Abstract
A polyphasic taxonomic study involving DNA-DNA hybridization, whole-cell protein electrophoresis, and 16S ribosomal DNA sequence analysis revealed that a group of Burkholderia cepacia-like organisms isolated from the rhizosphere or tissues of maize, wheat, and lupine belong to B. cepacia genomovar III, a genomic species associated with “cepacia syndrome” in cystic fibrosis patients. The present study also revealed considerable protein electrophoretic heterogeneity within this species and demonstrated that the B. cepacia complex consists of two independent phylogenetic lineages.
In a survey of nonnative plant rhizosphere bacteria conducted in La Côte Saint André (France) with maize and in Kapunda (South Australia, Australia) with wheat, high levels of two groups of Burkholderia strains were found. The first group was characterized by using a polyphasic approach and formed a new taxon, Burkholderia graminis (15). Strains of the second group (designated phenon B) were found to be closely related to the Burkholderia cepacia complex; large numbers of these strains were present on roots, and more recently, new isolates were also obtained from inside the tissues of wheat and lupine in Kapunda (Table 1). Here, characterization of this taxonomic group was revisited by including reference strains of the B. cepacia complex in DNA-DNA hybridization, whole-cell protein electrophoretic, and 16S ribosomal DNA (rDNA) sequence analyses.
TABLE 1.
Strains used in this study
| Strain | Other designationa | Reference or source | Ecology |
|---|---|---|---|
| Rhizosphere strains | |||
| AUS 13 | LMG 19240 | 15 | Continuous wheat plot (Kapunda, Australia) |
| AUS 26 | LMG 19247 | 15 | Continuous wheat plot (Kapunda, Australia) |
| AUS 27 | LMG 19238 | 15 | Continuous wheat plot (Kapunda, Australia) |
| AUS 30 | LMG 19245 | 15 | Continuous wheat plot (Kapunda, Australia) |
| AUS 12 | LMG 19243 | 15 | Wheat pasture rotation (Kapunda, Australia) |
| AUS 29 | LMG 19246 | 15 | Wheat pasture rotation (Kapunda, Australia) |
| AUS 31 | LMG 19244 | 15 | Wheat pasture rotation (Kapunda, Australia) |
| AUS 32 | LMG 19248 | 15 | Wheat pasture rotation (Kapunda, Australia) |
| AUS 34 | LMG 19242 | 15 | Wheat pasture rotation (Kapunda, Australia) |
| AUS 36 | LMG 19239 | 15 | Wheat pasture rotation (Kapunda, Australia) |
| AUS 37 | LMG 19241 | 15 | Wheat pasture rotation (Kapunda, Australia) |
| C3B1M | R-13371 | 15 | Maize rhizosphere (Côte Saint André, France) |
| m32 | R-13369 | 15 | Maize rhizosphere (Côte Saint André, France) |
| m35b | R-13370 | 15 | Maize rhizosphere (Côte Saint André, France) |
| Endophytes | |||
| WS11.7 | LMG 19232 | K. Ophel-Keller | Wheat shoot endophyte (Kapunda, Australia) |
| WS9.1 | LMG 19231 | K. Ophel-Keller | Wheat shoot endophyte (Kapunda, Australia) |
| WR2.5 | LMG 19237 | K. Ophel-Keller | Wheat root endophyte (Kapunda, Australia) |
| WR2.6 | LMG 19230 | K. Ophel-Keller | Wheat root endophyte (Kapunda, Australia) |
| LS2.4 | LMG 19233 | K. Ophel-Keller | Lupine shoot endophyte (Kapunda, Australia) |
| LS12.9 | LMG 19234 | K. Ophel-Keller | Lupine shoot endophyte (Kapunda, Australia) |
| LR1.4 | LMG 19235 | K. Ophel Keller | Lupine root endophyte (Kapunda, Australia) |
| LR14.9 | LMG 19236 | K. Ophel-Keller | Lupine root endophyte (Kapunda, Australia) |
| Clinical isolates | |||
| 1–36 | R-9061 | C. Segonds | Cystic fibrosis patient (Clermont-Ferrand, France) |
| 751 | R-11750 | C. Segonds | Non-cystic fibrosis hemoculture, (Bordeaux, France) |
| Reference strains | |||
| B. cepacia (I)b | ATCC 25416Tc | 1 | Onion sour skin |
| B. cepacia (I) | LMG 6964 | Haywood, 1965 | Tomato |
| B. multivorans (II) | 1–45 | 11 | Cystic fibrosis patient (France) |
| B. cepacia (III) | LMG 13053 | 3 | Cystic fibrosis sputum (Belgium) |
| B. cepacia (III) | LMG 16661 | 13 | Cystic fibrosis sputum (United Kingdom) |
| B. cepacia (III) | LMG 12614 | 13 | Cystic fibrosis sputum (United Kingdom) |
| B. cepacia (III) | LMG 12615 | 13 | Cystic fibrosis sputum (United Kingdom) |
| B. cepacia (III) | LMG 16659 | 13 | Cystic fibrosis sputum (United Kingdom) |
| B. cepacia (III) | LMG 6988 | Leg wound, (Sweden, 1972) | |
| B. vietnamiensis (V) | LMG 10929T | 5 | Rice rhizosphere |
| B. pyrrocinia | ATCC 15958T | 7 | Soil |
| B. graminis | ATCC 700544T | 15 | Maize rhizosphere |
ATCC, American Type Culture Collection, Manassas, Va.; LMG, Culture Collection, Laboratory of Microbiology, State University of Ghent, Ghent, Belgium.
The roman numerals in parentheses indicate genomovars.
T = type strain.
Total DNA-DNA hybridization analyses were performed by using two methods, one involving tritiated reference DNAs (Table 2) and one involving photobiotin-labeled probes (Table 3). In a preliminary study, the two methods showed good correlation. For instance, the levels of hybridization of strain AUS 27 DNA with DNA of strain LMG 12614 were 65% when tritiated DNA was used and 63% when photobiotin-labeled DNA was used.
TABLE 2.
Levels of total DNA hybridization with radioactively labeled DNAs of eight reference strains
| Source of unlabeled DNA | % Hybridization with labeled DNA of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| AUS 27 | C3B1M | 1–36 | B. cepacia ATCC 25416T | B. vietnamiensis LMG 10929T | B. multivorans 1–45 | B. pyrrocinia ATCC 15958T | B. graminis ATCC 700544T | |
| AUS 27 | 100 | 69 (3.2)a | 60 (4.3) | 51 | 43 (10) | 50 | 60 | 15 |
| AUS 37 | 100 | 71 | 50 | |||||
| AUS 14 | 100 | 36 | 40 | 39 | 55 | |||
| AUS 26 | 99 | |||||||
| AUS 32 | 98 | 48 | ||||||
| AUS 34 | 92 | 66 | 45 | |||||
| AUS 13 | 92 | 61 | 46 | |||||
| AUS 30 | 91 | 64 | 44 | 53 | 15 | |||
| AUS 31 | 90 (2.8) | 41 | 39 | |||||
| AUS 12 | 90 | |||||||
| AUS 29 | 90 | |||||||
| AUS 36 | 89 | |||||||
| C3B1M | 70 (3.8) | 100 | 63 (3.1) | 57 | 44 (5.6) | 40 | 56 | 13 |
| m32 | 75 | 98 | ||||||
| PHQB17 | 65 | 76 | 15 | |||||
| 751 | 72 (4.2) | 43 | ||||||
| LMG 12614 | 65 (4.2) | 42 | ||||||
| LMG 6988 | 58 (4.9) | 35 | ||||||
| 1–36 | 63 (4.6) | 100 | 46 | 38 (7.3) | 44 | 50 | ||
| LMG 16661 | 76 | 75 (0.8) | 45 | 41 (5.8) | 48 | |||
| 1–47 | 66 (4.8) | 68 (4) | 48 | 44 (9) | 40 | 49 | ||
| ATCC 25416T | 56 (6.5) | 52 (5.9) | 52 (6.5) | 100 | 39 (8) | 41 | 53 | |
| LMG 6964 | 56 (6.4) | 54 (7.8) | 55 (6.8) | 66 | 43 | 44 | 55 | |
| ATCC 15958T | 62 (9.2) | 55 (9.6) | 62 (6.2) | 51 | 43 (6.7) | 41 | 100 | |
| m35b | 47 | 48 | 44 | 40 | 48 | 43 | 46 | |
The values in parentheses are ΔTm values (in degrees Centigrade).
TABLE 3.
DNA-DNA hybridization of environmental strains with genomovar III representatives
| Source of unlabeled DNA | % Hybridization with labeled DNA of:
|
||||
|---|---|---|---|---|---|
| LMG 12614 | LMG 13053 | LMG 16659 | WS11.7 | AUS 27 | |
| LMG 12614 | 100 | ||||
| LMG 13053 | 79 | 100 | |||
| LMG 16659 | 67 | 91 | 100 | ||
| WS11.7 | 68 | 82 | 71 | 100 | |
| AUS 27 | 63 | 82 | 71 | 99 | 100 |
For hybridization we used photobiotin-labeled probes at 50°C. Each value is the average of the values from at least two hybridization experiments.
In the first experiments we used tritiated reference DNAs of eight isolates, including two rhizosphere isolates (AUS 27 and C3B1M), one recent cystic fibrosis isolate (1–36), and reference strains of B. cepacia genomovar I (ATCC 25416T), Burkholderia vietnamiensis (LMG 10929T), Burkholderia multivorans (1–45), Burkholderia pyrrocinia (ATCC 15958T), and B. graminis (ATCC 700544T). DNAs from 15 of our rhizosphere isolates, reference strains belonging to B. cepacia genomovar I (ATCC 25416T and LMG 6964) and genomovar III (LMG 12614, LMG 16661, and LMG 6988), and B. pyrrocinia (ATCC 15958T), and three recent cystic fibrosis isolates (strains 751, 1–36, and 1–47) were hybridized with these radioactively labeled DNAs. When hybridized with labeled DNA of strain AUS 27, all rhizosphere isolates except m35b showed levels of DNA-DNA hybridization greater than 65% and differences in melting temperatures (ΔTm values) less than 5°C, indicating that they belong to the same genomic species (12). When they were hybridized with labeled DNA of strain C3B1M, slightly lower values (as low as 61%) were obtained, indicating a certain degree of genomic heterogeneity in this species. Strain m35b showed significant but low levels of hybridization (40 to 48%) with all reference strains and thus does not belong to any of the genomovars examined. The possibility that this strain could belong to Burkholderia stabilis was not eliminated and will be tested further. B. cepacia genomovar III reference strains exhibited levels of hybridization of 58 to 76% with labeled DNA of strain AUS 27, indicating that the rhizosphere isolates belong to B. cepacia genomovar III. Reference strains of the other B. cepacia genomovars and of B. pyrrocinia exhibited levels of DNA-DNA hybridization between 39 and 60%, values which are in complete agreement with values reported previously (13). The levels of hybridization with DNA of the B. graminis type strain were much lower (13 to 15%). The three recent cystic fibrosis isolates (strains 1–36, 751, and 1–47) showed levels of hybridization between 63 and 76% with AUS 27 DNA with ΔTm values less than 5°C. These data show unambiguously that these three isolates also belong to the same genomic species as AUS 27 (i.e., B. cepacia genomovar III).
A second group of DNA-DNA hybridization experiments (Table 3) was performed in order to substantiate the relationships among B. cepacia genomovar III strains. In addition, a representative endophytic isolate was included. Values between 63 and 82% were obtained, which confirmed that all of these isolates belong to a single genomic species.
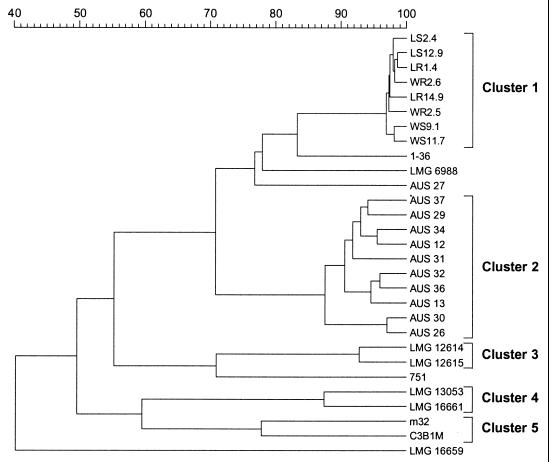
Whole-cell protein extracts were prepared from 48-h cultures of all of the B. cepacia genomovar III strains and several additional endophytic isolates. Data for the reference strains were obtained from previous studies (3a, 13, 14). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analyses were performed as described previously (13). Protein profiles were analyzed by using the GelCompar software package (version 4.2; Applied Maths, Kortrijk, Belgium). Levels of similarity between the patterns were computed by using the Pearson product moment correlation coefficient and were expressed as percentages of similarity for convenience. Considerable heterogeneity was apparent, and the strains grouped into two main protein electrophoretic clusters comprising the endophytic isolates (cluster 1) and all of the Australian rhizosphere isolates except isolate AUS 27 (cluster 2), three small clusters comprising two isolates each (clusters 3 to 5), and several isolates with distinct positions in the dendrogram (Fig. 1). Cluster 3 comprises two reference strains (LMG 12614 and LMG 12615) and represents cluster xii described previously (13). Cluster 4 also comprises two reference strains (LMG 13053 and LMG 16661) and corresponds to cluster xi described previously (13). Finally, cluster 5 comprises two French rhizosphere isolates (C3B1M and m32). In spite of this protein electrophoretic heterogeneity, DNA-DNA hybridization data (Tables 2 and 3) demonstrated that all of the isolates shown in Fig. 1 represent a single genomic species; most of the isolates belong to clusters 2 through 5, and one strain, strain W11.7, is a cluster 1 reference isolate. The protein electrophoretic homogeneity of the other cluster 1 isolates indicates that they are members of the genomic species as well.
FIG. 1.
Dendrogram derived from unweighed pair group average linkage of correlation coefficients (expressed for convenience as percentages of similarity) for the whole-cell protein patterns of the B. cepacia genomovar III strains studied.
To investigate the phyletic relatedness of the genomovar III isolates, almost complete 16S rDNA sequences of the following two endophytic isolates and two genomovar III strains were obtained: LMG 12615, LMG 12614, LS2.4 and WS11.7. These sequences and selected GenBank 16S rDNA sequences of 19 representative strains of the B. cepacia complex were aligned. A total of 1351 16S rDNA sites were then selected, and sites involving indels (insertions or deletions) were excluded from further analysis. Evolutionary distances (representing the percentages of transversion type differences between sequence pairs) were computed by the method of Jukes and Cantor (9). A phylogenetic tree was inferred by using the neighbor-joining method (10), and bootstrapping was performed (9). This phylogenetic analysis divided the 16S rDNA sequences of the B. cepacia complex into two major clusters: (i) a lineage containing the B. vietnamiensis (genomovar V), B. multivorans (genomovar II), and LMG 18941 (genomovar VI) DNA sequences, and (ii) a group containing the B. stabilis (genomovar IV), B. pyrrocinia, B. cepacia genomovar I (ATCC 25416T and ATCC 17759), B. cepacia genomovar III (LMG 12614, LMG 12615, WS11.7, LS2.4, and C3B1M), and unclassified strain m35b sequences. This division was strongly supported by 98% of the bootstrap replicates and should be obtained with any other phylogenetic markers that match classical bacterial evolutionary patterns.
Public health implications.
Our 14 isolates represent environmental niches ranging from the rhizosphere to the inner tissues of wheat, lupine, and maize and were obtained in France and in South Australia. These plants are cultivated all over the world, and it is likely that our isolates represent very common bacteria.
We demonstrate here that a significant proportion of these maize-, wheat-, and lupine- associated bacteria are actually members of B. cepacia genomovar III. This B. cepacia genomovar is particularly relevant for cystic fibrosis as most strains associated with the “cepacia syndrome” belong to it. This syndrome is characterized by a dramatic necrotizing pneumonia that results in rapid death of the patient (6, 8). Recent deadly outbreaks which occurred in many parts of the world have been attributed to strains of genomovar III, suggesting that high transmissibility could be a characteristic of B. cepacia genomovar III in the B. cepacia complex (13). It has been suggested that the environment is a source of new isolates, but so far no clear evidence of this has been obtained. Attempts to recover B. cepacia isolates similar to clinical isolates from soils and other environmental sites have failed, most likely because of the selective agents used (antibiotics). The environmental strains studied here were isolated by using the PCAT medium (2), whose selective power is based solely on the metabolism of unusual sources of carbon and nitrogen (viz., azelaic acid and tryptamine). Thus, our Australian and French isolates represent the first collection of genuinely environmental B. cepacia genomovar III strains. This collection opens the way for comparisons of closely related strains of environmental and clinical origin, which could provide some clue about the properties acquired by hospital-adapted strains and their pathogenicity characteristics or genes. It could also help refine strategies for avoiding acquisition of new Burkholderia strains in cystic fibrosis treatment centers. Most of the soil isolates have been deposited in the BCCM/LMG Bacteria Collection (University of Ghent, Ghent, Belgium) under the accession numbers shown in Table 1.
Nucleotide sequence accession numbers.
The nucleotide sequences of strains LMG 12615, LMG 12614, LS2.4, and WS11.7 have been deposited in the GenBank database under accession numbers AF311969, AF311970, AF311971, and AF311972, respectively.
Acknowledgments
K. Ophel-Keller (Commonwealth Scientific and Industrial Research Organisation, Adelaide, South Australia, Australia) isolated the endophyte strains which were kindly sent to us by C. Lodewyckx from M. Mergeay's laboratory (SCK/CEN, Mol, Belgium). We thank G. Chabanon and C. Segonds for sending us cystic fibrosis patient isolates of Burkholderia. F. Fontaine efficiently contributed to the 16S rDNA sequencing of several strains.
J.B. was supported by an OECD fellowship and a CNRS-CSIRO collaborative program. P.V. is indebted to the Fund for Scientific Research-Flanders (Belgium) for a position as a postdoctoral fellow. T.C. acknowledges support received from the Vlaams Instituut voor Bevordering van Wetenschappelijk-technologisch Onderzoek in de Industrie (Belgium) in the form of a bursary for advanced study. We acknowledge the financial support provided by the Cystic Fibrosis Trust (United Kingdom) (grant RS15). We acknowledge the financial support of Centre National de la Recherche Scientifique for the research performed at UMR CNRS 5557.
REFERENCES
- 1.Ballard R W, Palleroni N J, Stanier R Y, Mandel M. Taxonomy of the aerobic pseudomonads Pseudomonas cepacia, P. marginata, P. alliicola and P. caryophylli. J Gen Microbiol. 1970;60:199–214. doi: 10.1099/00221287-60-2-199. [DOI] [PubMed] [Google Scholar]
- 2.Burbage D A, Sasser M. A medium selective for Pseudomonas cepacia. Phytopathol Abstr. 1982;72:706. [Google Scholar]
- 3.Coenye T, Schouls L M, Govan J R W, Kersters K, Vandamme P. Identification of Burkholderia species and genomovars from cystic fibrosis patients by AFLP fingerprinting. Int J Syst Bacteriol. 1999;49:1657–1666. doi: 10.1099/00207713-49-4-1657. [DOI] [PubMed] [Google Scholar]
- 3a.Coenye, T., S. Laevens, A. Willems, M. Ohlén, W. Hannant, J. R. W. Govan, M. Gillis, E. Falsen, and P. Vandamme.Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 4.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 5.Gillis M, Tran Van V, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and the proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 6.Govan J R W, Hughes J, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 7.Imanaka H, Kousaka M, Tamura G, Arima K. Studies on pyrrolnitrin, a new antibiotic. Taxonomy studies on pyrrolnitrin-producing strain. J Antibiot. 1965;18:205–206. [PubMed] [Google Scholar]
- 8.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 9.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. III. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 10.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 11.Segonds C, Heulin T, Marty N, Chabanon G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J Clin Microbiol. 1999;37:2201–2208. doi: 10.1128/jcm.37.7.2201-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 13.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Gowan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 14.Vandamme P, Mahenthiralingam E, Holmes B, Coenye T, Hoste B, De Vos P, Henry D, Speert D P. Identification and population structure of Burkholderia stabilis sp. nov (formerly Burkholderia cepacia genomovar IV) J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viallard V, Poirier I, Cournoyer B, Haurat J, Wiebkin S, Ophel-Keller K, Balandreau J. Burkholderia graminis sp. nov., a novel species of rhizospheric Burkholderia and reassessment of Pseudomonas phenazinium, P. pyrrocinia and P. glathei into Burkholderia. Int J Syst Bacteriol. 1998;48:549–563. doi: 10.1099/00207713-48-2-549. [DOI] [PubMed] [Google Scholar]