Abstract
A PCR-based method for the detection of Salmonella spp. in food was developed. The method, set up on typical salami from the Italian region of Marche, is sensitive and specific and shows excellent correlation with the conventional method of reference when naturally contaminated foods are analyzed. Moreover, it can be easily performed within a maximum of 12 h from food sampling, thus allowing prompt detection of Salmonella spp. in the food stocks analyzed.
Salmonella spp. are facultative, intracellular parasites that invade the mucous membrane and are transmitted to humans mainly through meat, eggs, and poultry products (4). The conventional method used for Salmonella detection in food relies on preenrichment in buffered peptone water (BPW) and enrichment in selective media, followed by isolation on differential media and serological confirmation (6). The main limitation of this method is that it is time-consuming and requires 5 or 6 days during which the food stocks being analyzed are forbidden to be sold. Thus, several research groups have been working to optimize innovative and more rapid methods for the detection of Salmonella spp. (1, 2, 5, 7, 8). One of these, validated by the Association Française de Normalisation, allows the detection of Salmonella spp. in food within 24 h (5).
In this report, we propose a more rapid and simpler method that, relying on a 6-h nonselective enrichment in BPW followed by cell breaking and PCR, allows the detection of Salmonella spp. within a maximum of 12 h from the receipt of food samples.
All of the experiments were carried out on ciauscolo, which is a typical salami from the Italian region of Marche. Ciauscolo is a product at high risk of Salmonella contamination because it is made of finely chopped, raw pork and is matured for 15 days at temperatures gradually decreasing from 27 to 15°C, after which the salami is ready to be consumed.
To determine the lowest level of contamination that could be detected with this method, the experiments were conducted as described below. Ten grams of ciauscolo was homogenized with 90 ml of BPW (Oxoid, Basingstoke, England) by means of a Stomacher (400 Circulator PBI) at 260 rpm for 1 min, heat-treated at 85°C for 5 min, and inoculated as follows with Salmonella enterica serovar Enteritidis or Salmonella enterica serovar Typhimurium (provided by Istituto Superiore di Sanità, Rome, Italy). Aliquots of 1 ml of tenfold serial dilutions (10−4 to 10−9) of an overnight culture of Salmonella isolates containing 1 × 108 CFU ml−1, as confirmed by plate counting on brilliant green agar (Oxoid), were inoculated in 100 ml of each homogenized sample. Uninoculated heat-treated ciauscolo homogenates were used as a control. Immediately after the inoculation and after 2, 4, 6, and 8 h of nonselective enrichment in BPW at 37°C, 200 μl of each sample was combined with 10 μl of a 10-mg ml−1 proteinase K solution (Sigma Chemical Co., St. Louis, Mo.), incubated at 65°C for 1 h, taken to 99°C for 10 min to inactivate the proteinase K, and cooled down at 4°C (3). PCRs were performed in 50-μl reaction mixtures containing 5 μl of the crude extract and 10 mM Tris-HCl (pH 8.3)–50 mM KCl–1.5 mM MgCl2–2 pg of each primer μl−1–1.25 IU of Taq DNA polymerase (Roche Diagnostics GmbH, Mannheim, Germany)–0.125 mM each deoxynucleoside triphosphate. The PCRs were carried out in a Gene Amp PCR System 9700 (The Perkin-Elmer Corp., Norwalk, Conn.) under the following conditions: heat denaturation at 95°C for 5 min, followed by 35 cycles (90 s at 95°C, 60 s at 62°C, and 90 s at 72°C), and an elongation step of 7 min at 72°C. The primers used were Salm3 (5′-GCTGCGCGCGAACGGCGAAG-3′) and Salm4 (5′-TCCCGGCAGAGTTCCCATT-3′), which amplify a 389-bp fragment within the conserved invA gene sequence of Salmonella spp. (3). Salm3 and Salm4 proved to be highly specific for Salmonella spp. in the experimental conditions used in our study. In fact, these primers produced the characteristic 389-bp fragment when tested on 17 different Salmonella serovars. The results of the amplification were compared with a positive control represented by an “in-house” standard containing 106 S. enterica serovar Enteritidis or S. enterica serovar Typhimurium cells ml−1 and a negative control, in which the template DNA was replaced with sterile distilled water. Thirty microliters of each PCR product was analyzed by electrophoresis on a 2% Tris-borate-EDTA agarose gel stained with ethidium bromide. The gel images were visualized by means of a Bio-Rad Gel DOC 1000 and acquired with Multi-Analyst software (Bio-Rad Laboratories, Richmond, Calif.). All of the experiments were conducted in triplicate with both of the Salmonella serovars tested.
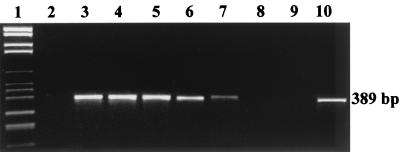
The method proposed proved to be sensitive and rapid. In fact, even though Salmonella spp. were undetectable immediately after the inoculation and after 2 h of incubation in BPW (data not shown), the 100-ml homogenates inoculated with 104, 103, and 102 CFU of Salmonella cells showed the 389-bp amplicon after 4 h of enrichment. After 6 h, Salmonella was also detectable in the samples inoculated with 1 ml of the 10−7 and 10−8 dilutions, virtually inoculated with 10 cells and 1 cell per 100 ml of homogenate, respectively. No signal was obtained from the homogenized samples inoculated with the 10−9 dilutions, which very likely did not contain Salmonella spp. (Fig. 1). As expected, S. enterica serovar Enteritidis and S. enterica serovar Typhimurium gave similar results, thus indicating that the method presented can detect different Salmonella serovars. Furthermore, the sensitivity of the method was not increased after 8 h of enrichment on BPW (data not shown), thus allowing definition of the optimal enrichment time as 6 h. This result represents an important advance in respect to other methods which, because they prescribe longer enrichment times, slow down the detection of Salmonella spp. in food samples (3, 7, 8). Moreover, taking into account that, after the nonselective enrichment in BPW, about 5 to 6 h is required for the preparation of the crude extract, PCR, and electrophoresis, the detection of Salmonella spp. with this method is completed within a maximum of 12 h from food sampling. Another main advantage of this method is that the interpretation of the results is immediate. In fact, as Salm3 and Salm4 do not amplify any nonspecific PCR products in the experimental conditions utilized, the presence or absence of Salmonella spp. in food is unequivocally reflected by the presence or absence of the 389-bp amplicon on the gel.
FIG. 1.
Agarose gel electrophoresis of PCR-amplified DNA extracted from food homogenates after inoculation with S. enterica serovar Enteritidis and incubation for 6 h at 37°C in BPW. Lanes: 1, DNA Molecular Weight Marker VI (Roche); 2, uninoculated food homogenate; 3 to 8, food samples inoculated respectively with 104, 103, 102, 10, 1, and 0 CFU per 100 ml of food homogenate; 9, PCR negative control; 10, pure culture of S. enterica serovar Enteritidis (positive control). The data shown are representative of three independent experiments.
The detection of Salmonella cells may be easier in inoculated than in naturally contaminated samples. In fact, while in the first case fresh and active cultures of this pathogen are inoculated in pasteurized samples, in the second case Salmonella cells, besides competing with the composite microflora responsible for salami fermentation, may be stressed and less viable as a consequence of the temperatures involved in the salami maturation process (8). Thus, in order to test the capability of this method to detect Salmonella in naturally contaminated food, 10 mature ciauscolo, coming from four separate lots, were sampled after 15 days of maturation, subjected to 6 h of enrichment in BPW, and processed as already described. In parallel, all of the samples were analyzed by means of the conventional method (6).
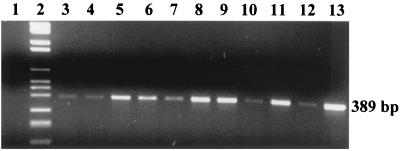
Surprisingly, Salmonella cells were detected in all of the samples, with both the conventional method and the 12-h PCR-based test (Fig. 2). This result, besides highlighting the fact that ciauscolo is a product at high risk of Salmonella contamination, indicates that the method proposed shows excellent correlation with the conventional method of reference (6) and confirms its specificity when naturally contaminated foods are analyzed.
FIG. 2.
Agarose gel electrophoresis of the PCR-amplified DNA extracted from the homogenates of 10 different naturally contaminated ciauscolo, incubated for 6 h at 37°C in BPW. Lanes: 1, PCR negative control; 2, DNA Molecular Weight Marker VI (Roche); 3 to 12, ciauscolo homogenates 1 to 10, respectively; 13, pure culture of S. enterica serovar Enteritidis (positive control). The data shown are representative of three independent experiments.
REFERENCES
- 1.Bailey J L. Detection of Salmonella cells within 24 to 26 hours in poultry samples with the polymerase chain reaction BAX system. J Food Prot. 1998;61:792–795. doi: 10.4315/0362-028x-61.7.792. [DOI] [PubMed] [Google Scholar]
- 2.Bennett A R, Greenwood D, Tennant C, Banks J G, Betts R P. Rapid and definitive detection of Salmonella in foods by PCR. Lett Appl Microbiol. 1998;26:437–441. doi: 10.1046/j.1472-765x.1998.00368.x. [DOI] [PubMed] [Google Scholar]
- 3.Cocolin L, Manzano M, Cantoni C, Comi G. Use of polymerase chain reaction and restriction enzyme analysis to directly detect and identify Salmonella typhimurium in food. J Appl Microbiol. 1998;85:673–677. doi: 10.1111/j.1365-2672.1998.00575.x. [DOI] [PubMed] [Google Scholar]
- 4.D'Aoust J Y. Pathogenicity of foodborne Salmonella. Int J Food Microbiol. 1991;12:14–70. doi: 10.1016/0168-1605(91)90045-q. [DOI] [PubMed] [Google Scholar]
- 5.Fach P, Dilasser F, Grout J, Tache J. Evaluation of a polymerase chain reaction-based test for detecting Salmonella spp. in food samples: Probelia Salmonella spp. J Food Prot. 1999;62:1387–1393. doi: 10.4315/0362-028x-62.12.1387. [DOI] [PubMed] [Google Scholar]
- 6.International Organization for Standardization. Microbiology—general guidance on methods for the detection of Salmonella. (Revision of 2nd ed., ISO, 6579, 1990.) Geneva, Switzerland: International Organization for Standardization; 1991. [Google Scholar]
- 7.Manzano M, Cocolin L, Astori G, Pipan C, Botta G A, Cantoni C, Comi G. Development of a PCR microplate-capture hybridization method for simple, fast and sensitive detection of Salmonella serovars in food. Mol Cell Probes. 1998;11:459–462. doi: 10.1006/mcpr.1998.0176. [DOI] [PubMed] [Google Scholar]
- 8.Waage A S, Vardund T, Lund V, Kapperud G. Detection of low numbers of Salmonella in environmental water, sewage and food samples by a nested polymerase chain reaction assay. J Appl Microbiol. 1999;87:418–428. doi: 10.1046/j.1365-2672.1999.00835.x. [DOI] [PubMed] [Google Scholar]