Abstract
Bacterial strains were isolated from samples of Japanese rice vinegar (komesu) and unpolished rice vinegar (kurosu) fermented by the traditional static method. Fermentations have never been inoculated with a pure culture since they were started in 1907. A total of 178 isolates were divided into groups A and B on the basis of enterobacterial repetitive intergenic consensus-PCR and random amplified polymorphic DNA fingerprinting analyses. The 16S ribosomal DNA sequences of strains belonging to each group showed similarities of more than 99% with Acetobacter pasteurianus. Group A strains overwhelmingly dominated all stages of fermentation of both types of vinegar. Our results indicate that appropriate strains of acetic acid bacteria have spontaneously established almost pure cultures during nearly a century of komesu and kurosu fermentation.
The rice vinegars komesu and kurosu are produced from polished and unpolished rice, respectively, by the same process (saccharification of rice, alcohol fermentation, and oxidation of ethanol to acetic acid). Both of these vinegars are traditional seasonings that have long been used in Japan, China, and Asian countries. Komesu is colorless and has a plain taste and thus is used for sushi cooking, while kurosu is black and contains more amino acids and vitamins than komesu and thus is used as a healthy drink. With increasing public interest in health, the effective health-related elements of the traditional vinegars have come under scrutiny.
Vinegar is produced industrially by two main methods, a slow process involving static surface acetic acid fermentation and a fast submerged fermentation process. Generally, static fermentation is employed in traditional vinegar production. This technique is not costly in terms of plant investment, and the quality of the product is good, although a rather long time is required to complete the fermentation. An alcoholic liquid with vinegar (moromi) is fermented in appropriate containers fitted with covers, and this is considered a good way to prevent bacterial contamination during static fermentation. In a few days, a crepe pellicle of acetic acid bacteria covers the surface of the moromi, after which the fermentation proceeds and finishes about 1 month later. In this process, no strict sterilization measures are used. No purified strain is inoculated after the start of vinegar fermentation, which in some cases has been continued without inoculation of a pure culture for more than 100 years.
The acetic acid bacteria consist of two genera, Acetobacter and Gluconobacter. Strains of Acetobacter are generally involved in vinegar production (12). Identification of the species and characterization of the dominant strains in static acetic acid fermentation are desirable in order to stabilize the fermentation and improve the strain (7). In recent years the enterobacterial repetitive intergenic consensus (ERIC)-PCR method and random amplified polymorphic DNA (RAPD) fingerprinting have been applied to taxonomic grouping of bacteria, including enterobacteria (3, 16), Acetobacter sp. (15), lactic acid bacteria (17), and rhizobia (4, 6). In this study, we investigated acetic acid bacterial strains isolated from samples obtained during commercial production of komesu and kurosu by using the ERIC-PCR and RAPD methods. The isolates were identified and characterized genetically and physiologically. Changes in the flora during the manufacturing process were also examined.
All of the TN strains isolated in this study and Acetobacter pasteurianus ATCC 33445T were cultivated aerobically at 28°C in potato medium containing (per liter) 20.0 g of glycerol, 10 g of Polypeptone, 10 g of yeast extract, 5 g of glucose, and 100 ml of potato extract. Escherichia coli JM109 (Takara Shuzo Co., Ltd., Ohtsu, Shiga, Japan) was grown in Luria-Bertani medium. Samples used for isolation of bacteria were taken from komesu and kurosu commercially produced by Tamanoi Vinegar Co., Ltd. (Nara, Japan). The samples were obtained from microbial films on the moromi surface in the early, middle, and late phases of acetic acid fermentation (and in the last fermentation period in the case of komesu). The initial moromi contained about 3 to 3.5% acetic acid and 4 to 4.5% ethyl alcohol and had a pH of 3.3. Acetic acid bacteria were isolated on isolation medium agar (1% glucose, 1% glycerol, 0.2% yeast extract, 0.2% Polypeptone, 10% potato extract, 1% acetic acid, 2% ethanol, 0.9% agar) and were grown at 30°C for 4 days.
Total DNA was extracted by the method of Ohmori et al. (10). DNA for the ERIC-PCR template was prepared by the rapid method described by Nuswantara et al. (9). The PCR conditions used with oligonucleotide primers ERIC1R (3′-CACTTAGGGGTCCTCGAATGTA-5′) and ERIC2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) were those described by Versalovic et al. (16). PCR were carried out by using a programmable temperature control system (PC-700; Astec, Fukuoka, Japan). RAPD Analysis Beads and Primers were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). Electrophoresis was carried out on a horizontal 0.7, 1.5, or 2% agarose gel (SeaKem GTG agarose; FMC BioProducts, Rockland, Maine) in electrophoresis buffer (0.04 M Tris-acetate, 0.001 M EDTA) at a constant voltage of 100 V. The gel was stained with ethidium bromide and photographed with a UV transilluminator.
16S ribosomal DNA (rDNA) was amplified in vitro (PCR) by using the method of Both et al. (1) in combination with oligonucleotide primers complementary to highly conserved regions of bacterial rRNA genes. The 5′- and 3′-terminal primers used were GAGTTTGAT(C/T)(C/A)TGGCTCA (positions 9 to 26, according to the E. coli numbering system [2]) and CA(G/T)AAAGGAGGTGATCC (positions 1545 to 1529), respectively (11). Double-stranded PCR products were ligated at the HincII site of pUC19. The DNA was transferred into E. coli JM109 competent cells (Takara Shuzo). Sequencing was performed with an ABI PRISM 310 genetic analyzer (PE Biosystems Japan) according to the manufacturer's instructions. DNA sequences were processed with GENETYX-MAC, version 10.1 (Software Development Co., Tokyo, Japan) for multialignment analysis.
The G+C contents of DNA were determined by high-performance liquid chromatography as described previously (8, 14). The nucleotides obtained by treatment with P1 nuclease and alkaline phosphatase were applied to a high-performance liquid chromatograph; a Hitachi L7000 analyzer equipped with a Cosmosil 5C18-AR column (4.6 by 150 mm; Nacalai Teque, Inc., Kyoto, Japan) was used for this analysis.
Gram staining, catalase production tests, oxidase tests, tests for acid production from glucose under aerobic or anaerobic conditions, and tests for acetate and lactate oxidation to CO2 were conducted with all isolates. Tests for assimilation of ammoniacal nitrogen, growth on carbon sources, acid production from carbon sources, formation of ketogluconic acids from d-glucose (13), and determination of the ubiquinone system were conducted with representative isolates as described in Bergey's Manual of Systematic Bacteriology (5). Ubiquinone 10 was obtained from Sigma Chemical Co. (St. Louis, Mo.). Ubiquinone 9 was isolated and purified from Acetobacter aceti IFO3281. Acetobacter strains were observed with a Hitachi S-3500N scanning electron microscope equipped with a field emission gun and operated at 10 kV. In acetic acid fermentation tests, acetic acid bacterial mats were floated on 50 ml of a medium containing 1.0% d-glucose, 1.0% glycerol, 0.2% Polypeptone, 0.2% yeast extract, 10% potato extract, 1.0% acetic acid, and 4.0% ethanol in 100-ml vials, and surface fermentation was continued at 30°C for 2 weeks. Acetic acid contents were determined by titration with 0.1 N NaOH against phenolphthalein.
A total of 178 bacterial strains were obtained from the moromi of komesu and kurosu in static surface acetic acid fermentations. Surface bacterial mats of moromi were suspended in sterile water, and bacteria were isolated on isolation medium plates incubated at 30°C for a few days. In static fermentations, a portion of the surface mat bacteria from the early phase of fermentation is used as a starter for the next fermentation batch. Both komesu and kurosu are fermented for about 1 month at room temperature, which can range from 10 to 30°C. The final concentration of acetic acid is about 6 to 6.5%, and the pH is 3.1. The residual alcohol content is usually 0.1%.
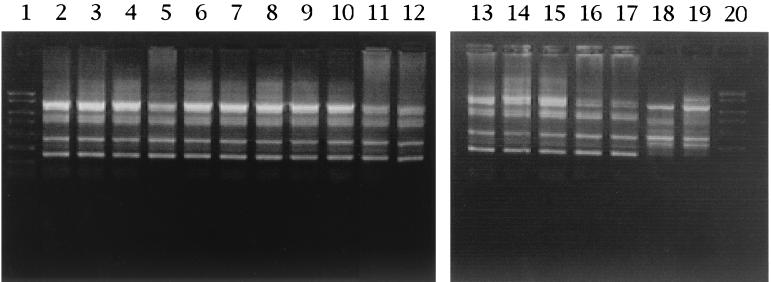
DNA prepared from pure cultures of all isolates obtained from samples taken at different stages of fermentation were amplified by ERIC-PCR (Fig. 1). On the basis of the amplification profiles, the bacterial strains were divided into two groups, groups A and B. Most of the isolates produced ERIC bands at 1,350, 1,150, 860, 770, 390, and 220 bp, and these isolates were placed in group A. The rest of the isolates, including TN-1 and TN-2, produced ERIC bands at 1,150, 710, 640, 500, 390, and 310 bp and were placed in group B. These strains were isolated mainly after fermentation had finished. All of the isolates from kurosu had the same profiles as group A isolates (data not shown).
FIG. 1.
ERIC profiles of selected acetic acid bacterial strains from rice vinegar. Lanes 1 and 20, PCR markers (Novagen); Lanes 2 to 19, strains TN-106, TN-6, TN-9, TN-12, TN-70, TN-71, TN-21, TN-23, TN-27, TN-117, TN-118, TN-119, TN-96, TN-98, TN-193, TN-195, TN-1, and TN-2, respectively.
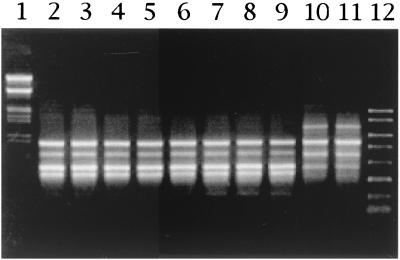
Intraspecific variation was demonstrated by differential amplification of certain DNA fragments displaying polymorphism in the following strains: TN-1, TN-2, TN-6, TN-9, TN-12, TN-21, TN-23, TN-27, TN-70, and TN-71 (Fig. 2). These strains were arbitrarily selected from groups A and B. On average, the PCR products obtained with RAPD analysis primer 6 generated fingerprints consisting seven or eight fragments that varied in length from 150 bp to 2.0 kbp. These strains all produced RAPD bands at 810, 600, and 430 bp. However, TN-1 and TN-2 produced an additional band at about 1.3 kb, whereas the other strains produced an additional band at about 340 bp. This result shows that strains TN-1 and TN-2 (group B) are different from other strains (group A) and is consistent with the ERIC-PCR findings.
FIG. 2.
RAPD profiles of selected acetic acid bacterial strains from rice vinegar. Lane 1, lambda HindIII-EcoRI fragments used as DNA size markers; lanes 2 to 11, strains TN-6, TN-9, TN-12, TN-70, TN-71, TN-21, TN-23, TN-27, TN-1, and TN-2, respectively; lane 12, PCR markers (Novagen).
The 16S rDNA of strain TN-27 in group A and strains TN-1 and TN-2 in group B were sequenced. The level of similarity between the sequence of strain TN-27 and the sequence of the type strain of A. pasteurianus, LMD 22.1 (EMBL data library accession no. X71863), was 99.5%, and the difference corresponded to 8 base changes. The 16S rDNA sequences of strains TN-1 and TN-2 were also very similar (99.5%) to that of A. pasteurianus LMD 22.1. The TN-1 and TN-2 sequences differed at 7 and 8 bases, respectively, from the sequence of the A. pasteurianus strain. Levels of similarity between group A and B strains of 99.9% were found. Since the level of similarity between each TN strain and A. aceti NCIB 8621 (EMBL data library accession no. X74066) was only 96.9%, the TN strains apparently belong to A. pasteurianus.
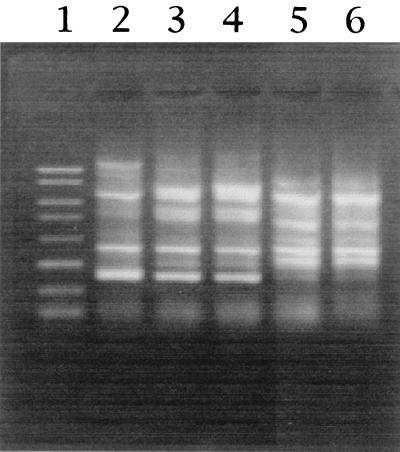
The taxonomic characteristics of two strains belonging to group A, two strains belonging to group B, and the type strain of A. pasteurianus, ATCC 33445, were examined. All of the strains were gram-negative rods that were catalase positive and oxidase negative. The results of the oxidation-fermentation test showed oxidation. The strains were able to oxidize lactate and acetate. The G+C contents of the chromosomal DNA ranged from 53.6 to 54.3 mol%, values which are within the range of values for A. pasteurianus according to Bergey's Manual of Systematic Bacteriology (5). No formation of ketogluconic acids was detected by thin-layer chromatography analysis. The strains produced ubiquinone 9. No assimilation of ammoniacal nitrogen was detected. All isolates utilized d-glucose, galactose, d-mannose, glycerol, l-sorbose, and inositol as carbon sources. The group A strains could assimilate methanol, whereas the group B isolates could not. With regard to acid production from carbon sources, the group A isolates produced acids from methanol, but the group B isolates did not. For the most part, these characteristics were consistent with those of A. pasteurianus, although differences were found in growth on and acid formation from carbon sources. We also observed that the colonies of group A strains differed from those of group B strains. Colonies of members of both groups were pale brownish or pinkish, had regular edges, and were 5 mm in diameter. However, the colonies of group A strains were umbonate, whereas those of group B strains were flat. Group A strains produced acid from d-arabinose after about 20 days, but the group B strains and A. pasterurianus did not. Thus, the group A strains may constitute a novel subgroup within A. pasteurianus, although they were classified as A. pasteurianus based on the results of the 16S rDNA sequence analysis. When we compared the ERIC-PCR patterns of A. pasteurianus ATCC 33445T and the four strains isolated, the patterns obtained for both group A and B isolates were different from the ATCC 33445T pattern, which had ERIC bands at 2,300, 1,200, 400, and 225 bp (Fig. 3).
FIG. 3.
Comparison of ERIC profiles of A. pasteurianus ATCC 33445T and TN strains. Lane 1, PCR markers (Novagen); lanes 2 to 6, strains ATCC 33445T, TN-27, TN-136, TN-1, and TN-2, respectively.
We observed cell shape and the surfaces of isolated bacteria by using a scanning electron microscope. In spite of their classification in the same species on the basis of 16S rDNA sequence data, group A and B organisms appeared to be slightly different at the cell level. Cells of group A strain TN-27 had a swollen appearance, and they were 0.8 to 0.9 by 1.2 to 1.3 μm. The cell surface was coated with an unknown material. During preparation of the specimens, the cells were observed to be more conglomerated than those of the group B bacterium, which was thought to indicate a difference in cell surface construction. The cells of group B strain TN-1 were rod shaped compared to those of TN-27 and they were 0.6 to 0.7 by 1.6 to 1.8 μm.
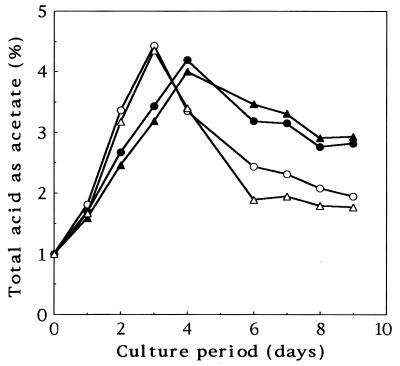
Representative group A and B strains were tested in a small-scale acetic acid fermentation experiment (Fig. 4). Under the conditions employed, the group B strains seemed to be slightly superior in terms of acetic acid formation ability. These strains consumed acetic acid promptly, a phenomenon referred to as “overoxidation”. Members of the genus Acetobacter reportedly are able to oxidize acetate into carbon dioxide and water. Group A strains TN-27 and TN-136 consumed acetic acid at rates of 0.121 and 0.0790 mmol/h, respectively. In comparison, group B strains TN-1 and TN-2 consumed acetic acid at rates of 0.229 and 0.266 mmol/h, respectively. These results showed that the strains differed in the ability to overoxidize acetic acid and that group B strains tended to oxidize acetic acid more strongly than group A strains.
FIG. 4.
Time courses of acid production by isolated acetic acid bacteria. Acetic acid bacteria were grown statically on 50 ml of medium containing 1.0% d-glucose, 1.0% glycerol, 0.2% Polypeptone, 0.2% yeast extract, 10% potato extract, 1.0% acetic acid, and 4.0% ethanol in 100-ml vials at 30°C for 2 weeks. Acetic acid contents were determined by titration with 0.1 N NaOH against phenolphthalein. The data are averages based on three trials. Symbols: ●, TN-27; ▴, TN-136; ○, TN-1; ▵, TN-2.
We investigated the changes of the flora in komesu and kurosu surface fermentations used for commercial production by using the ERIC-PCR method (Table 1). It is valuable to classify acetic acid bacteria isolated from the vinegar fermentation process at the species level in order to be able to assess and control vinegar fermentation. The fermentation was divided into three periods, early (1 to 10 days), middle (11 to 20 days), and late (21 to 32 days). Samples were taken from moromi pellicles during each period, and colonies of acetic acid bacteria were randomly picked from each sample. In addition, in the case of komesu, the moromi was kept after the end of acetic acid fermentation (that is, after the residual alcohol content became less than 0.1%), and acetic acid bacteria were isolated from the postfermented samples as described above. Group A strains accounted for 100% of the fermentation flora throughout acetic acid fermentation of both types of vinegar. This appears to indicate that in the course of vinegar production by the traditional fermentation method, appropriate bacterial strains have been selected spontaneously to give almost pure cultures without any sterilization or purified strain inoculation for almost 100 years. In komesu, two group B strains (8%) were found in the postfermentation period. Group B strains did not always appear after the end of acetic acid fermentation, and they appeared only once every several batches. During acetic acid fermentation, the pellicle is an almost pure culture of group A strains. It is thought that group B strains would lower the quality of fermentation since they have a stronger propensity to oxidize acetic acid. The traditional practice has been to end the acetic acid fermentation before the residual alcohol completely disappears in order to prevent inferior vinegar fermentation. The results of this study have proved that this traditional idea is scientifically valid. Even if it is due to the low pH of the acetic acid fermentation, we were very surprised to find that an almost pure culture of acetic acid bacteria has been kept for such a long time. We should emphasize that all the processes of fermentation were well controlled and maintained, including the use of a pure water supply and cleaning of the containers and room.
TABLE 1.
Flora changes in rice and unpolished rice vinegar fermentations
| Source | Fermentation period | No. of isolates (appearance rate [%])
|
|
|---|---|---|---|
| Group A | Group B | ||
| Rice vinegar (komesu) | Early (1 to 10 days) | 49 (100) | NDa |
| Middle (11 to 20 days) | 28 (100) | ND | |
| Late (21 to 32 days) | 26 (100) | ND | |
| Postfermentation (more than 32 days) | 23 (98) | 2 (8.0) | |
| Unpolished rice vinegar (kurosu)b | Early (1 to 10 days) | 16 (100) | ND |
| Middle (11 to 20 days) | 22 (100) | ND | |
| Late (21 to 32 days) | 12 (100) | ND | |
ND, not detected.
Unpolished rice vinegar production has no postfermentation stage.
REFERENCES
- 1.Both B, Krupp G, Stackebrandt E. Direct sequencing of double-stranded polymerase chain reaction-amplified 16S rDNA. Anal Biochem. 1991;199:216–218. doi: 10.1016/0003-2697(91)90092-8. [DOI] [PubMed] [Google Scholar]
- 2.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 3.Cocconcelli P S, Porro D, Galandini S, Senini L. Development of RAPD protocol for typing of strains of lactic acid bacteria and enterococci. Lett Appl Microbiol. 1995;21:376–379. doi: 10.1111/j.1472-765x.1995.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 4.de Bruijn F J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Ley J, Swings J, Gossele M. Genus I. Acetobacter Beijerinck. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. p. 268. [Google Scholar]
- 6.Dooley J J, Harrison S P, Mytton L R, Dye M, Cresswell A, Skot L, Beeching J R. Phylogenetic grouping and identification of Rhizobium isolates on the basis of random amplified polymorphic DNA profiles. Can J Microbiol. 1993;39:665–673. doi: 10.1139/m93-096. [DOI] [PubMed] [Google Scholar]
- 7.Fleet G H. Microorganisms in food ecosystems. Int J Food Microbiol. 1999;50:101–117. doi: 10.1016/s0168-1605(99)00080-x. [DOI] [PubMed] [Google Scholar]
- 8.Mesbah M, Premachandran U, Whitman W B. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol. 1989;39:159–167. [Google Scholar]
- 9.Nuswantara S, Fujie M, Sukiman H I, Yamashita M, Yamada T, Murooka Y. Phylogeny of bacterial symbionts of the leguminous tree Acacia mangium. J Ferment Bioeng. 1997;84:511–518. [Google Scholar]
- 10.Ohmori S, Uozumi T, Beppu T. Loss of acetic acid resistance and ethanol oxidizing ability in an Acetobacter strain. Agric Biol Chem. 1982;46:381–389. [Google Scholar]
- 11.Sievers M, Wolfgang L, Teuber M. Phylogenetic positioning of Acetobacter, Gluconobacter, Rhodopila and Acidiphilium species as a branch of acidophilic bacteria in the α-subclass of Proteobacteria based on 16S ribosomal DNA sequences. Syst Appl Microbiol. 1994;17:189–196. [Google Scholar]
- 12.Sokollek S J, Hertel C, Hammes W P. Cultivation and preservation of vinegar bacteria. J Biotechnol. 1998;60:195–206. [Google Scholar]
- 13.Sugisawa T, Hoshino T, Masuda S, Nomura S, Setoguchi Y, Tazoe M, Shinjoh M, Someha S, Fujiwara A. Microbial production of 2-keto-l-gulonic acid from l-sorbose and d-sorbitol by Gluconobacter melanogenus. Agric Biol Chem. 1990;54:1201–1209. [Google Scholar]
- 14.Tamaoka J, Komagata K. Determination of DNA base composition by reversed-phase high-performance liquid chromatogrophy. FEMS Microbiol Lett. 1984;25:125–128. [Google Scholar]
- 15.Trcek J, Ramus J, Raspor P. Phenotypic characterization and RAPD-PCR profiling of Acetobacteri sp. isolated from spirit vinegar production. Food Technol Biotechnol. 1997;35:63–67. [Google Scholar]
- 16.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward L J, Timmins M J. Differentiation of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus by polymerase chain reaction. Lett Appl Microbiol. 1999;29:90–92. doi: 10.1046/j.1365-2672.1999.00586.x. [DOI] [PubMed] [Google Scholar]