Abstract
Real-time PCR was used to quantify populations of ammonia-oxidizing bacteria representing the β subdivision of the class Proteobacteria in samples of arable soil, both nitrogen fertilized and unfertilized, from Mellby, Sweden. Primers and probes targeting a 16S ribosomal DNA region of the ammonia-oxidizing bacteria were designed and used. In the fertilized soil there were ∼6.2 × 107 ammonia-oxidizing bacteria per g of soil, three times more than the number of bacteria in the unfertilized soil. The lytic efficiency of bead beating in these soils was investigated by using populations of free or loosely attached bacteria, bacteria tightly bound to particles, and bacteria in nonfractionated samples. The shapes of the curves generated in these tests showed that the concentration of template DNA released at various times remained constant after 10 to 100 s of bead beating.
Quantification of microbial populations is important in many aspects of microbial ecology. The development of molecular biological methods involving PCR has led to new techniques that are not limited by the culturability of the microorganisms. Since it is known that only a small proportion of the bacteria in soils can be cultivated under standard laboratory conditions, the PCR-based quantification methods have found many applications. Three PCR-based methods, limiting-dilution PCR (27, 31), kinetic PCR (3, 14), and competitive PCR (10, 11, 23), have been used for quantitative analysis of DNA having different origins. However, kinetic PCR and limiting-dilution PCR often have the disadvantage of relying on endpoint measurements of the amount of DNA produced, which makes it difficult to deduce the initial concentration of template DNA. In competitive PCR it may be difficult to achieve the same affinity of the primers for the target and competitive molecules, which complicates quantification. A modified PCR technique, real-time PCR (13), measures the DNA concentration continuously during amplification, which enables the initial template concentration to be determined and the cell numbers to be more accurately deduced without the use of a competing molecule.
Quantification of ammonia-oxidizing bacteria (AOB), which are responsible for the oxidation of ammonia to nitrite in the nitrification process, has been attempted by using several different methods. These include the most-probable-number technique (6, 7, 19), in situ hybridization (29), a competitive enzyme-linked immunosorbent assay using monoclonal antibodies (28), and competitive PCR (17, 26, 30) based on traditional methods of amplification. However, all of these methods have significant disadvantages. Thus, a reliable and reproducible method for quantifying AOB would be valuable for evaluating correlations between microbial activities and cell numbers, the effects of different treatments on cell density, and population changes in time and space. Determination of DNA concentrations with real-time PCR overcomes some of the problems associated with traditional PCR. The real-time PCR technique is based on continuously monitoring fluorescence throughout the reaction. This is made possible by adding a dually labeled fluorescent probe that hybridizes to the template in each cycle. The fluorescent emission from one of the dyes, the reporter, is quenched by the emission from the other dye. Cleavage of the probe, mediated by the 5′-to-3′ nuclease activity of the polymerase which acts only on template-annealed probes, increases the emission from the reporter dye. Quantification of DNA by real-time PCR is based on measurements obtained during the early exponential phase, when amplification of the PCR product is first detected and the amount of the amplified product is proportional to the concentration of the template DNA (13).
In the present study we used real-time quantitative PCR to evaluate the efficiency of lysing AOB by bead beating as the initial step for extracting the DNA of these organisms from arable soil samples. The numbers of AOB were studied in two bacterial fractions and compared to the numbers in nonfractionated soil samples in order to investigate the influence of adherence to soil particles on lysability and cell density. Furthermore, the numbers of cells in both nitrogen-fertilized soil and unfertilized soil were studied in order to investigate the effect of fertilization on the density of AOB in the soil. The soils which we chose were sandy loams with good oxygen diffusion, which was assumed to promote nitrifying activity. Thus, both of the soils investigated should have been favorable environments for nitrification, but they were likely to differ in the availability of substrates for AOB.
The fertilized and unfertilized soil samples were collected in August 1999 in Mellby, Sweden (Table 1). Multiple (15 to 20) samples were randomly collected from each plot (40 by 40 m) at depths of 0 to 30 cm and subsequently pooled to give composite samples and thoroughly mixed by sieving (grid size, 6.3 mm). Two 10-ml mixed soil samples (0.3 g [dry weight] per ml of TE buffer [10 mM Tris-HCl, 1 mM EDTA; pH 7.5]) were kept on ice while they were homogenized with a hand-held blender (DIAX 900; Homogenizer tool G6; Heidolph, Kelheim, Germany) for 5 min. Density gradient centrifugation was performed by the method described by Bakken and Lindahl (4). Twelve milliliters of Nycodenz (0.8 g ml of TE buffer−1; Nycomed Pharma A/S, Oslo, Norway) was placed under 20 ml of each homogenized soil suspension, and the samples were subsequently centrifuged at 2,000 × g (swing-out rotor) for 2 h. Two fractions were collected: the visible band, just above the Nycodenz, containing the free or loosely attached (FLA) bacterial fraction and the pellet containing the tightly bound (TB) bacterial fraction. After the two separated fractions were washed twice in TE buffer, subsamples of the FLA fraction, the TB fraction, and nonfractionated samples were shaken in a mini bead beater (BioSpec Products, Inc., Bartlesville, Okla.) at 5,000 rpm for various times. DNA was extracted with a FastDNA SPIN kit for soil (Bio 101, Inc., La Jolla, Calif.); 1 g of 0.1-mm-diameter glass beads (BioSpec Products) was used per sample. No MT buffer (supplied with the FastDNA SPIN kit) was added to the samples that were not bead beaten (the time-zero samples). To optimize the concentrations of AOB-specific primers and probes and to construct standard curves, DNA extractions were performed (QIAamp tissue kit; QIAGEN GmbH, Hilden, Germany) with four pure cultures of AOB grown in liquid medium (8, 21).
TABLE 1.
Characteristics of the soil samples (depth, 0 to 30 cm) collected from Mellby, Halland, southwest Sweden
| Soil | pH(KCl) | Moisture content (%) | Organic matter content (%) | NH4+-N content (kg ha−1) | NO3−-N content (kg ha−1) | Amt of manure applied (kg of N yr−1 ha−1)a | Amt of commercial fertilizer applied (kg of N yr−1 ha−1)b |
|---|---|---|---|---|---|---|---|
| Unfertilized | 5.6 | 13 | 5.7 | 4.5 | 2.3 | 0 | 0 |
| Fertilized | 5.7 | 15 | 7.1 | 16.4 | 38.7 | 180 | 45 |
73% NH4+-N.
50% NH4+-N plus 50% NO3−-N.
For quantification of AOB by real-time PCR, amplification was performed in 25-μl reaction mixtures by using buffers supplied with a TaqMan Universal PCR Master Mix kit (PE Applied Biosystems, Foster City, Calif.) in MicroAmp Optical 96-well reaction plates with optical caps (PE Applied Biosystems). The template DNA in the reaction mixtures was amplified and monitored with an ABI Prism SDS 7700 instrument (PE Applied Biosystems). The primers, probes, and concentrations used are listed in Table 2. Primers CTO 189fA/B and CTO 189fC were used at a 2:1 ratio. Linearized plasmid pUC18 (24), together with RT2f, RT2r, and TMP2 (Table 2), was used as an internal control (0.7 pg of pUC18 DNA per reaction mixture) in each well containing an environmental sample to correct for possible interference from contaminating substances (e.g., humic substances) in the DNA samples extracted from the soil. The conditions used for the amplifying reactions were as follows: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The lowest dilution of the DNA extracts from the TB fractions and the nonfractionated samples that gave real-time PCR products of both AOB DNA and internal standard DNA was 1:1,000. No dilution of the DNA extracts from the FLA bacterial fractions was necessary.
TABLE 2.
Primers and probes used in real-time PCR targeted against 16S ribosomal DNA of β-subgroup AOB and the pUC18 plasmid
| Primer or probe | Nucleotide sequence (5′-3′) | Sequence position | Initial concn in PCR mixture (nM) | Target | Reference |
|---|---|---|---|---|---|
| Primers | |||||
| CTO 189fA/B | GGAGRAAAGCAGGGGATCG | 189-207b | 300 | AOB | 18 |
| CTO 189fC | GGAGGAAAGTAGGGGATCG | 189-207b | 300 | AOB | 18 |
| RT1r | CGTCCTCTCAGACCARCTACTG | 283-304b | 300 | AOB | This studyd |
| RT2f | CTCCCGGCATCCGCTA | 597-622c | 40 | pUC18 | This studyd |
| RT2r | CGCGCGTTTCGGTGAT | 668-683c | 40 | pUC18 | This studyd |
| R-Q probesa | |||||
| TMP1 (5′-FAM and 3′-TAMRA) | CAACTAGCTAATCAGRCATCRGCCGCTC | 226-253b | 125 | AOB | This studyd |
| TMP2 (5′-CR6G and 3′- TAMRA) | ACGGTGAAAACCTCTGACACATGCAGCT | 639-666c | 125 | pUC18 | This studyd |
The AOB-specific PCR primers were selected to amplify a 116-bp DNA fragment in the V2 region (12) of the 16S ribosomal DNA. The concentrations of the primers and probes (Table 2) were empirically optimized in order to minimize the Ct values (i.e., the threshold cycles in which exponential amplification of PCR products was first detected). The Ct values were determined on the basis of the mean baseline signals during the early cycles of amplification. In this way the two amplifications (i.e., DNA amplification of the AOB and DNA amplification of the internal standard) were both optimized to achieve an amplification efficiency close to one (i.e., doubling of the amplification product in each cycle). The amplification efficiencies were determined by serial dilution of AOB DNA extracted from the pure cultures and pUC18 DNA, followed by real-time PCR.
The use of a standard curve based on known concentrations of DNA makes it theoretically possible to quantify DNA from any source. In this case, standard curves constructed after real-time PCR amplification of eight different DNA concentrations ranging from 0.6 pg to 10 ng of DNA/well were used. DNA extracted from Nitrosomonas europaea NCIMB 11850, Nitrosospira sp. strain B6 (15), Nitrosospira sp. strain 40 KI (15), and Nitrosospira multiformis NCIMB 11849, representing different clusters of AOB (18), generated slopes of −3.56, −3.59, −3.55, and −3.63 ΔCt ng of DNA−1, respectively. The R2 values were greater than 0.99 for all of the curves. The similarity of the slopes confirms that primers CTO 189fA/B, CTO 189fC, and RT1r and probe TMP1 are well suited for amplification of different groups of AOB with real-time PCR. Furthermore, the similarity indicates that the 16S rRNA gene is an appropriate target molecule for this application. Quantification of AOB DNA extracted from the two types of soil (the nonfractionated samples and the FLA and TB bacterial fractions), assumed to contain a mixed flora of AOB, was based on a mean slope value (−3.58 ± 0.04 ΔCt ng of DNA−1) derived from the four standard curves.
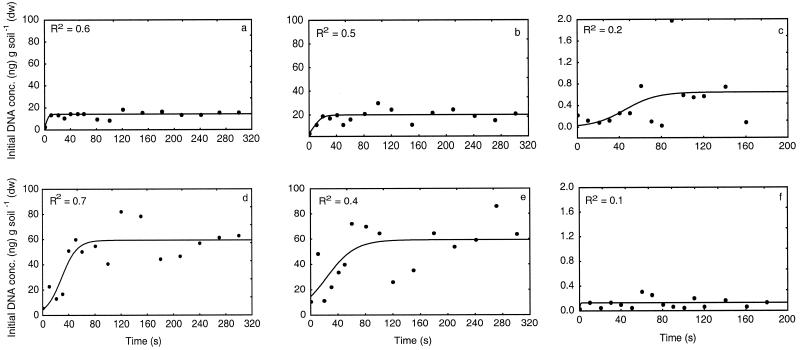
The effect of the duration of bead beating on the efficiency of AOB DNA extraction was studied by varying the bead-beating times between 0 and 300 s and measuring the DNA released. The majority of the DNA extracted from soil samples subjected to beat beating for 300 s was shown to be between 1,000 and 5,000 bp long (results not shown). Therefore, fragmentation of the DNA due to excessive bead beating was unlikely to bias the real-time PCR results, since the length of the amplicon is only 116 bp. Since we expected the curve estimated from the initial amount of DNA released during the bead-beating time course to be similar to that of a microbial growthlike function, we used a nonlinear regression, y = aa/[1 + bb × exp(cc × x)] (where a, b, and c are constants estimated for each function), and the least-squares method for estimation (Fig. 1). The software STATISTICA (version 5.5; StatSoft Inc., Tulsa, Okla.) was used for the analysis. Since soil contains numerous microniches, the scattering of the data can be explained by the heterogeneity in the samples. Furthermore, the soil samples used in this study were composite samples, which added to the heterogeneity.
FIG. 1.
Initial concentrations of DNA from AOB as determined by real-time PCR with DNA extracted from unfertilized soil (a to c) and fertilized soil (d to f) during bead beating. DNA concentrations were determined for nonfractionated samples (a and d), the TB fraction (b and e), and the FLA fraction (c and f). The curves were estimated by nonlinear regression, y = aa/[1 + bb × exp (cc × x)], where a, b, and c are constants estimated for each function. Each value plotted is based on three real-time PCR amplifications. dw, dry weight.
After the results were assessed, the bead-beating duration was standardized to 100 s in further analyses. The shapes of the DNA release time curves were similar for the nonfractionated samples and the TB bacterial fractions from both soil types (Fig. 1). A plateau was reached between 10 and 100 s, which indicated the times at which the majority of the lysable cells had been disrupted. The cells in the FLA bacterial fractions were probably lysed instantly, explaining the low R2 values for these curves (Fig. 1c and f). If the extremely high value in Fig. 1c is regarded as an outlier, the curves in Fig. 1c and f have similar shapes. For the unfertilized soil, bead beating for 30 s was sufficient for optimal lysis of the AOB in the nonfractionated sample and the bacterial fraction tightly bound to soil particles. Longer bead beating was required to reach the plateau phase for the nonfractionated sample and the TB fraction derived from the fertilized soil than for corresponding fractions from the unfertilized soil. This may have been because there were differences in the AOB populations in the two soils and/or because the cell density was greater in the fertilized soil. However, since the DNA released in these experiments was not impaired by longer-than-necessary bead beating, the same duration (100 s, the time required to ensure that nearly all lysable cells had released their DNA) was used for all samples in further analyses. Furthermore, none of the curves started from zero, indicating that there were small amounts of free AOB DNA in the samples even before the bead beating. This could have been due to lysis during storage of the soils, homogenization, and density gradient centrifugation of the samples and/or to spontaneous lysis.
The estimates of bacterial cell numbers were based on the following assumptions: that the genome size is 3 Mb (32); that there are approximately 1.3 genomes/cell; and that all AOB have only one rrn operon per genome, as shown for all the strains studied so far (2). An analysis of variance (STATISTICA) was performed with the estimated cell numbers. Quantification of the AOB in the unfertilized soil gave a significantly different population size for the FLA fraction (1.2 × 106 ± 1.7 × 105 cells g [dry weight] of soil−1; mean ± standard error) than for the TB fraction (1.6 × 107 ± 1.4 × 106 cells g [dry weight] of soil−1) and the nonfractionated sample (1.8 × 107 ± 2.0 × 106 cells g [dry weight] of soil−1). In the fertilized soil the AOB population sizes were significantly different for the FLA fraction (2.7 × 106 ± 1.8 × 106 cells g [dry weight] of soil−1), the TB fraction (4.5 × 107 ± 3.0 × 106 cells g [dry weight] of soil−1), and the nonfractionated sample (6.2 × 107 ± 5.7 × 106 cells g [dry weight] of soil−1). The amounts of AOB in the two soils (nonfractionated samples) suggested there were two to three times as many bacteria in the fertilized soil as in the unfertilized soil, a difference also valid for the bacterial fractions measured separately. These results corroborate earlier findings (6) suggesting that increasing the amount of substrate by nitrogen fertilization increases the number of AOB in the soil. The lower percentage of AOB in the TB fraction of the fertilized soil than in the unfertilized soil probably reflects the fact that newly grown AOB in substrate-rich environments are less strongly attached to particles (1).
The numbers of AOB found in the two soils in this study are 1 or 2 orders of magnitude higher than the numbers found in most previous studies of arable soils (6, 20, 23). Comparisons should be treated with caution since the earlier studies involved soils from different sites with variable characteristics and different methods were used. However, Mendum et al. (23) performed competitive PCR by using sequences of both the amoA gene and the 16S rRNA gene, and the numbers of 16S rRNA gene copies in fertilized soil that they found are on the same order of magnitude as the numbers which we found in our study. AOB have been shown to possess only one rRNA gene copy per genome (2), which makes using the 16S rRNA gene more valid for quantification of AOB than for quantification of most other organisms, which often have variable numbers of rrn operons per genome, which biases quantification (9). The amoA gene has been found only in organisms capable of ammonia oxidation, but more than one copy may be present in each genome and the gene copy numbers in different AOB species may be different (16, 22, 25). If a selective set of primers directed against the amoA gene is used, the amplification reaction is very specific, but quantification is uncertain since the exact number of gene copies per genome is unknown.
To our knowledge, this study demonstrates for the first time the potential of the real-time PCR technique for quantifying AOB in soil. The assay is a fast and reliable method, and we assume that it has great potential for quantification of bacteria in other types of soil. The number of AOB in certain environments is relatively low (∼104 to 106 cells g of soil−1), making real-time PCR, which is highly sensitive, a convenient method for enumeration of these organisms. Furthermore, the rapidity of real-time PCR and its capacity to handle high sample throughput make it less time-consuming and more convenient than other quantitative PCR methods. Real-time PCR can also be used to determine a broad range of starting molecule template concentrations spanning at least 5 orders of magnitude (13). Thus, it is also suitable for quantifying soil bacteria with population dynamics radically different than those of AOB.
Acknowledgments
The four isolates of AOB were a generous gift from Å. Aakra, Agricultural University of Norway, Ås. The soil was kindly provided by Å. Kasimir-Klemedtsson, IVL, Gothenburg, Sweden. We are grateful to P. Milberg, Linköping University, Linköping, Sweden, for help with the statistical analyses. We also thank H.-J. Monstein, University Hospital, Linköping, Sweden, and C. Tebbe, Federal Agricultural Research Centre, Braunschweig, Germany, for valuable comments on the manuscript.
This study was supported by grants from the Swedish Council for Forestry and Agricultural Research (contracts 23.0215/94 and 711.0790/96).
REFERENCES
- 1.Aakra Å, Hesselsøe M, Bakken L R. Surface attachment of ammonia-oxidising bacteria in soil. Microb Ecol. 2000;39:222–235. doi: 10.1007/s002480000006. [DOI] [PubMed] [Google Scholar]
- 2.Aakra Å, Utåker J B, Nes I F. RFLP of rRNA genes and sequencing of the 16S–23S rDNA intergenic spacer region of ammonia-oxidizing bacteria: a phylogenetic approach. Int J Syst Bacteriol. 1999;49:123–130. doi: 10.1099/00207713-49-1-123. [DOI] [PubMed] [Google Scholar]
- 3.Alard P, Lantz O, Sebagh M, Calvo C F, Weill D, Chavanel G, Senik A, Charpentier B. A versatile ELISA-PCR assay for mRNA quantitation from a few cells. BioTechniques. 1993;15:730–737. [PubMed] [Google Scholar]
- 4.Bakken L R, Lindahl V. Recovery of bacterial cells from soil. In: Trevors J T, van Elsas J D, editors. Nucleic acids in the environment. Methods and applications. Berlin, Germany: Springer-Verlag; 1995. pp. 9–27. [Google Scholar]
- 5.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Bruns M A, Stephen J R, Kowalchuck G A, Prosser J I, Paul E A. Comparative diversity of ammonia oxidizer 16S rRNA gene sequences in native, tilled, and successional soils. Appl Environ Microbiol. 1999;65:2994–3000. doi: 10.1128/aem.65.7.2994-3000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnol M, Ineson P. Environmental factors controlling NO3− leaching, N2O emissions and numbers of NH4+ oxidisers in a coniferous forest soil. Soil Biol Biochem. 1999;31:979–990. [Google Scholar]
- 8.Donaldson J M, Henderson G S. A dilute medium to determine population size of ammonium oxidizers in soil. Soil Sci Soc Am J. 1989;53:1608–1611. [Google Scholar]
- 9.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felske A, Akkermans A D L, De Vos W M. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl Environ Microbiol. 1998;64:4581–4587. doi: 10.1128/aem.64.11.4581-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilliland G, Perrin S, Blanchard K, Bunn H F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray M W, Sankoff D, Cedergren R J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984;12:5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 14.Higuchi R, Dollinger G, Walsh P S, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Bio/Technology. 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Q Q. Ph.D. thesis. Ås, Norway: Agricultural University of Norway; 1996. [Google Scholar]
- 16.Klotz M G, Norton J M. Multiple copies of ammonia monooxygenase (amo) operons have evolved under biased AT/GC mutational pressure in ammonia-oxidizing autotrophic bacteria. FEMS Microbiol Lett. 1998;168:303–311. doi: 10.1111/j.1574-6968.1998.tb13288.x. [DOI] [PubMed] [Google Scholar]
- 17.Kowalchuk G A, Naoumenko Z S, Derikx P J L, Felske A, Stephen J R, Arkhipchenko I A. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl Environ Microbiol. 1999;65:396–403. doi: 10.1128/aem.65.2.396-403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowalchuk G A, Stephen J R, De Boer W, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kowalchuk G A, Stienstra A W, Heilig G H J, Stephen J R, Woldendorp J W. Molecular analysis of ammonia-oxidising bacteria in soil of successional grasslands of the Drentsche A (The Netherlands) FEMS Microbiol Ecol. 2000;31:207–215. doi: 10.1111/j.1574-6941.2000.tb00685.x. [DOI] [PubMed] [Google Scholar]
- 20.Laanbroek H J, Gerards S. Effects of organic manure on nitrification in arable soils. Biol Fertil Soils. 1991;12:147–153. [Google Scholar]
- 21.MacDonald R M, Spokes J R. A selective and diagnostic medium for ammonia oxidising bacteria. FEMS Microbiol Lett. 1980;8:143–145. [Google Scholar]
- 22.McTavish H, Fuchs J, Hooper A. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol. 1993;175:2436–2444. doi: 10.1128/jb.175.8.2436-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendum T A, Sockett R E, Hirsch P R. Use of molecular and isotopic techniques to monitor the response of autotrophic ammonia-oxidizing populations of the β subdivision of the class Proteobacteria in arable soils to nitrogen fertilizer. Appl Environ Microbiol. 1999;65:4155–4162. doi: 10.1128/aem.65.9.4155-4162.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 25.Norton J M, Low J M, Klotz M G. The gene encoding ammonia monooxygenase subunit A exists in three nearly identical copies in Nitrosospira sp. NpAV FEMS Microbiol Lett. 1996;139:181–188. doi: 10.1111/j.1574-6968.1996.tb08200.x. [DOI] [PubMed] [Google Scholar]
- 26.Phillips C J, Paul E A, Prosser J I. Quantitative analysis of ammonia oxidising bacteria using competitive PCR. FEMS Microbiol Ecol. 2000;32:167–175. doi: 10.1111/j.1574-6941.2000.tb00710.x. [DOI] [PubMed] [Google Scholar]
- 27.Pillai S D, Josephson K L, Bailey R L, Gerba C P, Pepper I L. Rapid method for processing soil samples for polymerase chain reaction amplification of specific gene sequences. Appl Environ Microbiol. 1991;57:2283–2286. doi: 10.1128/aem.57.8.2283-2286.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandén B, Grunditz C, Hansson Y, Dalhammar G. Quantification and characterisation of Nitrosomonas and Nitrobacter using monoclonal antibodies. Water Sci Technol. 1994;29:1–6. [Google Scholar]
- 29.Schramm A, de Beer D, van den Heuvel J C, Ottengraf S, Amann R. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol. 1999;65:3690–3696. doi: 10.1128/aem.65.8.3690-3696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephen J R, Chang Y-J, Macnaughton S J, Kowalchuck G A, Leung K T, Flemming C A, White D C. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol. 1999;65:95–101. doi: 10.1128/aem.65.1.95-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sykes P J, Neoh S H, Brisco M J, Hughes E, Condon J, Morley A A. Quantitation of targets for PCR by use of limiting dilution. BioTechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- 32.Utåker J B. Ph.D. thesis. Ås, Norway: Agricultural University of Norway; 1997. [Google Scholar]