Abstract
Mucus, a biopolymer hydrogel that covers all wet epithelia of the body, is a potential site for infection by pathogenic bacteria. Mucus can bind small molecules and influence bacterial physiology, two factors that may affect the efficacy of antibiotics. In spite of this, the impact of mucus on antibiotic activity has not been thoroughly characterized. We examined the activity of polymyxin and fluoroquinolone antibiotics against the opportunistic pathogen Pseudomonas aeruginosa in native mucus and purified mucin biopolymer environments. We found that mucus reduces the effectiveness of polymyxins and fluoroquinolones against P. aeruginosa. Mucin biopolymers MUC5AC, MUC2, and MUC5B are primary contributors to this reduction. Our findings highlight that the biomaterial environmental context should be considered when evaluating antibiotics in vitro.
Keywords: mucus, mucin, antibiotics, efficacy, Pseudomonas aeruginosa
Graphical Abstract
Treatment of antimicrobial-resistant infections is a major public health challenge that makes accurate in vitro evaluation of antibiotic activity critically important. Many studies have investigated how alterations in microbial genomes and transcriptomes reduce the activity of antibiotics.1,2 However, fewer studies have explored the contributions of the microbial environment to antimicrobial activity. Although previous studies have examined the impact of environmental factors such as oxygen availability,3 microbial byproducts,4 negatively charged bacterial polysaccharides,5 and culture media6 on efficacy, we focus on the fact that antibiotics used to treat bacterial infections in the human body must frequently act in the mucus layer.
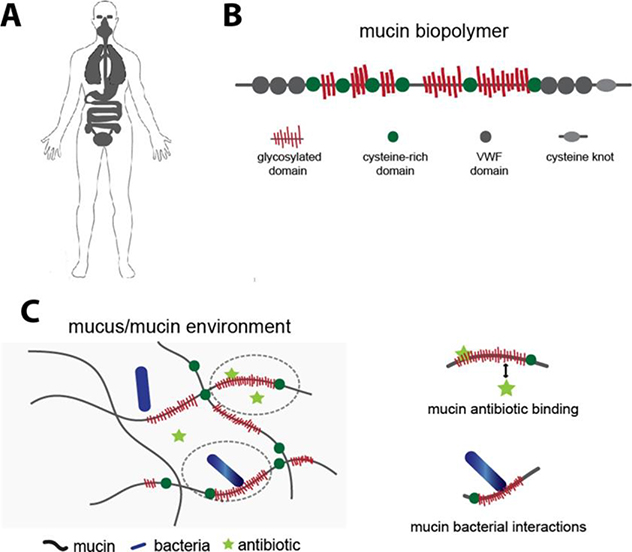
Mucus is a hydrogel that coats and protects all wet epithelia including the eyes and the respiratory, gastrointestinal, and cervicovaginal tracts (Figure 1A). It is a selectively permeable layer that permits the passage of some substrates while restricting others, protecting the underlying epithelial surface. The primary gel-forming components of mucus are high-molecular weight polyanionic glycoprotein polymers called mucins (Figure 1B). Purified mucins exhibit important features of native mucus, including characteristic viscoelastic and selective barrier properties7,8 and specific interactions with mucosal microbes.9,10 Hence, purified mucins may serve as a simplified model environment for the study of mucus. Mucins present an abundance of potential electrostatic and hydrophobic binding sites for small molecules;11,12 mucin-antibiotic binding may reduce the activity of antibiotics by sequestering them. Mucin can also modulate the physiology of bacteria,13–16 which may alter their susceptibility to antibiotics (Figure 1C). These considerations suggest that including mucins in the evaluation of antimicrobial activity may improve the predictability of in vitro analysis of antibiotic function.
Figure 1.
(A) Mucus covers all wet epithelia including the eyes, respiratory tract, gastrointestinal tract, and female reproductive tract, and is one of the primary arenas for microbes in the body. (B) Mucus contains mucins: glycoprotein polymers composed of densely glycosylated domains, hydrophobic cysteine rich domains, von Willebrand factor (VWF)-like domains and cysteine knots, with the number and spatial distribution of each of these domains varying depending on the specific mucin (MUC5AC, MUC2, MUC5B) (C) Mucins may interact with antibiotics or bacteria via electrostatic and/or weak hydrophobic interactions to alter antibiotic effectiveness.
Reduction of antimicrobial activity in sputum17,18 or mucin,19 and antibiotic-mucus/mucin binding19–23 have been observed for a limited number of antibiotics. These previous studies provide an important but incomplete picture of the role played by mucus and mucin on antibiotic efficacy, and that of mucin-antibiotic binding. In order to properly evaluate the relationship between mucin-antibiotic binding and antibiotic activity in mucus, the two must be tested in the same conditions. In the work presented here, we examined the impact of native mucus from mucosal layers across the body on the activity of antibiotics against the model mucosal bacterium Pseudomonas aeruginosa. P. aeruginosa is an opportunistic pathogen that colonizes the respiratory and gastrointestinal mucosa of immune-compromised individuals as well as the mucosa of individuals with diseases such as chronic obstructive pulmonary disease and cystic fibrosis.24 We focused on two clinically relevant antipseudomonal antibiotic classes: fluoroquinolones and polymyxins. After observing that mucus inhibited the efficacy of these antibiotics, we evaluated the ability of purified mucins to mimic the effects of native mucus by comparing antibiotic activity in both environments. Our experimental design enabled direct comparison of native mucus with mucins purified from the same native mucus source. Finally, we explored the mechanism by which mucus influences antibiotic efficacy of polymyxin and fluoroquinolone antibiotics by examining mucin-antibiotic binding in the same experimental conditions.
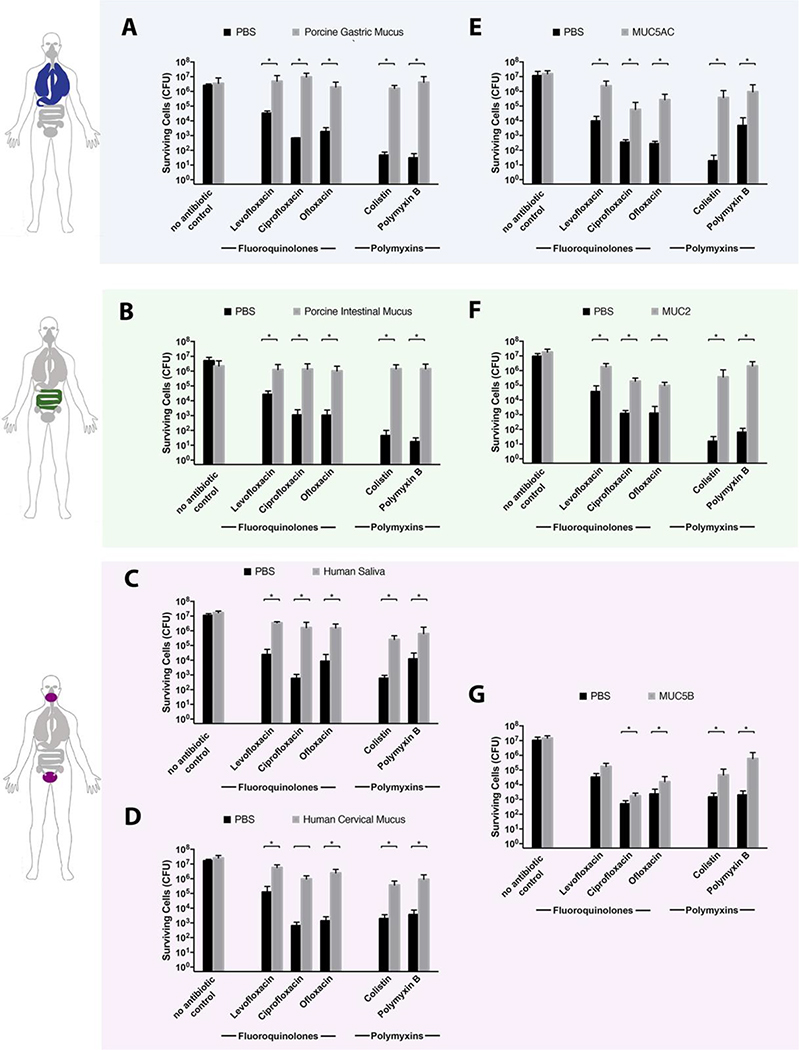
We first evaluated antibiotic activity in native mucus using a modified minimum bactericidal concentration for planktonic bacteria (MBC-P) assay.25 For each antibiotic tested, the MBC-P was determined in phosphate-buffered saline (PBS) after 2h of antibiotic exposure. P. aeruginosa cells were exposed to one-half the MBC-P in phosphate-buffered saline (PBS) alone or in native mucus, and the number of surviving cells was quantified by colony forming units (CFUs) after 2 h. Porcine intestinal and gastric mucus, which are easy to source and routinely employed in studies of drug delivery,26,27 were used to model the human gastrointestinal environment. Native human saliva and cervical mucus were tested in order to measure the effects of these environments on antibiotic activity. All mucus types significantly reduced the effectiveness of polymyxin and fluoroquinolone antibiotics relative to the buffer-only controls (Figure 2A–D). When cells were exposed to polymyxin antibiotics in gastric (Figure 2A) or intestinal (Figure 2B) mucus, we observed at least a 49,000-fold increase in cell survival compared to antibiotic exposure in PBS alone. For fluoroquinolones, gastric (Figure 2A) and intestinal (Figure 2B) mucus provided a 75-fold or greater increase in cells surviving antibiotic exposure versus PBS alone. Native human saliva (Figure 2C) and cervical mucus (Figure 2D) reduced P. aeruginosa killing by all antibiotics tested relative to buffer alone, with at least 50-fold and 180-fold increases in cell survival for polymyxin and fluoroquinolone antibiotics, respectively. For all mucus samples tested, cells incubated for 2 h in mucus alone (Figure 2A–D, no antibiotic control condition) showed no difference in cell counts between the buffer-only and mucus conditions, confirming that the increase in bacterial survival observed with mucus present was due to a reduction in antibiotic activity rather than an increase in bacterial growth stimulated by mucus. These data suggest that the reduction in antibiotic efficacy in its presence may be a general property of mucus, independent of anatomical source.
Figure 2.
Mucus and mucins reduce the efficacy of polymyxin and fluoroquinolone antibiotics against P. aeruginosa. PAO1 was exposed to polymyxin or fluoroquinolone antibiotics (polymyxin B, 8 μg/mL; colistin, 8 μg/mL; ofloxacin, 1 μg/mL; ciprofloxacin, 0.25 μg/mL; or levofloxacin, 0.25 μg/mL) in mucin-free buffer (PBS), and in native mucus from different surfaces in the body: (A) native gastric mucus, (B) native intestinal mucus, (C) saliva, and (D) native cervical mucus. Cells were also exposed to antibiotics in purified mucins (0.5% w/v dissolved in PBS): (E) MUC5AC, the primary mucin present in the human lungs and stomach; (F) MUC2, the primary mucin found in the intestines; and (G) MUC5B, a mucin primarily found in saliva and the cervix. Cells were exposed to antibiotics for 2 h at 37 °C and surviving cells were quantified by serial dilution and plating. All mucus and mucin samples increased the number of surviving cells. No antibiotic control condition for each mucus and mucin type demonstrated that the presence of mucus or mucin does not lead to a substantial increase in PAO1 growth. All error bars represent standard deviation of biological replicates (n ≥ 3). (*) indicate a significant increase in the number of surviving cells after antibiotic exposure in mucus/mucin compared to that in mucus/mucin-free buffer (PBS), as determined by the t test (P < 0.05).
We hypothesized that the increased survival of bacteria after antibiotic exposure in mucus environments may be due to a component of mucus common across all these niches: mucin proteins. We tested whether three distinct mucins, MUC2, MUC5AC, and MUC5B, also reduced the activity of polymyxins and fluoroquinolones against P. aeruginosa, as observed in native mucus samples. MUC2 is the predominant mucin in the intestines and thus may serve as a simplified model of the human intestinal mucus environment.28 MUC5AC is predominant in the stomach and lungs;28 although MUC5B is the primary mucin found in the oral cavity and female reproductive tract, it is also found in the lungs.28,29 MUC2 and MUC5AC were first purified from native porcine intestinal and gastric mucus samples, which were used to evaluate antibiotic efficacy. Similarly, MUC5B was purified from human saliva. Purification of mucins directly from these sources—rather than using commercially available mucins—enabled meaningful comparisons with the results from native mucus. Additionally, it has been reported that compared to commercially available mucins, proteins purified in this manner better recapitulate the properties of native mucus.30
Purified MUC5AC (Figure 2E), MUC2 (Figure 2F), and MUC5B (Figure 2G) reduced the efficacy of all antibiotics tested relative to mucus-free buffer controls. When cells were exposed to polymyxin antibiotics in MUC5AC (Figure 2E) or MUC2 (Figure 2F), we observed a 10,000-fold or greater increase in cell survival compared to antibiotic exposure in PBS alone and a 140-fold increase for fluoroquinolone antibiotics (Figure 2E,F). In MUC5B, we observed a 30-fold or greater increase in cells surviving polymyxin antibiotic exposure (Figure 2G) versus buffer. For fluoroquinolones in MUC5B, we measured 30-fold, 4-fold, and 12-fold increases in cells surviving exposure to ofloxacin, ciprofloxacin, or levofloxacin, respectively (Figure 2G). Cell counts after incubation for 2 h without antibiotics in buffer or mucin solution were not substantially different (Figure 2E–G, no antibiotic control condition), demonstrating that the presence of mucins does not increase cell growth but reduces antibiotic activity. Our data suggest that mucins are primary, though not necessarily singular, contributors to the reduction in antibiotic efficacy observed in mucus. These data also highlight the potential for purified mucins as a three-dimensional model biomaterial for more accurate evaluation of antibiotic activity.
We were further interested in understanding the mechanism by which mucins and mucus affect antibiotic efficacy. Methylcellulose, a polymer often used as a mucin mimetic because of similarities in its viscoelastic properties,31 did not impact antibiotic activity to the same degree for all antibiotics tested (Figure 3A), suggesting that the mucin-mediated reduction in antibiotic efficacy is not due to macromolecular crowding. Alternatively, we hypothesized that mucin–antibiotic binding could reduce the free concentration of antibiotic available to kill bacterial cells. We performed equilibrium dialysis to evaluate whether mucins were binding antibiotics. We prepared two chambers separated by a 12 kDa molecular weight cutoff membrane that permitted the passage of antibiotics, but not mucins (Figure 3B). One chamber contained mucins (0.5% w/v dissolved in PBS) and the other PBS; the initial concentration of antibiotic was equal in both. The system equilibrated for 4 h before the final concentration of free antibiotic in the buffer chamber was quantified by mass spectrometry. Buffer composition and mucin concentration were maintained between the antibiotic efficacy experiments and these assays to ensure consistency in ionic strength, pH, and the number of potential mucin binding sites, which are experimental conditions expected to affect binding. It was expected that if an antibiotic bound to mucin, the relative concentration of antibiotic would increase in the mucin chamber and decrease in the buffer chamber. This was quantified by calculating the uptake ratio, defined as the mucin chamber:-buffer chamber antibiotic concentration ratio. Uptake ratios greater than one indicate binding. For MUC5AC and MUC2, we found that the average uptake ratios for colistin and polymyxin were greater than or equal to 2.4, and for MUC5B, 1.4 or more. These data suggest that polymyxin antibiotics bind mucins in the conditions used to evaluate antibiotic efficacy (Figure 3C), with stronger binding to MUC5AC and MUC2 compared with MUC5B. This binding is likely electrostatic in nature (cationic polymyxins and polyanionic mucins) and may account for the reduction in polymyxin activity in mucus and mucins (Figure 2).
Figure 3.
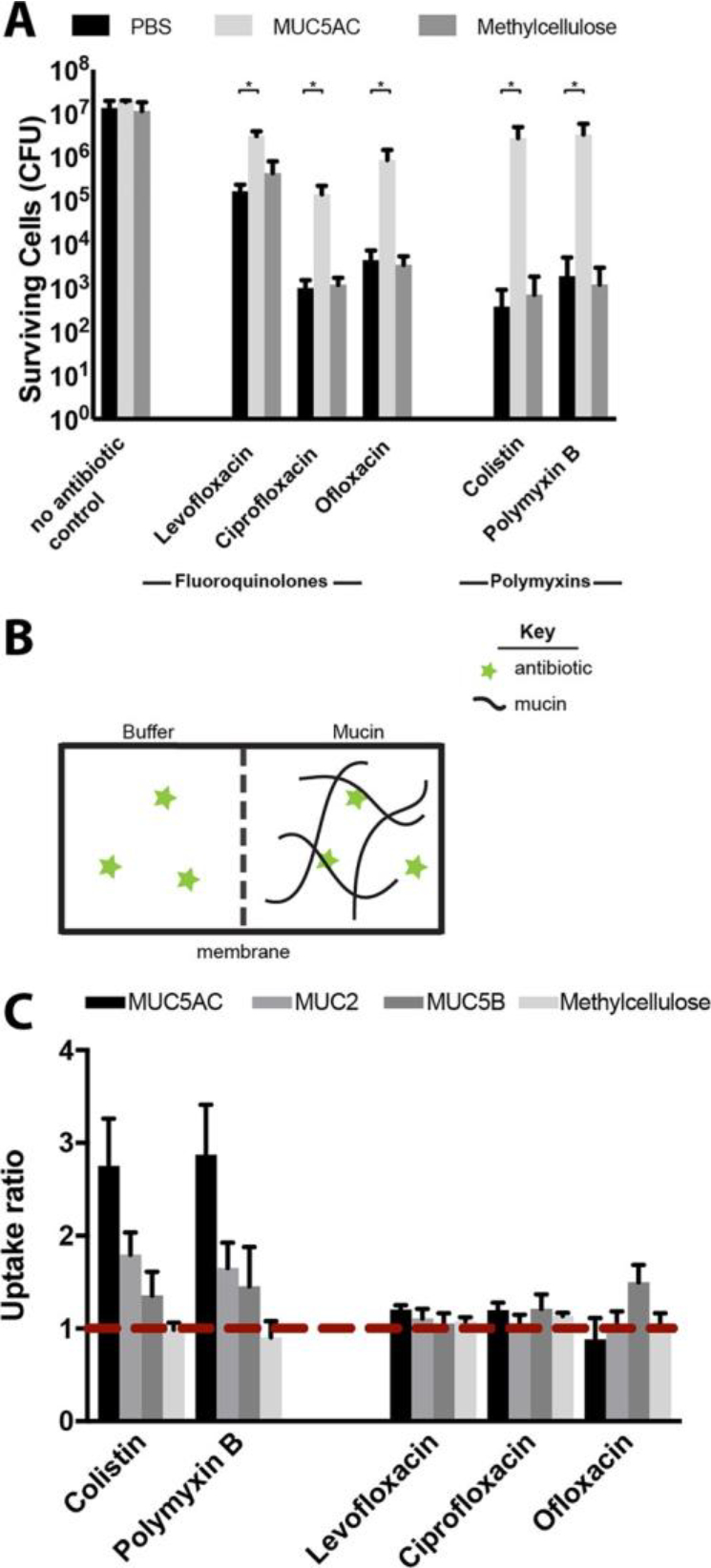
Evaluating the mechanism of mucus/mucin reduction of antibiotic efficacy. Although MUC5AC (0.5% w/v) protected bacterial cells from killing compared to buffer alone, (A) methylcellulose (0.5% w/v) did not, suggesting that neither mechanical cues nor macromolecular crowding are responsible for the protection observed. All error bars represent standard deviation of biological replicates (n ≥ 3). (*) indicates a significant increase in the number of surviving cells after antibiotic exposure in mucus compared to that in mucin-free buffer (PBS), as determined by the t test (P < 0.01) (B) Schematic illustration of experimental set up for equilibrium dialysis experiments to evaluate polymyxin and fluoroquinolone binding to mucin (C) Uptake ratio from equilibrium dialysis for each antibiotic with MUC5AC, MUC2, MUC5B and methylcellulose, in the conditions used to evaluate antibiotic efficacy. Ratios greater than 1 (red dotted line) indicate binding. Colistin and polymyxin B both bind to MUC5AC, MUC2, and MUC5B but not to methylcellulose. Uptake ratios of fluoroquinolone antibiotics to biopolymers are close to 1, consistent with weak or no binding. Error bars represent standard error of biological replicates (n ≥ 3) (see Methods for standard error calculation).
Despite the reduction in activity observed for fluoroquinolone antibiotics in mucus and mucins (Figure 2), we found that the uptake ratios for levofloxacin, ciprofloxacin, and ofloxacin were close to 1, suggesting weak or no binding of fluoroquinolones to mucins in the conditions used to evaluate antimicrobial efficacy (Figure 3C). This is consistent with the fact that fluoroquinolones, with no net charge and moderate hydrophobicity, are unlikely to bind to mucin through electrostatic or hydrophobic means. These results suggest that mechanisms other than mucin–antibiotic binding contribute to the reduction in fluoroquinolone activity observed in mucus/mucins. To pursue this hypothesis, we performed a modified antibiotic efficacy assay in which we exposed P. aeruginosa to antibiotics in a system with two connected chambers separated by a 12 kDa MWCO membrane that allowed the selective diffusion of only antibiotics between them. As in the equilibrium dialysis experiments, we first added mucin, buffer, and antibiotic to the chambers and allowed the system to equilibrate for 4 h before adding the bacteria. Cells were exposed to antibiotic for 2 h and then quantified by serial dilution and CFU counting (Figure S1).
This modified assay enabled us to compare antibiotic efficacy in two conditions with mucins: one in which cells and mucin were in the same chamber (Figure 4ii), and one in which cells were separated from mucins by a membrane (Figure 4iii). Figure 4 shows that exposure to ciprofloxacin with mucin and cells in the same chamber (Figure 4ii) significantly increased bacterial survival relative to a mucus-free buffer control (Figure 4i), consistent with our initial efficacy experiments (Figure 2). However, when cells were separated from mucin by a membrane and exposed to ciprofloxacin, no such increase was observed (Figure 4iii). For a mucin-binding antibiotic, we would expect to see a reduction in antibiotic activity even when mucins are separate from the cells, because mucin–antibiotic binding would reduce the free antibiotic concentration. Indeed, for colistin, which binds mucin strongly, this is exactly what was observed (Figure S2). These data support our hypothesis that for fluoroquinolone antibiotics such as ciprofloxacin, mucin may be mediating the reduction in antibiotic efficacy via a mechanism other than mucin–antibiotic binding. We hypothesize that possible mechanisms include mucin modulation of bacterial physiology or mucin reduction of antibiotic uptake by the cells; however, further studies will be required to elucidate the exact mechanism(s).
Figure 4.
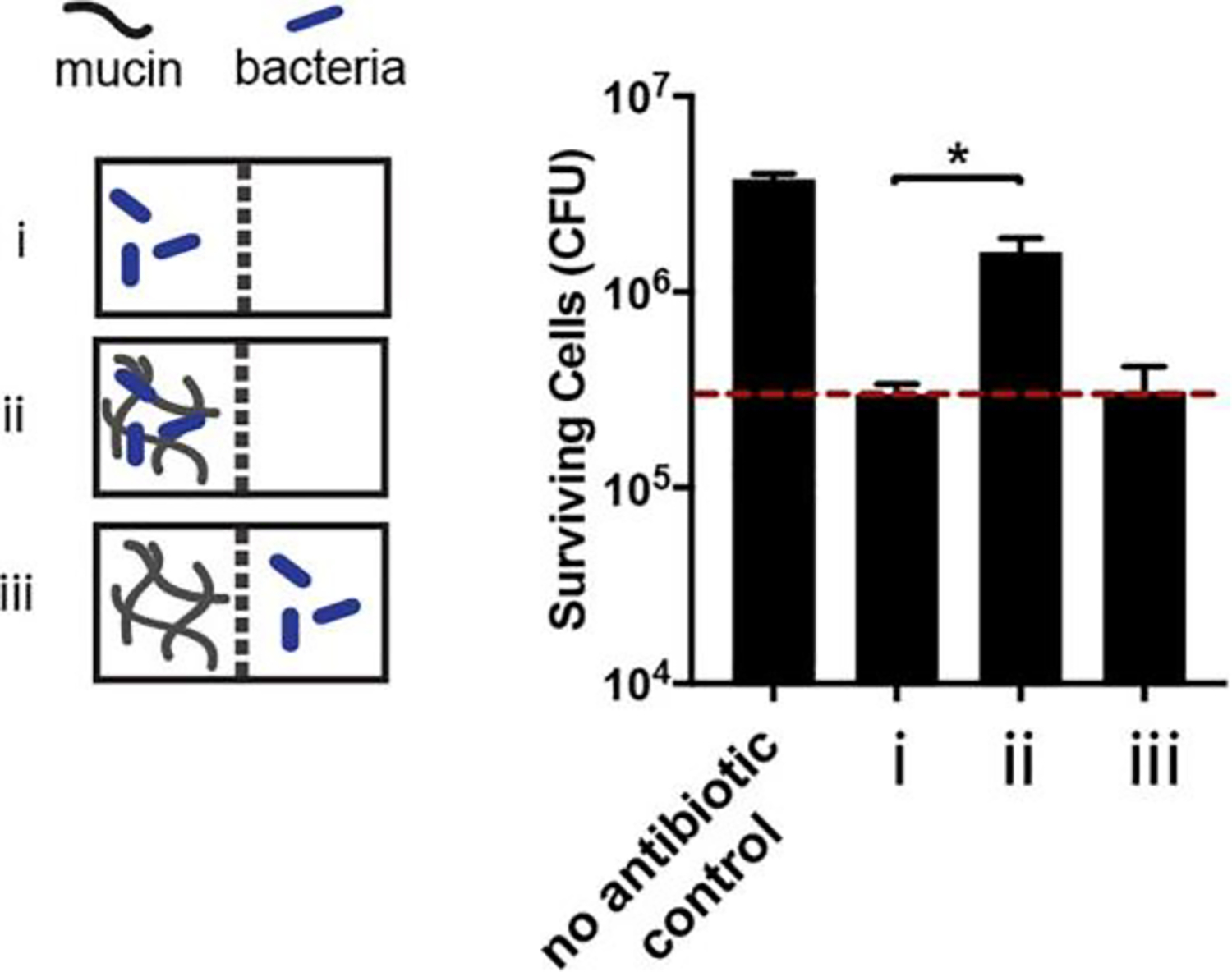
Two chambers were separated by a 12 kDa MWCO membrane that allowed for the free diffusion of antibiotics, but not mucin or cells, between the chambers. As in the equilibrium dialysis experiments, mucin, buffer, and antibiotic were added to the chambers (Figure S1) and the system was left to equilibrate for 4 h before adding P. aeruginosa cells. Bacteria were exposed to ciprofloxacin for 2 h in three different conditions: (i) buffer with no mucin, (ii) mucin, and (iii) buffer with mucin in the connected chamber, separated from the cells by the membrane. In condition iii, mucin was not in direct contact with the bacteria, but could still interact with the antibiotic. Ciprofloxacin efficacy was reduced relative to the mucin-free control in condition ii, recapitulating the result from Figure 2 in this modified system. However, in condition iii (bacteria were separated from mucin by a membrane and exposed to ciprofloxacin in buffer), no increase in surviving cells was observed. This suggests that mucin is not able to bind and reduce the antibiotic concentration of ciprofloxacin and that a mechanism other than mucin-antibiotic binding accounts for the observed reduction in ciprofloxacin efficacy. Dotted red line marks the number of surviving cells in the mucin free control (i). All error bars represent standard deviation of biological replicates (n ≥ 3). (*) indicates a significant increase in the number of surviving cells after antibiotic exposure compared to that in mucin-free buffer, as determined by the t test (P < 0.0001).
CONCLUSION
We examined the impact of native mucus from surfaces throughout the body on the efficacy of antibiotics against P.aeruginosa, a formidable opportunistic pathogen. Our data show that mucus substantially diminishes the activity of polymyxin and fluoroquinolone antibiotics against P. aeruginosa. Mucin biopolymers, the gel-forming components of mucus, are primary contributors to this effect and are strong candidates for the construction of a model environment that would enable more accurate in vitroantimicrobial evaluation. We determined that antibiotic binding by mucin likely plays a role in the reduced effectiveness of polymyxin antibiotics, but that mucin may reduce the activity of fluoroquinolones via alternative mechanisms. While this work focused on two relevant classes of antibiotics and P. aeruginosa, mucus and mucins are likely to impact other antibiotic classes and other mucosal microbes. Our findings highlight the importance of considering the biomaterial environment for in vitro evaluation of antibiotics, particularly for mucosal pathogens. Understanding the mechanism(s) by which mucus reduces antibiotic activity is important for devising strategies to maximize therapeutic effect. Thinking broadly, this work highlights important considerations for the future design of antimicrobial releasing polymers and materials, as our data suggest that researchers should consider not only the ability of these polymers and materials to bind and release antimicrobials but also whether interactions between bacteria and the polymer/material may reduce the efficacy of the antimicrobial.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nicole Billings, Erica Shapiro Frenkel, Wesley Chen, Gerardo Cárcamo-Oyarce, and Caroline Wagner for helpful comments on the manuscript.
Funding
This work was supported by the National Institutes of Health under Award NIH R01-EB017755, the National Science Foundation under award NSF Career PHY-1454673, and the MRSEC Program of the National Science Foundation under award DMR-14-19807. J.W. and T.S. were supported in part by the National Science Foundation Graduate Research Fellowship under Grant 1122374. T.S was supported by the Siebel Scholarship and the MIT Collamore-Rogers Fellowship. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsbiomaterials.8b01054.
Materials and Methods and Supplemental Figures and Tables (PDF)
REFERENCES
- (1).Kapoor G; Saigal S; Elongavan A Action and Resistance Mechanisms of Antibiotics: A Guide for Clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33 (3), 300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Breidenstein EBM; de la Fuente-Núñez C; Hancock REW Pseudomonas Aeruginosa: All Roads Lead to Resistance. Trends Microbiol. 2011, 19 (8), 419–426. [DOI] [PubMed] [Google Scholar]
- (3).Schaible B; Taylor CT; Schaffer K Hypoxia Increases Antibiotic Resistance in Pseudomonas Aeruginosa through Altering the Composition of Multidrug Efflux Pumps. Antimicrob. Agents Chemother. 2012, 56 (4), 2114–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Radlinski L; Rowe SE; Kartchner LB; Maile R; Cairns BA; Vitko NP; Gode CJ; Lachiewicz AM; Wolfgang MC; Conlon BP Pseudomonas Aeruginosa Exoproducts Determine Antibiotic Efficacy against Staphylococcus Aureus. PLoS Biol. 2017, 15 (11), No. e2003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Tseng BS; Zhang W; Harrison JJ; Quach TP; Song JL; Penterman J; Singh PK; Chopp DL; Packman AI; Parsek MR The Extracellular Matrix Protects Pseudomonas Aeruginosa Biofilms by Limiting the Penetration of Tobramycin. Environ. Microbiol. 2013, 15 (10), 2865–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Crabbé A; Liu Y; Matthijs N; Rigole P; Fuente-Nùñez CDL; Davis R; Ledesma MA; Sarker S; Houdt RV; Hancock REW; et al. Antimicrobial Efficacy against Pseudomonas Aeruginosa Biofilm Formation in a Three-Dimensional Lung Epithelial Model and the Influence of Fetal Bovine Serum. Sci. Rep. 2017, 7, 43321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Celli J; Gregor B; Turner B; Afdhal NH; Bansil R; Erramilli S Viscoelastic Properties and Dynamics of Porcine Gastric Mucin. Biomacromolecules 2005, 6 (3), 1329–1333. [DOI] [PubMed] [Google Scholar]
- (8).Li LD; Crouzier T; Sarkar A; Dunphy L; Han J; Ribbeck K Spatial Configuration and Composition of Charge Modulates Transport into a Mucin Hydrogel Barrier. Biophys. J. 2013, 105 (6), 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Celli JP; Turner BS; Afdhal NH; Keates S; Ghiran I; Kelly CP; Ewoldt RH; McKinley GH; So P; Erramilli S; et al. Helicobacter Pylori Moves through Mucus by Reducing Mucin Viscoelasticity. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (34), 14321–14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Caldara M; Friedlander RS; Kavanaugh NL; Aizenberg J; Foster KR; Ribbeck K Mucin Biopolymers Prevent Bacterial Aggregation by Retaining Cells in the Free-Swimming State. Curr. Biol. 2012, 22 (24), 2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Bansil R; Turner BS Mucin Structure, Aggregation, Physiological Functions and Biomedical Applications. Curr. Opin. Colloid Interface Sci. 2006, 11 (2–3), 164–170. [Google Scholar]
- (12).Witten J; Samad T; Ribbeck K Selective Permeability of Mucus Barriers. Curr. Opin. Biotechnol. 2018, 52, 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Caldara M; Friedlander RS; Kavanaugh NL; Aizenberg J; Foster KR; Ribbeck K Mucin Biopolymers Prevent Bacterial Aggregation by Retaining Cells in the Free-Swimming State. Curr. Biol. 2012, 22 (24), 2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Frenkel ES; Ribbeck K Salivary Mucins Protect Surfaces from Colonization by Cariogenic Bacteria. Appl. Environ. Microbiol. 2015, 81, 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kavanaugh NL; Zhang AQ; Nobile CJ; Johnson AD; Ribbeck K Mucins Suppress Virulence Traits of Candida Albicans. mBio 2014, 5 (6), e01911–e01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Frenkel ES; Ribbeck K Salivary Mucins Promote the Coexistence of Competing Oral Bacterial Species. ISME J. 2017, 11 (5), 1286–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Mendelman PM; Smith AL; Levy J; Weber A; Ramsey B; Davis RL Aminoglycoside Penetration, Inactivation, and Efficacy in Cystic Fibrosis Sputum. Am. Rev. Respir. Dis. 1985, 132 (4), 761–765. [DOI] [PubMed] [Google Scholar]
- (18).King P; Lomovskaya O; Griffith DC; Burns JL; Dudley MN In Vitro Pharmacodynamics of Levofloxacin and Other Aerosolized Antibiotics under Multiple Conditions Relevant to Chronic Pulmonary Infection in Cystic Fibrosis. Antimicrob. Agents Chemother. 2010, 54 (1), 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Huang JX; Blaskovich MAT; Pelingon R; Ramu S; Kavanagh A; Elliott AG; Butler MS; Montgomery AB; Cooper MA Mucin Binding Reduces Colistin Antimicrobial Activity. Antimicrob. Agents Chemother. 2015, 59, 5925–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Bhat PG; Flanagan DR; Donovan MD The Limiting Role of Mucus in Drug Absorption: Drug Permeation through Mucus Solution. Int. J. Pharm. 1995, 126 (1–2), 179–187. [Google Scholar]
- (21).Niibuchi J-J; Aramaki Y; Tsuchiya S Binding of Antibiotics to Rat Intestinal Mucin. Int. J. Pharm. 1986, 30 (2), 181–187. [Google Scholar]
- (22).Ramphal R; Lhermitte M; Filliat M; Roussel P The Binding of Anti-Pseudomonal Antibiotics to Macromolecules from Cystic Fibrosis Sputum. J. Antimicrob. Chemother. 1988, 22 (4), 483–490. [DOI] [PubMed] [Google Scholar]
- (23).Hunt BE; Weber A; Berger A; Ramsey B; Smith AL Macromolecular Mechanisms of Sputum Inhibition of Tobramycin Activity. Antimicrob. Agents Chemother. 1995, 39 (1), 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Klockgether J; Tümmler B Recent Advances in Understanding Pseudomonas aeruginosa as a Pathogen. F1000Research 2017, 6, 1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mah T-F; Pitts B; Pellock B; Walker GC; Stewart PS; O’Toole GA A Genetic Basis for Pseudomonas Aeruginosa Biofilm Antibiotic Resistance. Nature 2003, 426 (6964), 306–310. [DOI] [PubMed] [Google Scholar]
- (26).Crater JS; Carrier RL Barrier Properties of Gastrointestinal Mucus to Nanoparticle Transport. Macromol. Biosci. 2010, 10 (12), 1473–1483. [DOI] [PubMed] [Google Scholar]
- (27).Boegh M; Nielsen HM Mucus as a Barrier to Drug Delivery – Understanding and Mimicking the Barrier Properties. Basic Clin. Pharmacol. Toxicol. 2015, 116 (3), 179–186. [DOI] [PubMed] [Google Scholar]
- (28).McGuckin MA; Thornton DJ; Whitsett JA Mucins and Mucus. In Mucosal Immunology, fourth ed.; Mestecky J, Strober W, Russell MW, Kelsall BL, Cheroutre H, Lambrecht BN, Eds.; Academic Press: Boston, 2015; Chapter 14, pp 231–250. [Google Scholar]
- (29).Andersch-Björkman Y; Thomsson KA; Larsson JMH; Ekerhovd E; Hansson GC Large Scale Identification of Proteins, Mucins, and Their O-Glycosylation in the Endocervical Mucus during the Menstrual Cycle. Mol. Cell. Proteomics 2007, 6 (4), 708–716. [DOI] [PubMed] [Google Scholar]
- (30).Celli JP; Turner BS; Afdhal NH; Ewoldt RH; McKinley GH; Bansil R; Erramilli S Rheology of Gastric Mucin Exhibits a pH-Dependent Sol-Gel Transition. Biomacromolecules 2007, 8 (5), 1580–1586. [DOI] [PubMed] [Google Scholar]
- (31).Ivic A; Onyeaka H; Girling A; Brewis IA; Ola B; Hammadieh N; Papaioannou S; Barratt CLR Critical Evaluation of Methylcellulose as an Alternative Medium in Sperm Migration Tests. Hum. Reprod. 2002, 17 (1), 143–149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.