Abstract
The synthesis of polyglutamic acid (PGA) was repressed by exogenous glutamate in strains of Bacillus licheniformis but not in strains of Bacillus subtilis, indicating a clear difference in the regulation of synthesis of capsular slime in these two species. Although extracellular γ-glutamyltranspeptidase (GGT) activity was always present in PGA-producing cultures of B. licheniformis under various growth conditions, there was no correlation between the quantity of PGA and enzyme activity. Moreover, the synthesis of PGA in the absence of detectable GGT activity in B. subtilis S317 indicated that this enzyme was not involved in PGA biosynthesis in this bacterium. Glutamate repression of PGA biosynthesis may offer a simple means of preventing unwanted slime production in industrial fermentations using B. licheniformis.
Strains of Bacillus licheniformis and Bacillus subtilis may synthesize a water-soluble, viscous slime material containing d- and l-glutamic acid residues. This polyglutamic acid (PGA) is polymerized via amide linkages between the α-amino and γ-carboxylic groups of the amino acid residues. PGA is the principal component of “Itohiki-natto,” a traditional Japanese food prepared from steamed soybean by the biological action of PGA-producing strains of B. subtilis, which are generally referred to as “Bacillus natto” or B. subtilis (natto) (17). PGA has other biotechnological applications in cosmetics, medicines, and foods. However, the synthesis of even small amounts of PGA can be a problem in the fermentation industry, most notably in the production of extracellular enzymes from bacilli, where PGA accumulation causes increased viscosity of the fermentation broth, reduced enzyme yield, uncontrollable foaming, and complications in product recovery. The unpredictable nature of PGA synthesis is particularly troublesome.
Cultural conditions affecting PGA biosynthesis have been studied using various poorly identified strains of B. licheniformis and B. subtilis in complex, ill-defined media, and these studies have resulted in conflicting conclusions. In general, strains have been classified into two categories: (i) those that require exogenous l-glutamate for PGA synthesis, for example, B. subtilis strains IFO 3335 (6) and F-2-01 and B. licheniformis 9945 (16) (although Birrer et al. reported that strain 9945 does not require exogenous glutamate for PGA synthesis [2]), and (ii) strains that do not require exogenous glutamate (often described as de novo synthesis), such as B. licheniformis A35 (3) and B. subtilis TAM-4 (10).
The mechanisms of PGA biosynthesis in B. licheniformis and B. subtilis have been elusive until recently. A membranous synthetase complex from B. licheniformis which catalyzes the activation, racemization, and polymerization of l-glutamate into exclusively poly-d-glutamate has been partially characterized (5), but B. subtilis (natto) has been studied more extensively in this context. A gene originally thought to code for γ-glutamyltranspeptidase (GGT) (8) but later referred to as a PGA “stimulating factor” (7) has been cloned and sequenced, although its role in PGA synthesis is unknown. More recently, an unrelated gene coding for an extracellular GGT has been cloned and sequenced from B. subtilis (natto) (12) which is essentially identical to a ggt gene previously cloned from B. subtilis (18). However, the recent cloning and characterization of three genes, pgsA, -B, and -C, that are homologs of the Bacillus anthracis cap genes (1) casts doubt on the relevance of the ggt genes and GGT in PGA synthesis.
In this study we show that PGA synthesis is repressed by exogenous l-glutamate in strains of B. licheniformis but not in B. subtilis strains. Moreover, a role for extracellular GGT in PGA production has been shown to be unlikely through the poor correlation between enzyme activity and PGA yield and the finding that B. subtilis S317 synthesizes large amounts of PGA in the absence of detectable GGT activity.
Analytical procedures.
B. licheniformis and B. subtilis strains (Table 1) were grown in basal salts comprising (per liter) glucose (20 g), K2HPO4 (14 g), KH2PO4 (6 g), MgSO4 · 7H2O (0.2 g), and 1 ml of trace element solution (FeSO4 · 7H2O, CaCl2 · 2H2O, MnSO4 · 4H2O, ZnCl2; 1 mM each) at 37°C. Nitrogen sources were added as described in Tables 1 and 2. Medium (20 ml) in 100-ml flasks was inoculated with 100 μl of a frozen spore suspension in 20% glycerol and was shaken at 250 rpm. All data are average results from three separate flask cultures with less than 12% variation. PGA was determined in culture supernatants after clarifying cultures by centrifugation at room temperature. The supernatant was dialyzed against water for 24 h at 4°C and hydrolyzed with an equal volume of 6 M HCl at 100°C overnight. The hydrolysate was neutralized with 6 M NaOH, and the quantity of glutamic acid in the solution was determined using ninhydrin with glutamic acid as a standard. Glutamate was confirmed as the sole product in the hydrolysate by thin-layer chromatography on Silica Gel-60 plates (Merck) using n-butanol–acetic acid–water (12:3:5). The plates were dried and sprayed with acetone containing 0.2% ninhydrin and 2% collidine to visualize the amino acids (data not shown). The amount of PGA is given as glutamic acid equivalent to the difference between the hydrolyzed and unhydrolyzed samples. The degree of polymerization of PGA was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The polymer was purified from culture supernatants using ethanol precipitation and deoxyribonuclease I treatment (10) before SDS-PAGE in a 10% gel. The protein standards (ovalbumin, 45 kDa; serum albumin, 66.2 kDa; phosphorylase b, 97 kDa) were stained with Coomassie brilliant blue and, after destaining in 7% acetic acid-10% methanol, the gel was stained for PGA with 0.5% methylene blue in 3% acetic acid and destained in water.
TABLE 1.
PGA synthesis and GGT activity in cultures of some strains of B. licheniformis and B. subtilisa
| Strainb | PGA (μg/ml) in mediumc:
|
GGT (U/ml) in mediumc:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | |
| B. licheniformis | ||||||||
| DSMZ 13T | 0 | 0 | 450 | 0 | 0 | 0 | 2.6 | 0.9 |
| NCIMB 9375T | 540 | 0 | 620 | 0 | 1.7 | 6.4 | 30.0 | 5.6 |
| ATCC 9945a (A) | 0 | 0 | 260 | 0 | 0 | 0 | 4.9 | 7.9 |
| ATCC 9945a (B) | 510 | 0 | 420 | 60 | 3.8 | 0 | 3.8 | 3.4 |
| NCIMB 6346 | 370 | 0 | 790 | 0 | 2.2 | 2.8 | 3.2 | 5.4 |
| NCTC 8223 | 530 | 0 | 420 | 0 | 2.5 | 0 | 3.5 | 1.5 |
| S170 | 0 | 0 | 410 | 170 | 0 | 2.6 | 6.5 | 3.2 |
| S173 | 260 | 0 | 390 | 90 | 3.6 | 17.3 | 3.9 | 4.4 |
| S178 | 0 | 0 | 1,470 | 0 | 0 | 0 | 5.2 | 0.7 |
| B. subtilis | ||||||||
| NCIMB 3610T | 0 | 360 | 300 | 300 | 0 | 1.1 | 1.0 | 3.0 |
| NRS 264 | 0 | 1,900 | 0 | 0 | 0 | 1.7 | 3.2 | 1.1 |
| S031 | 0 | 120 | 0 | 0 | 0 | 0.3 | 0 | 0 |
| S317 | 0 | 0 | 390 | 280 | 0 | 0 | 0 | 0 |
| NRRL B-1490 | 0 | 0 | 0 | 0 | 0 | 0 | 3.0 | 0.2 |
| 1E2 (pLS11) | 0 | 0 | 20 | 0 | 0 | 0 | 2.4 | 1.6 |
| 1E3 (pLS12) | 0 | 340 | 220 | 340 | 0 | 0.6 | 6.2 | 11.3 |
PGA and GGT activities are normalized to an OD660 of 1.0. The limit of PGA detection was 20 μg/ml and that of GGT activity was 0.3 U/ml.
Strains were from our culture collection and have been described previously (13) with the exception of strains 1E2 and 1E3 from the Bacillus Genetic Stock Center (Ohio State University). DSMZ 13T and NCIMB 9375T are independent cultures of the type strain and two strains of ATCC 9945a were used (see text for details).
Cultures were grown in basal salts medium containing the following (per liter): medium 1, (NH4)2SO4 (4 g); medium 2, sodium glutamate (4 g); medium 3, casein hydrolysate (4 g); medium 4, casein hydrolysate (4 g) plus sodium glutamate (4 g) for B. licheniformis strains or (NH4)2SO4 (4 g) for B. subtilis strains.
TABLE 2.
Effect of different nitrogen sources on PGA synthesis and GGT activity in cultures of B. licheniformis S173
| Nitrogen source (4 g/liter) | Growth (OD660) | PGA (μg/ml) | GGT (U/ml) |
|---|---|---|---|
| NH4Cl | 3.7 | 1,270 | 13.4 |
| (NH4)2SO4 | 3.7 | 1,060 | 10.9 |
| NH4NO3 | 4.8 | 1,180 | 7.9 |
| NaNO3 | 1.0 | 50 | 3.1 |
| Arginine | 2.8 | 790 | 22.4 |
| Aspartate | 3.0 | 580 | 35.3 |
| Glutamine | 2.9 | 910 | 10.8 |
| Glutamate | 2.5 | 0 | 12.1 |
| (NH4)2SO4 + glutamate | 2.7 | 0 | 0 |
| Glutamine + glutamate | 4.3 | 370 | 13.3 |
GGT activity was negligible in cell extracts prepared by lysis through a French press (data not shown) and was therefore measured only in culture supernatants prepared by centrifugation at room temperature. The enzyme was assayed using γ-glutamyl-p-nitroanilide as substrate (18). One unit of activity is the amount of enzyme which liberated 1 mM p-nitroaniline (detected as A410)/min/ml of culture supernatant at 37°C. All data are averages from at least two replicate determinations.
Effect of glutamate on PGA biosynthesis in B. licheniformis and B. subtilis.
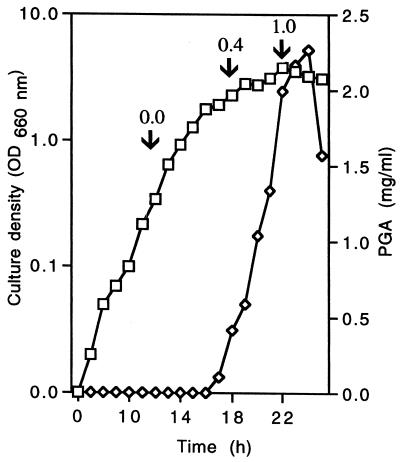
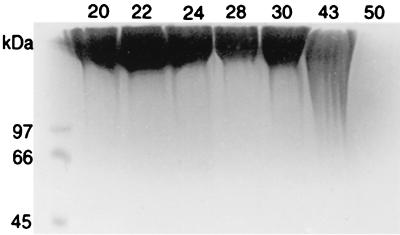
The effect of exogenous l-glutamate on PGA accumulation and GGT activity in various strains of B. licheniformis and B. subtilis is shown in Table 1. The typical culture density (optical density at 650 mm [OD650]) after incubation for 24 h was between 2.4 and 3.5 with (NH4)2SO4 as the nitrogen source, between 1.0 and 3.5 with glutamate as the nitrogen source, and between 2.0 and 4.0 with casein hydrolysate. PGA accumulation was determined at the beginning of stationary phase, when the yield was likely to be maximal (Fig. 1). This is a considerably shorter incubation period than that used by others, which varied between 48 and 96 h (2, 3, 5), but this probably reflects the simplicity and low concentrations of medium components used here. Although depolymerization of PGA ensues as the bacterium enters stationary phase, this is a slow process (2, 4, 6), and electrophoresis of precipitated PGA showed that there was no substantial degradation until incubation beyond 30 h (Fig. 2).
FIG. 1.
Growth of B. licheniformis S173 and PGA biosynthesis in medium containing casein hydrolysate as the nitrogen source. Symbols: □, OD660; ◊, PGA accumulation. At the times indicated by the arrows, the cultures were equally divided into two flasks and sodium glutamate was added to 4 g/liter to one of the flasks. Incubation of both flasks continued for 26 h, and the final yield of PGA after incubation in the presence of sodium glutamate is given in milligrams per milliliter above the arrows.
FIG. 2.
Molecular size of extracellular PGA from B. licheniformis S 173 as a function of culture age (numbers above lanes represent hours since inoculation). PGA was precipitated from culture supernatant and examined by SDS-PAGE (see text for details).
B. licheniformis strains varied in PGA synthesis when supplied with (NH4)2SO4 alone as the nitrogen source, but all strains synthesized PGA when casein hydrolysate was provided as the nitrogen source. Glutamate invariably repressed PGA synthesis in B. licheniformis strains growing with casein hydrolysate or (NH4)2SO4. The same organism from different sources (DSMZ 13T and NCIMB 9375T are independent cultures of the type strains) and the same organism stored under different conditions (ATCC 9945a [A, B]) showed greatly different levels of PGA synthesis when grown with (NH4)2SO4 as the nitrogen source. In both cases, the productive strains had been lyophilized on receipt and were not used in the laboratory for some 15 years, while the nonproductive strains had been subcultured routinely. This supports the view that strain “degeneration” occurs upon repeated subculture on solid media (2, 16). We can therefore categorize B. licheniformis strains as capable of de novo synthesis of PGA in the absence of exogenous glutamate and as susceptible to glutamate inhibition of PGA synthesis.
In contrast, the B. subtilis strains produced PGA only when growing on glutamate or casein hydrolysate (Table 1). Given the lack of synthesis when (NH4)2SO4 was used as the nitrogen source, we examined the effects of adding (NH4)2SO4 to cultures growing on casein hydrolysate to see if it repressed PGA production, but it had little effect. B. subtilis 1E2 and 1E3 contain plasmids which have been implicated in PGA production (9), but unlike typical B. subtilis (natto) strains, they do not require biotin for growth (17) and they do not synthesize large amounts of PGA (Table 1). B. subtilis strains are distinguishable from B. licheniformis strains in PGA physiology and are best categorized as organisms in which (NH4)2SO4 cannot support PGA synthesis and glutamate does not repress its production.
The synthesis of extracellular GGT showed no correlation with PGA production in B. licheniformis strains, for example, under glutamate-repressing conditions (medium 2 in Table 1); GGT synthesis varied between 0 and 17 U/ml although there was no detectable PGA in any of these cultures. The situation was similar with the strains of B. subtilis, of which S317 was particularly interesting because it synthesized large amounts of PGA in the absence of detectable GGT. This appears to be a natural GGT mutant and indicates that extracellular GGT is not involved in PGA biosynthesis in this bacterium. ggt is not an essential gene in B. subtilis, and mutants grow and sporulate normally (18); therefore, the role of GGT in B. subtilis physiology remains obscure, but it seems likely that PGA is synthesized exclusively by the pgs genes (1).
Factors affecting PGA synthesis in B. licheniformis S173.
Given the industrial importance of B. licheniformis in the extracellular enzyme industry (14), we examined the synthesis of PGA in B. licheniformis S173 in more detail. PGA synthesis was estimated after growth in basal salts medium containing (NH4)2SO4 (4 g/liter) as the nitrogen source with various carbon sources. Glucose, maltose, sucrose, and xylose supported similar levels of PGA synthesis (0.6 to 0.9 mg/ml). Citric acid stimulated maximal synthesis (1.0 mg/ml), and with galactose as the carbon source, PGA was undetectable. Citrate has been noted previously for supporting high levels of PGA synthesis in B. licheniformis 9945a (11) and B. subtilis IFO 3335 (6), which suggests that the requirement for the tricarboxylic acid cycle for metabolism of citrate enhances the production of α-ketoglutarate as the direct precursor for glutamate and PGA (4).
The effects of different nitrogen sources on PGA production by B. licheniformis S173 were also investigated. We discovered that the ammonium component of (NH4)2SO4 is important, since (NH4)2NO3 supported the same high yield of PGA as (NH4)2SO4 but NaNO3 failed to result in appreciable PGA accumulation (Table 2). Arginine, aspartate, and glutamine as the sole nitrogen source supported PGA synthesis; only glutamate repressed PGA biosynthesis, and it did so whether (NH4)2SO4 or glutamine was provided as the additional nitrogen source (Table 2).
The effect of glutamate on PGA synthesis was most pronounced when glutamate was added to early exponential phase cultures of B. licheniformis S173, in which it totally abolished PGA synthesis (Fig. 1). The addition of glutamate at late exponential phase also inhibited PGA synthesis, but when glutamate was added to stationary phase cells it reduced the final level of PGA, presumably by abolishing further synthesis and accelerating depolymerization. Glutamate halted PGA biosynthesis at the point at which the amino acid was added to the culture, indicating that glutamate represses the expression of the PGA biosynthetic enzyme genes in young cultures and prevents further synthesis of PGA in cultures in which the biosynthetic machinery has been previously synthesized.
We observed that strains of Bacillus amyloliquefaciens, a close relative of B. subtilis and B. licheniformis, also produced PGA on solid media but only when grown anaerobically with KNO3 as the terminal electron donor (data not shown). Given that B. licheniformis reduces nitrate under anaerobic conditions (15), we examined the synthesis of PGA in anaerobic cultures growing in basal salts medium containing glucose as the carbon source, (NH4)2SO4 as the nitrogen source, and KNO3 (3 g/liter) as the terminal electron acceptor. Growth was limited under these conditions (final OD660 was 0.89 after 24 h), but PGA accumulation was relatively high (0.284 mg/ml) and similar to that of aerobically grown cultures when the differences in growth are taken into account. B. licheniformis S173, like B. licheniformis strain A35 (3), produces PGA under anaerobic conditions, but unlike B. amyloliquefaciens it is not dependent on anaerobiosis for PGA synthesis.
In conclusion, we have shown that PGA synthesis in strains of B. licheniformis and B. subtilis responds differently to the presence of exogenous glutamate. The unpredictable synthesis of PGA by industrial strains of B. licheniformis used for extracellular enzyme production can cause difficulties in downstream processing and product recovery. The simple expedient of modifying the glutamate content of the medium could reduce this problem.
Acknowledgments
M.K. thanks the Royal Society for a postdoctoral fellowship.
REFERENCES
- 1.Ashiuchi M, Soda K, Misono H. A poly-gamma-glutamate system of Bacillus subtilis IFO 3336: gene cloning and biochemical analysis of poly-gamma-glutamate produced by Escherichia coli clone cells. Biochem Biophys Res Commun. 1999;263:6–12. doi: 10.1006/bbrc.1999.1298. [DOI] [PubMed] [Google Scholar]
- 2.Birrer G A, Cromwick A M, Gross R A. γ-Poly(glutamic acid) formation by Bacillus licheniformis 9945a: physiological and biochemical studies. Int J Biol Macromol. 1994;16:265–275. doi: 10.1016/0141-8130(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 3.Cheng C, Asada Y, Aida T. Production of γ-polyglutamic acid by Bacillus licheniformis A35 under denitrifying conditions. Agric Biol Chem. 1989;53:2369–2375. [Google Scholar]
- 4.Cromwick A M, Gross R A. Effects of manganese (II) on Bacillus licheniformis ATCC 9945A physiology and γ-poly(glutamic acid) formation. Int J Biol Macromol. 1995;17:259–267. doi: 10.1016/0141-8130(95)98153-p. [DOI] [PubMed] [Google Scholar]
- 5.Gardner J M, Troy F A. Chemistry and biosynthesis of the poly(γ-D-glutamyl) capsule in Bacillus licheniformis. Activation, racemization and polymerization of glutamic acid by a membranous polyglutamyl synthetase complex. J Biol Chem. 1979;254:6262–6269. [PubMed] [Google Scholar]
- 6.Goto A, Kunioka M. Biosynthesis and hydrolysis of poly(γ-glutamic) acid from Bacillus subtilis IFO 3335. Biosci Biotechnol Biochem. 1992;56:1031–1035. doi: 10.1271/bbb.56.1031. [DOI] [PubMed] [Google Scholar]
- 7.Hara T, Nagatomo S, Iwamoto N, Tadakoro Y, Kaneko S, Orima M, Ogata S. Cloning and nucleotide sequence of γ-polyglutamate production stimulating factor on Bacillus subtilis (natto) plasmid pUH1. J Fac Agric Kyushi Univ. 1994;39:43–51. [Google Scholar]
- 8.Hara T, Nagatomo S, Ogata S, Ueda S. The DNA sequence of γ-glutamyltranspeptidase gene of Bacillus subtilis (natto) plasmid pUH1. Appl Microbiol Biotechnol. 1992;37:211–215. doi: 10.1007/BF00178173. [DOI] [PubMed] [Google Scholar]
- 9.Hara T, Nakajima K, Saito H, Ishizaki A, Ogata S, Ueda S. Sequence analysis of replication origin of plasmid pLS11 of Bacillus subtilis IFO 3022. Biosci Biotechnol Biochem. 1992;56:223–227. doi: 10.1271/bbb.56.223. [DOI] [PubMed] [Google Scholar]
- 10.Ito Y, Tanaka T, Ohmachi T, Asada Y. Glutamic acid independent production of poly(γ-glutamic acid) by Bacillus subtilis TAM-4. Biosci Biotechnol Biochem. 1996;60:1239–1242. [Google Scholar]
- 11.Leonard C G, Housewright R D, Thorne C B. Effects of some metallic ions on glutamyl polypeptide synthesis by Bacillus subtilis. J Bacteriol. 1958;76:499–503. doi: 10.1128/jb.76.5.499-503.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogawa Y, Sugiura D, Motai H, Yuasa K, Tahara Y. DNA sequence of Bacillus subtilis (natto) NR-1 γ-glutamyltranspeptidase gene, ggt. Biosci Biotechnol Biochem. 1997;61:1596–1600. doi: 10.1271/bbb.61.1596. [DOI] [PubMed] [Google Scholar]
- 13.Priest F G, Goodfellow M, Todd C. A numerical classification of the genus Bacillus. J Gen Microbiol. 1988;134:1847–1882. doi: 10.1099/00221287-134-7-1847. [DOI] [PubMed] [Google Scholar]
- 14.Priest F G, Harwood C R. Bacillus species. In: Hui Y H, Kachatourians G G, editors. Food biotechnology: microorganisms. New York, N.Y: VCH; 1994. pp. 377–421. [Google Scholar]
- 15.Shariati P, Mitchell W J, Boyd A, Priest F G. Anaerobic metabolism in Bacillus licheniformis NCIB 6346. Microbiology. 1995;141:1117–1124. doi: 10.1099/13500872-141-5-1117. [DOI] [PubMed] [Google Scholar]
- 16.Thorne C B, Gomez C G, Noyes H G, Housewright R D. Production of glutamyl polypeptide by Bacillus subtilis. J Bacteriol. 1954;68:307–315. doi: 10.1128/jb.68.3.307-315.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda S. Utilization of soybean as natto, a traditional Japanese food. In: Maruo B, Yoshikawa H, editors. Bacillus subtilis: molecular biology and industrial application. Amsterdam, The Netherlands: Elsevier; 1989. pp. 143–161. [Google Scholar]
- 18.Xu K, Strauch M A. Identification, sequence, and expression of the gene encoding γ-glutamyltranspeptidase in Bacillus subtilis. J Bacteriol. 1996;178:4319–4322. doi: 10.1128/jb.178.14.4319-4322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]