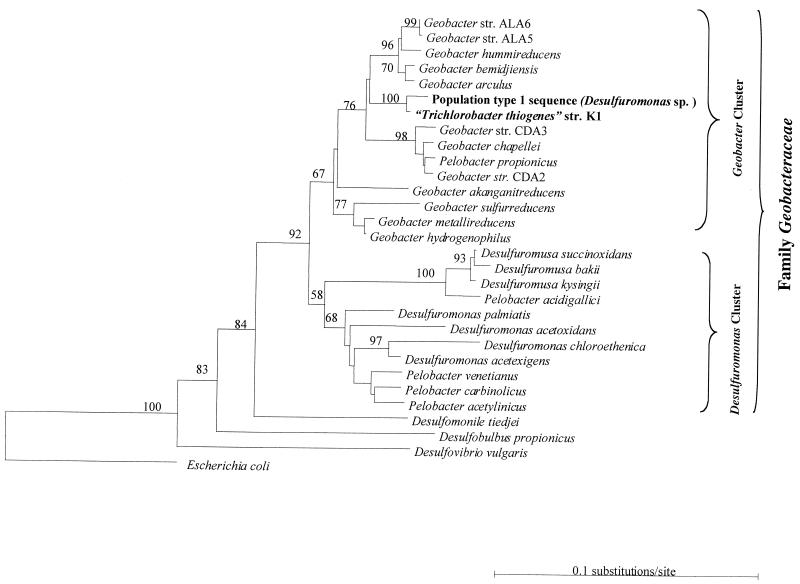
In a recent paper by De Wever et al. (2), it is proposed that the recently isolated microorganism strain K1 be assigned to a new genus within the delta Proteobacteria. Those authors state that the 16S ribosomal DNA (rDNA) sequence of strain K1 places it within a “cluster of mixed taxonomic affiliation” within the delta Proteobacteria. From this statement it is apparent that De Wever et al. (2) are unaware of several previous phylogenetic analyses of this group within the delta Proteobacteria, most notably, a study by Lonergan et al. (5). As De Wever et al. (2) note, the 16S rDNA sequence of strain K1 is nearly identical (99% sequence identity) to a 16S rDNA sequence recovered from a bioreactor that they misidentify as “environmental sp. 2” but that is actually an environmental sequence first described as a population type 1 sequence (Desulfuromonas-like sp.) (1) and listed by both GenBank and the Ribosomal Database Project (RDP) II (9) release 7.1 as Desulfuromonas sp. (GenBank accession number M80618). If De Wever et al. (2) had known about the study by Lonergan et al. (5), they would have realized that detailed analysis of this sequence has demonstrated that it rests squarely within the Geobacter cluster of the family Geobacteraceae (5). Our own analysis of the strain K1 sequence confirms not only that the overall sequence of strain K1 is closely related to organisms in the Geobacter cluster (Fig. 1) but also that the sequence contains the signature secondary structures characteristic of the Geobacter cluster (5).
FIG. 1.
Phylogenetic tree inferred from 16S rRNA sequences showing the phylogenetic placement of “Trichlorobacter thiogenes” strain K1. Phylogenetic relationships shown here were inferred by using neighbor joining and Kimura two-parameter genetic distances in TREECON (10). Bootstrap values above 60 are shown adjacent to branch nodes and were calculated from 100 resampled data sets using neighbor joining. The scale bar shows the number of expected nucleotide substitutions per site per unit of branch length. Operational taxonomic units shown on the tree were obtained from GenBank and the RDP databases. A similar tree topology was generated for trees constructed using maximum-likelihood and maximum-parsimony methods (data not shown).
At this time, the Geobacter cluster contains only one organism, Pelobacter propionicus, that does not have the genus designation Geobacter (Fig. 1). There are four more species of Pelobacter within the Geobacteraceae family, but these other Pelobacter species are interspersed throughout two other genera outside the Geobacter cluster. Thus, it is clear that the genus Pelobacter is not phylogenetically coherent and that organisms in the Geobacter cluster cannot be renamed Pelobacter as this would result in organisms of different phylogenetic clusters being placed within the same genus. It has been suggested that the simplest way to avoid confusion with the phylogeny is to place P. propionicus in the genus Geobacter (5).
Therefore, designation of strain K1 as a new genus within the Geobacter cluster, as suggested by De Wever et al. (2), creates havoc within an otherwise logical grouping of organisms, comprised of predominantly a single genus within a phylogenetically coherent cluster. Designation of a new genus might be warranted if strain K1 had some unique physiological characteristics that distinguished it from previously described Geobacter species, but this does not appear to be the case. Strain K1 uses acetate as an electron donor for the reduction of S0 and fumarate, and it has been suggested that it might also use Fe(III) as an electron acceptor (2). The oxidation of acetate coupled to the reduction of S0 and Fe(III) is one of the defining physiological characteristics of Geobacter species, and many Geobacter species can also use fumarate as an electron acceptor for acetate oxidation (6, 7, 8). Strain K1 does reductively dechlorinate trichloroacetic acid, a physiological capacity not previously reported for Geobacter species. However, to our knowledge, no other organisms in the Geobacter cluster have been evaluated for the ability to carry out this reaction. Therefore, it is not clear that strain K1 is unique among organisms in the Geobacter cluster in this ability. Furthermore, De Wever et al. (2) suggest that at least part of the reductive dechlorination observed with strain K1 is the result of strain K1 reducing S0 to sulfide, with the subsequent reduction of the trichloroacetic acid by sulfide. Since all organisms in the Geobacter cluster have the ability to reduce S0 to sulfide, it is likely that all Geobacter species have the ability to dechlorinate trichloroacetic acid via this electron shuttling mechanism.
Furthermore, even if all the organisms in the Geobacter cluster other than strain K1 were found to not be able to reductively dechlorinate trichloroacetic acid, precedence suggests that the designation of a new genus would not be warranted. When the first organism in the Desulfuromonas cluster of the Geobacteraceae found to have the ability to carry out reductive dechlorination was described (4), it was not assigned to a new genus; rather, it was designated a new species in the genus Desulfuromonas (3). This is consistent with the concept of not cluttering phylogenetically coherent groups with multiple genus designations.
In summary, designating strain K1 as a new genus in the Geobacter cluster at this time advances neither clarity in phylogeny or understanding of physiology. It is suggested that, once the characterization of the physiology of strain K1 is completed, it be designated a new species in the genus Geobacter.
REFERENCES
- 1.Amann R I, Stromley J, Devereux R, Key R, Stahl D A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992;58:614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Wever H, Cole J R, Fettig M R, Hogan D A, Tiedje J M. Reductive dehalogenation of trichloroacetic acid by Trichlorobacter thiogenesgen. nov., sp. nov. Appl Environ Microbiol. 2000;66:2297–2301. doi: 10.1128/aem.66.6.2297-2301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krumholz L R. Desulfuromonas chloroethenicasp. nov. uses tetrachloroethylene and trichloroethylene as electron acceptors. Int J Syst Bacteriol. 1997;47:1262–1263. [Google Scholar]
- 4.Krumholz L R, Sharp R, Fishbain S S. A freshwater anaerobe coupling acetate oxidation to tetrachloroethylene dehalogenation. Appl Environ Microbiol. 1996;62:4108–4113. doi: 10.1128/aem.62.11.4108-4113.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lonergan D J, Jenter H, Coates J D, Phillips E J P, Schmidt T, Lovley D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lovley D R. Fe(III) and Mn(IV) reducing prokaryotes. In: Dworkin M, Falkow S, Rosenberg M, Stackebrandt E, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 2000. pp. 1–47. [Google Scholar]
- 7.Lovley D R. Fe(III) and Mn(IV) reduction. In: Lovley D R, editor. Environmental microbe-metal interactions. Washington, D.C.: ASM Press; 2000. pp. 3–30. [Google Scholar]
- 8.Lovley D R, Coates J D, Saffarini D A, Lonergan D J. Dissimilatory iron reduction. In: Winkelman G, Carrano C J, editors. Iron and related transition metals in microbial metabolism. Geneva, Switzerland: Harwood Academic Publishers; 1997. pp. 187–215. [Google Scholar]
- 9.Maidak B L, Cole J R, Charles J, Parker T, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van de Peer Y, De Wachter Y. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]