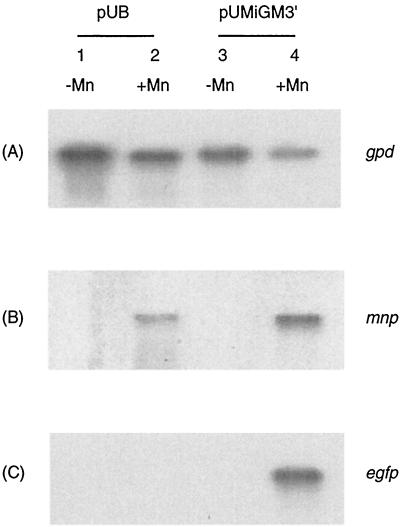
Total RNA from 5-day-old stationary cultures of transformant T-5 carrying pUMiGM3′ in HCLN media with 180 μM MnSO4 or with no Mn was probed with the egfp gene and mnp1 cDNA (Fig. 7). Both the mnp1 and egfp mRNAs were observed only in the cultures with added Mn2+. The same RNAs were also probed with the constitutively expressed gpd gene to show the quality and quantity of the total RNA sample. These data suggest that the expression of both the endogenous mnp- and exogenous gfp-encoding mRNAs of transformants carrying pUMiGM3′ is most likely affected by Mn at the transcriptional level in transformants carrying pUMiGM3′ (8, 15, 17).
FIG. 7.
Effect of Mn supplementation on the expression of MiGM3′ at the RNA level. Nitrogen-limited cultures of T-5 were grown in the presence and absence of Mn. RNA was extracted, separated by electrophoresis, transferred to a membrane, and probed as described in the text. (A) Northern blots probed with the gpd gene; (B) Northern blots probed with the mnp1 gene, showing RNA from endogenous genes; (C) Northern blots probed with the gfp gene. Lanes 1 and 2 contain RNA from transformant T-1 carrying the control plasmid pUB; lanes 3 and 4 contain RNA from transformant T-5 carrying pUMiGM3′; lanes 2 and 4 contain RNA from cells growing in the presence of 180 μM Mn; lanes 1 and 3 contain RNA from cells grown in the absence of exogenous Mn.
DISCUSSION
The activities of LiP and MnP, two major components of the lignin-degrading system, are detectable in extracellular culture media only during the secondary metabolic phase of growth (19, 30, 31, 33). Our earlier studies indicated that MnP activity and mnp RNA are observed only in nitrogen-limited cultures in the presence of Mn(II) (7, 8, 15, 19).
To further examine the regulation of mnp gene expression, a suitable gene reporter system is required. The gfp gene from A. victoria has been used in numerous studies as a reporter of gene expression and protein localization (11, 12) and has been expressed in several fungal systems (11, 26, 34, 46). The commercial A. victoria egfp gene contains no introns. In contrast, all sequenced genes from the basidiomycetes S. commune and P. chrysosporium contain introns (34) (GenBank), suggesting that introns may play an important role in gene expression in these fungi. Recently, it was demonstrated that an intronless hydrophobin gene was very weakly expressed in S. commune; however, high protein expression was observed when a native or synthetic intron was introduced into this intronless hydrophobin gene 3′ of the translation stop codon (34).
In the present study, a native intron from the P. chrysosporium gpd gene, which has characteristics of introns similar to those of other P. chrysosporium genes, was inserted at the 5′ end of the egfp gene; other egfp constructs contained no introns. Unlike in other studies of the effect of introns on gene expression in fungi and other eukaryotic cells (27, 34), we inserted the intron within the coding region of the egfp gene instead of in the 5′ or 3′ UTR. GPD is a constitutively expressed enzyme which plays an important role in glycolysis. The P. chrysosporium gpd gene has important features: its first exon is extremely short (6 bp, including the start codon), and this is followed by a short 55-bp intron. This intron has characteristics of introns in other P. chrysosporium genes. These features enable the insertion of the intron into the coding region of the reporter gene, egfp, with minimal extra amino acids fused to the reporter. When exon 1, intron 1, and 9 bp of exon 2 of gpd are fused to the ATG-less egfp, the resulting protein has only four extra amino acids compared to wild-type GFP. Our results show that egfp with an introduced intron exhibits high expression in P. chrysosporium under either gpd or mnp1 promoter control but that the intronless egfp does not. Steady-state RNA levels are clearly higher in the transformants carrying constructs with a 5′ intron than in those carrying constructs with no introns. Since the absence of an intron apparently does not affect transcriptional initiation of the S. commune hydrophobin genes (34), the difference in RNA levels of intron-containing versus intronless genes probably occurs at the posttranscriptional level, affecting factors such as RNA stability.
Based on an earlier survey (25), introns located 3′ downstream of the stop codon are rare, while roughly one-third of 328 vertebrate genes surveyed contained introns in the 5′ noncoding region. A study using mammalian cells suggested that introns increase the efficiency of RNA 3′ processing and the accumulation of cytoplasmic RNA (27). More recent work using Xenopus oocytes suggests that the location of an inserted intron strongly affects the translation of the cytoplasmic RNA (25, 35). Comparisons of sequences of the mnp, lip, gpd, and quinone reductase genes of P. chrysosporium found in the GenBank database indicate that more introns are found in the 5′ half of these genes than in the 3′ half. These observations suggest that the 5′ introns may have a role in gene expression, RNA processing, or RNA translation.
Fluorescence micrography of P. chrysosporium mycelia transformed with the pUGiGM3′ (5′ intron) construct showed that the GFP produced was intracellular and probably cytoplasmic. Significant green fluorescence was not observed in the extracellular medium or in the conidiospores (data not shown). Control cultures transformed with pUGGM3′ did not exhibit green fluorescence, suggesting that GFP may be a good marker for viewing P. chrysosporium in wood or in a consortium of other fungi. The spotty fluorescence of some mycelial fragments is probably due to the growth conditions on coverslips and/or the age of the particular hypha and was not observed in all micrographs.
Transformation of P. chrysosporium with plasmid DNA results in the ectopic integration of the DNA into the fungal genome (3, 5), where the site of integration is not controlled. It is recognized that, in fungal transformations, the site of integration can affect the expression of the gene. This may be a reason for the different GFP expression levels for the individual transformants carrying pUGiGM3′ or pUMiGM3′. Most importantly, egfp faithfully reports the function of the two promoters used in these experiments. When the gpd promoter is used, GFP production is observed under primary metabolic conditions. When endogenous mnp genes are used as an internal control, all the transformants carrying pUMiGM3′ produce GFP only during secondary metabolic growth triggered by nutrient nitrogen source limitation and only under Mn2+ supplementation. Expression of egfp in transformants with pUMiGM3′ parallels that of endogenous mnp. Northern blots further indicate that gfp mRNA in these transformants is considerably reduced when cultures are grown in the absence of Mn2+ compared with the levels in cultures grown in the presence of Mn2+, confirming that the Mn effect is probably at the level of gene transcription (8, 15, 17).
The role of Mn in regulating the expression of a reporter gene can best be examined in an induction experiment, where the effect of Mn is independent of other variables such as nutrient nitrogen or O2 levels in the flasks. Our results show that the addition of Mn to cultures grown in the absence of Mn for 5 days leads to the simultaneous appearance of both GFP and endogenous MnP. In addition, adding Mn to cells grown in HCHN medium does not lead to the appearance of either MnP activity of GFP fluorescence.
In conclusion, we have prepared four different egfp constructs to determine whether GFP may be used as a reporter of promoter function in P. chrysosporium. We demonstrate that, if an intron is placed at the 5′ end of the egfp gene, egfp is an effective reporter of either gpd or mnp promoter function, yielding high levels of regulated protein expression, but that the absence of an intron in the egfp gene results in very reduced levels of expression. This result suggests that the presence or absence of an intron may be an important determinant of RNA stability and/or RNA processing in this system. We also conclude that 1,500 bp of the mnp promoter sequence regulates egfp reporter expression in a manner similar to the manner of regulation of endogenous mnp genes by Mn, nutrient nitrogen levels, and metabolic phase of growth. We plan to use the 5′ intron-containing egfp construct to examine the effects of cis-acting sequences on the regulation of mnp by Mn and other factors.
ACKNOWLEDGMENT
This research was supported by grant MCB-9723725 from the National Science Foundation.
REFERENCES
- 1.Akileswaran L, Alic M, Clark E K, Hornick J L, Gold M H. Isolation and transformation of uracil auxotrophs of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Curr Genet. 1993;23:351–356. doi: 10.1007/BF00310898. [DOI] [PubMed] [Google Scholar]
- 2.Alic M, Akileswaran L, Gold M H. Characterization of the gene encoding manganese peroxidase isozyme 3 from Phanerochaete chrysosporium. Biochim Biophys Acta. 1997;1338:1–7. doi: 10.1016/s0167-4838(96)00235-x. [DOI] [PubMed] [Google Scholar]
- 3.Alic M, Kornegay J R, Pribnow D, Gold M H. Transformation by complementation of an adenine auxotroph of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1989;55:406–411. doi: 10.1128/aem.55.2.406-411.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alic M, Letzring C, Gold M H. Mating system and basidiospore formation in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1987;53:1464–1469. doi: 10.1128/aem.53.7.1464-1469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alic M, Mayfield M B, Akileswaran L, Gold M H. Homologous transformation of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Curr Genet. 1991;19:491–494. doi: 10.1007/BF00310898. [DOI] [PubMed] [Google Scholar]
- 6.Bao W, Fukushima Y, Jensen K A, Jr, Moen M A, Hammel K E. Oxidative degradation of non-phenolic lignin during lipid peroxidation by fungal manganese peroxidase. FEBS Lett. 1994;354:297–300. doi: 10.1016/0014-5793(94)01146-x. [DOI] [PubMed] [Google Scholar]
- 7.Brown J A, Alic M, Gold M H. Manganese peroxidase gene transcription in Phanerochaete chrysosporium: activation by manganese. J Bacteriol. 1991;173:4101–4106. doi: 10.1128/jb.173.13.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J A, Glenn J K, Gold M H. Manganese regulates expression of manganese peroxidase by Phanerochaete chrysosporium. J Bacteriol. 1990;172:3125–3130. doi: 10.1128/jb.172.6.3125-3130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown J A, Li D, Alic M, Gold M H. Heat shock induction of manganese peroxidase gene transcription in Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:4295–4299. doi: 10.1128/aem.59.12.4295-4299.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bumpus J A, Aust S D. Biodegradation of environmental pollutants by the white rot fungus Phanerochaete chrysosporium: involvement of the lignin-degrading system. Bioessays. 1987;6:166–170. [Google Scholar]
- 11.Cormack B. Green fluorescent protein as a reporter of transcription and protein localization in fungi. Curr Opin Microbiol. 1998;1:406–410. doi: 10.1016/s1369-5274(98)80057-x. [DOI] [PubMed] [Google Scholar]
- 12.Cubitt A B, Heim R H, Adams S R, Boyd A E, Gross L A, Tsien R Y. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 13.Cullen D. Recent advances on the molecular genetics of lignolytic fungi. J Biotechnol. 1997;53:273–289. doi: 10.1016/s0168-1656(97)01684-2. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson K- E L, Blanchette R A, Ander P. Microbial and enzymatic degradation of wood and wood components. Berlin, Germany: Springer-Verlag; 1990. [Google Scholar]
- 15.Gettemy J M, Ma B, Alic R, Gold M H. Reverse transcription PCR analysis of the regulation of the manganese peroxidase gene family. Appl Environ Microbiol. 1998;64:569–574. doi: 10.1128/aem.64.2.569-574.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glenn J K, Gold M H. Purification and characterization of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete, Phanerochaete chrysosporium. Arch Biochem Biophys. 1985;242:329–341. doi: 10.1016/0003-9861(85)90217-6. [DOI] [PubMed] [Google Scholar]
- 17.Godfrey B J, Akileswaran L, Gold M H. A reporter gene construct for studying the regulation of manganese peroxidase gene expression. Appl Environ Microbiol. 1994;60:1353–1358. doi: 10.1128/aem.60.4.1353-1358.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godfrey B J, Mayfield M B, Brown J A, Gold M H. Characterization of a gene encoding a manganese peroxidase from Phanerochaete chrysosporium. Gene. 1990;93:119–124. doi: 10.1016/0378-1119(90)90144-g. [DOI] [PubMed] [Google Scholar]
- 19.Gold M H, Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev. 1993;57:605–622. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gold M H, Wariishi H, Valli K. Extracellular peroxidases involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium. ACS Symp Ser. 1989;389:127–140. [Google Scholar]
- 21.Gold M H, Youngs H L, Sollewijn Gelpke M D. Manganese peroxidases. Met Ions Biol Syst. 1999;37:558–586. [PubMed] [Google Scholar]
- 22.Hammel K E. Oxidation of aromatic pollutants by lignin-degrading fungi and their extracellular peroxidases. Met Ions Biol syst. 1992;28:41–60. [Google Scholar]
- 23.Harmsen M C, Schuren F H J, Moukha S M, van Zuilen C M, Punt P J, Wessels J G H. Sequence analysis of the glyceraldehyde-3-phosphate dehydrogenase genes from the basidiomycetes Schizophyllum commune, Phanerochaete chrysosporium and Agaricus bisporus. Curr Genet. 1992;22:447–454. doi: 10.1007/BF00326409. [DOI] [PubMed] [Google Scholar]
- 24.Harvey P J, Schoemaker H E, Bowen R M, Palmer J M. Single-electron transfer processes and the reaction mechanism of enzymic degradation of lignin. FEBS Lett. 1985;183:13–16. [Google Scholar]
- 25.Hawkins J D. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9905. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henriksen A L S, Even S, Muller C, Punt P J, van den Hondel C A M J J, Nielsen J. Study of the glucoamylase promoter in Aspergillus niger using green fluorescent protein. Microbiology. 1999;145:729–734. doi: 10.1099/13500872-145-3-729. [DOI] [PubMed] [Google Scholar]
- 27.Huang M T F, Gorman C M. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kersten P J, Kirk T K. Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J Bacteriol. 1987;169:2195–2205. doi: 10.1128/jb.169.5.2195-2201.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kersten P J, Tien M, Kalyanaraman B, Kirk T K. The ligninase of Phanerochaete chrysosporium generates cation radicals from methoxybenzenes. J Biol Chem. 1985;260:2609–2612. [PubMed] [Google Scholar]
- 30.Kirk T K, Farrell R L. Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 31.Kirk T K, Schultz E, Connors W J, Lorenz L F, Zeikus J G. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch Microbiol. 1978;177:277–285. [Google Scholar]
- 32.Li D, Alic M, Brown J A, Gold M H. Regulation of manganese peroxidase gene transcription by hydrogen peroxide, chemical stress, and molecular oxygen. Appl Environ Microbiol. 1995;61:341–345. doi: 10.1128/aem.61.1.341-345.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D, Alic M, Gold M H. Nitrogen regulation of lignin peroxidase gene transcription. Appl Environ Microbiol. 1994;60:3447–3449. doi: 10.1128/aem.60.9.3447-3449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lugones L G, Scholtmeijer K, Klootwijk R, Wessels J G H. Introns are necessary for mRNA accumulation in Schizophyllum commune. Mol Microbiol. 1999;32:681–689. doi: 10.1046/j.1365-2958.1999.01373.x. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto K, Wassarman K M, Wolffe A P. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayfield M B, Kishi K, Alic M, Gold M H. Homologous expression of recombinant manganese peroxidase in Phanerochaete chrysosporium. Appl Environ Microbiol. 1994;60:4303–4309. doi: 10.1128/aem.60.12.4303-4309.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naylor L. Reporter gene technology: the future looks bright. Biochem Pharmacol. 1999;58:749–757. doi: 10.1016/s0006-2952(99)00096-9. [DOI] [PubMed] [Google Scholar]
- 38.Pease E A, Andrawis A, Tien M. Manganese-dependent peroxidase from Phanerochaete chrysosporium: primary structure deduced from cDNA sequence. J Biol Chem. 1989;264:13531–13535. [PubMed] [Google Scholar]
- 39.Prasher D C, Eckenrode V K, Ward W W, Prendergast F G, Cormier M J. Primary structure of the Aequora victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 40.Pribnow D, Mayfield M B, Nipper V J, Brown J A, Gold M H. Characterization of a cDNA encoding a manganese peroxidase, from the lignin-degrading basidiomycete Phanerochaete chrysosporium. J Biol Chem. 1989;264:5036–5040. [PubMed] [Google Scholar]
- 41.Raeder U, Broda P. Comparison of the lignin-degrading white rot fungi Phanerochaete chrysosporium and Sporotrichum pulverulentum at the DNA level. Curr Genet. 1984;8:499–506. doi: 10.1007/BF00410436. [DOI] [PubMed] [Google Scholar]
- 42.Reddy G V B, Gold M H. A two-component tetrachlorohydroquinone reductive dehalogenase system from the lignin-degrading basidiomycete Phanerochaete chrysosporium. Biochem Biophys Res Commun. 1999;257:901–905. doi: 10.1006/bbrc.1999.0561. [DOI] [PubMed] [Google Scholar]
- 43.Reddy G V B, Sollewijn Gelpke M D, Gold M H. Degradation of 2,4,6-trichlorophenol by Phanerochaete chrysosporium: involvement of reductive dechlorination. J Bacteriol. 1998;180:5159–5164. doi: 10.1128/jb.180.19.5159-5164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith P K, Krohn R I, Hermanson G T. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 45.Sollewijn Gelpke M D, Mayfield-Gambill M, Cereghino G P L, Gold M H. Homologous expression of recombinant lignin peroxidase in Phanerochaete chrysosporium. Appl Environ Microbiol. 1999;65:1670–1674. doi: 10.1128/aem.65.4.1670-1674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spellig T, Bottin A, Kahmann R. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol Gen Genet. 1996;252:503–509. doi: 10.1007/BF02172396. [DOI] [PubMed] [Google Scholar]
- 47.Sundaramoorthy M, Kishi K, Gold M H, Poulos T L. The crystal structure of manganese peroxidase from Phanerochaete chrysosporium at 2.06-Å resolution. J Biol Chem. 1994;269:32759–32767. [PubMed] [Google Scholar]
- 48.Sundaramoorthy M, Kishi K, Gold M H, Poulos T L. Crystal structures of substrate binding site mutants of manganese peroxidase. J Biol Chem. 1997;272:17574–17580. doi: 10.1074/jbc.272.28.17574. [DOI] [PubMed] [Google Scholar]
- 49.Tien M, Tu C-P D. Cloning and sequencing of a cDNA for a ligninase from Phanerochaete chrysosporium. Nature. 1987;326:520–523. doi: 10.1038/326520a0. [DOI] [PubMed] [Google Scholar]
- 50.Wariishi H, Valli K, Renganathan V, Gold M H. Thiol-mediated oxidation of nonphenolic lignin model compounds by manganese peroxidase of Phanerochaete chrysosporium. J Biol Chem. 1989;264:14185–14191. [PubMed] [Google Scholar]
- 51.Wariishi H, Valli K, Gold M H. Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J Biol Chem. 1992;267:23688–23695. [PubMed] [Google Scholar]